Abstract
The analysis of biotransformations that occur in lysosomes and other endocytic organelles is critical to studies on intracellular degradation, nutrient recycling and lysosomal storage disorders. Such analyses require bioactive organelle preparations that are devoid of other contaminating organelles. Commonly used differential centrifugation techniques produce impure fractions and may not compatible with micro-scale separation platforms. Density gradient centrifugation procedures reduce the level of impurities but may compromise bioactivity. Here we report on simple magnetic setup and a procedure that produce highly enriched bioactive organelles based on their magnetic capture as they traveled through open tubes. Following capture, in-line laser-induced fluorecence detection (LIF) determined for the first time that each magnetically retained individual endocytic organelles have an acidic pH. Unlike bulk measurements, this method was suitable to describe the distributions of pH values in endocytic organelles from L6 rat myoblasts treated with dextran-coated iron oxide nanoparticles (for magnetic retention) and fluorescein/TMRM-conjugated dextran (for pH measurements by LIF). Their individual pH values ranged from 4 to 6, which is typical of bioactive endocytic organelles. These analytical procedures are of high relevance to evaluate lysosomal-related degradation pathways in aging, storage disorders and drug development.
Introduction
Lysosomes are organelles that degrade endocytosed material1 such as proteins,2, 3 lipids,4 fatty acids,5 and carbohydrates.3 These organelles play important roles in drug metabolism, biotransformation, reactive oxygen species production, autophagy, and lysosomal storage disorders.6–9 Lack of lysosomal function can lead to many diseases (e.g. Fabry’s,10 Gaucher’s,11 and Niemann-Pick disease).8 Lysosomes are also important in Alzheimer’s disease,12 because it has been shown that degradation of β-amyloid inside lysosomes may lead to novel disease treatments.
Investigating such roles and biological function of lysosomes usually involves studies done on enriched isolated lysosomes, devoid of other contaminating organelle types. For instance, proteomic studies aiming at identifying lysosomal proteomes usually require extensive lysosomal isolation procedures prior to proteomic analysis.13–15 Another example is the detection of reactive oxygen species in lysosomes that requires these organelles to be functional and free of contaminating mitochondria.6, 16, 17 Lastly, many xenobiotics, such as penicillin G, tilmicosin, and vitamin E may be accumulated and biotransformed in the lysosomes, but confirmation that these processes occur in the lysosomes requires enriched functional lysosomes, devoid of contaminants in which similar processes may occur.18–20 In the absence of high enrichment of a specific organelle, direct links cannot be made.
The most common methods to isolate and enrich lysosomes are differential centrifugation and density-gradient centrifugation. Differential centrifugation separates organelles based upon their sedimentation rate.21 It is relatively quick (~ 2 hours), but peroxisomes and mitochondria are present as contaminants. Density-gradient centrifugation in self forming gradients of colloidal particles separates organelles directly22, 23 or after loading of the lysosomes with compounds such as dextran,24 iron,25 and gold26 to change their sedimentation rate. These methods often require multiple centrifugation and sample cleanup steps that can lead to a lengthy experimental time (~6 hours) and the potential dissipation of the acidic pH within lysosomes and other endocytic organelles. 27 Free flow electrophoresis has been shown to separate lysosomes based on their electrophoretic mobility, however it requires specialized instrumentation.28
Magnetic isolation based on antibody-coated paramagnetic beads, which is commonly used to isolate proteins and cells,29–32 is another approach to isolate organelles such mitochondria, peroxisomes, and autophagosomes.33–35 Magnetic isolation of lysosomes after endocytosis of dextran-coated superparamagnetic iron oxide nanoparticles (FeDex) has also been reported35–37 and used to investigate pharmacological properties in bulk.37, 38 These studies did not evaluate directly whether such organelles retained an acidic pH during the isolation procedure. Magnetic isolation of particles in microfluidic channels39 and capillary40 have been used to isolate and concentrate particles used in protein digestions39, triglyceride analysis41 and immunoassays,42 but have not been applied to organelle studies.
Here, we report on the analysis of the pH of individual magnetically-enriched endocytic organelles after their release from a magnetic retention in an open tube. The post-nuclear fraction of L6 cells that had endocytosed both FeDex and fluorescein/TMRM-conjugated dextran was delivered through a fused silica capillary. This capillary passed through a custom-built on-line magnetic isolation device and ended at a post-column dual-channel laser induced fluorescence detector (LIF). For each individual organelle detected, the ratio of its fluorescence signals at each channel was used to determine its pH. Acidic pH values of the magnetically retained individual organelles suggested that magnetic isolation in an open tube is suitable to prepare functional endocytic organelles. Future magnetic purification of acidic organelles in open tubes would be highly beneficial to investigate the role of lysosomal activity in aging, storage disorders and metabolism.
Experimental
Reagents
Dulbecco’s Modified Eagle Medium (DMEM) high glucose cell medium was obtained from Gibco. Bovine calf serum was obtained from Omega Scientific (Tarsana, CA). Gentamycin, iron (III) chloride hexahydrate, dextran from Leuconostoc mesenteroides, 1,10-phenanthroline, hydroxylamine hydrochloride, poly-L-lysine, magnesium chloride hexahydrate, protease inhibitor, dimethyl sulfoxide, 2,6-dicholoroindophenol, sodium azide, succinic acid disodium salt, Triton X-100, titanium oxysulfate sulfuric acid complex hydrate, cacodylate, glutaraldehyde, osmium tetroxide, polyvinyl alcohol, propylene oxide, 3-sn-phosphatidyl-L-serine, L-α-phosphatidylcholine, L-α-phosphatidylethanolamine, iron (II) chloride tetrahydrate, imidazole, and dichloroindophenol (DCIP) were obtained from Sigma-Aldrich (St. Louis, MO). Ammonium hydroxide, hydrochloric acid, potassium hydroxide, and hydrogen peroxide were obtained from Mallinckrodt (Phillipsburgh, NJ). Fluorescein, tetramethylrhodamine (TMRM), para-nitrophenylphosphate (pNPP), fluorescein/TMRM-conjugated dextran (70,000 MW) (FTD), LysoTracker Red, and AlexaFluor488-conjugated dextran (10,000 MW, referred thereafter as AlexaFluor488-dextran) were obtained from Invitrogen (Carlsbad, CA). Sodium acetate trihydrate, 4,6-diamidino-2-phenylindole, and sulfuric acid were obtained from Fisher Scientific (Pittsburgh, PA). Bovine serum albumin was obtained from Roche (Basel, Switzerland). Ethanol was obtained from Pharmco-AAPER (Brookfield, CT). Phosphate buffered saline (10× PBS) is obtained from BioRad (Hercules, CA). UltraPure brand sucrose (99.9%) was obtained from MP Biomedicals, Inc. (Illkirch, France). Iron (96.0% pure) was obtained from VWR (Radnor, PA). Water was purified with a Millipore Synergy UV System (18.2 m3/cm, Bedford, MA).
A nigericin stock solution (1 mM) was made by mixing 5 mg nigericin with 6.89 mL water. Potassium phosphate solution (used with nigericin; 900 mM) was made by adding 3.92 g of the reagent to 20 mL of water. The pH was then adjusted to 6.1 with concentrated NaOH and brought to 25 mL. Buffer N, used for cell handling for enzyme assay procedures, was made by adding 27.01 g mannose (300 mM) and 240 mg MgCl2 (5.0 mM) to 50.0 mL of 10× PBS. The solution was then diluted to 450 mL with water. The pH was adjusted to 7.4 with concentrated NaOH and brought to 500 mL. Buffer M, used for cell handling for magnetic isolation procedures for determination of the pH of the retained organelles, was made by adding 19.12 g mannitol (210 mM), 11.98 g sucrose (70 mM), 596 mg 4-(2-hydroxyetheyl)-1-piperazineethanesulfonic acid (5 mM), and 731 mg ethylenediaminetetraacetic acid (5 mM) to 450 mL water. The pH was adjusted to 7.4 with concentrated NaOH and brought to 500 mL. The catalase assay solution was made the day of the experiment by adding 50 mg bovine serum albumin, 5 mL 0.2 M imidizole (pH 7.6), 1 mL 10 % w/v Triton X-100, 100 μL of 37 % H2O2, and was brought to 50 mL. The catalase stop buffer was made in a fume hood by adding 2.25 g TiOSO4 to 300 mL 2 N H2SO4. Once dissolved, the solution was brought to room temperature. A 200mL aliquot of the solution was added to 200 mL of 2N H2SO4. This solution was then diluted to 1 L. Unless noted otherwise, solutions were diluted with deionized water.
The synthesis of FeDex was prepared as described previously,36 except for preparation of AlexaFluor 488-FeDex, that used a mixture of 5 mg of AlexaFluor488 dextran and 2 g of dextran. After synthesis, the particles were dialyzed against water in a Fisher 4.8 nm seamless cellulose membrane for 48 hours at 4 °C. The dialysis water was exchanged after 24 hours. FeDex was filtered with a 0.22-μM filter under vacuum to remove large aggregates and impurities. FeDex (~ 4 mg/mL) was stored at 4 °C in water and used within 2 months.
FeDex particles were characterized by transmission electron microscopy (TEM), x-ray diffraction, light scattering and hysteresis loop measurements. For TEM characterization, FeDex particles were imaged on lacey carbon/formvar copper 200 mesh grids (Ted Pella Inc, Redding CA) on a JEOL 1200 transmission electron microscope. TEM does not display the dextran coating and revealed iron cores with irregular morphology with long-axis length of 7.4 ± 2.2 nm (average ± standard deviation, n=30; range: 4.3 – 14.4 nm), which are similar to those previously observed (8.7 ± 2.1 nm).36 For x-ray diffraction, 80 mg FeDex were analyzed in a Pananalytical X’Pert PRO MRD x-ray diffraction instrument. Using the Scherrer equation the calculated crystalline volume was 16.5 ± 8 (average ± standard deviation, n=7), which is consistent with the dimensions observed by TEM. For light scattering, 4.0 mg/mL FeDex in water were analyzed in a Brookhaven Instruments 90Plus/BI-MAS particle sizing instrument. The average particle size was 61.2 ± 0.7 (average ± standard deviation, n = 3 trials). This is a hydrodynamic determination that includes the total particle dimensions, including the thickness of the dextran coating. Hysteresis loop measurements were done in a Princeton Measurements Alternating Gradient Magnetometer (23° C, 0T – 2.2T, 1 × 10−11 Am2, Princeton, NJ). The FeDex particles were superparamagnetic and were at 97% of their maximum magnetic moment (160.7 μAm2) when exposed to the maximum magnetic field obtained with the magnets used here.
Cell Culture and Homogenate Preparation with Differential Centrifugation
The rat myoblast L6 cells (ATCC, Manassas, VA) were maintained in DMEM supplemented with 10% bovine serum and 0.01% gentamycin at 37 °C and 5% CO2. Cells were subcultured upon reaching 95% confluence in a 1:40 ratio. L6 cells were treated with nanoparticles FeDex (2.0 mg/mL) for 30 minutes in DMEM. Trypsin in PBS (0.5%) was used to release cells from T-flasks for subculture or collection of cells. Cells used for determination of individual organelle pH values were treated with fluorescein/TMRM-conjugated dextran (2 mg/mL) for 18 hours before cell homogenization.
In most of the studies described here, after their release by trypsinization cells were harvested by centrifugation at 1,000g for 10 minutes and washed twice with 3 mL of buffer N. This was repeated a second time. Cells were then treated with 1% (v/v in DMSO) protease inhibitor cocktail and homogenized in a Dounce homogenizer (0.0025″ clearance, Kontes Glass, Vineland, NJ) with 100 strokes. About 10% of the whole cell homogenate volume was retained for enzyme assays. The remaining 90% of homogenate was centrifuged at 600g for 10 minutes to remove the nuclear fraction and unlysed whole cells. The resulting supernatant was the post nuclear fraction (PNF) and contained lysosomes, mitochondria, peroxisomes, endosomes, and cytosol. For differential centrifugation studies, the PNF was centrifuged at 13,000g for 10 minutes to form a pellet that contained lysosomes among other organelles. The supernatant containing mainly cytosol and cytosolic proteins was removed with a pipette. The pellet was diluted with 1.1 mL of buffer N and then used in enzyme assays. In addition the whole cell homogenate fraction was diluted and characterized in a similar fashion.
Fluorescent Microscopy of L6 Cells
Cells seeded in a LabTek 4 chambered coverslip were labeled with Lysotracker Red (50 μM) for 45 minutes at 37 °C. After 15 minutes, cells were labeled with AlexaFluor488-FeDex (3 mg iron/mL) for the remaining 30 minutes at 37 °C. After labeling, the tray was washed twice by removing media, adding PBS with a pipette, removing the first wash solution, and replacing with 300 μL of PBS.
All images were taken with an Olympus IX81 inverted microscope (Melville, NY) equipped with an Olympus IX2-DSU disk scanning unit and a 60× (NA 1.45) oil immersion objective. A mercury lamp (Addison, TX) was used as the excitation source. AlexaFluor488 and Lysotracker Red fluorescence were detected respectively with filter sets GFP (excitation 460–500 nm; dichoric 505 nm; emission 510–560 nm) and RFP (excitation 510–560 nm; dichoric 565 nm; emission 572–649 nm) (Chroma (Rockingham, VT). Imaging was done with a C9100-01 CCD camera (Hamamatsu, Bridgewater, NJ). CImaging Simple PCI 5.3 (Compix Inc., Cranberry Township, PA) software was used to control the microscope, image acquisition and processing. The levels of background fluorescence, AlexaFluor 488 cross talk, and Lysotracker Red cross talk were determined by observing unlabeled controls and samples labeled with only AlexaFluor488-FeDex or Lysotracker Red, respectively. The values of the crosstalk were undetectable and 3.6 ± 1.1 % (average ± standard deviation), respectively (See Section S-1 Supplementary Material).
Image J-W Version 1.43s (National Institutes of Health) was used to calculate the Manders colocalization coefficients (M1 and M2) and the ICQ correlation coefficient (Section S-1, Supplementary Material). M1 and M2 range from zero to one representing total to total lack of colocalization of fluorophores.43 ICQ values range from 0.5 to −0.5 representing same to opposite correlation, respectively, of the fluorescence intensities of the two fluorophores.
Enzyme Assays
Enzyme assays specific to lysosomes, mitochondria, and peroxisomes were used to determine the abundance of these organelle types. The initial change of absorbance versus time was the enzymatic activity. After correcting for dilutions, the total specific activity (S.A.) was calculated as the ratio of enzymatic activity to protein concentration in the original sample. The relative activity (R.A.) of a sample was calculated as the ratio of its S.A. relative to the S.A. of the whole cell homogenate. The yield of a method was calculated as ratio of the total enzymatic activity in a recovered fraction over the total enzymatic activity of the PNF used in the fraction isolation procedure (i.e., differential centrifugation or magnetic isolation). Lysosomal abundance was determined with the pNPP assay.44 First 150 μL of 25 mM pNPP and then 150 μL of the sample were added to a well of a 96-well plate (F96, Nunc, Rochester, NY). Absorbance readings at 405 nm (ε = 1.80 × 104 cm−1 M−1)45 were taken every 8 seconds for 15 minutes. Measurements were done in triplicate. LOD’s for enzymatic assays were determined by calculating the enzymatic activity and protein concentration equivalent to three standard deviations above baseline. The LOD of the pNPP assay was 700 pmol pNPP/min/mg protein. Mitochondrial abundance was determined with the succinate dehydrogenase assay.46 The following reagents were added sequentially to a well of a 96-well plate: 130 μL of buffer N, 25 μL of 40 mM sodium azide, 25 μL of 200 mM succinate, and 25 μL aliquot of 0.050 mM DCIP. Then, 80 μL of the sample were added and thoroughly mixed. Absorbance readings at 600 nm (ε = 1.88 × 104 cm−1 M−1)47 were taken every 8 seconds for 15 minutes. The LOD of the succinate dehydrogenase assay was 1.0 nmol succinate/min/mg protein. Peroxisomal abundance was determined with the catalase assay.48 A 50 μL aliquot of organelle sample was mixed with a 12 μL aliquot of the catalase assay solution. Samples were incubated for 8 minutes and then treated with a 205 μL aliquot catalase stop solution to quench the reaction and allows for remaining hydrogen peroxide to react with titanium (IV) oxysulfate. A 250 μL aliquot of the total solution was added to a well in a 96-well plate and read at 405 nm (ε = 3.97 × 105 cm−1 M−1).49 The LOD of the catalase assay was 7.6 nmol/min/mg protein. Protein concentrations were determined using the bicinchoninic acid protein assay (BCA kit, ThermoScientific, Wilmington, MA) according to the manufacturer’s instructions.
On-line magnetic isolation device
Neodymium borate magnets (2.0 × 2.0 sq. in.; magnetized through their 0.5-inch thickness; K&J Magnetics, Inc., Jamison, PA) were modeled with Vizimag 3.18 software (Figure 1). The relative permeability of each magnet was set to 1.10 and the magnetic strength was set to 1.32 Tesla to match the manufacturer’s values. The best configuration found had four magnets per pole (Figure 1A). For 1-cm gap, the predicted field was 817 mT. We adopted such configuration and positioned each set of magnets on an aluminum mount equipped with a high-precision micrometer to adjust the gap between zero and ten centimeters (Figure 1B). The magnetic field was measured with a hand held Gaussmeter (GM08, Hirst, Cornwell, United Kingdom) that fitted when the gap between the magnets was 1 cm or larger (Figure 1C). For 1-cm gap the measured magnetic field was 758 mT.
Figure 1.
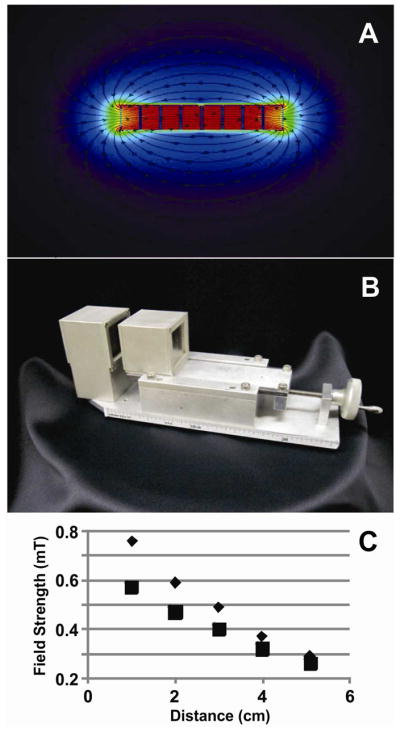
Characterization of magnet configuration. (A) Magnetic simulations of a cavity defined by two sets of four magnets each. This configuration shows a field strength of 817 mT when the gap is 1 cm. The simulation was done with Vizimag software. (B) On-line magnetic isolation device. (C) Field strength measurements at the center of the magnet cavity. Triangles and squares represent the experimental and the simulation data, respectively. Simulation data fitted the equation y = 0.0058x2 – 0.11x + 0.67 and predicted magnetic field strengths of 650 and 670 mT for gaps equivalent to those used for PEEK tubing and fused-silica capillary, respectively. Experimental data fitted the equation y = 0.012x2 – 0.19x + 0.93 and predicted magnetic field strengths of 900 and 930 mT for gaps equivalent to those used for PEEK tubing and fused-silica capillary, respectively.
Magnetic Isolation Procedures for Enzymatic Assays
A syringe pump (Model number 210, KDS Scientific, Holliston, MA) delivered ~ 100 μL of PNF from a 1-mL Hamilton RN syringe to PEEK tubing (#1531, IDEX Health and Science, Oak Harbor, WA) passing through the on-line magnetic isolation device. The syringe needle (22 g) was connected to the tubing with an adapter (VISF-2 and U-411, Chrom Tech, Inc, Apple Valley, MN). The PEEK tubing was positioned between the two magnet poles and held in place by reducing the gap until to the magnets slightly squeezed the tubing. The length of the tubing in the magnetic zone was 5.1 centimeters, which represents a volume of 2.6 μL within the tubing.
Following delivery of 15 μL of PNF through the tubing for loading of magnetic material into the magnetic zone, 25 μL of buffer N were delivered to wash off unretained material. The eluted volumes from the loading and wash steps contained unretained material. Upon completion of the wash step at 1.3 cm/min linear flow rate, the PEEK tubing was removed from the magnet gap. Then the previously retained magnetic material was eluted with at least 125 μL of buffer N. This elution step took under 9.5 minutes. The collected material was analyzed with the enzymatic assays described above. The linear velocity for the loading and elution steps was 21.9 cm/min. The linear velocities used for the wash step were 0, 0.6, and 1.32 cm/min, but only 1.32 cm/min was adequate to remove mitochondria and peroxisomes (Figure S2 in the Supplementary Material).
Magnetic Isolation Procedures for determination of the pH of the retained organelles
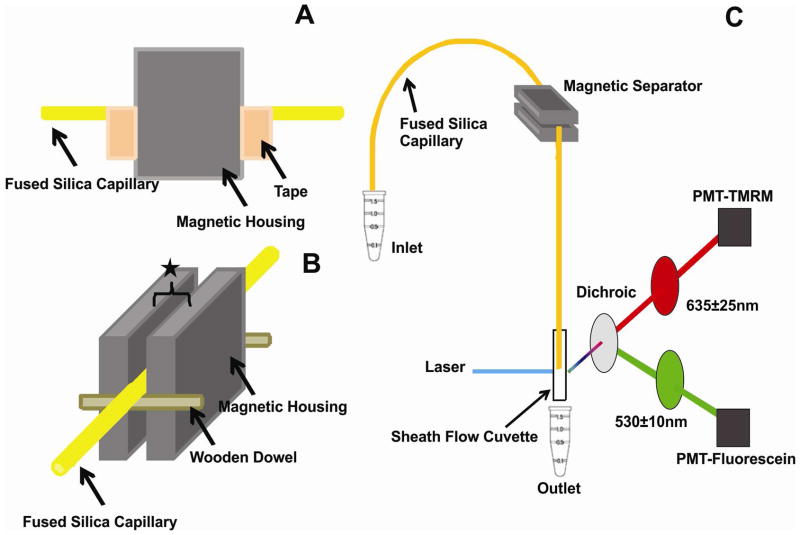
To detect the pH of organelles, we used the setup shown in Figure 2. Fused-silica capillary replaced the PEEK tubing described above. A fused silica capillary (49-μm I.D., 150-μm O.D., 80-cm long, coated with 2% polyvinyl alcohol50) was immersed in a vial containing either the PNF or buffer N and pressurized at 4 psi with an argon tank. The linear flow velocity in the capillary was 7.7 cm/min. The magnetic separator was placed 40 cm from the injection end. The capillary position was fixed within the magnet gap because two pieces of tape at each side of the magnet cavity prevented it from moving (Figure 2A), wooden dowels confined it to the middle of magnet gap (Figure 2B), and the gap was adjusted to match the capillary O.D. A capillary length of 5 cm was within the magnet gap and define an internal capillary volume of ~ 100 nL where magnetic capture occurred. When loading the capillary with PNF the plug length was ~ 5 cm long, matching the length of the capillary within the magnet gap. After loading the PNF, one capillary volume (1.6 μL) of buffer M was used to displace the PNF to the magnet gap and to wash off unretained material at a linear flow rate of 7.7 cm/min. Then, the capillary was removed from the magnet gap and retained organelles were mobilized by the pressure driven-flow. Upon completion of this step the capillary was flushed with methanol to reduce carry over into the next experimental run.
Figure 2.
Detail of the on-line magnetic isolation device. (A) Tape ensured that the capillary did not shift along the capillary’s length in the magnetic separator. (B) Wooden dowels attached to the magnetic housing kept the capillary in the middle of the magnetic housing. The star represents the gap adjusted to hold snug the PEEK tubing (gap ~ 1.59 mm) or the fused-silica capillary (gap ~ 0.150 mm). (C) An LIF detector placed ~ 40 cm after the device. Detection and excitation conditions are described in the Experimental section.
The detection end of the capillary was connected to a post-column LIF detector that has been previously described (Figure 2C).51 Briefly, the 488-nm argon-ion laser line was used for excitation. A 560 nm dichroic mirror (XF2016, Omega Optical, Brattleboro, VT) was used to reflect green fluorescence. A 635 ± 25 nm filter (XF3015, Omega Optical,) was used to select tetramethylrhodamine (TMRM) fluorescence and a 530 ± 10 nm filter (XF3017, Omega Optical) was used to select fluorescein and AlexaFluor488 fluorescence, respectively. Photomultiplier tubes were biased at 1,000V. The LOD of fluorescein was ~ 9 zmol. Fluorescently labeled organelles were detected as individual events as they eluted out during washing of the non-retained material and elution of the magnetically retained material. Events also occurred during the washing with buffer M and before elution of the fluorescently labeled organelles. These random false positives occurred at a low rate (0.049 ± 0.004 events/s). The non-retained fluorescent organelles (e.g. endocytic organelles with low FeDex contents but detectable levels of fluorescein/TMRM-conjugated dextran) appeared first at a rate of 0.17 ± 0.14 events/s. The retained fluorescent organelles (e.g. endocytic organelles with both FeDex and fluorescein/TMRM-conjugated dextran) appeared after ~100 s delay after removal of the capillary from the magnet. The yield of retained organelles was calculated as the ratio of the number of events in the retained zone to the number of events in both the unretained and retained zone.
Fluorescent data was analyzed using Igor Pro (WaveMetrics, Inc. Lake Oswego, OR). Broad bands corresponding to freely diffusing species were eliminated with a median filter. Individual organelle peak analysis was done with the custom-written program “coincidence analysis”.52 The average peak base width for peaks used in coincidence analysis was 24 ± 3 ms (n = 10,374) for TMRM peaks and 32 ± 3 ms (n = 10959) for fluorescein peaks. Peak intensities were corrected for spectral crosstalking (i.e, red-to-green ~ 16% and green-to-red ~ 6%). Peaks detected at the same time (± 20 msec) by the two PMTs of the LIF detector were used to calculate pH as previously reported53 using a pH calibration curve of FTD-loaded liposomes with different internal pH’s (Section S-3, Supplementary Material). Peak overlap between individual events defined by statistical overlap theory54 was not significant for all the magnetically retained organelles and for most of the non-retained organelles. For histogram distributions of individual pH values the bin width (2.0 pH units) was determined as the cubic root of total number of retained organelles, plus one.55 This value is larger than the error of individual pH values calculated by error propagation (Section S-3, Supplementary Material). Because pH distributions are non-parametric, previously described Q-Q plots56 were used for comparison of distributions.
Safety Considerations
Biosafety level 1 was observed in all procedures involving L6 cells. Biological waste was treated with bleach prior to disposal. Used cell culture supplies were autoclaved prior to disposal.
Results & Discussion
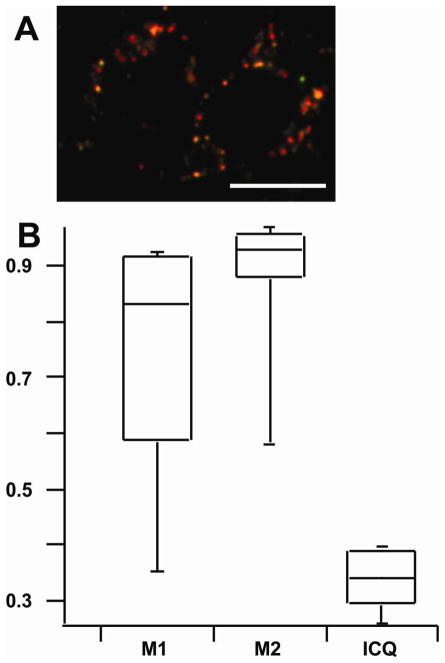
Fluorescence confocal microscopy measurements confirmed endocytosis of FeDex by L6 cells and was used to assess localization of FeDex in endocytic organelles based on the colocalization of LysoTracker Red and AlexaFluor488-bound FeDex (Figure 3 and Figure S1, Supplementary Material). LysoTracker Red accumulates in acidic organelles because the dye’s amine functional groups become protonated in acidic environments, which causes retention within the lumen of acidic organelles.57, 58 Based on the M2 Manders coefficient, 93% AlexaFluor488 fluorescence was colocalized with Lysotraker Red fluorescence. The remaining 7% may correspond to AlexaFluor 488 found in the cytosol (i.e., released from the lysosomes upon dextran degradation) or organelles with undetectable levels of LysoTracker Red (e.g., early endosomes). Based on the M1 Manders coefficient, 74% of the LysoTracker Red fluorescence was colocalized with AlexaFluor488 fluorescence. This is not surprising because endocytosis of AlexaFluor488-bound FeDex is expected to distribute to endocytic organelles (late endosomes, lysosomes)59, 60 excluding other non-endocytic acidic organelles (e.g. Golgi). ICQ suggested a positive correlation in the abundances of AlexaFluor488-bound FeDex and LysoTracker Red (ICQ = 0.34). Based on the fluorescence microscopy results and considering dextran’s role as a lysosomal and endocytic organelle marker,60, 61 we concluded that FeDex particles were mainly localized in late endocytic organelles, including lysosomes.
Figure 3.
Intracellular colocalization of FeDex with acidic organelles. (A) Overlay of confocal fluorescent images of L6 myocytes after endocytosis of AlexaFluor488-FeDex (Green) and treatment with LysoTracker Red (Red). Scale bar = 10 μm. (B) Boxplots of colocalization statistics of 45 cells. The M1 statistic represents the fraction of red fluorescence pixels with green colocalization. The M2 statistic represents the fraction of green fluorescence pixels with red colocalization. The ICQ statistic is > 0 indicating that intensities of both fluorophores are correlated. The ICQ statistic ranges from −0.5 to 0.5. See the Experimental and Section S-1 for more details.
Subcellular composition of magnetically isolated material
Organelle specific enzyme assays are commonly used to determine composition, enrichment (i.e. R.A), and yields of subcellular fractions.18 We utilized the pNPP, succinate dehydrogenase, and catalase assays to compare the subcellular fractions prepared by (i) magnetically retaining organelles from PNF flowing through PEEK tubing or (ii) differential centrifugation. While the subcellular fraction prepared by differential centrifugation was positive for all the enzymatic markers tested here (i.e., lysosomes, mitochondria, peroxisomes), the fraction that was magnetically isolated had only lysosomal markers present (R.A. = 1.7 ± 0.5, average ± Std. Dev., n=3 independent tests); other markers were below their limits of detection (Table 1). The R.A. for differential centrifugation for lysosomes was 1.2 ± 0.6 (average ± Std. Dev., n=3 independent tests). While the magnetic isolation procedure appears to have a higher yield than differential centrifugation, these values are not statistically different (p=0.05). Density gradient centrifugation has also been used for purification of lysosomal fractions.22, 23 Unfortunately, this procedure did not provide sufficient material to perform the organelle marker assays used here (data not shown).
Table 1.
Enrichment and composition of organelle fractions. The abundance of each organelle type was determined using organelle-specific enzymatic markers. The relative abundance (R.A.) for each fraction was calculated a described in the Experimental. The para-nitrophenylphosphatase assay was used to determine lysosomal abundance (LOD = 700 pmol p-nitrophenol/min/mg protein). The succinate dehydrogenase assay was used to determine mitochondrial abundance (LOD = 1.0 nmol DCIP/min/mg protein) as used for mitochondria. The catalase assay was used to determine peroxisomal abundance (LOD = 7.6 nmol Ti(IV)peroxysulfate/min/mg protein).
| Organelle | Differential Centrifugation (R.A.) | Magnetic Isolation (R.A.) |
|---|---|---|
| Lysosome | 1.2 ± 0.6 | 1.7 ± 0.5 |
| Mitochondria | 1.4 ± 0.5 | <0.2 |
| Peroxisome | 1.7 ± 0.4 | <0.1 |
The lysosomal yield of the magnetic isolation procedure was 18 ± 6% (average ± Std. Dev., n = 3 independent tests). The yield for differential centrifugation was 42 ± 10 (average ± Std. Dev., n = 3 independent tests). While the magnetic isolation procedure appears to have a lower yield than differential centrifugation, these values are not statistically different (p=0.05). When the goal is to isolate large amounts of material, the yield could be further improved by coiling the tubing within the magnet gap,62 design of other magnet geometries,39 and further optimization isolation procedure. On the other hand, the current design is highly suitable when working with limited amounts of material (e.g., 5 μL of PNF).
Determination of the individual pH of magnetically isolated organelles
Lysosomes and other late endocytic organelles require an acidic pH to maintain biological activity.1 For lysosomes to be enriched and to be of possible use for metabolic and biotransformation in vitro studies, their functionality must be retained. The first step to assess their biofunctionality is the determination of their pH. Determining the pH value of isolated organelles in bulk does not describe the ranges in pH values found in isolated endocytic organelles at the different stages of their maturation. Knowing the distribution of pH values is critical to associate pH with the overall bioactivity of the isolated organelles. Detection of the pH of individual organelles addresses this issue. We previously reported an LIF detection scheme to measure the pH of individual organelles.53 In order to use this detection scheme, here we combine magnetic purification with the analysis of pH of individual organelles based on LIF detection. Organelles that were magnetically isolated and then detected had both FeDex nanoparticles and FTD. Since FTD has two fluorophores (Alexa 488 and TMRM) each with distinct pH dependence, While FeDex allows for magnetic retention, the ratio of fluorescence signals of FTD (Alexa 488 and TMR) are used calculate the pH of individual organelles (see Section S-3, Supplementary Material).
The instrumental setup used for pH determinations consisted a fused silica capillary (50 μm. I.D.), which replaced the PEEK tubing, connected to a post-column LIF detector (Figure 2C). The LIF detection of individual organelles produced narrow events (i.e., 24 and 32 ms wide at their base for fluorescein and TMRM detection, respectively), with a separate fluorescence signal detected at each PMT for each fluorophore (Figure 4B). While the PNF was being loaded and before it reached the detector, only a small number of random events were detected (regions (i) and (ii) in Figure 4A). When the PNF material began reaching the LIF detector, organelles began to appear (region (iii) in Figure 4A). These events are the non-retained organelles and decreased in frequency as washing with Buffer M proceeded. Once the frequency of appearance of events decreased to 0–5% of that of the most crowded region, the capillary was removed from the magnet gap. As expected, the frequency of events increased, indicating that the magnetically retained events were reaching the LIF detector (region (iv) in Figure 4A). This region lasted only 50–60 seconds and had a low number of events (~ 20), consistent with the small volume of capillary exposed to the magnetic field (Figure 2A). The combined results of three runs had 83 events (yield ~ 2.6% of the total number of events) detected after removal of the capillary from the magnetic field. The yield was lower than that calculated based on organelle markers because here we used a higher linear flow rate than that used for the PEEK tubing-based experiments (7.7 cm/min for fused silica capillary; 1.3 cm/min for PEEK tubing). As indicated in Figure S-2 (Supplementary Material), the recovery decreased as the flow rate increases. The numbers of non-specific absorption and random events (12% and 4% of the detected events in region (iv), respectively) were not a problem because the non-specific absorption events are also endocytic organelles with pH values indicative of their bioactivity; the random events were rare and did not affect the interpretation of the pH results. Thus, we used the detected events in region (iv) to assess the pH of the magnetically isolated organelles.
Figure 4.
LIF detection of individual organelles loaded with FTD. (A) Detection of fluorescein (upper trace) and TMRM (lower trace). Both traces were modified with a median filter to remove broad bands corresponding to free fluorophores (c.f. Figure S-4 in the Supplementary Material).58 The four regions shown in this plot include: (i) Detection while PNF was loaded and before organelles reached the LIF detection; (ii) dead time during the exchange of PNF for buffer M; (iii) detection of events from organelles that were not magnetically retained; (iv) detection of events that were magnetically retained. Before detection of events in region (iv) the frequency of events had been decreased to 0 – 2.3% of the event density observed at the beginning of region (iii). (B) Expanded view from 4A indicates that events are narrow as expected from the rapid passage of organelles through the LIF laser and they contain FTD because they are detected simultaneously in both channels.
As a reference, we determined the distribution of the pH of individual endocytic organelles in the PNF of L6 cells, treated only with FTD and prepared by differential centrifugation (Figure 5A). The observed range of individual pH was 4–6, which is consistent with the expected pH of lysosomes and late endocytic organelles.63 We also determined a distribution of pH of individual endocytic organelles in the PNF prepared from L6 cells treated with nigericin and FTD (Figure 5B). Nigericin dissipates the luminal pH in acidic organelles that acquire the pH of their surroundings.64 The Q-Q plot of the distributions shown in Figure 5B confirmed that nigericin-treated organelles had an overall increase in pH (Figure S-5A, Supplementary Material).
Figure 5.
Individual organelle pH Distributions. (A) The pH of organelles in a PNF from L6 cells not treated with FeDex (B) Same as in A, except for treatment with nigericin prior to cell homogenization. (C) The pH of magnetically retained organelles. (D) Q-Q plot of pH distributions of magnetically retained versus unretained organelles in the same run. Markers joined by solid lines represent increasing quantiles (10%, 20%, 30%, 40%, 50%, 60%, 70%, 80%, 90%). The dashed line (y=x) represents hypothetical identical distributions.
We also investigated the effect of FeDex on the LIF determination of pHs of individual organelles. The presence of FeDex caused a systematic increase in pH (1.7 ± 0.1 pH units) relative those organelles from cells treated only with FTD (Section S-4, Table S-1, Figure S-5B, Supplementary Material). This may be the result from scattering or fluorescent quenching by the iron oxide nanoparticle.65 This shift was used as a correction factor for pH values of individual organelles obtained from FeDex-treated L6 cells. After correction, both the pH of the retained (Figure 5C) and the eluted organelles (Figure S-5C, Supplementary Material) were consistent with previous literature reports.63 Most importantly, the determination of the individual pH of magnetically retained organelles revealed how pH distributions shifted relative to the distribution of the organelles in the PNF (Figures 5C and 5A, respectively). This slight shift may indeed correspond to uncorrected biases caused by the FeDex particles or the additional time needed to carry out the magnetic isolation (~30 min) during which the pH gradient may begin to dissipate. On the other hand, a Q-Q plot comparing the individual pH values of magnetically retained and non-retained organelles in the same PNF revealed that their pH distributions are practically unbiased by the magnetic isolation procedure (Figure 5D).
CONCLUSIONS
The magnetic isolation system described here was effective at isolating enriched lysosomes and endocytic organelles containing FeDex particles. Based on organelle enzyme markers, common contaminants in differential centrifugation preparations such as mitochondria and peroxisomes were not observed. Laser-induced fluorescence detection of the individual pH of magnetically retained organelles demonstrated that magnetic isolation does not influence the distribution of pH values relative to non-retained organelles. We found a pH for isolated organelles of 4 – 6, which is characteristic of lysosomes and endocytic organelles retaining acidic pH.
Future applications of the work described here include evaluation of different subpopulations of magnetically enriched organelles. This could be achieved with on-line magnetic capture of organelles followed by capillary electrophoresis coupled to LIF detection53 or isolectric focusing of the organelles based upon either free flow electrophoresis66 or within a capillary with LIF detection67. Future improvements also includes designing alternate open tube configurations (e.g. coiled62 or parallel lines37) or microfabricating magnetic geometries that are more conducive to attaining higher organelle yields.68 The advantages of newly proposed configurations could be easily explored using physical modeling using software packages such as ANSYS (fluid dynamics) and Maxwell (magnetism).69 The device could also be used to magnetically isolate other subcellular compartments using magnetic beads coated with organelle-specific antibodies.29–35 The bioactivity of such organelles could be evaluated using previously reported70 or novel individual organelle assays. Lastly, the work described here has a great potential to become the basis for future magnetic purifications of endocytic organelles that are highly needed to investigate the role of lysosomal function in aging, storage disorders and metabolism.
Supplementary Material
Acknowledgments
NIH grant RO1 AG020866 supported this work. C.S. was supported by NIH grant T32 GM8347. T.E.M. was carried out at the Institute of Technology Characterization Facility, University of Minnesota, member of the NSF-funded Materials Research Facilities Network. Hysteresis loop experiments were performed at the University of Minnesota Institute of Rock Magnetism. The authors would like to thank the Lee Penn laboratory for access to their x-ray diffraction equipment and the Christy Haynes laboratory for access to light scattering equipment.
References
- 1.Mukherjee S, Ghosh RN, Maxfield FR. Physiol Rev. 1997;77:759–803. doi: 10.1152/physrev.1997.77.3.759. [DOI] [PubMed] [Google Scholar]
- 2.Vellodi A. Br J Haematol. 2005;128:413–431. doi: 10.1111/j.1365-2141.2004.05293.x. [DOI] [PubMed] [Google Scholar]
- 3.Winchester B. Glycobiology. 2005;15:1R–15R. doi: 10.1093/glycob/cwi041. [DOI] [PubMed] [Google Scholar]
- 4.Kolter T, Sandhoff K. FEBS Lett. 2010;584:1700–1712. doi: 10.1016/j.febslet.2009.10.021. [DOI] [PubMed] [Google Scholar]
- 5.Tanaka M, Ilo T, Tabata T. J Biochem. 1987;101:619–624. doi: 10.1093/jb/101.3.619. [DOI] [PubMed] [Google Scholar]
- 6.Zhao M, Antunes F, Eaton JW, Brunk UT. Eur J Biochem. 2003;270:3778–3786. doi: 10.1046/j.1432-1033.2003.03765.x. [DOI] [PubMed] [Google Scholar]
- 7.Hsu KF, Wu CL, Huang SC, Wu CM, Hsiao JR, Yo YT, Chen YH, Shiau AL, Chou CY. Autophagy. 2009;5:451–460. doi: 10.4161/auto.5.4.7666. [DOI] [PubMed] [Google Scholar]
- 8.Dietz HC. N Engl J Med. 2010;363:852–863. doi: 10.1056/NEJMra0907180. [DOI] [PubMed] [Google Scholar]
- 9.Kohler A, Pluta HJ. Mar Environ Res. 1995;39:255–260. [Google Scholar]
- 10.Schiffmann R, Murray GJ, Treco D, Daniel P, Sellos-Moura M, Myers M, Quirk JM, Zirzow GC, Borowski M, Loveday K, Anderson T, Gillespie F, Oliver KL, Jeffries NO, Doo E, Liang TJ, Kreps C, Gunter K, Frei K, Crutchfield K, Selden RF, Brady RO. Proc Natl Acad Sci U S A. 2000;97:365–370. doi: 10.1073/pnas.97.1.365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Grabowski GA, Leslie N, Wenstrup R. Blood Rev. 1998;12:115–133. doi: 10.1016/s0268-960x(98)90023-6. [DOI] [PubMed] [Google Scholar]
- 12.Pimplikar SW, Nixon RA, Robakis NK, Shen J, Tsai LH. J Neurosci. 2010;30:14946–14954. doi: 10.1523/JNEUROSCI.4305-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Della Valle MC, Sleat DE, Zheng HY, Moore DF, Jadot M, Lobel P. Mol Cell Proteomics. 2011:10. doi: 10.1074/mcp.M110.006403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Trotter MWB, Sadowski PG, Dunkley TPJ, Groen AJ, Lilley KS. Proteomics. 2010;10:4213–4219. doi: 10.1002/pmic.201000359. [DOI] [PubMed] [Google Scholar]
- 15.de Araujo MEG, Huber LA, Stasyk T. Expert Rev Proteomics. 2011;8:303–307. doi: 10.1586/epr.11.27. [DOI] [PubMed] [Google Scholar]
- 16.Eder AR, Arriaga EA. Chem Res Toxicol. 2006;19:1151–1159. doi: 10.1021/tx060083i. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pourahmad J, Ross S, O’Brien PJ. Adv Free Radical Biol Med. 2001;30:89–97. doi: 10.1016/s0891-5849(00)00450-0. [DOI] [PubMed] [Google Scholar]
- 18.Renard C, Vanderhaeghe HJ, Claes PJ, Zenebergh A, Tulkens PM. Antimicrob Agents Chemother. 1987;31:410–416. doi: 10.1128/aac.31.3.410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Agoston M, Orsi F, Feher E, Hagymasi K, Orosz Z, Blazovics A, Feher J, Vereckei A. Toxicology. 2003;190:231–241. doi: 10.1016/s0300-483x(03)00188-4. [DOI] [PubMed] [Google Scholar]
- 20.Wang YH, Arriaga EA. Cancer Lett (Shannon, Irel) 2008;262:123–132. doi: 10.1016/j.canlet.2007.11.034. [DOI] [PubMed] [Google Scholar]
- 21.Claude A. J Exp Med. 1946;84:51–59. [PubMed] [Google Scholar]
- 22.Cuervo AM, Dice JF, Knecht E. J Biol Chem. 1997;272:5606–5615. doi: 10.1074/jbc.272.9.5606. [DOI] [PubMed] [Google Scholar]
- 23.Sipos G, Fuller RS. Guide to Yeast Genetics and Molecular and Cell Biology, Pt C. 2002;351:351–365. [Google Scholar]
- 24.Arai K, Kanaseki T, Ohkuma S. J Biochem. 1991;110:541–547. doi: 10.1093/oxfordjournals.jbchem.a123616. [DOI] [PubMed] [Google Scholar]
- 25.Arborgh B, Ericsson J, Claumann H. FEBS Letters. 1973;32:190–194. doi: 10.1016/0014-5793(73)80769-0. [DOI] [PubMed] [Google Scholar]
- 26.Henning R, Plattner H. Biochim Biophys Acta. 1974;354:114–120. doi: 10.1016/0304-4165(74)90059-2. [DOI] [PubMed] [Google Scholar]
- 27.Alberts B, Johnson A, Lewis J, Raff M, Roberts K, Walter P. Molecular Biology of the Cell. 4. Garland Science; New York: 2002. [Google Scholar]
- 28.Marsh M. Methods Cell Biol. 1989;31:319–334. doi: 10.1016/s0091-679x(08)61617-9. [DOI] [PubMed] [Google Scholar]
- 29.Franzreb M, Siemann-Herzberg M, Hobley TJ, Thomas ORT. Appl Microbiol Biotechnol. 2006;70:505–516. doi: 10.1007/s00253-006-0344-3. [DOI] [PubMed] [Google Scholar]
- 30.Yavuz CT, Prakash A, Mayo JT, Colvin VL. Chem Eng Sci. 2009;64:2510–2521. [Google Scholar]
- 31.Thiel A, Scheffold A, Radbruch A. Immunotechnology. 1998;4:89–96. doi: 10.1016/s1380-2933(98)00010-4. [DOI] [PubMed] [Google Scholar]
- 32.Tomasello E, Desmoulins PO, Chemin K, Guia S, Cremer H, Ortaldo J, Love P, Kaiserlian D, Vivier E. Immunity. 2000;13:355–364. doi: 10.1016/s1074-7613(00)00035-2. [DOI] [PubMed] [Google Scholar]
- 33.Hornig-Do HT, Gunther G, Bust M, Lehnartz P, Bosio A, Wiesner RJ. Anal Biochem. 2009;389:1–5. doi: 10.1016/j.ab.2009.02.040. [DOI] [PubMed] [Google Scholar]
- 34.Gao WT, Kang JH, Liao Y, Ding WX, Gambotto AA, Watkins SC, Liu YJ, Stolz DB, Yin XM. J Biol Chem. 2010;285:1371–1383. doi: 10.1074/jbc.M109.054197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Luers GH, Hartig R, Mohr H, Hausmann M, Fahimi HD, Cremer C, Volkl A. Electrophoresis. 1998;19:1205–1210. doi: 10.1002/elps.1150190722. [DOI] [PubMed] [Google Scholar]
- 36.Rodriguezparis JM, Nolta KV, Steck TL. J Biol Chem. 1993;268:9110–9116. [PubMed] [Google Scholar]
- 37.Diettrich O, Mills K, Johnson AW, Hasilik A, Winchester BG. FEBS Lett. 1998;441:369–372. doi: 10.1016/s0014-5793(98)01578-6. [DOI] [PubMed] [Google Scholar]
- 38.Duvvuri M, Krise JP. Mol Pharmaceutics. 2005;2:440–448. doi: 10.1021/mp050043s. [DOI] [PubMed] [Google Scholar]
- 39.Slovakova M, Minc N, Bilkova Z, Smadja C, Faigle W, Futterer C, Taverna M, Viovy JL. Lab Chip. 2005;5:935–942. doi: 10.1039/b504861c. [DOI] [PubMed] [Google Scholar]
- 40.Gassner AL, Morandini J, Josserand J, Girault HH. Anal Methods. 2011;3:614–621. doi: 10.1039/c0ay00596g. [DOI] [PubMed] [Google Scholar]
- 41.Chen SP, Yu XD, Xu JJ, Chen HY. Analyst. 2010;135:2979–2986. doi: 10.1039/c0an00231c. [DOI] [PubMed] [Google Scholar]
- 42.Chen HX, Busnel JM, Gassner AL, Peltre G, Zhang XX, Girault HH. Electrophoresis. 2008;29:3414–3421. doi: 10.1002/elps.200800106. [DOI] [PubMed] [Google Scholar]
- 43.Adler J, Parmryd I. Cytometry Part A. 2010;77A:733–742. doi: 10.1002/cyto.a.20896. [DOI] [PubMed] [Google Scholar]
- 44.Takeuchi T, Pang M, Amano K, Koide J, Abe T. Clin Exp Immunol. 1997;109:20–26. doi: 10.1046/j.1365-2249.1997.4371334.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhang ZY, Clemens JC, Schubert HL, Stuckey JA, Fischer MWF, Hume DM, Saper MA, Dixon JE. J Biol Chem. 1992;267:23759–23766. [PubMed] [Google Scholar]
- 46.Masters B, Bilimoria M, Kamin H. J Biol Chem. 1965;240:4081–4088. [PubMed] [Google Scholar]
- 47.Bissbort SH, Vermaak WJH, Elias J, Bester MJ, Dhatt GS, Pum JKW. Clin Chim Acta. 2001;303:139–145. doi: 10.1016/s0009-8981(00)00388-0. [DOI] [PubMed] [Google Scholar]
- 48.Csanyi L. Anal Chem. 1970;42:680–682. [Google Scholar]
- 49.Aviram I, Shaklai N. Arch Biochem Biophys. 1981;212:329–337. doi: 10.1016/0003-9861(81)90373-8. [DOI] [PubMed] [Google Scholar]
- 50.Whiting CE, Arriaga EA. Electrophoresis. 2006;27:4523–4531. doi: 10.1002/elps.200600249. [DOI] [PubMed] [Google Scholar]
- 51.Duffy CF, Gafoor S, Richards DP, Admadzadeh H, O’Kennedy R, Arriaga EA. Anal Chem. 2001;73:1855–1861. doi: 10.1021/ac0010330. [DOI] [PubMed] [Google Scholar]
- 52.Andreyev D, Arriaga EA. Anal Chem. 2007;79:5474–5478. doi: 10.1021/ac070770u. [DOI] [PubMed] [Google Scholar]
- 53.Chen Y, Arriaga EA. Anal Chem. 2006;78:820–826. doi: 10.1021/ac051513x. [DOI] [PubMed] [Google Scholar]
- 54.Davis JM, Arriaga EA. J Chromatogr, A. 2009;1216:6335–6342. doi: 10.1016/j.chroma.2009.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Terrell GR, Scott DW. J Am Stat Assoc. 1985;80:209–214. [Google Scholar]
- 56.Whiting CE, Arriaga EA. J Chromatogr, A. 2007;1157:446–453. doi: 10.1016/j.chroma.2007.04.065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Lemieux B, Percival MD, Falgueyret JP. Anal Biochem. 2004;327:247–251. doi: 10.1016/j.ab.2004.01.010. [DOI] [PubMed] [Google Scholar]
- 58.Fuller KM, Arriaga EA. Anal Chem. 2003;75:2123–2130. doi: 10.1021/ac026476d. [DOI] [PubMed] [Google Scholar]
- 59.Shi H, He XX, Yuan Y, Wang KM, Liu D. Anal Chem. 2010;82:2213–2220. doi: 10.1021/ac902417s. [DOI] [PubMed] [Google Scholar]
- 60.Duzgunes N, Majumdar S, Goren MB. Methods Enzymol. 1993;221:234–238. doi: 10.1016/0076-6879(93)21020-9. [DOI] [PubMed] [Google Scholar]
- 61.Sallusto F, Cella M, Danieli C, Lanzavecchia A. J Exp Med. 1995;182:389–400. doi: 10.1084/jem.182.2.389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Beveridge JS, Stephens JR, Latham AH, Williams ME. Anal Chem. 2009;81:9618–9624. doi: 10.1021/ac9016456. [DOI] [PubMed] [Google Scholar]
- 63.Geisow MJ, Hart PD, Young MR. J Cell Biol. 1981;89:645–652. doi: 10.1083/jcb.89.3.645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Schindler M, Grabski S, Hoff E, Simon SM. Biochemistry. 1996;35:2811–2817. doi: 10.1021/bi952234e. [DOI] [PubMed] [Google Scholar]
- 65.Sathe TR, Agrawal A, Nie SM. Anal Chem. 2006;78:5627–5632. doi: 10.1021/ac0610309. [DOI] [PubMed] [Google Scholar]
- 66.Ellinger I, Klapper H, Fuchs R. Electrophoresis. 1998;19:1154–1161. doi: 10.1002/elps.1150190716. [DOI] [PubMed] [Google Scholar]
- 67.Wolken GG, Kostal V, Arriaga EA. Anal Chem. 2011;83:612–618. doi: 10.1021/ac102712r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Ostergaard S, Blankenstein G, Dirac H, Leistiko O. J Magn Magn Mater. 1999;194:156–162. [Google Scholar]
- 69.Gleich B, Hellwig N, Bridell H, Jurgons R, Seliger C, Alexiou C, Wolf B, Weyh T. IEEE Trans Nanotechnol. 2007;6:164–170. [Google Scholar]
- 70.Meany DL, Poe BG, Navratil M, Moraes CT, Arriaga EA. Adv Free Radical Biol Med. 2006;41:950–959. doi: 10.1016/j.freeradbiomed.2006.06.003. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.