Abstract
Cofilin is a key actin-binding protein that is critical for controlling the assembly of actin within the cell. Here we present the results of molecular docking and dynamics studies using a muscle actin filament and human cofilin I. Guided by extensive mutagenesis results as well as other biophysical and structural studies, we arrive at a model for cofilin bound to the actin filament. This predicted structure agrees very well with cryo-electron microscopy results for cofilin-decorated filaments, provides molecular insight into how the known F- and G-actin sites on cofilin interact with the filament, and also suggests new interaction sites that may play a role in cofilin binding. The resulting atomic-scale model also helps us understand the molecular function and regulation of cofilin and provides testable data for future experimental and simulation work.
Keywords: ADF/cofilin, cytoskeleton, molecular docking, mutagenesis
INTRODUCTION
The regulation of actin polymerization is vital for cellular function, particularly in processes such as cell division and migration where rapid reorganization of the cytoskeleton is required. Cofilin plays a key role in this regulatory process. Cofilin was first implicated for its function as a filament severing protein 1; 2, but it has subsequently been shown to have a much broader physiological role (see 3 for a recent review). In order to understand the molecular function of this protein, the past two decades have seen an extensive array of biochemical and genetic studies 4; 5; 6; 7; 8; 9, x-ray, NMR and cryo-EM structural work 10; 11; 12; 13, numerous biophysical studies 14; 15; 16; 17; 18; 19, as well as computer simulation work 20; 21; 22; 23; 24. Collectively, these studies have each given us critical pieces of information about the interaction of cofilin with the actin filament, however a detailed understanding of cofilin binding is still lacking.
One complicating factor in this system is that cofilin binds both to monomeric or G-actin as well as polymerized actin filaments or F-actin. The generation of numerous cofilin and actin mutations has provided molecular details on how these proteins interact with each other 4; 5; 6; 7; 8; 9. Through these studies it has been determined that the G- and F-actin binding modes have some common features; however there are some intrinsic differences and a number of amino acids appear to only affect one of these two interactions. Along with this mutagenesis work, structural studies have provided significant insight into the interaction of cofilin with actin. Here again, cryo-EM work has characterized the filament interactions, such as the twist induced by cofilin binding 13 as well as changes in the orientation of the protomer 11; 25, and our knowledge of the interaction with G-actin with cofilin is informed by co-crystal structures with two ADF/cofilin homologs: gelsolin segment 1 26 and more recently, twinfilin 27. These structural data are invaluable, however they are not without caveats. The cryo-EM model from Galkin et al. 25 resulted from manually fitting atomic structures into the ~23 Å density map, and the crystal structures are with the actin monomer and not the filament. Thus we still lack a detailed understanding of the direct molecular interactions between cofilin and F-actin.
Here we present the results of molecular docking and molecular dynamics studies aimed at deriving a molecular model for the cofilin/F-actin complex. Recent work from Pfaendtner et al. 20 illustrated the utility of such types of simulations in studying this system. Using the cofilactin structure derived from electron microscopy by Galkin et al.25, the authors simulated the dynamics of the cofilin-bound filament and thus were able to gain insight into the dynamics and mechanics of this system. The methods we employ in this work are similar, however our objective here is very different, namely to derive an independent structural model for the cofilin-bound filament that agrees with the biochemical and structural data detailed above.
RESULTS
Sequence/Structure Alignment
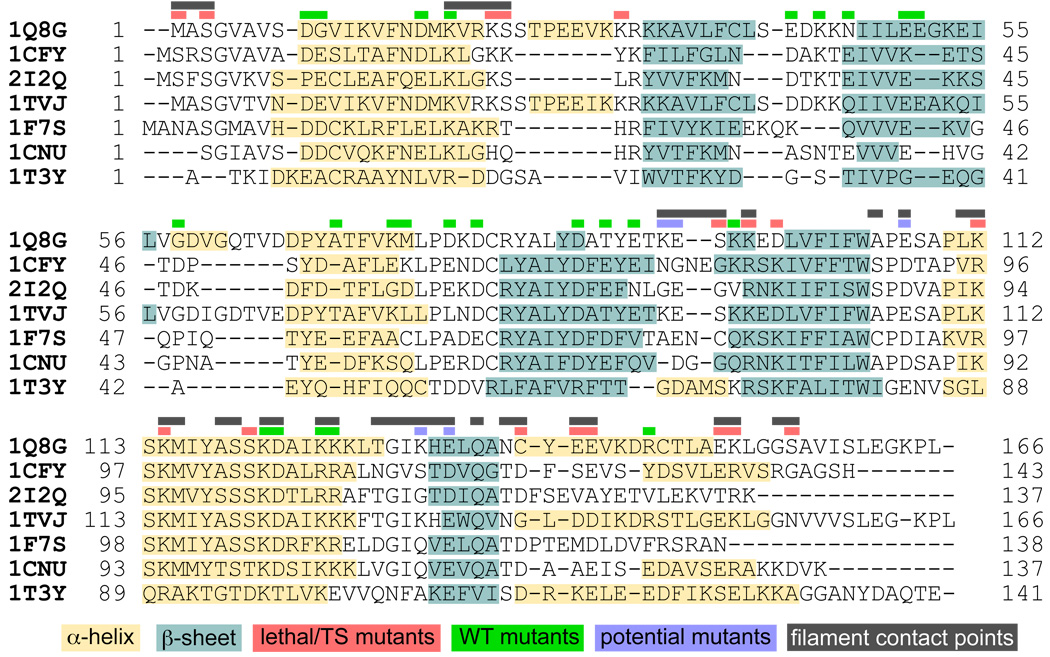
In order to correlate all of the published experimental mutagenesis results, our first task was to generate an accurate alignment of the ADF/cofilin sequences. Structural conservation within this family is high, although sequence conservation is variable depending on which proteins are chosen (average of 32% across the sequences presented here). Further, there are many insertions and deletions that occur, and this presents real challenges for alignment methods that only consider sequence similarity. Structure-based methods produce better sequence alignments in situations like this, but automated methods that consider both structure and sequence still face difficulties 28. For this reason, we started with a sequence/structure-based alignment using MultiSeq 29 and manually adjusted the gaps based on the secondary and tertiary structure. This method brings more regions of the proteins into register and we see a much higher degree of correlation between both the secondary structure elements as well as the individual mutagenesis results (see Figure 1). Obviously the alignment of human and yeast cofilin is of primary interest in this effort since these two systems formed the basis for the majority of the experimental work (Figure 2). The difficulties of aligning the yeast and human cofilin sequences have been noted in the past 30. Our manually curated alignment solves these ambiguities, and we now see an improved alignment, for example, between K22-R32/G58-D66 in human cofilin with the regions K23-K26/D47-S49 in the yeast protein.
Figure 1.
A structure-based sequence alignment for seven ADF/cofilin proteins and homologs (see Methods for details). Shaded regions of the sequences indicated secondary structure elements and colored bars above the sequence denote regions of contact in our model and the location of lethal mutations (red), mutations with a WT phenotype (green) and novel interaction sites suggested by our model (blue).
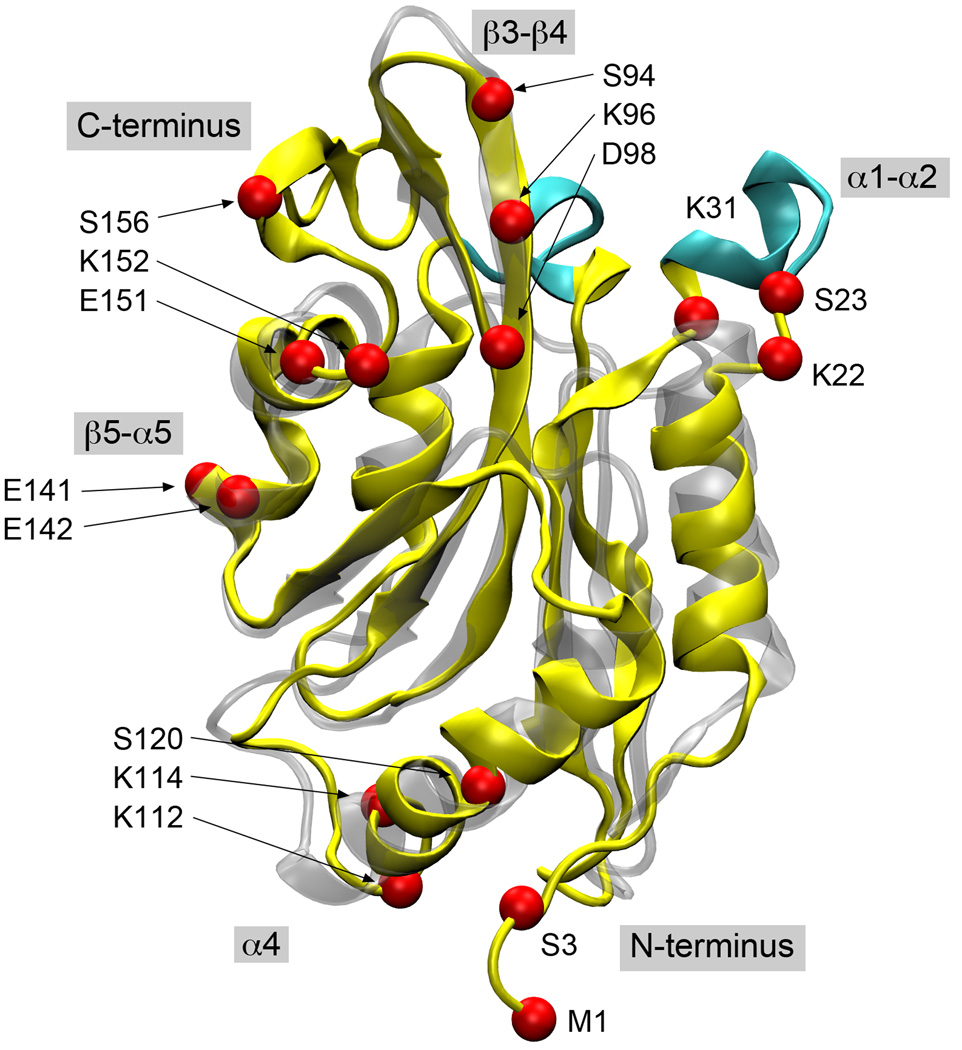
Figure 2.
A structural alignment of human cofilin (yellow) and yeast cofilin (silver). The secondary structure elements discussed in the text are the shaded labels and the locations of lethal point mutations explained by our docking model are shown as red spheres and labeled based on the human cofilin sequence. The major loop insertions in the human protein are highlighted in cyan.
Model for the Cofilin/F-actin Complex
As detailed in the Methods, we performed several iterations of molecular docking, using both the mutagenesis results and the stability of the cofilin/F-actin complex in molecular dynamics (MD) simulations as quality metrics for a given model. The final model showed very good interaction with the filament and appeared to maintain a stable complex with the filament when subjected to MD simulation. The protein-protein binding interfaces explain mutations on both the cofilin and actin sides. Figure 3 shows details of our cofilin/F-actin model where mutations known to affect cofilin binding are shown in red and new contact residues predicted by the model are blue spheres. This model was used as a template to produce a fully decorated cofilactin filament consisting of 8 actin protomers and 6 cofilin molecules. MD simulations were then performed using this cofilactin model and we analyzed buried surface area as well as the statistics of atomic contacts, salt bridges and hydrogen bonds formed during the course of the simulation. We find an average of ~700 Å2 of buried surface area per bound cofilin as well as 4–5 salt bridges at any given point in the simulation. The molecular details of these interactions will be discussed in more detail in the following section.
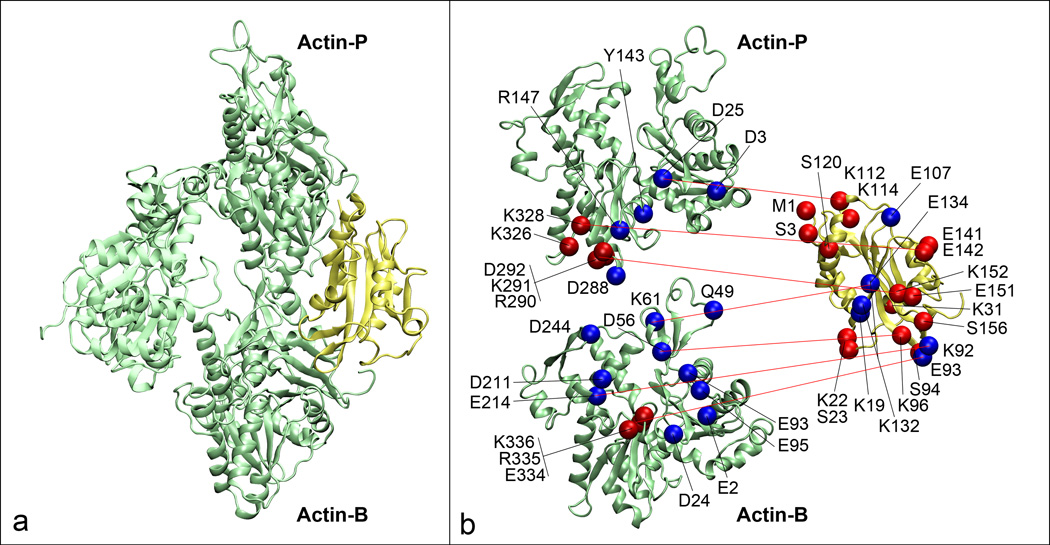
Figure 3.
Molecular details of the interaction between cofilin and the actin filament. (a) An actin trimer (green) where we have labeled the two actin protomers, Actin-P and Actin-B that contact cofilin (yellow). (b) Molecular details where lethal mutations for both proteins are shown as red spheres while other contact points predicted by the model are shown in blue and red lines indicate contact points. The two actin protomers have been spread apart to more clearly show the interactions.
Analysis of Known Cofilin Mutations
Having a detailed molecular model allows us to perform equally detailed analysis of the interactions between cofilin and the actin filament. In the following section we look at some of the key interactions between cofilin and actin and how these compare with experimental findings. We have adopted the convention where Actin-B/Actin-P refers to the actin protomer directly adjacent to the bound cofilin on barbed/pointed side. Further, the amino acid numbers and the locations of secondary structure elements all use the human cofilin sequence/structure as the reference (see Figure 2). In the sections below we work from the N-terminus to the C-terminus of cofilin, and each subsection has a heading corresponding to the region or key secondary structure elements that we analyze.
N terminus and Ser3
The deletion of the first five residues in the N-terminus (cof1-28) prevents binding to both G- and F-actin 4; 6. Similarly, the mutation of a known phosphorylation site, S3, to D/E also eliminates binding 4; 5; 8. We see significant interactions with this region of cofilin in our model with M1 making contact 100% of the time, often with D3 on Actin-P, and S3 contacting actin about 78% of the time, with no specific hydrogen bonding interactions.
Alpha 1/Alpha 2
The cof1-8 yeast mutation K23A/K24A/Y25A aligns to K22/S23/K31 in human cofilin. This is a temperature sensitive mutation, however it has not been tested in vitro for F-actin binding 4. K22 has 71% contact with the filament and forms intermittent salt bridges with D24 and E2 of Actin-B while S23 has 63% contact. Interestingly, the cof1-8 mutation occurs around a loop region where human cofilin has a short α-helix inserted. This point will be covered later in the Discussion.
Beta 3/Beta 4
The R80A/K82A mutation in yeast (cof1-16) is lethal and weakens binding to F-actin 4. Further, both the S94D mutation 6 and the K96Q mutation 8 appear to abrogate binding of human cofilin to the actin filament. In many sequence alignments, R80/K82 in yeast are aligned with K96/D98 in human cofilin 12; 27; 31; 32 however based on function and interactions we see, it seems more appropriate to equate these amino acids with S94/K96 in human cofilin even though they are not aligned. We see sustained interaction between both S94 and K96 with Actin-B, with K96 on cofilin forming a persistent salt bridge with D56 on actin, while D98 on cofilin has only 34% contact and no observable hydrogen bonding or salt bridge interactions. Alternatively, it may be that only R80A is necessary to lose F-actin binding.
Alpha 4
This alpha helix has the highest level of sequence conservation within the protein and it makes several key contacts with the actin filament. Yeast R96A/K98A (cof1-17) is a lethal mutation 4 that aligns with human K112/K114. Just two turns along this same helix, S120A also decreases F-actin binding as compared to WT cofilin 6. We see very good interactions between K112 and D25 on Actin-P with a salt bridge formed about 82% of the time. K114 also forms consistent contacts with Actin-P and makes periodic ionic bonds with E334. S120 only makes some contact with the filament (13% of the time), but S119 has 100% contact in our model. D122 makes 85% contact with both Q49 on Actin-B and Y143 on Actin-P.
Beta 5/Alpha 5 Loop
The yeast D123A/E126A (cof1-20) mutation is lethal and results in weaker filament binding 4. These yeast residues align with C139/E142, however the human protein has an additional glutamate at position 141. C139 does make consistent contacts in our model, but both E141 and E142 make salt bridges: E141 with K326 and K328 on Actin-P, and E142 with R147 and K328, also on Actin-P. It seems highly probable that E141 is matching the interactions of D123 in yeast while E142 matches those of E126, but this discrepancy underlines the difficulties of making simple assignments based on sequence alignments.
C-terminus
Yeast E134A/R135A/R138A (cof1-22) is a temperature sensitive mutation that leads to reduced F-actin binding 4. Based solely on sequence, these three yeast residues would align with E151/K152/G155 12; 27; 31; 32 and indeed we see good interactions between E151 and K291 on Actin-P, and K152 with D292 also on Actin-P. G155 obviously appears as a weak candidate for an interaction site based on our knowledge of protein biochemistry, however the neighboring residue S156 seems like a much more promising equivalent since it makes contacts more than 75% of the time and forms hydrogen bonds with residues D211 and D244 on Actin-P.
WT mutations
In addition to the many mutations that affect F-actin binding, there are numerous cofilin mutations that appear to have WT binding. As seen in Figure 1, the vast majority of these mutations fall outside of our contact regions with the filament. In this sense they do support our binding model as negative controls, but mutation of contact residues does not always have an effect on binding 33 and thus we cannot place too much weight in this correspondence.
New Putative Cofilin Contacts
Although there has been extensive mutagenesis performed on cofilin, our model has several points of interaction that appear to be novel, namely K19, K92, E93, E107, K132 and E134. K19 and K22 are part of a strongly conserved KXXK motif that forms consistent contacts with the filament. K22 was previously identified, but K19, which aligns to K20A in yeast, looked to be like wild type (cof1-6 D18A/K20A), but was never directly tested for filament binding. In our model K19 forms a salt bridge with E93 on Actin-B, and the E93A/E95A mutation on actin eliminates cofilin binding 9. Similarly, we see interactions between K92 and E214 on Actin-B, as well as E93 with R335/K336 on Actin-B. Although K92 and E93 have not been mutated, the E334A/R335A/K336A mutation on actin abrogates cofilin binding 9. E107 on cofilin interacts with both R147 and K328 on Actin-P, and the K326A/K328A actin mutation affects cofilin binding 9, but we also see E141/E142 on cofilin interacting with this same site. Finally, K132 and E134 show interactions with D56/E93 and K61 respectively, both on Actin-B. Interestingly, these two residues flank H133 that was tested for its ability to confer pH dependent severing 24, however these interaction sites have not been directly tested.
Unmatched Cofilin Mutations
Our model matches the majority of mutagenesis results, however there are a few cofilin mutations that are not readily explained. Yeast D10A/E11A (cof1-5) and D34A/K36A/E38A (cof1-9) align to human D9/G10 and E42/K44/N46 respectively, and both are temperature sensitive mutations 4. Neither region on cofilin shows contact with the filament in our model, but these mutations also show no change in F-actin binding in in vitro assays suggesting some other mechanism may be at play. Consistent with this idea, a recent study found that the cof1-5 and cof1-9 alleles bound to actin but were defective in binding Srv2 34, a key cofilin regulator, and this likely explains the phenotype observed in earlier experiments. Another mutation, D68A/E70A/E72A in yeast (cof1-14), is lethal and lowers the stability cofilin, but only slightly decreases F-actin binding 4. This mutation maps to D86/T88/E90 on human cofilin, which does not make contact in our model, but eliminating the internal salt bridge that is seen between D86 and R32 could explain the lowered protein stability.
Analysis of Actin Mutations
Apart from changes to cofilin, there has also been mutagenesis performed with actin 35. This collection of mutations were tested for cofilin activity with a mixture of results 9. Some of the mutations (act1-107, act1-130, act1-127, act1-128 and act1-108) failed to show interaction with cofilin or any other actin binding protein, suggesting that they may be altering the structure or stability of the actin. However, three mutations, act1-103, act1-106 and act1-126, exhibited binding defects specific for cofilin. The act1-103 mutation is E334A/R335A/K336A, and as described above we see ionic bonds formed between both R335 and K336 with E93 on cofilin. Similarly, the act1-106 mutation involves R290A/K291A/E292A, and we observe salt bridges formed between K291 and E151 on cofilin as well as E292 with K152 on cofilin. Finally, the act1-126 mutation site has multiple interactions. This mutation is K326A/K328A and we see several interchangeable salt bridges between K326A and E141/E151 on cofilin plus K328 and E107/E141/E142 on cofilin. Several salt bridges are also formed with E93 on Actin-B and D288 in Actin-P, but since these amino acids are part of the act1-130 and act1-107 mutations, respectively, we cannot draw any firm conclusions. As a potential new contact site, we see significant interactions between R147 on Actin-P with both E107 and E142 on cofilin.
Comparison with Structural Data
Our model complex obviously matches well with mutagenesis results, but it also is in very good agreement with available structure information. Figure 4 shows a comparison of our cofilin/F-actin complex with the cryo-EM density from Galkin et al. 25 as well the twinfilin/G-actin crystal structure 27. In Figure 4a we see that our molecular model fits extremely well within the envelope of the EM density map. This is perhaps only weak support for our model given the limited resolution of the density map, however failure to agree with this data would raise serious questions about the validity of our model. It is also interesting that although is a complex with monomeric actin, the twinfilin/G-actin complex also shows good correspondence with our docked model when put in the context of the full filament (Figure 4b). There are some minor clashes between twinfilin and the Actin-B protomer, but since this crystal is with G-actin, this is not surprising. The contacts with Actin-P are very similar in nature and the contact points of twinfilin match well with the G-sites on cofilin. This is particularly true for the contacts on α4 where the interactions we see with K112, K114, S119 and S120 are largely recapitulated in the crystal complex. This strongly supports the idea that twinfilin and cofilin interact with actin through similar mechanisms.
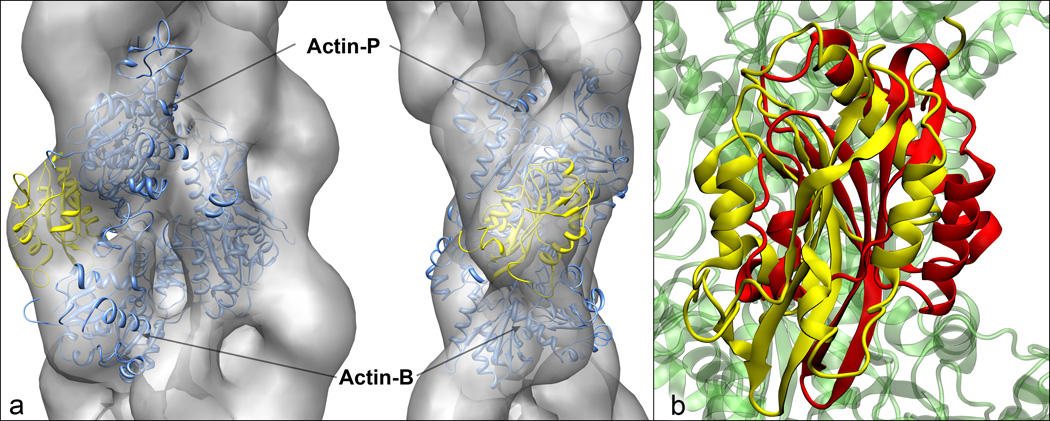
Figure 4.
Comparison of our cofilin/F-actin complex with (a) the EM volume reconstruction of Galkin et al. 25 and (b) the twinfilin/G-actin structure of Paavilainen et al.27. For the EM comparison in (a), our model depicts the actin trimer in blue and cofilin in yellow, and the position and orientation was determined by optimizing the fit between our model and the electron density map using Chimera 41. In (b), our model cofilin is shown in yellow and twinfilin in red, and the relative orientation was determined by fitting the G-actin in the twinfilin structure to our Actin-P protomer.
DISCUSSION
We have developed a molecular model of human cofilin bound to F-actin by utilizing a combination of published experimental data and iterative docking and molecular dynamics studies. Our model explains numerous mutagenesis results on both cofilin and actin, and the binding site matches extremely well with available structural data.
One feature that our results highlight is the challenge associated with aligning the sequences of various cofilin homologs. Sequence conservation is moderate within this family of proteins (~32%), but apart from amino acid substitutions, there are numerous insertions and deletions in the protein, making it very difficult to translate point mutations from one system to another. Human and yeast cofilin are great examples of this since the human protein has two insertions in the N-terminal portion of the protein (Figure 2). Based on the protein structures, we would place these insertions such that the region K22-R32 in human cofilin would align with K23-K26 in yeast, and G58 to D66 in human would correspond to D47-S49 in yeast. Since the function of these two proteins is very similar, one might expect that these two insertions points are not critical for filament binding, however this does not appear to be the case. First, the cof1-8 allele (K23A/K24A/Y25A) spans the first insertion site and produces a temperature sensitive phenotype in yeast 4. Second, an attempt to insert fluorescent proteins into the yeast protein produced a loss of function phenotype at the first insertion site, but resulted in a function protein at the second site 30. Although our manual alignment fixes discrepancies in these insertions, there is still some ambiguity in interpreting mutations made in other regions of the protein, particularly in some of the central beta sheets such as S94 or in highly dynamic regions of the proteins such as E142. In this sense, a given sequence alignment does not provide a one-to-one correspondence, but simply is a guide to understanding the relationship between sequence, structure and function.
A key interaction point between cofilin and the filament is the DNase-I loop (D-loop) of the actin protomer (residues 40–51). Cryo-EM observations 25, cross-linking studies 36, and molecular dynamics simulations 20 have provided significant evidence that the D-loop, as well as other portions of subdomain-2 (residues 33–69), become disordered upon cofilin binding, thus breaking the contacts between protomers along the long-pitch helix. We see significant interactions with subdomain-2 in our model. The α-helix containing E93/E95 in Actin-B makes sustained ionic bonds with cofilin, and this was one of the regions of actin that was found to be remodeled in previous MD simulations 20. We also see hydrogen bonding interactions with Q49 at the C-terminal end of the D-loop and D56/K61 directly below the D-loop. These interactions appear to alter the dynamics and conformation of the D-loop enough that its interactions with subdomain-1 of Actin-P are reduced by about 10% over the course of our MD simulation, consistent with the prior results of Pfaendtner et al. 20. The length of our simulation is clearly a limiting factor and we have not had sufficient time to observe the twisting of the filament that would further disrupt these interactions, but this is still consistent with the previous observations mentioned above.
The phosphorylation of S3 on cofilin is a key method of regulating cofilin activity. In our model S3 makes consistent contacts with the actin filament, but we do not see any specific hydrogen bonding interactions. Interestingly, through a series of MD simulations Frantz et al. found that the N-terminus of cofilin is significantly rearranged in the phosphorylated form and pS3 forms salt bridges with K126/K127 on cofilin 24. We see that the α-helix containing K126/K127 makes significant contact with the filament, and although mutation of these two basic residues (R109A/R110A in yeast cof1-19) has no discernable phenotype in yeast 4, the interaction of pS3 with K126/K127 would cause the N-terminus of the cofilin to block several other interaction points along the helix (e.g. K112, K114, S120) and this would likely be enough to eliminate filament binding.
One outstanding question that remains is the basis of pH regulation of cofilin and the fact that mammalian cofilin has a pH-dependent severing activity while yeast cofilin does not 1; 37. Human cofilin has a single histidine, H133, and it appeared to be a prime candidate for this regulation since it aligns to a threonine in yeast and was at a filament contact site, however the H133A mutant has been shown to have pH-sensitive severing activity that is unchanged from the WT protein 24. We have performed pKa analysis in the hopes of finding another potential regulatory site, but we could not identify other regulatory site and this question will need to be left for future studies.
In summary, we have derived an independent model for cofilin bound to the actin filament that agrees well with structural and biochemical data, and further provides insight into filament remodeling and regulation by phosphorylation.
MATERIALS AND METHODS
Secondary Structure Alignment
Multiple sequence alignments (MSA) using standard alignment tools do not account for secondary or tertiary structure and this has been shown to lead to incorrect alignments for ADF/cofilin family proteins 30. Here, a secondary structure MSA was built from a selection of relevant ADF/cofilin family proteins as well as ADF Homology Domain (ADF-H) homologs. The VMD plugin tool, MultiSeq 29, was used for the initial alignment of sequences. The structures used were for human cofilin (PDB 1Q8G, GI:50513339, also the seed structure), S. cerevisiae cofilin (PDB 1CFY, GI: 2098523), S. pombe cofilin (PDB 2I2Q, GI:118138074), chicken cofilin (PDB 1TVJ, GI:78100779), A. thaliana ADF (PDB 1F7S, GI:11513711), A. polyphaga actotrophin (PDB 1CNU, GI:5107573), and human coactosin (PDB 1T3Y, GI:75765235). Manual corrections to the alignment were necessary particularly at the anchor points near the loops (between human R21 to R32 as well as V57 to D65), which would not be done correctly based on sequence alone. Porcine cofilin was used in some mutagenesis work, but since there is no solved structure and it has nearly 100% sequence identity with human cofilin (with only a single C108S substitution), we did not include it in our alignment.
Docking and Refinement Simulations
In order to prepare for molecular docking studies, we first constructed an 8-protomer actin filament using the recent model from Oda et al. (PDB 2ZWH) 38. The filament was simulated using molecular dynamics (MD) as detailed in the following section. A series of 6 filament structures were then extracted at 0, 5, 10, 15, 20 and 25 ns for use in docking studies. For cofilin, instead of performing additional MD simulations, we were able to use 10 models from the NMR structure of human cofilin I (PDB 1Q8G) 12.
Docking was carried out using RosettaDock 39. Each combination of filament and cofilin structure was used resulting in 60 different pairs. An initial unbiased docking using default parameters and random rotation of both binding partners was performed; thus the cofilin was given full freedom to bind anywhere along the actin filament. All bound complexes were ranked by their docking score and their general correspondence to the cryo-EM structure of the decorated filament, and the top 20 scoring models were chosen as the starting structures for refinement dockings. In this next round of docking, more restrictive parameters (e.g. allowing 10 A translation, 30° rotation from the starting structure) were applied. We generated a total of 30,000 model structures and the top cofilin models that had the most buried surface area and the greatest number of F- and G- sites (S3, K96/K98, K112/K114, C139/E142 and E151/K152) in contact with the actin filament were chosen for each of the six binding sites along the filament. The 25 ns filament snapshot had the best representation of these top-binding models and became the basis for our fully decorated filament.
In order to test the stability of the models, the fully decorated filament was simulated using MD for more than 80 ns. Using contact analysis and RMSD, two of the six models appeared to be more stable than their counterparts (RMSD of around 3 A) and maintained better contact with the actin filament. These two conformations ended up with very similar structures and contacts, and formed the basis for the final model, and this structure was replicated at all six binding sites on the filament. This new, decorated filament was used for analysis and further MD simulation as detailed below
Molecular Dynamics Simulations
The bare and decorated filaments were simulated with all atom molecular dynamics. Simulations were performed in an isothermal-adiabatic (NpT) ensemble with the CHARMM27 force field and TIP3P water model, under periodic boundary conditions. Each actin protomer had ADP as the bound nucleotide. Bonded interactions were computed every 2 fs using SHAKE, short-ranged non-bonded electrostatic and van der Waals (with a 10 A cutoff and smooth switching function beginning at 8.5 A, and pair-list distance of 11.5 A). Long-ranged, full electrostatic interactions were computed every 4 fs with the Particle Mesh Ewald method with grid points at least 1/A in each dimension. Using Langevin Dynamics, NpT conditions were maintained at 1 atm and temperature of 300 K with a damping coefficient of 1/ps and a Langevin piston oscillation period of 200 fs and decay of 100 fs for the production run (200 fs and 500 fs respectively for both heating and equilibration). Following minimization, the system was heated in 50 K steps with 40 ps per step and restraints on the Cα atoms. At 300 K, the restraints were removed and the system was allowed to equilibrate for approximately 1 ns before the production run was started.
All molecular graphics were produced using VMD 40, except for Figure 4a which was generated with Chimera 41.
Highlights.
-
>
We construct an atomic model for the F-actin/cofilin complex
-
>
Resultant structure matches most known mutations and agrees with structural data
-
>
New amino acids that may be important to binding were identified
Supplementary Material
Acknowledgements
We would like to thank Ed Egeleman for sharing their EM data for the cofilin decorated actin filament. This work was supported in part by a grant from the National Institutes of Health to D.S. (GM067246).
Abbreviations
- Actin-B/Actin-P
actin protomer adjacent to bound cofilin on barbed/pointed side.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
SUPPLEMENTAL DATA
A pdb file for the structure shown in Figure 3 is provided.
REFERENCES
- 1.Hawkins M, Pope B, Maciver SK, Weeds AG. Human actin depolymerizing factor mediates a pH-sensitive destruction of actin filaments. Biochemistry. 1993;32:9985–9993. doi: 10.1021/bi00089a014. [DOI] [PubMed] [Google Scholar]
- 2.Hayden SM, Miller PS, Brauweiler A, Bamburg JR. Analysis of the interactions of actin depolymerizing factor with G- and F-actin. Biochemistry. 1993;32:9994–10004. doi: 10.1021/bi00089a015. [DOI] [PubMed] [Google Scholar]
- 3.Bernstein BW, Bamburg JR. ADF/cofilin: a functional node in cell biology. Trends Cell Biol. 2010;20:187–195. doi: 10.1016/j.tcb.2010.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lappalainen P, Fedorov EV, Fedorov AA, Almo SC, Drubin DG. Essential functions and actin-binding surfaces of yeast cofilin revealed by systematic mutagenesis. Embo J. 1997;16:5520–5530. doi: 10.1093/emboj/16.18.5520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Moriyama K, Iida K, Yahara I. Phosphorylation of Ser-3 of cofilin regulates its essential function on actin. Genes Cells. 1996;1:73–86. doi: 10.1046/j.1365-2443.1996.05005.x. [DOI] [PubMed] [Google Scholar]
- 6.Moriyama K, Yahara I. The actin-severing activity of cofilin is exerted by the interplay of three distinct sites on cofilin and essential for cell viability. Biochem J. 2002;365:147–155. doi: 10.1042/bj20020231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Moriyama K, Yonezawa N, Sakai H, Yahara I, Nishida E. Mutational analysis of an actin-binding site of cofilin and characterization of chimeric proteins between cofilin and destrin. J Biol Chem. 1992;267:7240–7244. [PubMed] [Google Scholar]
- 8.Pope BJ, Gonsior SM, Yeoh S, McGough A, Weeds AG. Uncoupling actin filament fragmentation by cofilin from increased subunit turnover. J Mol Biol. 2000;298:649–661. doi: 10.1006/jmbi.2000.3688. [DOI] [PubMed] [Google Scholar]
- 9.Rodal AA, Tetreault JW, Lappalainen P, Drubin DG, Amberg DC. Aip1p interacts with cofilin to disassemble actin filaments. J Cell Biol. 1999;145:1251–1264. doi: 10.1083/jcb.145.6.1251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fedorov AA, Lappalainen P, Fedorov EV, Drubin DG, Almo SC. Structure determination of yeast cofilin. Nat Struct Biol. 1997;4:366–369. doi: 10.1038/nsb0597-366. [DOI] [PubMed] [Google Scholar]
- 11.Galkin VE, Orlova A, Lukoyanova N, Wriggers W, Egelman EH. Actin depolymerizing factor stabilizes an existing state of F-actin and can change the tilt of F-actin subunits. J Cell Biol. 2001;153:75–86. doi: 10.1083/jcb.153.1.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pope BJ, Zierler-Gould KM, Kuhne R, Weeds AG, Ball LJ. Solution structure of human cofilin: actin binding, pH sensitivity, and relationship to actin-depolymerizing factor. J Biol Chem. 2004;279:4840–4848. doi: 10.1074/jbc.M310148200. [DOI] [PubMed] [Google Scholar]
- 13.McGough A, Pope B, Chiu W, Weeds A. Cofilin changes the twist of F-actin: implications for actin filament dynamics and cellular function. J Cell Biol. 1997;138:771–781. doi: 10.1083/jcb.138.4.771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Andrianantoandro E, Pollard TD. Mechanism of actin filament turnover by severing and nucleation at different concentrations of ADF/cofilin. Mol Cell. 2006;24:13–23. doi: 10.1016/j.molcel.2006.08.006. [DOI] [PubMed] [Google Scholar]
- 15.Blanchoin L, Pollard TD. Mechanism of interaction of Acanthamoeba actophorin (ADF/Cofilin) with actin filaments. J Biol Chem. 1999;274:15538–15546. doi: 10.1074/jbc.274.22.15538. [DOI] [PubMed] [Google Scholar]
- 16.De La Cruz EM. Cofilin binding to muscle and non-muscle actin filaments: isoform-dependent cooperative interactions. J Mol Biol. 2005;346:557–564. doi: 10.1016/j.jmb.2004.11.065. [DOI] [PubMed] [Google Scholar]
- 17.Grintsevich EE, Benchaar SA, Warshaviak D, Boontheung P, Halgand F, Whitelegge JP, Faull KF, Loo RR, Sept D, Loo JA, Reisler E. Mapping the cofilin binding site on yeast G-actin by chemical cross-linking. J Mol Biol. 2008;377:395–409. doi: 10.1016/j.jmb.2007.12.073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.McCullough BR, Blanchoin L, Martiel JL, De la Cruz EM. Cofilin increases the bending flexibility of actin filaments: implications for severing and cell mechanics. J Mol Biol. 2008;381:550–558. doi: 10.1016/j.jmb.2008.05.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Michelot A, Berro J, Guerin C, Boujemaa-Paterski R, Staiger CJ, Martiel JL, Blanchoin L. Actin-filament stochastic dynamics mediated by ADF/cofilin. Curr Biol. 2007;17:825–833. doi: 10.1016/j.cub.2007.04.037. [DOI] [PubMed] [Google Scholar]
- 20.Pfaendtner J, De La Cruz EM, Voth GA. Actin filament remodeling by actin depolymerization factor/cofilin. Proc Natl Acad Sci U S A. 2010;107:7299–7304. doi: 10.1073/pnas.0911675107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wriggers W, Tang JX, Azuma T, Marks PW, Janmey PA. Cofilin and gelsolin segment-1: molecular dynamics simulation and biochemical analysis predict a similar actin binding mode. J Mol Biol. 1998;282:921–932. doi: 10.1006/jmbi.1998.2048. [DOI] [PubMed] [Google Scholar]
- 22.Carlsson AE. Stimulation of actin polymerization by filament severing. Biophys J. 2006;90:413–422. doi: 10.1529/biophysj.105.069765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.De La Cruz EM, Sept D. The kinetics of cooperative cofilin binding reveals two states of the cofilin-actin filament. Biophys J. 2010;98:1893–1901. doi: 10.1016/j.bpj.2010.01.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Frantz C, Barreiro G, Dominguez L, Chen X, Eddy R, Condeelis J, Kelly MJ, Jacobson MP, Barber DL. Cofilin is a pH sensor for actin free barbed end formation: role of phosphoinositide binding. J Cell Biol. 2008;183:865–879. doi: 10.1083/jcb.200804161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Galkin VE, Orlova A, VanLoock MS, Shvetsov A, Reisler E, Egelman EH. ADF/cofilin use an intrinsic mode of F-actin instability to disrupt actin filaments. J Cell Biol. 2003;163:1057–1066. doi: 10.1083/jcb.200308144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.McLaughlin PJ, Gooch JT, Mannherz HG, Weeds AG. Structure of gelsolin segment 1-actin complex and the mechanism of filament severing. Nature. 1993;364:685–692. doi: 10.1038/364685a0. [DOI] [PubMed] [Google Scholar]
- 27.Paavilainen VO, Oksanen E, Goldman A, Lappalainen P. Structure of the actin-depolymerizing factor homology domain in complex with actin. J Cell Biol. 2008;182:51–59. doi: 10.1083/jcb.200803100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kim C, Lee B. Accuracy of structure-based sequence alignment of automatic methods. BMC Bioinformatics. 2007;8:355. doi: 10.1186/1471-2105-8-355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Roberts E, Eargle J, Wright D, Luthey-Schulten Z. MultiSeq: unifying sequence and structure data for evolutionary analysis. BMC Bioinformatics. 2006;7:382. doi: 10.1186/1471-2105-7-382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lin MC, Galletta BJ, Sept D, Cooper JA. Overlapping and distinct functions for cofilin, coronin and Aip1 in actin dynamics in vivo. J Cell Sci. 2010;123:1329–1342. doi: 10.1242/jcs.065698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bamburg JR, McGough A, Ono S. Putting a new twist on actin: ADF/cofilins modulate actin dynamics. Trends Cell Biol. 1999;9:364–370. doi: 10.1016/s0962-8924(99)01619-0. [DOI] [PubMed] [Google Scholar]
- 32.Gorbatyuk VY, Nosworthy NJ, Robson SA, Bains NP, Maciejewski MW, Dos Remedios CG, King GF. Mapping the phosphoinositide-binding site on chick cofilin explains how PIP2 regulates the cofilin-actin interaction. Mol Cell. 2006;24:511–522. doi: 10.1016/j.molcel.2006.10.007. [DOI] [PubMed] [Google Scholar]
- 33.Bogan AA, Thorn KS. Anatomy of hot spots in protein interfaces. J Mol Biol. 1998;280:1–9. doi: 10.1006/jmbi.1998.1843. [DOI] [PubMed] [Google Scholar]
- 34.Quintero-Monzon O, Jonasson EM, Bertling E, Talarico L, Chaudhry F, Sihvo M, Lappalainen P, Goode BL. Reconstitution and dissection of the 600-kDa Srv2/CAP complex: roles for oligomerization and cofilin-actin binding in driving actin turnover. J Biol Chem. 2009;284:10923–10934. doi: 10.1074/jbc.M808760200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wertman KF, Drubin DG, Botstein D. Systematic mutational analysis of the yeast ACT1 gene. Genetics. 1992;132:337–350. doi: 10.1093/genetics/132.2.337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kim E, Wriggers W, Phillips M, Kokabi K, Rubenstein PA, Reisler E. Cross-linking constraints on F-actin structure. J Mol Biol. 2000;299:421–429. doi: 10.1006/jmbi.2000.3727. [DOI] [PubMed] [Google Scholar]
- 37.Pavlov D, Muhlrad A, Cooper J, Wear M, Reisler E. Severing of F-actin by yeast cofilin is pH-independent. Cell Motil Cytoskeleton. 2006;63:533–542. doi: 10.1002/cm.20142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Oda T, Iwasa M, Aihara T, Maeda Y, Narita A. The nature of the globular-to fibrous-actin transition. Nature. 2009;457:441–445. doi: 10.1038/nature07685. [DOI] [PubMed] [Google Scholar]
- 39.Gray JJ, Moughon S, Wang C, Schueler-Furman O, Kuhlman B, Rohl CA, Baker D. Protein-protein docking with simultaneous optimization of rigid-body displacement and side-chain conformations. J Mol Biol. 2003;331:281–299. doi: 10.1016/s0022-2836(03)00670-3. [DOI] [PubMed] [Google Scholar]
- 40.Humphrey W, Dalke A, Schulten K. VMD: Visual molecular dynamics. Journal of Molecular Graphics. 1996;14:33–38. doi: 10.1016/0263-7855(96)00018-5. [DOI] [PubMed] [Google Scholar]
- 41.Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE. UCSF Chimera--a visualization system for exploratory research and analysis. J Comput Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.