Abstract
The matricellular glycoprotein Secreted Protein Acidic and Rich in Cysteine (SPARC) plays an important role in the regulation of cell adhesion and proliferation as well as in tumorigenesis and metastasis. Earlier, we reported that, in addition to its potent anti-angiogenic functions, SPARC also induces apoptosis in medulloblastoma cells, mediated by autophagy. We therefore sought to investigate the underlying molecular mechanism through which SPARC inhibits migration and invasion of Daoy medulloblastoma cells, both in vitro and in vivo. For this study, we used SPARC-overexpressing stable Daoy medulloblastoma cells. SPARC overexpression in Daoy medulloblastoma cells inhibited migration and invasion in vitro. Additionally, SPARC overexpression significantly suppressed the activity of Rho, Rac and Cdc42, which all regulate the actin cytoskeleton. This suppression was accompanied by an increase in the phosphorylation of Src at TYR-416, which led to a loss of actin stress fibers and focal contacts and a decrease in the phosphorylation level of cofilin. The reduced phosphorylation level of cofilin, which is indicative of receding Rho function, in turn led to inhibition of active Rho A. To confirm the role of SPARC in inhibition of migration and invasion of Daoy medulloblastoma cells, we transfected parental and SPARC-overexpressing Daoy cells with a plasmid vector carrying siRNA against SPARC. Transfection with SPARC siRNA reversed Src-mediated disruption of the cytoskeleton organization as well as dephosphorylation of cofilin and activation of Rho A. Taken together, these results establish SPARC as an effector of Src-induced cytoskeleton disruption in Daoy medulloblastoma cells, which subsequently led to decreased migration and invasion.
Keywords: SPARC, Src, Migration, Invasion, Rho, Rac, Cdc42
1. Introduction
Medulloblastoma is the most prevalent malignant brain tumor of childhood, accounting for up to 20% of pediatric brain tumors arising in the cerebellum [1,2]. These tumors are known to metastasize outside the central nervous system [3–6] and are likely to spread into bones, bone marrow, lymph nodes, liver, and lungs [7,8]. Compared with other childhood tumors, the median survival rate for medulloblastoma is poor, with a five-year survival rate of less than 50%. Surgical resection followed by craniospinal irradiation and chemotherapy is the therapy of choice. Although there are encouraging results, intermittent late recurrence of these tumors is well documented, even after the most rigorous treatments [9,10]. Most of these incidences emerge within 18 months of treatment, with a mean survival period of 2 years [4]. The highly invasive nature of this neoplasm is a major reason for the low survival rate of patients with tumor recurrence.
Metastasis is a complex process mainly dependent on cell adhesion to the extracellular matrix (ECM) and basement membrane, and it involves various cell signaling pathways that permit cancer cells to restructure the ECM, leading to cancer cell invasion, migration, and establishment at a new site. Cell invasion is mediated by both extracellular and intracellular factors. Invasion is dependent on the specific, dynamic interaction of different cell surface receptors with the ECM [11–13].
The matricellular proteins are extracellular proteins, which do not contribute structurally to the extracellular milieu as the classical extracellular matrix proteins do but rather modulate interactions between the extracellular matrix and cells. One of these matricellular proteins is Secreted Protein Acidic and Rich in Cysteine (SPARC), also known as osteonectin or BM-40; SPARC is a 34 kDa, calcium-binding glycoprotein shown to associate with the cell membrane and membrane receptors [14]. SPARC is known to not only modulate cell–cell and cell–matrix interactions, but also to influence de-adhesive and growth regulatory properties [15]. In cancers, SPARC may elicit different actions, showing the complexity of the protein [16,17]. In certain types of cancers, such as melanomas and gliomas, SPARC is associated with a highly aggressive tumor phenotype, whereas in others, mainly ovarian, neuroblastomas, colorectal cancers and PNET tumors, SPARC may function as a tumor suppressor [15,18]. The capacity of SPARC to bind to several resident proteins of the ECM, to modulate growth factor efficacy, to affect the expression of matrix metalloproteinases (MMPs) [18], and to alter cell shape as a counter-adhesive factor supports the idea that SPARC acts to regulate cell interaction with the extracellular environment during development and in response to injury in vivo. SPARC can lead to a cytoskeletal rearrangement essential for cell transmigration by binding to VCAM-1, a surface molecule mediating leukocyte adhesion [19], and it can also alter cell shape by an important counter-adhesive function achieved by dissolution of focal adhesion complexes and reorganization of actin stress fibers [20].
Organization of actin filaments is controlled by the Rho family of small GTPases, Rho, Rac, and Cdc42[21]. Among them, Rho regulates the assembly of actin stress fibers and focal contacts through activation of downstream effectors, like Src. Src activation results in tyrosine phosphorylation of numerous substrates and simultaneous activation of several signaling pathways, including the Ras/MEK and PI3K pathways [22]. ERK activity has been implicated in cell migration and directly phosphorylates myosin light chain kinase, suggesting a role for ERK in the regulation of cytoskeleton and focal contact dynamics [23,24]. Viral Src-induced cytoskeleton disruption results from inactivation of Rho-mediated stress fiber assembly, and MEK contributes to this inactivation. Stimulation of a Rho-GAP (GTPase-activating protein) activity was reported upon v-Src activation in chicken embryo fibroblasts, suggesting that v-Src-induced cytoskeleton disruption is linked to inhibition of Rho activity [25].
PI3-K is a heterodimer made up of a regulatory subunit (p85) and a catalytic subunit (110 kDa). The capability of the regulatory subunit to associate with other proteins links PI3-K to a distinct signaling mechanism required for proliferation, adhesion, and motility[26]. It has been shown to act downstream of FAK in cell migration and survival. PI3-K and small Rho GTPases, such as Cdc42 and Rac1, are key effectors that regulate cell migration by means of dynamic changes in the actin cytoskeleton [27,28]. Rho-Rac-Cdc42 is also known to affect matrix metalloproteinase-9 (MMP-9) expression and activity [29]. MMP-9 expression is associated with migration and invasion of medulloblastoma in vitro and in vivo. In an earlier study, we demonstrated that SPARC overexpression inhibits MMP-9 expression in medulloblastoma cells[18]. Here, we have assessed the effect of SPARC overexpression in medulloblastoma cells in relation to migration and invasion.
2. Materials and Methods
2.1 Daoy cell culture
Daoy cells were obtained from ATCC (Manassas, VA) and cultured in Advanced MEM supplemented with 5% fetal bovine serum, 2 mM/L L-glutamine, 100 units/mL penicillin, and 100 µg/mL streptomycin. Cells were maintained in a humidified atmosphere containing 5% CO2 at 37°C.
2.2 Construction of pcDNA3.1-SPARC and transfection of Daoy cells
An 1100-bp cDNA fragment of human SPARC was amplified by PCR using synthetic primers and sub-cloned into a pcDNA3.1 vector (Invitrogen, San Diego, CA) in the “sense” orientation. Daoy cells were transfected with either a full length cDNA SPARC containing vector or with an empty vector using FuGene HD (Roche, Indianapolis, IN) as described earlier [30]. Stable transfectants were selected with cloning cylinders after 3–4 weeks in medium containing G418. Wild-type Daoy cells were termed Daoy parental (Daoy-P) and the stable cell lines expressing SPARC were designated as Daoy-SP; Daoy-EV is the cell line transfected stably with the empty vector.
2.3 siRNA design and transient transfection
SPARC siRNA sequences were designed with the help of a siRNA designer program (Imgenex, Sorrento Valley, CA). The siRNA was complementary to an exonic sequence of the target mRNA and compatible with the pcDNA3.1 vector (Invitrogen, San Diego, CA). The following siRNA sequence was used to construct the SPARC siRNA and designated as SP-siRNA 5'-TCGAGGGTGTGCAGCAATGACAACAAGAGTCGTCGTTGTTGTCATTGCTGCACACCG-3’. A control vector containing siRNA with a scrambled sequence was constructed and designated as control siRNA. We used the following scrambled sequence: 5'-CACGGAGGTTGCAAAGAATAATCGATTATTCTTTGCAA CCTCCGTGC-3’.
2.4 Plasmids and transfection
The plasmids encoding fusion proteins for the glutathione S-transferase (GST)-rhotekin Rho binding domain fusion proteins (pGEX-TRBD) were a kind gift from Dr Martin A. Schwartz (Cardiovascular Research Center, Mellon Prostate Cancer Institute, Departments of Microbiology and Biomedical Engineering, University of Virginia, Charlottesville, VA). pGEX-Rac1 (Addgene Plasmid 12200), pGEX-Cdc42 (Addgene Plasmid 12201), and siRNA against SPARC were used in this study. FuGene HD (Roche, Indianapolis, IN; 1 µg plasmid: 3 µL of FuGene HD) was used according to the manufacturer’s instructions. Following transfection, the cells were cultured in Advanced MEM-5% FBS for 24 hours, rinsed once with PBS, and cultured for an additional 16 hours in serum-free DMEM/F12 50/50. Conditioned medium and cell lysates were collected, and MMP-9 and SPARC levels were determined by gelatin zymography and western blot analysis, respectively.
2.5 Immunocytochemistry
We used a previously described protocol with minor changes [31]. Briefly, the cells were cultured on 8-well chamber slides and fixed with 4% paraformaldehyde (w/v) in PBS, permeabilized with 0.1% Triton X-100 (w/v) in PBS and blocked with 1% BSA (w/v) in PBS for 1 hour at 4°C. Cells were incubated overnight at 4°C with anti-SPARC antibody followed by corresponding HRP-conjugated secondary antibody for 1 hour, and protein expression was detected using 3,3-diaminobenzidine solution (Sigma, St. Louis, MO). Sections were counterstained with hematoxylin, and negative control slides were obtained by nonspecific IgG. Sections were washed and mounted with anti-fade mounting solution (Invitrogen, San Diego, CA) and analyzed with an inverted microscope.
2.6 Western blotting
Western blot analysis was performed as described previously [32]. Briefly, 36 hours after seeding, the Daoy-P, Daoy-EV, and Daoy-SP cells were collected and lysed in RIPA buffer. Equal amounts of protein were resolved on SDS-PAGE and transferred onto a PVDF membrane. The blot was blocked with 5% non-fat dry milk and probed overnight with primary antibodies followed by HRP-conjugated secondary antibodies. An ECL system was used to detect chemiluminescent signals. All blots were re-probed with GAPDH antibody to confirm equal loading.
2.7 RT-PCR
Daoy-P, Daoy-EV and Daoy-SP cells were grown for 36 hours. Total RNA was extracted from these cells and cDNA synthesized using poly-dT primers as described earlier [33]. PCR was performed using the following primers: SPARC, 5’-GGAAGAAACTGTGGCAGAGG-3’ (sense) and 5’-ATTGCTGCACACCTTCTCAA-3’ (antisense); GAPDH, 5’-TGAAGGTCGGAGTCAACGGATTTGGT-3’ (sense) and 5’-CATGTGGGCCATGAGGTCCACCAC-3’ (antisense). Quantification of SPARC mRNA levels was determined based on densitometry.
2.8 GTPase activity assays
pGEX-Rac1, pGEX-Cdc42, and pGex-TRBD plasmids were expressed in Escherichia coli, and the fusion proteins GST-Rac1, GST-Cdc42, and GST-TRBD were affinity purified. Glutathione agarose beads containing 20–30 µg of GST-TRBD or GST-PAK PBD were incubated with lysates of Daoy-P, Daoy-EV, or Daoy-SP for 60 minutes. The proteins that were precipitated on the agarose beads were then washed three times with wash buffer and eluted by incubating in loading buffer at 95°C for 10 minutes. The samples were subjected to SDS-PAGE and immunoblotted with corresponding antibodies.
2.9 Cell adhesion assays
Cell adhesion assay on different matrices was performed as described earlier [34]. Briefly, 1×104 cells were plated on a 96-well plate previously coated with fibronectin (2 µg/mL), laminin (2µg/mL), vitronectin (2 µg/mL), or bovine serum albumin (100 µg/mL). After 1 hour, the matrix was removed and DAOY or Daoy-SP cells were seeded and allowed to attach on the coated matrices for an additional 1 hour. The attached cells were washed with PBS three times. Cells that adhered to the ECM components were quantified by staining with Hema-3 stain (Fisher Diagnostics, VA). Images of the stained cells from different fields were taken under a light microscope (Olympus IX-71).
2.10 Matrigel invasion assay
Matrigel invasion was carried out as described previously [34]. Briefly, 2×105 (Daoy and Daoy-SP) cells were plated on Matrigel-coated transwell inserts and placed in a 12-well plate containing serum medium. After overnight incubation, invaded cells (towards the bottom of the membrane) were stained with Hema-3 stain. The invading cells on the reverse side of the filter were stained, photographed and counted. Five different fields per filter were analyzed, and all experiments were done in triplicate.
2.11 Wound healing migration
Wound healing migration assay was performed as described earlier [35]. Briefly, the cells (Daoy and Daoy-SP) were grown to full confluence to form a monolayer, a scratch was made as described previously [36], and the cells were allowed to migrate towards each other. With careful monitoring using a light microscope, we measured and quantified the distance the cells migrated over the indicated time periods.
2.12 Intracranial tumor model and immunohistochemistry
The Institutional Animal Care and Use Committee at the University of Illinois College of Medicine-Peoria approved all experimental procedures involving the use of animals. A small, hand-controlled twist drill was used to make the hole in the skull, and a specially devised screwdriver was used to thread and secure the screw into the hole. After a recovery period of 5 days, these animals were divided into three groups each containing 6 animals. Nude mice were anesthetized and Daoy cells [(1×105 cells/5 µl PBS), Daoy-P, Daoy-EV, and Daoy-SP] were stereotactically implanted as described previously [32]. Animals were monitored for 180 days, which was the designated termination point of the experiment. Animals that lost >20% of body weight or had trouble ambulating, feeding, or grooming were sacrificed. For histological analysis, brains were snap frozen and maintained at −70 °C until sectioning. Tumor volume was assessed as described previously [37]. Briefly, all brains were serially sectioned and every eighth 8 µm section was deparaffinized and incubated for 2 hours at 4°C with human specific MHC Class-I IgG (10 µg/mL) (mAb W6/32, Serotec, Inc., Raleigh, NC) followed by a multilink secondary antibody conjugated to biotin and HRP, followed by incubation with streptavidin, and then 3,3'-diaminobenzidine (DAB) substrate (Sigma, St. Louis, MO). Digital images were imported into Adobe Photoshop. Stained areas in each section were quantified as a pixel number, and pixel numbers for all sections from each brain were summed to obtain a total pixel number. Excised brains were fixed in 10% buffered formalin and embedded in paraffin. Tissue sections (5 µm thick) were obtained from the paraffin blocks and were stained with hematoxylin and eosin using standard histologic techniques. For immunohistochemical analysis, sections were incubated with mAb (1 hour, RT) followed by the appropriate secondary antibody. For HRP-conjugated secondary antibodies, we used DAB solution as the chromogen. Nuclei were counterstained with either hematoxylin or DAPI. Negative control slides were obtained by nonspecific IgG. Sections were mounted with mounting solution and analyzed with an inverted microscope.
2.13 Statistical analysis
All data are expressed as mean ± SD. Statistical analysis was performed using the student’s t test or a one-way analysis of variance (ANOVA). A p value of less than 0.05 was considered statistically significant. All experiments were performed in triplicate with consistent results.
3. Results
3.1 Overexpression of SPARC in Daoy cells
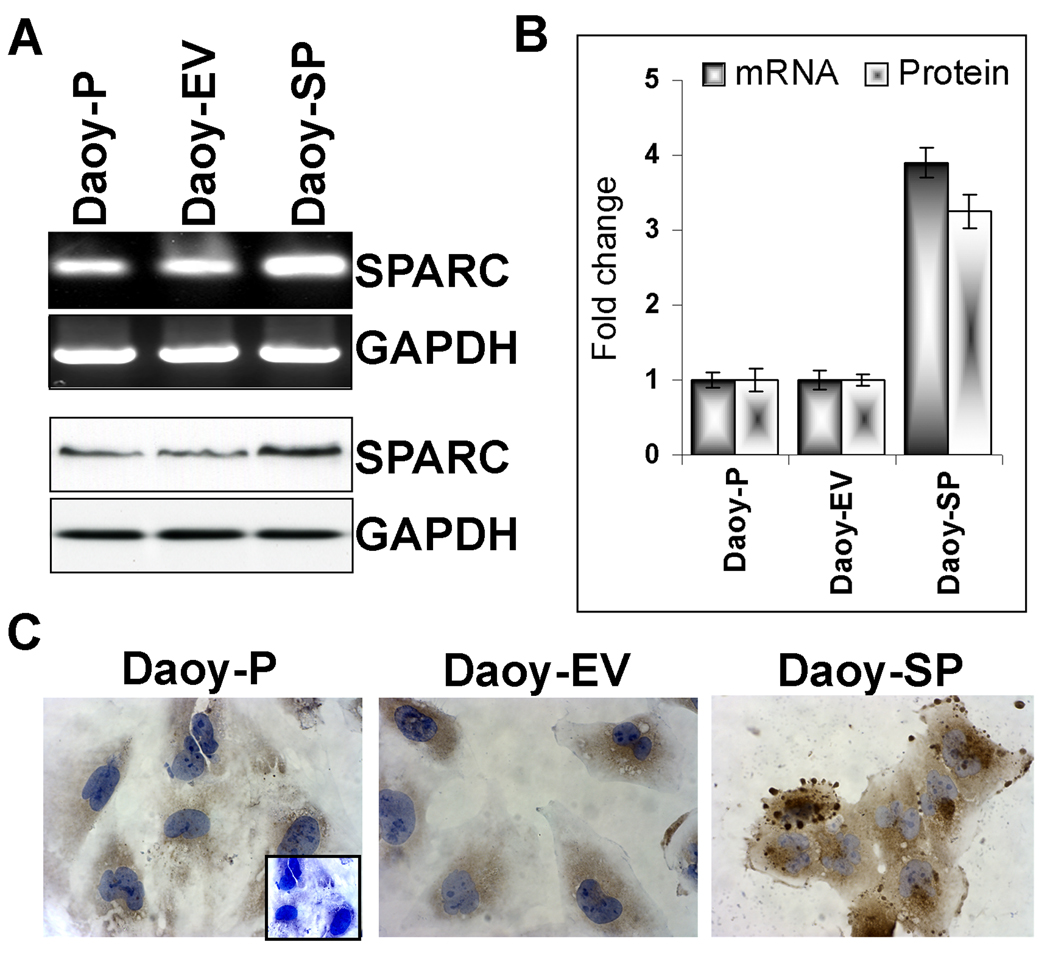
SPARC is a known to influence migration, and modulate growth factor-induced cell proliferation by modulating cellular interaction by interacting with extracellular matrix and by its abrogation of focal adhesions. To experiment with a genetic approach to induce SPARC expression and observe its effects on medulloblastoma tumor cell invasion and migration, we sub-cloned human SPARC cDNA into the pcDNA3.1 mammalian expression vector and transfected it into Daoy parental (Daoy-P) cells. The stable cell lines expressing SPARC were designated as Daoy-SP while the stable cell line expressing the empty pcDNA3.1 vector was designated as Daoy-EV. We tested clones randomly for mRNA expression of SPARC transcript (data not shown) and selected three stable SPARC-overexpressing clones [18]. Figure 1A indicates that SPARC transcript levels were increased in the Daoy-SP cells compared with those of parental and vector controls. There was about a 3-fold increase in mRNA transcript levels in the Daoy-SP clone (Figure 1A, top panel) (p<0.01 vs. controls; Fig. 1B). To confirm that this upregulation of SPARC mRNA translated into increased levels of SPARC protein, we next carried out western blot and immunocytochemical analyses for SPARC expression in the Daoy-SP clones (Fig. 1A, bottom panel). We found a 3- to 4-fold increase in SPARC expression in the Daoy-SP cells as compared to parental and empty vector controls (p<0.01; Fig. 1B). Immunocytochemical analysis for SPARC using specific antibody showed intense staining in Daoy-SP cells when compared to Daoy-P and Daoy-EV controls (Fig. 1C).
Figure 1. Development of Daoy medulloblastoma stable cell line for SPARC overexpression.
Daoy cells were stably transfected with plasmid containing full-length SPARC cDNA or empty vector. (A) Total RNA was extracted using Trizol reagent, and RT-PCR was performed for assessment of SPARC mRNA transcript level. GAPDH served as a control for RNA quality. SPARC protein levels were determined in total cell lysates by western blot analysis using a SPARC-specific antibody. GAPDH was used to confirm equal loading of cell lysates. (B) Protein and mRNA transcripts levels were quantified by densitometric analysis as shown in the corresponding bar graph. Columns, mean of triplicate experiments; bars, SE; *p<0.01, significant difference from Daoy-P control cells. (C) Immunocytochemical analysis for SPARC in different SPARC-overexpressing clones. Mouse IgG was used as negative control. Representing images are shown taken from 10 different microscopic fields of three independent experiments.
3.2 SPARC overexpression affects medulloblastoma cell adhesion to extracellular matrix protein–coated plates and rounding index
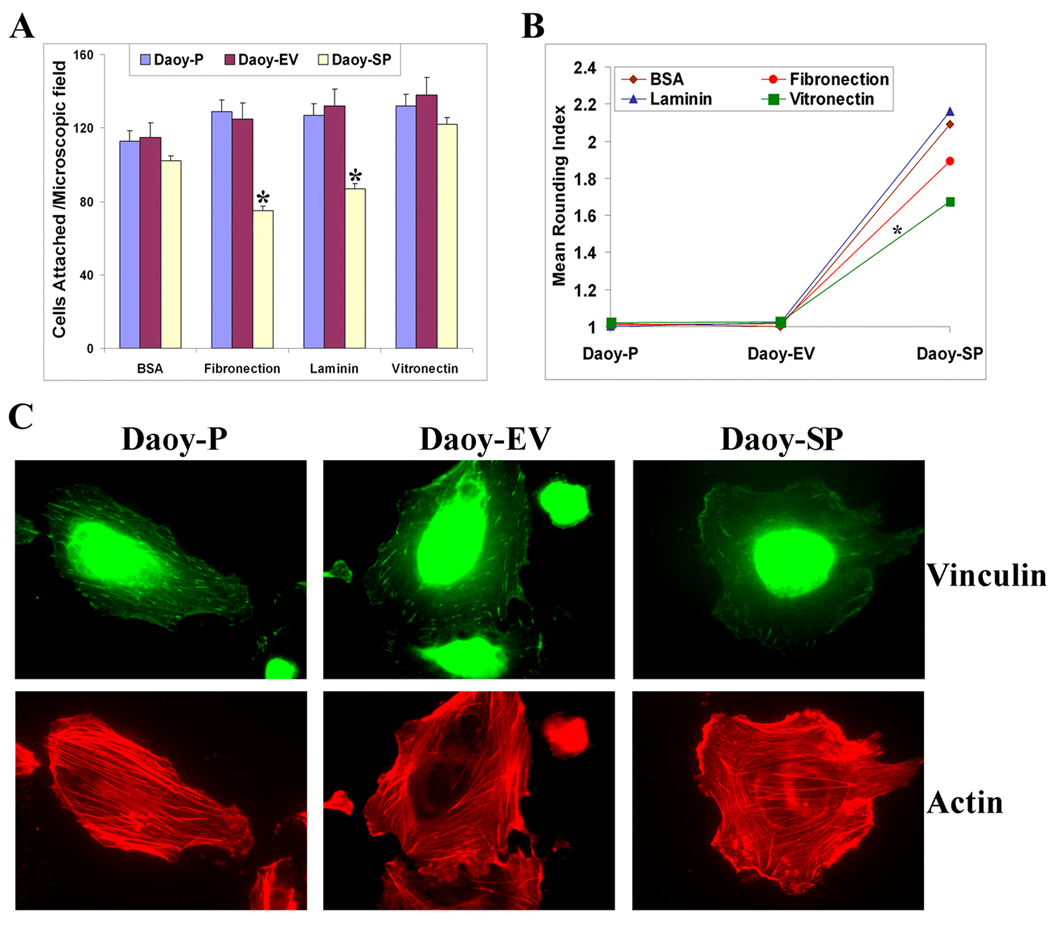
Earlier studies show that SPARC and several SPARC peptides promoted a partial detachment (rounding) of endothelial cells and inhibited the spreading of newly plated cells [38,39]. To investigate the effect of SPARC on cell-matrix adhesion of medulloblastoma cells, we examined the effect of SPARC overexpression on Daoy cell attachment onto various extracellular matrix components. SPARC overexpression was associated with an overall decrease in adhesion to extracellular matrices; this effect was most evident when cells were allowed to adhere to vitronectin (Fig. 2A). The effect of SPARC on the Daoy medulloblastoma cell rounding index 1 hour after plating on a different matrix is shown in Figure 2B. Photographs were analyzed for cell shape and the shape of each cell was placed in one of three categories: (1) highly rounded cells were counted as type ‘a’ cells, (2) fully spread cells were counted as type ‘c’ cells, and (3) cells that exhibited morphological characteristics between ‘a’ and ‘c’ were counted as type ‘b’ cells. This data was quantified by a rounding index (RI) assay [40] formula as follows: RI=[(3Xa)+(2Xb)+(1Xc)]/(a+b+c). Daoy-SP cells exhibited an activity whereby nearly all spreading was inhibited in comparison to parental Daoy-P (RI = 1±0.12) and vector control Daoy-EV (RI = 1±0.21) cells (Fig. 2B) where the majority of cells had begun spreading or were already significantly spread. It was noted that there was no statistical difference between RI numbers of Daoy-P and Daoy-EV. In contrast, Daoy-SP cells exhibited RI numbers (RI = 1.6 – 2.2 ±0 .34) that were consistently higher than those of parental or vector control cells.
Figure 2. Overexpression of SPARC in Daoy medulloblastoma cells inhibits adhesion, increases rounding index, and disrupts actin cytoskeleton.
(A) Adhesion assays were done on vitronectin, fibronectin, and laminin. Daoy-P, Daoy-EV, and Daoy-SP cells (2×104/well), were seeded in eight-well chamber slides. After 2 hours, the medium was removed, attached cells were stained with Hema-3 reagent, and then cells were photographed using an inverted microscope. The total number of cells per microscopic field were counted and represented in a graphical manner. Columns, mean of three experiments; bars, SD. (B) Rounding index was calculated using the following criteria: (1) highly rounded cells were counted as type ‘a’ cells, (2) fully spread cells were counted as type ‘c’ cells, and (3) cells that exhibited morphological characteristics between ‘a’ and ‘c’ were counted as type ‘b’ cells. This data was quantified by a rounding index (RI) assay (Bassuk et al., 1996a) formula as follows: RI=[(3Xa)+(2Xb)+(1Xc)]/(a+b+c). All experiments were done in triplicate. p<0.05 is considered significant. (C) Immunocytochemical analysis for actin and vinculin in Daoy-P, Daoy-EV, and Daoy-SP cells. Representing images are shown taken from 10 different microscopic fields of three independent experiments.
3.3 Morphological changes in the actin stress fibers and disruption of vinculin points
Morphology of the cells was examined by indirect immunofluorescence after 24 hours of culture. Immunofluorescence labeling showed that the punctuated actin staining characteristic of Daoy-P and Daoy-EV cells was organized into thick bundles of actin stress fibers that were associated with vinculin-containing focal contacts. By contrast, Daoy-SP cells exhibited a disrupted actin cytoskeleton, despite some accumulation of actin at the cell-cell boundary. This suggests that SPARC overexpression in Daoy medulloblastoma cells disrupts actin cytoskeleton structure (Fig. 2C).
3.4 Expression of SPARC and its effects on proliferation, migration, and invasion of Daoy medulloblastoma cells
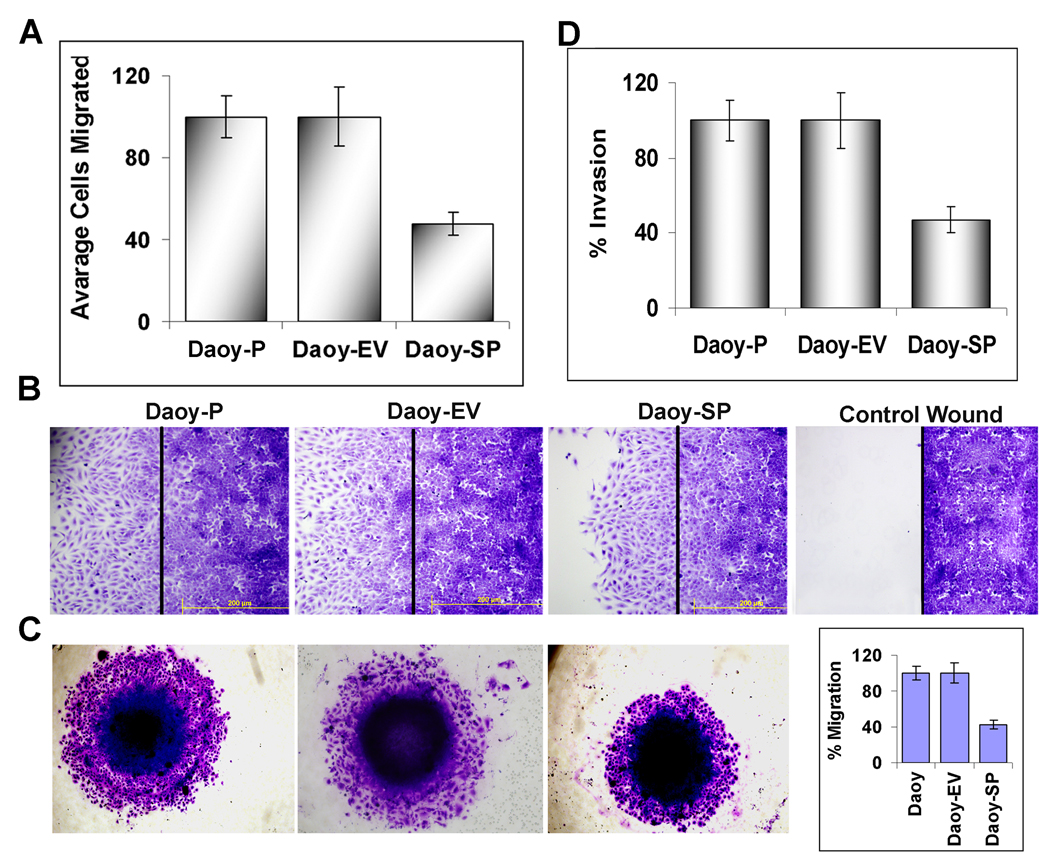
Earlier, we tested whether SPARC overexpression affected the growth of Daoy cells and compared the growth rates of SPARC-overexpressing cells (Daoy-SP) with those of the parental and empty vector controls (Daoy-P and Daoy-EV). A very minimal decrease in proliferation was observed at 24 hours (5–8%). At 48 hours, there was ~15% decrease in proliferation of Daoy-SP compared to Daoy-P and Daoy-EV cells [18]. To examine effects of SPARC on the migratory ability of medulloblastoma cells, we performed a migration assay using a modified Boyden chamber method. Migration of the SPARC-overexpressing Daoy cells was significantly reduced compared with the scrambled and empty vector control cells (Fig. 3A). Further, migration activity was also determined using a wound healing assay. The cells were allowed to migrate for 24 hours after scratching the cell layer. The overexpression of SPARC decreased the number of migrating cells scattered from the wound edges. In contrast, cells in the parental control and empty vector control collectively migrated and closed the gap (Fig. 3B). The areas covered by the migrating cells in the controls were significantly more extensive than those in the Daoy-SP group, indicating decreased cell migration with SPARC overexpression. Consequently, we evaluated whether SPARC overexpression suppresses migration and invasion of medulloblastoma cells. Figure 3B shows spheroids from parental and empty vector controls had a high number of cells that migrated from the spheroids into the surrounding area. However, Daoy-SP cells failed to migrate as much as controls (Fig. 3C). Daoy-SP cell shows decreased invasion when compare with parental or empty vector control Daoy cells in an in vitro matrigel invasion assay (Fig. 3D) suggesting that SPARC overexpression leading to decreased invasion in medulloblastoma.
Figure 3. SPARC overexpression in Daoy medulloblastoma cells decreases migration and invasion in vitro.
(A) Daoy-P, Daoy-EV, and Daoy-SP cells were allowed to migrate through the transwell membrane and the migrated cells were stained with Hema-3 solution and photographed. The total number of cells per microscopic field were counted and represented in a graphical manner. Columns, mean of three experiments; bars, SD. (B) Wound healing migration assay was carried out to determine the migratory ability of SPARC-overexpressing Daoy medulloblastoma cells. Daoy medulloblastoma cells were grown until a monolayer formed and then a scratch was made using a 200-µL pipette tip. After thorough washing with PBS, the cells were further incubated for 24 hours. The distance migrated by the cells was monitored over a period of time by observation under a light microscope. (C) SPARC overexpression inhibited the migration of Daoy medulloblastoma cells. Daoy-P, Daoy-EV, and Daoy-SP cell spheroids of approximately the same diameter were selected. Spheroids were then transferred to a 96-well plate and incubated for 48 hours to allow for migration. Finally, cell migration was observed, the medium was removed, and cells were stained using Hema-3 then photographed. The percent migration was calculated using Image J software (NIH) and represented in a graphical manner. (D) Transwells were pre-seeded with matrigel and allowed Daoy-P, Daoy-EV, and Daoy-SP cells to migrate through the transwell membrane and the migrated cells were stained with Hema-3 solution and photographed. The total number of cells per microscopic field were counted and represented in a graphical manner. Columns, mean of three experiments; bars, SD.
3.5 Overexpression of SPARC in Daoy medulloblastoma cells inhibits Rho-Rac-Cdc42 GTPase activity
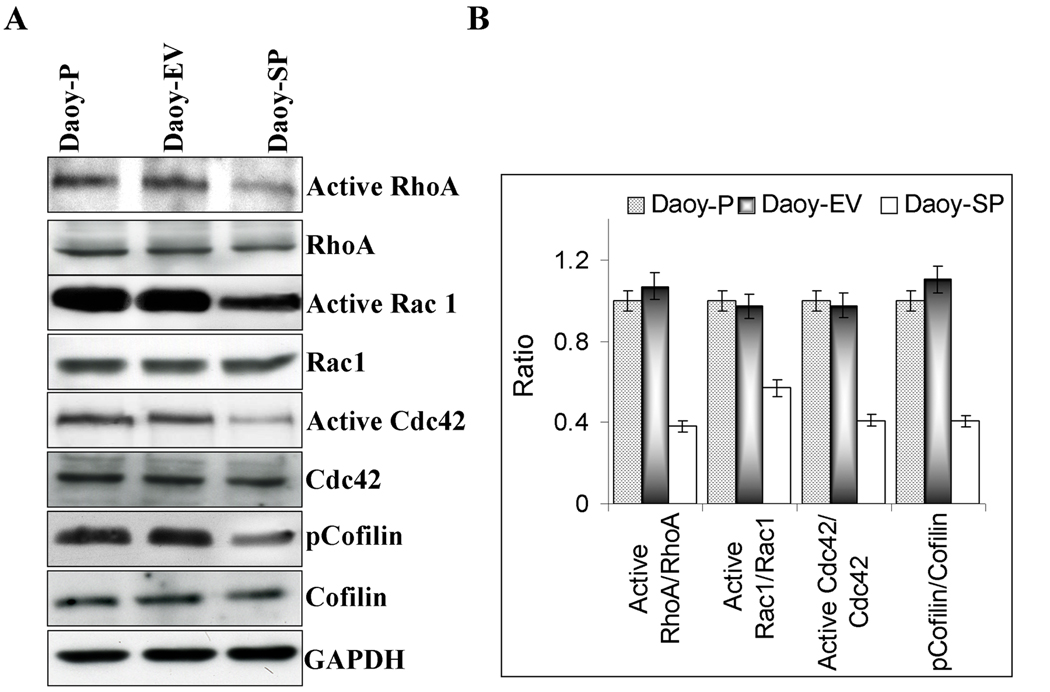
The actin cytoskeleton of animal cells maintains cellular shape and plays a pivotal role in cell motility. Rho and Rac, two members of the Ras-related superfamily of small GTPases, and Cdc42, another member of the Rho family, regulate the polymerization of actin to produce stress fibers and lamellipodia, respectively. To test the effect of SPARC overexpression on Rho-Rac-Cdc42 GTPase activity, Daoy-P, Daoy-EV and Daoy-SP cells were grown to 70% confluence and the cell lysates were assessed for Rho-Rac-Cdc42 activity. Stress fiber formation is controlled by the Rho family of small GTPases. More specifically, activation of the Rho-Rac-Cdc42 pathway leads to stress fiber assembly by both activation of actomyosin contractility and suppression of the actin-severing activity of cofilin [41]. A GTP pull-down assay was performed to measure the activity of endogenous Rho-Rac-Cdc42. There were decreased Rho-Rac-Cdc42 levels in Daoy-SP cells as compared to Daoy-P and Daoy-EV cells, suggesting that the loss of stress fibers in Daoy-SP cells is associated with a direct downregulation of Rho-Rac-Cdc42 activity (Fig. 4A). Next, we assessed the activation status of the Rho pathway by measuring the level of phosphorylation of cofilin, which is the terminal effector of this cascade [42]. Using a phosphospecific antibody, we found that cofilin phopshorylation decreased in Daoy-SP cells compared with parental Daoy-P or Daoy-EV cells (Fig. 4A). Taken together, these results suggest that activity of the Rho-Rac-Cdc42 pathway was decreased upon SPARC overexpression in Daoy medulloblastoma cells. Densitometric analysis revealed that there was a 50–65% decrease in Rho-Rac-Cdc42 activity and a 60% decrease in cofilin phopshorylation among Daoy-SP cells compared to the parental or empty vector control cells (Fig. 4B).
Figure 4. Overexpression of SPARC in Daoy medulloblastoma cells inhibits Rho-Rac-Cdc42 GTPase activity.
(A) Daoy-P, Daoy-EV, and Daoy-SP cells were grown to 70% confluence and the cell lysates were either incubated with the GST-TRBD fusion beads for 60 minutes or used for SDS-PAGE analysis. The lysates incubated with beads were then washed three times, resuspended in SDS loading buffer, and boiled for 10 min. SDS-PAGE and Western blot analyses were performed. (A) Western blot analysis for Active RhoA, total RhoA, active Rac1, total rac1, active Cdc42, total Cdc42, cofilin and pcofilin. (B) Densitometric analyses of western blots described in A. Results are expressed as the ratio between active and total levels of RhoA, Rac1, and cdc42. Phosphorylation is calculated as the ratio of the phosphorylated versus nonphosphorylated forms. Data are representative of three independent experiments (p<0.05).
3.6 SPARC overexpression activates Src and induces disruption of the RhoA pathway
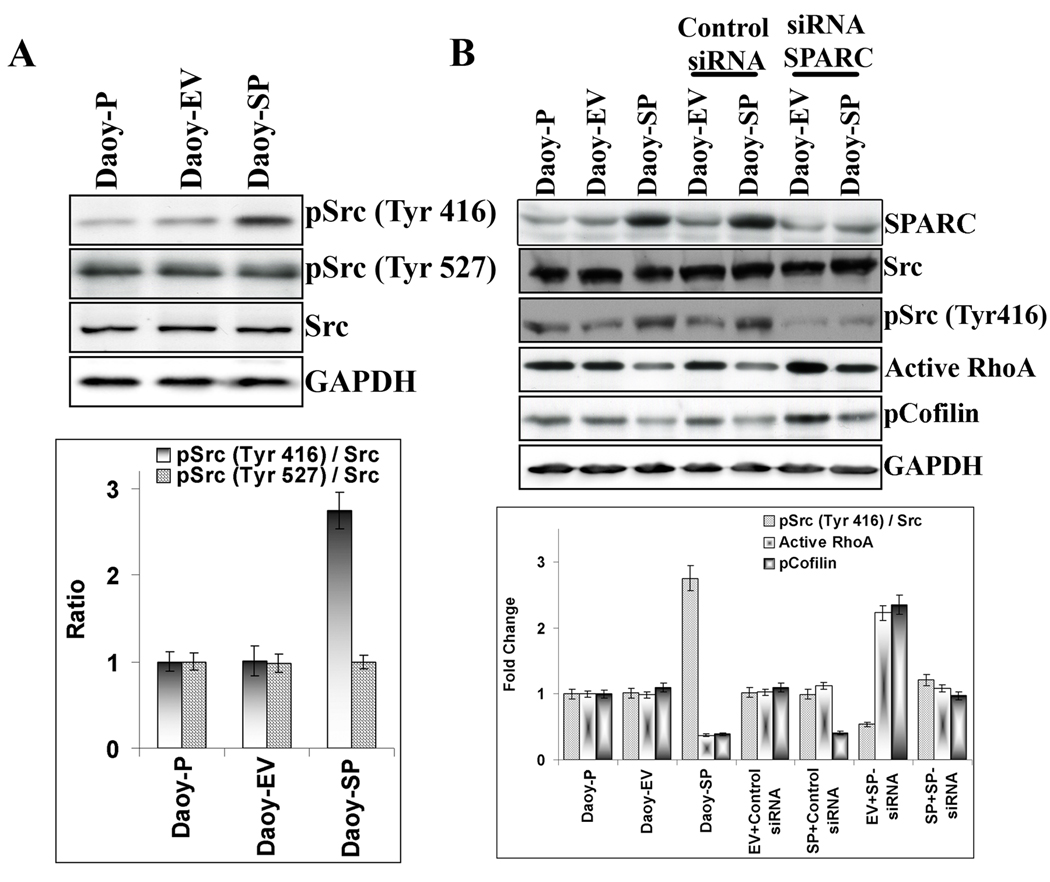
Cell–matrix adhesion and integrin engagement regulate the activity of Src family kinases [43,44]. In turn, Src was demonstrated to downregulate RhoA via tyrosine phosphorylation and activation of its upstream inhibitor p190RhoGAP [45]. Hence, we examined whether the decreased RhoA activity affected the Src signaling pathway. Src activity is regulated by tyrosine phosphorylation at two sites with opposing effects; the phosphorylation of Tyr-416 in the activation loop of the kinase domain upregulates enzyme activity whereas phopshorylation of Tyr-527 in the carboxy-terminal tail renders the enzyme less active. Analysis of Src phosphorylation in the Daoy-SP cells revealed approximately 3-fold higher phosphorylation levels of the activating site in Src (Src-pTyr-416) when compared to parental (Daoy-P) or vector control cells (Daoy-EV). However, SPARC overexpression in Daoy medulloblastoma cells did not alter the Src inactivating site phopshorylation at Tyr-527 (Fig. 5A). To confirm the activation of Src in these cells by SPARC overexpression, we further tested the effect of SPARC siRNA in these cell lines. SPARC siRNA inhibited Src phopshorylation at Tyr-416 in Daoy-SP cells comparable to the parental or empty vector controls, suggesting that SPARC overexpression is involved in Src activation. Further, increased Src activity inhibited RhoA activity in Daoy-SP cells (Fig. 5B).
Figure 5. Overexpression of SPARC in Daoy medulloblastoma cells activates Src activity.
(A) Daoy-P, Daoy-EV and Daoy-SP cells were grown to 70% confluence, and the cell lysates were used for SDS-PAGE analysis. Western blot analysis for Src and its phosphorylation was performed, and densitometric analyses of western blots described in (A) are shown. Phosphorylation is calculated as the ratio of the phosphorylated versus nonphosphorylated forms. Data are representative of three independent experiments (p<0.05). (B) Daoy-P, Daoy-EV, and Daoy-SP cells were transfected with either control siRNA or SPARC siRNA and incubated for 36 hours. The cell lysates were used for the western blot analysis of Src, pSrc and pcofilin. One set of cell lysates was incubated with the GST-TRBD fusion beads for 60 min. The beads incubated with lysates were then washed three times, resuspended in SDS loading buffer by boiling for 10 min and western blot analyses were performed for RhoA. Densitometric analyses of western blots described in (A). Data are representative of three independent experiments (p<0.05).
3.7 Overexpression of SPARC in medulloblastoma cells inhibits tumorigenicity and migration in nude mice
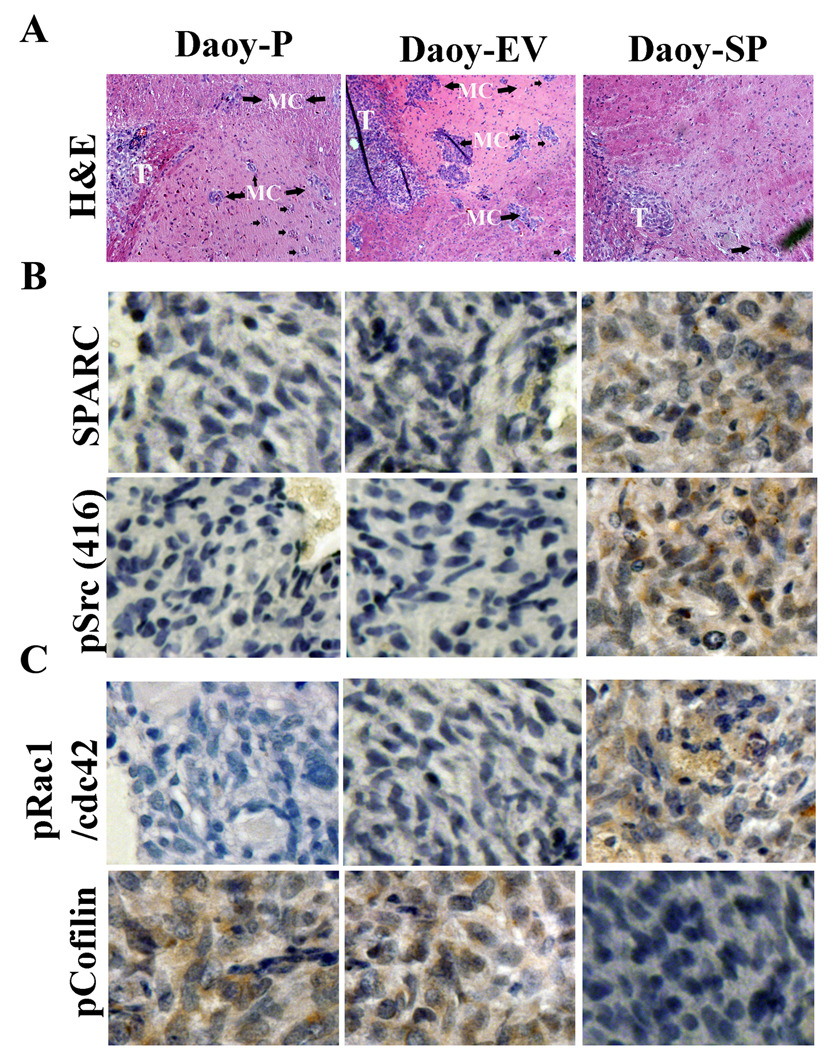
To assess the effect of SPARC overexpression on therapeutic efficacy, Daoy-P, Daoy-EV, and Daoy-SP cells were injected intracranially into nude mice. Mice injected with Daoy-P and Daoy-EV cells developed tumors, became symptomatic within 6 weeks, and were subsequently sacrificed. In contrast, mice injected with Daoy-SP cells survived for 180 days, which was the designated end point of the experiment. At this point, the animals were sacrificed and their brains were examined for tumor growth. Tumor volumes were evaluated by measuring the maximum cross-sectional areas stained for anti-human-specific MHC class-I IgG [western blot analysis of MHC class-I, data published previously [18] in digitalized sections of cerebellum/tumors. Earlier, we showed significant tumor growth in brains of mice implanted with Daoy-P and Daoy-EV cells whereas Daoy-SP cells showed a statistically significant decrease (>65%) in mean tumor volume [18]. H&E analysis revealed a significant number of cells had migrated from the core tumors in mice implanted with in Daoy-P and Daoy-EV cells. In contrast, very few cells migrated in Daoy-SP-implanted mice (Fig. 6A). To determine SPARC overexpression in vivo, brain sections were stained with a monoclonal antibody for human SPARC. Figure 6B indicates that tumor sections from Daoy-SP tumors showed more intense staining than Daoy-P and Daoy-EV tumors. To assess whether Src activation (Src-Tyr-416) mediated inhibition of migration, we analyzed the levels of Src activation in these tumor sections by Src-Tyr416 specific antibody. Daoy-SP tumor sections showed high staining for Src when compared to Daoy-P or Daoy-EV tumors (Fig. 6B). Further, we analyzed whether Src activation leads to inhibition of Rho activity by testing the levels of its downstream effectors, cofilin and phosphorylation of Rac1/cdc42 at Ser-71, which in turn deactivate RhoA. Cofilin staining was robust in Daoy-P and Daoy-EV tumors, but it was weaker in Daoy-SP tumors, suggesting that Rho activity was inhibited by Src activation in tumors; pRac1/Cdc42 showed intense staining in Daoy-SP tumors when compared to Daoy-P or Daoy-EV tumors (Fig. 6C). These results are consistent with a role for Src in the SPARC-mediated inhibition of migration observed in vitro.
Figure 6. SPARC overexpression inhibits medulloblastoma cell migration in vivo.
Medulloblastoma tumor sections from mice injected with Daoy-P, Daoy-EV, and Daoy-SP cells were analyzed as described in Materials and Methods. (A) H&E staining for the tumors showing migrated cells. T: tumor; MC: migrated cells from the implanted tumor. (B) Immunohistochemical analysis for SPARC, pSrc, and pcofilin was performed as described in Materials and Methods. All results are representative of multiple tumors taken from five separate mice in each treatment group.
4. Discussion
Tumor metastasis is a complex, multistep process that includes cell proliferation, attachment, migration, invasion, and sequential interactions between the tumor cells and the extracellular matrix (ECM). SPARC is an ECM molecule that influences tumor cell adhesion and migration. Some steps in tumor metastasis can be regulated by SPARC. In certain cancers, such as melanomas and gliomas, SPARC is associated with a highly aggressive tumor phenotype. In others, mainly ovarian, neuroblastomas, and colorectal cancers, SPARC may function as a tumor suppressor [15]. These opposing effects on cell growth, cell migration and tumor formation suggest that the functions of SPARC are cell specific and may be dependent on concentration as well as regulation of ECM components. Therefore, the focus of this investigation was to determine the role of SPARC in medulloblastoma cell migration. Our findings revealed that proliferation, adhesion, migration, invasion, and activity of MMP-9 (data published earlier [18], in medulloblastoma could be inhibited by SPARC. Additionally, SPARC activated the expression of Src by leading to decreased RhoA. These results proved that SPARC has the potential to prevent medulloblastoma cancer metastasis and tumor growth.
Using the Daoy cell line, stably transfected with SPARC cDNA, we determined the contribution of SPARC in medulloblastoma tumor migration and growth. Daoy-SP clones showed increased SPARC protein and gene transcript levels by about three-fold compared to Daoy-P and Daoy-EV stable clones, as determined by western blotting and real time PCR analyses. SPARC overexpression inhibited tumor cell migration as demonstrated by spheroid migration assay and wound healing migration assay and also inhibited invasion as demonstrated by Matrigel invasion assay in vitro. Previous studies have indicated that SPARC contributes to the regulation of tumor cell migration, even though its role appears to be cell-type specific. When the effects of exogenous SPARC on single cell movement and migration were investigated in pancreatic cell lines, pancreatic cell migration was inhibited in the presence of SPARC [46]. Another study shows that an inhibitory effect of SPARC on proliferation and migration has been found in breast and ovarian carcinoma cells [47]. Additionally, infection of MDA-231 breast carcinoma cells with viral constructs of osteonectin (SPARC) decreased the in vitro invasion of these cells through Matrigel [48].
SPARC is a part of a group of extracellular matrix proteins that regulate cell adhesion through a loss of focal adhesion plaques from spread cells. In this study, we illustrate that SPARC alters the morphology of cells grown on different matrix proteins and decreases migration, which is accompanied by decreased proliferation and also increased rounding indices of cells. We observed that SPARC induced Src activation, leading to inhibition of RhoA activity. We further show, through siRNA blockade, that knockdown of SPARC inhibits Src activation leading to increased RhoA activity, which in turn, increased cofilin phopshorylation. With these results, we demonstrate that siRNA inhibition of SPARC restores the lost migratory ability of Daoy-SP cells.
Previous reports show that SPARC reduced the number of bovine aortic endothelial (BAE) cells positive for focal adhesions [49]. Addition of SPARC to spread BAE cells caused a dose-dependent loss of focal adhesion-positive cells that was maximal at approximately 1 µg/mL (0.03 µM). Vinculin appeared diffuse and F-actin was redistributed to the periphery of cells incubated with SPARC, which is consistent with the loss of adhesion plaques as detected by interference reflection microscopy [20].
SPARC regulates several biological functions that could regulate tumor invasion [50]. In this study, we determined whether SPARC, a protein that modulates integrin-ECM interactions and affects cytoskeletal structure [51], also alters the ability of medulloblastoma cells to migrate. The data confirm that SPARC inhibits the migratory capacity of Daoy medulloblastoma cells both in vitro and in vivo.
Because our in vitro data indicated that SPARC inhibits migration and invasion via activation of Src, leading to decreased activity of RhoA/Rac/Cdc42, we reasoned both SPARC and Src should be expressed at the same time in tumor cells in vivo. To test this hypothesis, we grafted mice with Daoy-P, Daoy-EV, and Daoy-SP cells intracranially. The animals implanted with Daoy-SP tumor cells showed increased SPARC levels as compared to parental Daoy cells. These studies demonstrated that SPARC and Src are co-located in vivo and led to decreased cofilin phosphorylation. Other studies support our findings, since SPARC overexpression in ovarian cancer cell lines has been reported [52] to inhibit ovarian cancer cell chemotaxis, and fibronectin invasion induced by heated, mesothelial cell-conditioned medium. Bradshaw, et al. [53,54] showed that SPARC deficiency is associated with an accelerated closure of cutaneous wounds as well as decreased collagen content and enhanced contractibility of wounds in SPARC-deficient mice.
In summary, we have shown here that overexpression of endogenous SPARC decreases migration and the malignant phenotype of Daoy medulloblastoma cells. We hypothesized that overexpression of SPARC resulted in Src activation, leading to RhoA inactivation, which in turn caused the inhibition of cofilin that ultimately decreased migration in vitro and in vivo. On the basis of these observations, we conclude that endogenous overexpression of SPARC can decrease migration and tumor growth in medulloblastoma and therefore acts as a tumor suppressor in human Daoy medulloblastoma cells. Future studies in human medulloblastoma should focus on the design of treatment strategies that specifically target SPARC–Src interactions.
Acknowledgements
The authors wish to thank Shellee Abraham for manuscript preparation and Diana Meister and Sushma Jasti for manuscript review. We also thank Dr Martin A. Schwartz (Cardiovascular Research Center, Mellon Prostate Cancer Institute, Departments of Microbiology and Biomedical Engineering, University of Virginia, Charlottesville, VA) for providing pGEX-TRBD, Dr. Baroda S, for providing pGEX-Rac1, and pGEX-Cdc42.
Funding
This research was supported by a grant from National Institutes of Health, CA132853 (to S.S.L.) The contents are solely the responsibility of the authors and do not necessarily represent the official views of NIH.
Abbreviations
- SPARC
Secreted Protein Acidic and Rich in Cysteine
- ECM
Extra Cellular Matrix
- PI3K
Phosphatidylinositol 3-kinases
- FAK
Focal Adhesion Kinase
- MMP
matrix metalloproteinase
- PNET
Primitive neuroectodermal tumors
- VACM-1
vascular cell adhesion molecule 1
- MEK
Mitogen-activated protein kinase
- ERK
Extracellular signal-regulated kinases
- siRNA
short interfering ribonucleic acid
- GST
glutathione S-transferase
- HRP
horseradish peroxidase
- GAPDH
Glyceraldehyde 3-phosphate dehydrogenase
- TRBD
Rho binding domain
- MHC
major histocompatibility complex
- H&E
hematoxylin and eosin stain
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Reference List
- 1.Crawford JR, MacDonald TJ, Packer RJ. Lancet Neurol. 2007;6:1073–1085. doi: 10.1016/S1474-4422(07)70289-2. [DOI] [PubMed] [Google Scholar]
- 2.Howes TL, Buatti JM, Kirby PA, Carlisle TL, Ryken TC. J Neurooncol. 2006;80:191–194. doi: 10.1007/s11060-006-9175-4. [DOI] [PubMed] [Google Scholar]
- 3.Chan MY, Teo WY, Seow WT, Tan AM. Ann Acad Med Singapore. 2007;36:314–318. [PubMed] [Google Scholar]
- 4.Sun LM, Yeh SA, Wang CJ, Huang EY, Chen HC, Hsu HC, Lee SP. J Neurooncol. 2002;58:77–85. doi: 10.1023/a:1015865614640. [DOI] [PubMed] [Google Scholar]
- 5.Feltbower RG, Picton S, Bridges LR, Crooks DA, Glaser AW, McKinney PA. Pediatr Hematol Oncol. 2004;21:647–660. doi: 10.1080/08880010490501079. [DOI] [PubMed] [Google Scholar]
- 6.Ranger A, McDonald W, Bauman GS, Del MR. Can J Neurol Sci. 2009;36:631–637. doi: 10.1017/s0317167100008155. [DOI] [PubMed] [Google Scholar]
- 7.Mazloom A, Zangeneh AH, Paulino AC. Int J Radiat Oncol Biol Phys. 2010;78:72–78. doi: 10.1016/j.ijrobp.2009.07.1729. [DOI] [PubMed] [Google Scholar]
- 8.Lowenfels AB. Gastroenterology. 1989;96:551. doi: 10.1016/s0016-5085(89)91616-8. [DOI] [PubMed] [Google Scholar]
- 9.Chan AW, Tarbell NJ, Black PM, Louis DN, Frosch MP, Ancukiewicz M, Chapman P, Loeffler JS. Neurosurgery. 2000;47:623–631. doi: 10.1097/00006123-200009000-00018. [DOI] [PubMed] [Google Scholar]
- 10.Leo E, Schlegel PG, Lindemann A. J Neurooncol. 1997;32:149–154. doi: 10.1023/a:1005721510659. [DOI] [PubMed] [Google Scholar]
- 11.Painter KJ, Armstrong NJ, Sherratt JA. J Theor Biol. 2010;264:1057–1067. doi: 10.1016/j.jtbi.2010.03.033. [DOI] [PubMed] [Google Scholar]
- 12.Gupton SL, Waterman-Storer CM. Cell. 2006;125:1361–1374. doi: 10.1016/j.cell.2006.05.029. [DOI] [PubMed] [Google Scholar]
- 13.Ridley AJ, Schwartz MA, Burridge K, Firtel RA, Ginsberg MH, Borisy G, Parsons JT, Horwitz AR. Science. 2003;302:1704–1709. doi: 10.1126/science.1092053. [DOI] [PubMed] [Google Scholar]
- 14.Yan Q, Sage EH. J Histochem Cytochem. 1999;47:1495–1506. doi: 10.1177/002215549904701201. [DOI] [PubMed] [Google Scholar]
- 15.Tai IT, Tang MJ. Drug Resist Updat. 2008;11:231–246. doi: 10.1016/j.drup.2008.08.005. [DOI] [PubMed] [Google Scholar]
- 16.Clark CJ, Sage EH. J Cell Biochem. 2008;104:721–732. doi: 10.1002/jcb.21688. [DOI] [PubMed] [Google Scholar]
- 17.Podhajcer OL, Benedetti L, Girotti MR, Prada F, Salvatierra E, Llera AS. Cancer Metastasis Rev. 2008;27:523–537. doi: 10.1007/s10555-008-9135-x. [DOI] [PubMed] [Google Scholar]
- 18.Bhoopathi P, Chetty C, Gujrati M, Dinh DH, Rao JS, Lakka SS. Br J Cancer. 2010;102:530–540. doi: 10.1038/sj.bjc.6605538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kelly PN, Dakic A, Adams JM, Nutt SL, Strasser A. Science. 2007;317:337. doi: 10.1126/science.1142596. %20; [DOI] [PubMed] [Google Scholar]
- 20.Murphy-Ullrich JE, Lane TF, Pallero MA, Sage EH. J Cell Biochem. 1995;57:341–350. doi: 10.1002/jcb.240570218. [DOI] [PubMed] [Google Scholar]
- 21.Nobes CD, Hall A. Cell. 1995;81:53–62. doi: 10.1016/0092-8674(95)90370-4. [DOI] [PubMed] [Google Scholar]
- 22.Erpel T, Courtneidge SA. Curr Opin Cell Biol. 1995;7:176–182. doi: 10.1016/0955-0674(95)80025-5. [DOI] [PubMed] [Google Scholar]
- 23.Klemke RL, Cai S, Giannini AL, Gallagher PJ, de Lanerolle P, Cheresh DA. J Cell Biol. 1997;137:481–492. doi: 10.1083/jcb.137.2.481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nguyen DH, Catling AD, Webb DJ, Sankovic M, Walker LA, Somlyo AV, Weber MJ, Gonias SL. J Cell Biol. 1999;146:149–164. doi: 10.1083/jcb.146.1.149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pawlak G, Helfman DM. J Biol Chem. 2002;277:26927–26933. doi: 10.1074/jbc.M202261200. [DOI] [PubMed] [Google Scholar]
- 26.Chandrasekar N, Mohanam S, Gujrati M, Olivero WC, Dinh DH, Rao JS. Oncogene. 2003;22:392–400. doi: 10.1038/sj.onc.1206164. [DOI] [PubMed] [Google Scholar]
- 27.Jimenez C, Portela RA, Mellado M, Rodriguez-Frade JM, Collard J, Serrano A, Martinez A, Avila J, Carrera AC. J Cell Biol. 2000;151:249–262. doi: 10.1083/jcb.151.2.249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sturge J, Hamelin J, Jones GE. J Cell Sci. 2002;115:699–711. doi: 10.1242/jcs.115.4.699. [DOI] [PubMed] [Google Scholar]
- 29.Turner NA, O'Regan DJ, Ball SG, Porter KE. FASEB J. 2005;19:804–806. doi: 10.1096/fj.04-2852fje. [DOI] [PubMed] [Google Scholar]
- 30.Mohanam S, Jasti SL, Kondraganti SR, Chandrasekar N, Kin Y, Fuller GN, Lakka SS, Kyritsis AP, Dinh DH, Olivero WC, Gujrati M, Yung WK, et al. Clin Cancer Res. 2001;7:2519–2526. [PubMed] [Google Scholar]
- 31.Chetty C, Bhoopathi P, Lakka SS, Rao JS. Oncogene. 2007;26:7675–7683. doi: 10.1038/sj.onc.1210584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bhoopathi P, Chetty C, Kunigal S, Vanamala SK, Rao JS, Lakka SS. J Biol Chem. 2008;283:1545–1552. doi: 10.1074/jbc.M707931200. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 33.Chetty C, Lakka SS, Bhoopathi P, Kunigal S, Geiss R, Rao JS. Cancer Res. 2008;68:4736–4745. doi: 10.1158/0008-5472.CAN-07-6612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rao JS, Bhoopathi P, Chetty C, Gujrati M, Lakka SS. Cancer Res. 2007;67:4956–4964. doi: 10.1158/0008-5472.CAN-07-0380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nalla AK, Asuthkar S, Bhoopathi P, Gujrati M, Dinh DH, Rao JS. PLoS One. 2010;5:e13006. doi: 10.1371/journal.pone.0013006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jung JW, Hwang SY, Hwang JS, Oh ES, Park S, Han IO. Eur J Cancer. 2007;43:1214–1224. doi: 10.1016/j.ejca.2007.01.034. [DOI] [PubMed] [Google Scholar]
- 37.Ding Q, Grammer JR, Nelson MA, Guan JL, Stewart JE, Jr, Gladson CL. J Biol Chem. 2005;280:6802–6815. doi: 10.1074/jbc.M409180200. [DOI] [PubMed] [Google Scholar]
- 38.Lane TF, Sage EH. J Cell Biol. 1990;111:3065–3076. doi: 10.1083/jcb.111.6.3065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sage H, Vernon RB, Funk SE, Everitt EA, Angello J. J Cell Biol. 1989;109:341–356. doi: 10.1083/jcb.109.1.341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bassuk JA, Braun LP, Motamed K, Baneyx F, Sage EH. Int J Biochem Cell Biol. 1996;28:1031–1043. doi: 10.1016/1357-2725(96)00036-2. [DOI] [PubMed] [Google Scholar]
- 41.Bishop AL, Hall A. Biochem J. 2000;348(Pt 2):241–255. [PMC free article] [PubMed] [Google Scholar]
- 42.Schwartz M. J Cell Sci. 2004;117:5457–5458. doi: 10.1242/jcs.01582. [DOI] [PubMed] [Google Scholar]
- 43.Playford MP, Schaller MD. Oncogene. 2004;23:7928–7946. doi: 10.1038/sj.onc.1208080. [DOI] [PubMed] [Google Scholar]
- 44.Schwartz MA. Trends Cell Biol. 2001;11:466–470. doi: 10.1016/s0962-8924(01)02152-3. [DOI] [PubMed] [Google Scholar]
- 45.Arthur WT, Petch LA, Burridge K. Curr Biol. 2000;10:719–722. doi: 10.1016/s0960-9822(00)00537-6. [DOI] [PubMed] [Google Scholar]
- 46.Chen G, Tian X, Liu Z, Zhou S, Schmidt B, Henne-Bruns D, Bachem M, Kornmann M. Br J Cancer. 2010;102:188–195. doi: 10.1038/sj.bjc.6605440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Dhanesuan N, Sharp JA, Blick T, Price JT, Thompson EW. Breast Cancer Res Treat. 2002;75:73–85. doi: 10.1023/a:1016536725958. [DOI] [PubMed] [Google Scholar]
- 48.Koblinski JE, Kaplan-Singer BR, VanOsdol SJ, Wu M, Engbring JA, Wang S, Goldsmith CM, Piper JT, Vostal JG, Harms JF, Welch DR, Kleinman HK. Cancer Res. 2005;65:7370–7377. doi: 10.1158/0008-5472.CAN-05-0807. [DOI] [PubMed] [Google Scholar]
- 49.Murphy-Ullrich JE, Lightner VA, Aukhil I, Yan YZ, Erickson HP, Hook M. J Cell Biol. 1991;115:1127–1136. doi: 10.1083/jcb.115.4.1127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sato N, Fukushima N, Maehara N, Matsubayashi H, Koopmann J, Su GH, Hruban RH, Goggins M. Oncogene. 2003;22:5021–5030. doi: 10.1038/sj.onc.1206807. [DOI] [PubMed] [Google Scholar]
- 51.Golembieski WA, Thomas SL, Schultz CR, Yunker CK, McClung HM, Lemke N, Cazacu S, Barker T, Sage EH, Brodie C, Rempel SA. Glia. 2008;56:1061–1075. doi: 10.1002/glia.20679. [DOI] [PubMed] [Google Scholar]
- 52.Said NA, Najwer I, Socha MJ, Fulton DJ, Mok SC, Motamed K. Neoplasia. 2007;9:23–35. doi: 10.1593/neo.06658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Bradshaw AD, Reed MJ, Sage EH. J Histochem Cytochem. 2002;50:1–10. doi: 10.1177/002215540205000101. [DOI] [PubMed] [Google Scholar]
- 54.Bradshaw AD, Puolakkainen P, Dasgupta J, Davidson JM, Wight TN, Helene SE. J Invest Dermatol. 2003;120:949–955. doi: 10.1046/j.1523-1747.2003.12241.x. [DOI] [PubMed] [Google Scholar]