Abstract
In the AAA+ ClpXP protease, ClpX uses the energy of ATP binding and hydrolysis to unfold proteins before translocating them into ClpP for degradation. For proteins with C-terminal ssrA tags, ClpXP pulls on the tag to initiate unfolding and subsequent degradation. Here, we demonstrate that an initial step in ClpXP unfolding of the 11-stranded β barrel of superfolder GFP-ssrA involves extraction of the C-terminal β strand. The resulting 10-stranded intermediate is populated at low ATP concentrations, which stall ClpXP unfolding, and at high ATP concentrations, which support robust degradation. To determine if stable unfolding intermediates cause low-ATP stalling, we designed and characterized circularly permuted GFP variants. Notably, stalling was observed for a variant that formed a stable 10-stranded intermediate but not for one in which this intermediate was unstable. A stepwise degradation model in which the rates of terminal-strand extraction, strand refolding or recapture, and unfolding of the 10-stranded intermediate all depend on the rate of ATP hydrolysis by ClpXP accounts for the observed changes in degradation kinetics over a broad range of ATP concentrations. Our results suggest that the presence or absence of unfolding intermediates will play important roles in determining whether forced enzymatic unfolding requires a minimum rate of ATP hydrolysis.
Introduction
In cells ranging from bacteria to mammals, AAA+ proteases bind specific target proteins and then use cycles of ATP binding and hydrolysis to unfold them and to translocate the denatured polypeptide into a compartmental peptidase for degradation.1 Although these ATP-fueled machines can unfold substrates with diverse structures and stabilities, some proteins resist proteolysis or are only partially degraded.2–7 Inhibitory or slippery sequences, highly stable domains, or stable unfolding intermediates have all been proposed to play roles in helping proteins resist degradation.
The ClpXP protease of Escherichia coli consists of the AAA+ ClpX unfoldase and the associated ClpP compartmental peptidase.8–9 Peptide signals that bind in the axial pore of a hexameric ClpX ring, including the 11-residue ssrA tag, target substrates for ClpXP degradation.10–13 ATP-dependent translocation is thought to pull the peptide tag through the pore, eventually unfolding attached domains that cannot pass through the narrow channel in a native conformation (Fig. 1A). For stable proteins, unfolding is generally the rate-limiting step in ClpXP degradation, often requiring hydrolysis of hundreds of ATP molecules.14–15 Once the substrate is unfolded, ClpX translocates the unfolded polypeptide into the degradation chamber of ClpP in steps of 5–8 amino acids per power stroke.16–17 Because the ssrA tag is a C-terminal degradation signal, translocation of ssrA-tagged substrates begins at the C-terminus and proceeds to the N-terminus.
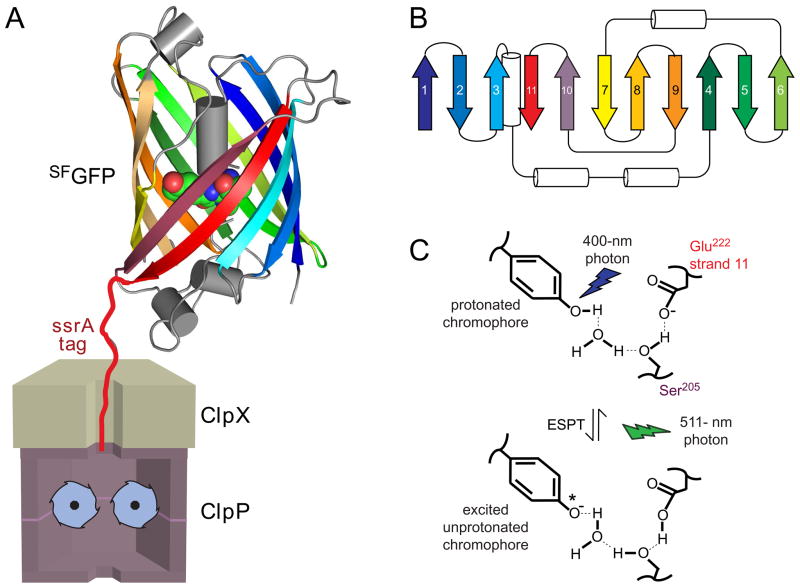
Fig. 1.
(A) Cartoon showing ClpXP after engaging the ssrA tag of SFGFP-ssrA but before unfolding, translocation, and degradation. The GFP chromophore is shown in CPK representation (nitrogen, blue; oxygen, red; carbon, green). (B) Diagram of the secondary structure of SFGFP-ssrA. β strands are the same color as in the structure in panel A. (C) Absorption of 400-nm light results in excited state proton transfer (ESPT) in which the phenolic proton moves to Glu222 on strand 11. Return to the ground state is accompanied by fluorescence emission at 511 nm. Mutation of Ser205 or Glu222 prevents ESPT and fluorescence after excitation with 400-nm light. Fluorescence arising from excitation of the deprotonated chromophore with 467-nm light does not depend on Ser205 or Gly222.
SsrA-tagged green fluorescent protein (GFP-ssrA) is an excellent model substrate for ClpXP, because unfolding and degradation can be monitored by loss of native fluorescence.14,18 Interestingly, however, ClpXP degradation of GFP-ssrA ceases or stalls at low ATP concentrations, whereas degradation of other stable proteins slows linearly as the rate of ATP hydrolysis decreases.7 To explain these observations, Martin et al. proposed that unfolding of GFP-ssrA by ClpX requires two steps: initial extraction of the C-terminal β strand, followed by unfolding of the resulting 10-stranded barrel.7 They also suggested that a thresh-hold rate of ATP hydrolysis was needed for degradation because capture of the strand-extracted intermediate required multiple rapid cycles of ATP hydrolysis to prevent refolding of the extracted strand.
The fluorescence properties of GFP depend on its folded structure. For example, denatured GFP displays very low fluorescence because of solvent quenching. In native GFP, by contrast, an 11-stranded β barrel shields the enclosed chromophore (residue 65–67), which contains a phenolic side chain that equilibrates slowly between protonated and unprotonated states (Fig. 1A, 1B, 1C).19–21. The unprotonated chromophore absorbs 467-nm light and emits 511-nm fluorescence (hereafter called 467-nm fluorescence). The protonated chromophore absorbs 400-nm light but also emits a 511-nm photon (hereafter called 400-nm fluorescence), because absorption transiently leads to deprotonation via an excited state proton transfer (ESPT) reaction (Fig. 1C).22–23 Importantly, the proton acceptor (Glu222) for this transfer reaction is on β-strand 11, near the C-terminus of GFP, and the Glu222→ Gln mutant displays normal 467-nm fluorescence but no 400-nm fluorescence.24 In the studies below, these fluorescence properties allow detection of a GFP species in which the β barrel is largely intact but the C-terminal β-strand is not.
In this paper, we use ssrA-tagged variants of superfolder GFP (SFGFP; ref. 25) to probe the mechanism of unfolding and degradation by ClpXP. Using fluorescence signals that monitor different protein species, we show that ClpXP produces a strand-extracted intermediate of SFGFP-ssrA, which is substantially populated both at low ATPase rates where degradation stalls, and at high ATPase rates where robust degradation occurs. The rates of appearance and disappearance of this intermediate suggest an on-pathway role in unfolding and degradation. SFGFP contains multiple stabilizing mutations,25 which allowed us to design, purify, and characterize circularly permuted variants in which different β strands of SFGFP contained the C-terminal ssrA tag. One of these variants showed ClpXP stalling at low ATPase rates, whereas another did not. We also engineered sites for thrombin cleavage between the C-terminal and penultimate strands of SFGFP-ssrA and a permuted variant, cleaved these proteins to produce split proteins, and then removed the C-terminal strand by ClpXP extraction to test if the resulting 10-stranded barrels maintained metastable structures. Stalling was only observed for substrates in which the strand-extracted protein remained stably folded. In combination, our experiments show that ClpXP unfolds GFP in a stepwise fashion and support a model in which the rates of terminal-strand extraction, strand refolding, and unfolding of the 10-stranded intermediate all depend on the rate of ATP hydrolysis.
Results
ClpXP extraction of terminal β strands in split-GFP variants
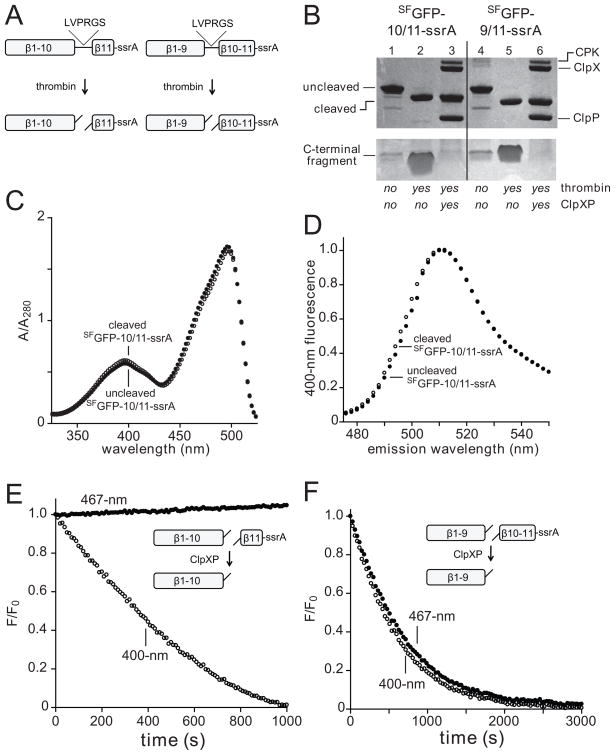
GFP lacking its 11th strand maintains a folded structure.7,26 We inserted a site for thrombin cleavage between strands 10 and 11 of SFGFP-ssrA (SFGFP-10/11-ssrA; Fig. 2A), incubated the purified protein with thrombin, and confirmed that cleavage had occurred by SDS-PAGE (Fig. 2B, lanes 1 and 2). This split protein had absorbance and fluorescence spectra similar to those of the uncleaved protein (Fig. 2C, 2D). Next, we incubated the split substrate with ClpXP and saturating ATP, and monitored fluorescence emission after excitation at 400 or 467 nm (hereafter, called 400-nm and 467-nm fluorescence). Importantly, we observed complete time-dependent loss of 400-nm fluorescence (initial rate ~1.5 min−1 enz−1) with a small increase in 467-nm fluorescence (Fig. 2E). SDS-PAGE confirmed that incubation of the split SFGFP-10/11-ssrA substrate with ClpXP destroyed the small fragment, corresponding to the ssrA-tagged 11th strand, but did not alter the large fragment, corresponding to the remaining structural elements of GFP (Fig. 2B, lane 3). Previous studies have shown that 467-nm fluorescence depends on shielding of the GFP chromophore from water by the β barrel,27 whereas 400-nm fluorescence requires excited state proton transfer (ESPT) from the chromophore to the side chain of Glu222 in strand 11 (Fig. 1C).22,24 In combination, these results indicate that ClpXP removes the ssrA-tagged 11th strand of the split substrate without denaturing the remaining 10-stranded structure.
Fig. 2.
(A) A thrombin cleavage sequence (LVPRGS) was inserted between strands 10/11 or 9/10 in the SFGFP-10/11-ssrA and SFGFP-9/10-ssrA proteins (NCBI accession codes JF951868 and JF951869, respectively), allowing creation of split proteins. (B) SDS-PAGE showing SFGFP-10/11-ssrA or SFGFP-9/10-ssrA (10 μM each) before and after thrombin cleavage and ClpXP (0.3 μM ClpX6; 0.9 μM ClpP14) extraction/degradation. The gel is a composite, with the lower portion taken from a gel containing 8-fold more sample than the upper portion. CPK, creatine phosphokinase. (C) Absorbance spectra of SFGFP-10/11-ssrA before (closed circles) or after (open circles) thrombin cleavage. (D) Fluorescence emission spectrum of SFGFP-10/11-ssrA before (closed circles) or after (open circles) thrombin cleavage. (E) Incubation of 10 μM thrombin-cleaved SFGFP-10/11-ssrA with 1 μM ClpXP (1 μM ClpX6; 2 μM ClpP14) and 4 mM ATP resulted in loss of 400-nm (open circles) but not 467-nm (closed circles) fluorescence. (F) Incubation of 10 μM thrombin-cleaved SFGFP-9/10-ssrA with 1 μM ClpXP and 4 mM ATP resulted in loss of 400-nm (open circles) and 467-nm (closed circles) fluorescence. The experiments in panels E and F contained an ATP-regeneration system.
We also constructed a SFGFP-9/10-ssrA variant, containing a thrombin-cleavage site between strands 9 and 10 (Fig. 2A), cleaved this protein with thrombin (Fig. 2B, lanes 4 and 5), and performed experiments similar to those described above. In this case, incubation of the 9/10-split substrate with ClpXP resulted in loss of both 400-nm and 467-nm fluorescence with an initial rate of ~0.5 min−1 enz−1 (Fig. 2F). We conclude that ClpXP extraction of the 11th strand of GFP leaves the β barrel largely intact, whereas extraction of both the 10th and the 11th strands leads to denaturation of the barrel, allowing solvent to quench the chromophore. Our finding that ClpXP extraction of strand 11 from the 10/11-split substrate occurred faster than extraction of strands 10 and 11 from the 9/10-split substrate is consistent with a sequential model of extraction of these elements of secondary structure in the intact native protein.
GFP fluorescence during ClpXP stalling supports terminal-strand extraction
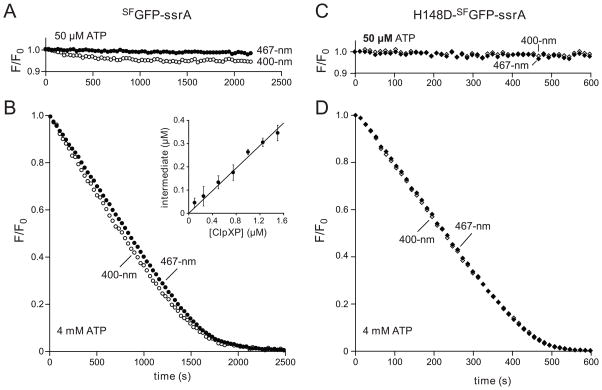
At the low ATPase rates that result in stalling, competition between ClpXP extraction and subsequent refolding of the eleventh strand of GFP was proposed to result in populations of the strand-extracted and native structures that depend on the rates of each reaction.7 This model predicts that stalling conditions should result in lower values of 400-nm fluorescence (a measure of intact GFP) as compared to 467-nm fluorescence (intact GFP plus the strand-extracted species). Indeed, using ClpXP (1 μM), SFGFP-ssrA (10 μM), and a low ATP concentration (50 μM), we observed a time-dependent decrease in 400-nm fluorescence but almost no change in 467-nm fluorescence (Fig. 3A). After ~1000 s, the 400-nm fluorescence stabilized, suggesting that equilibrium had been reached. We observed no change in 400-nm or 467-nm fluorescence in the absence of ClpXP but observed rapid loss of both signals when 4 mM ATP was added to the stalled reaction after 2500 s (data not shown).
Fig. 3.
(A) Changes in 400-nm fluorescence (open circles) or 467-nm fluorescence (closed circles) following incubation of SFGFP-ssrA (10 μM) with ClpXP (1 μM ClpX6; 2 μM ClpP14) and 50 μM ATP. (B) Same proteins as in panel A but using 4 mM ATP. The inset shows the concentration of the strand-extracted intermediate after 250 s as a function of ClpXP concentration from 4 mM ATP experiments like the one in the main panel. Values plotted are averages (n=4) ± 1 standard deviation. (C) Changes in 400-nm fluorescence (open diamonds) or 467-nm fluorescence (closed diamonds) following incubation of H148D-SFGFP-ssrA (10 μM; NCBI accession code JF951865) with ClpXP (1 μM ClpX6; 2 μM ClpP14) and 50 μM ATP. (D) Same proteins as in panel B but using 4 mM ATP. An ATP-regeneration system was used in all experiments.
To determine if the strand-extracted intermediate of SFGFP-ssrA accumulated under robust degradation conditions, we assayed changes in 400-nm and 467-nm fluorescence in a reaction containing 10 μM substrate, 1 μM ClpXP, and 4 mM ATP (Fig. 3B). As noted above, 467-nm fluorescence includes contributions from native GFP plus the intermediate, whereas 400-nm fluorescence depends only on the native GFP concentration. Thus, the concentration of the strand-extracted intermediate can be calculated as a function of the initial GFP concentration (GFP0), and the normalized 400-nm and 467-nm fluorescence: [I] = GFP0•(467F/467F0 − 400F/400F0). The concentration of the intermediate was substantial (~25% of the ClpXP concentration) during most of the degradation reaction (Fig. 3B). Moreover, when we varied the ClpXP concentration but kept the SFGFP-ssrA (10 μM) and ATP (4 mM) concentrations constant, the amount of the intermediate increased linearly with ClpXP concentration (Fig. 3B, inset). This result is expected for an enzyme-bound intermediate in degradation. An on-pathway intermediate would need to form at a faster rate than overall degradation. Indeed, the rate of ClpXP extraction of strand 11, calculated from the 10/11-split GFP experiment (Fig. 2E), was ~4-fold faster than the degradation rate in the experiment using 1 μM ClpXP and 4 mM ATP (Fig. 3B).
The His148→Asp GFP mutation on β-strand 7 provides an alternative ESPT acceptor and restores 400-nm fluorescence to Glu222→Gln GFP.28 Thus, H148D-SFGFP should not lose 400-nm fluorescence even upon extraction of Glu222 and strand 11 from the β barrel. Indeed, incubation of H148D-SFGFP-ssrA with ClpXP caused no change in 400-nm or 467-nm fluorescence using 50 μM ATP (Fig. 3C), which results in stalling, but caused concurrent loss of both signals during degradation using 4 mM ATP (Fig. 3D).
Stalling behavior of circularly permuted GFP variants
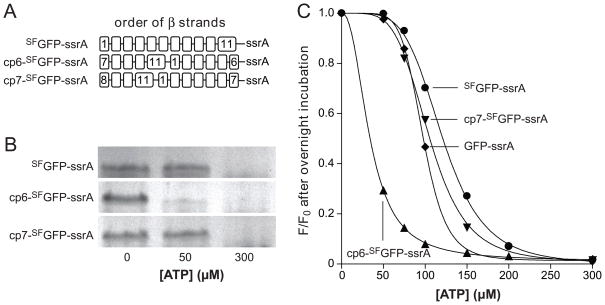
A circularly permuted GFP that lacks strand 7 (strand order 8-9-10-11-1-2-3-4-5-6) forms a 10-stranded fluorescent barrel.29 This result suggested that a circularly permuted ssrA-tagged variant in which strand 7 was at the C-terminus of the β barrel (cp7-SFGFP-ssrA) might also display ClpXP stalling behavior. Indeed, when we constructed and purified cp7-SFGFP-ssrA (Fig. 4A), ClpXP degraded this substrate at high but not low ATP concentrations as measured by SDS-PAGE (Fig. 4B) or by loss of 467-nm fluorescence (Fig. 4C).
Fig. 4.
(A) Cartoon representation of the order of β strands in SFGFP-ssrA and circularly permuted variants. (B) Permuted variants (1 μM) were incubated overnight with ClpXP (1.25 μM ClpX6; 2.5 μM ClpP14), the SspB adaptor (1 μM), and 0, 50, or 300 μM ATP before assaying degradation by SDS-PAGE. (C) End-point experiments like those in panel B were performed but degradation was assayed by reduced 467-nm fluorescence. GFP-ssrA (circles); SFGFP-ssrA (squares); cp6-SFGFP-ssrA (diamonds); cp7-SFGFP-ssrA (triangles). The lines are fits to a modified form of the Hill equation. In the panel B and C experiments, an ATP-regeneration system was used.
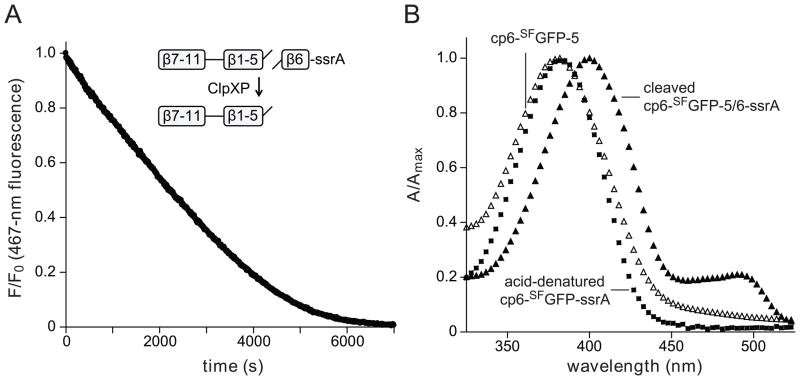
We also constructed a circularly permuted mutant with strand 6 at the C-terminus (cp6-SFGFP-ssrA; Fig. 4A). Strikingly, ClpXP degraded cp6-SFGFP-ssrA at low concentrations of ATP, which did not support degradation of SFGFP-ssrA or cp7-SFGFP-ssrA (Fig. 4B & 4C). We conclude that stalling is not an intrinsic property of the architecture of GFP, but depends instead on which strand is initially extracted by ClpX. To test the importance of strand 6 in barrel stability, we engineered a thrombin-cleavage site between the penultimate and terminal strands of cp6-SFGFP-ssrA to generate a cp6-SFGFP-5/6-ssrA variant. Following thrombin cleavage, ClpXP extraction of the terminal peptide of cp6-SFGFP-5/6-ssrA caused complete loss of native 467-nm fluorescence (Fig. 5A). After ClpXP strand extraction, the cp6-SFGFP-5 protein appeared to be aggregated and eluted in the void volume of an S200 gel-filtration column. Moreover, in comparison with the native split protein, the absorbance spectrum of the gel-filtered protein had a blue-shifted absorbance maximum near 395 nm and had lost an absorbance peak near 490 nm (Fig. 5B). These properties are similar to those of uncleaved cp6-SFGFP-ssrA after acid denaturation (Fig. 5B). Thus, ClpXP degradation of cp6-SFGFP-ssrA does not stall at low ATP concentrations, and extraction of the C-terminal strand of a closely related protein causes loss of barrel integrity.
Fig. 5.
(A) The cp6-SFGFP-5/6-ssrA protein (10 μM; NCBI accession code JF951870) was cleaved with thrombin and incubated with ClpXP (1 μM ClpX6; 2 μM ClpP14), 4 mM ATP, and an ATP-regeneration system. ClpXP extraction of the terminal β strand resulted in time-dependent loss of 467-nm fluorescence and 400-nm fluorescence (data not shown). The initial rate of this reaction (0.2 min−1 enz−1) was slow, but within error of the rate of ClpXP degradation of uncleaved cp6-SFGFP-5/6-ssrA (data not shown). (B) Absorbance spectra of thrombin-cleaved cp6-SFGFP-5/6-ssrA (closed triangles), the cp6-SFGFP-5 protein after ClpXP strand extraction and purification by S200 gel filtration (open triangles), and cp6-SFGFP-ssrA denatured by incubation at pH 2 (squares).
In combination, these results show a correlation between the stability of strand-extracted intermediates and the ability of different substrates to stall ClpXP degradation at low ATP concentrations. Specifically, the SFGFP-ssrA and cp7-SFGFP-ssrA proteins stall ClpXP and removal of their C-terminal strands (strands 11 and 7, respectively) results in 10-stranded barrels that are stable enough to maintain native fluorescence. By contrast, the cp6-SFGFP-ssrA protein did not stall ClpXP, and extraction of its C-terminal strand resulted in unfolding.
Stalling substrates have lower maximal rates of ClpXP degradation
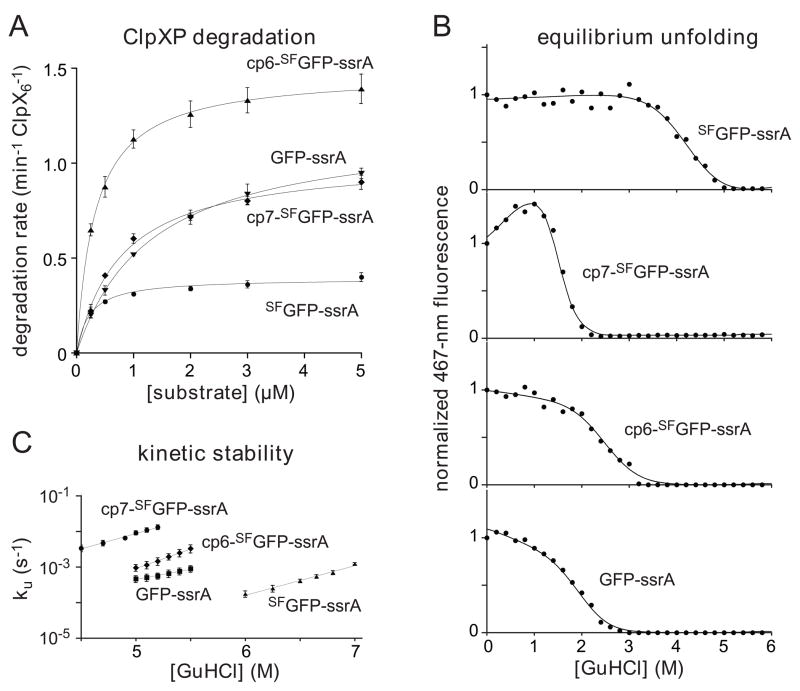
Using saturating ATP, we determined steady-state kinetic parameters for ClpXP degradation of different concentrations of GFP-ssrA, SFGFP-ssrA, cp6-SFGFP-ssrA, and cp7-SFGFP-ssrA (Fig. 6A; Table 1). Interestingly, cp6-SFGFP-ssrA, the only substrate which did not stall, also had the highest Vmax value. Although the “stalling” substrates displayed a range of Vmax values, with SFGFP-ssrA being the slowest, the overall correlation suggests that substrates whose degradation stalls at low ATPase rates are also more difficult to degrade at high ATPase rates.
Fig. 6.
(A) Michaelis-Menten plots of ClpXP degradation (0.1 μM ClpX6; 0.2 μM ClpP14) of ssrA-tagged variants of GFP and SFGFP in the presence of 4 mM ATP and an ATP-regeneration system. Initial degradation rates were calculated from changes in 467-nm fluorescence. The lines are fits to the equation rate = Vmax•[S]/(KM + [S]). Error bars (± 1 SD) based on four independent replicates. KM and Vmax values for each substrate are listed in Table 1. (B) Proteins (0.5 μM) were incubated with different concentrations of GuHCl for 2 weeks, and denaturation was assayed by 467-nm fluorescence. The solid lines are fits to a two-state unfolding model. cp7-SFGFP-ssrA has an unusual native baseline and higher meq value than the other proteins (Table 1). These properties could reflect an increase in the solvent accessibility of the unfolded protein and/or the presence of a populated unfolding intermediate. (C) Rates constants for unfolding (ku) were determined by single-exponential fits of changes in 467-nm fluorescence after jumps to different concentrations of GuHCl. The values plotted are averages of three independent experiments ± 1 standard deviation. The final protein concentration in each assay was 0.5 μM.
Table 1.
Properties of ssrA-tagged GFP substrates.
| degradation | ATP hydrolysis | protein stability | NCBI accession code | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| stalls* | KM μm | Vmax min−1 enz−1 | KM μm | Hill | Vmax min−1 enz−1 | G kcal/mol | meq kcal/mol M−1 | ku s−1 0 M GuHCl | mu kcal/mol M−1 | ||
| SFGFP- ssrA | yes | 0.36 | 0.39 | 114 | 1.85 | 159 | 6.9 | 1.6 | 1.6•10−9 | 1.1 | JF951864 |
| cp7- SFGFP- ssrA | yes | 0.79 | 1.02 | 131 | 1.85 | 203 | 4.4 | 3.0 | 4.2•10−7 | 1.2 | JF951867 |
| cp6- SFGFP- ssrA | no | 0.33 | 1.48 | 115 | 2.2 | 336 | 4.9 | 1.9 | 2.9•10−9 | 1.5 | JF951866 |
| GFP- ssrA | yes | 1.3 | 1.19 | 133 | 1.96 | 167 | 4.6 | 2.3 | 5.9•10−7 | 0.79 | JF951863 |
Stalling substrates displayed < 5% of the maximal degradation rate when the ATPase rate was 50% of maximal (Fig. 8B). Errors for KM and ku (± 50%); errors for other values (± 10%).
Equilibrium and kinetic stability
We determined the thermodynamic and kinetic stabilities of the GFP-ssrA, SFGFP-ssrA, cp6-SFGFP-ssrA, and cp7-SFGFP-ssrA proteins at different concentrations of GuHCl. The equilibrium stability of the SFGFP-ssrA protein was substantially greater than the stabilities of GFP-ssrA, cp6-SFGFP-ssrA, and cp7-SFGFP-ssrA (Fig. 5B; Table 1). At 5 M GuHCl, the order of kinetic stabilities from most to least stable was SFGFP-ssrA > GFP-ssrA > cp6-SFGFP-ssrA > cp7-SFGFP-ssrA (Fig. 5C); extrapolation to 0 M denaturant gave SFGFP-ssrA > cp6-SFGFP-ssrA > GFP-ssrA > cp7-SFGFP-ssrA (Table 1). These results show that neither ClpXP stalling nor the maximal rate of degradation correlate with the equilibrium or kinetic stabilities of the GFP variants, a result that is consistent with previous studies of different ClpXP substrates.4,30 Indeed, the single non-stalling substrate, cp6-SFGFP-ssrA, was degraded at the fastest rate but had stabilities intermediate between those of the non-stalling substrates.
Tests for sequence-dependent effects on unfolding
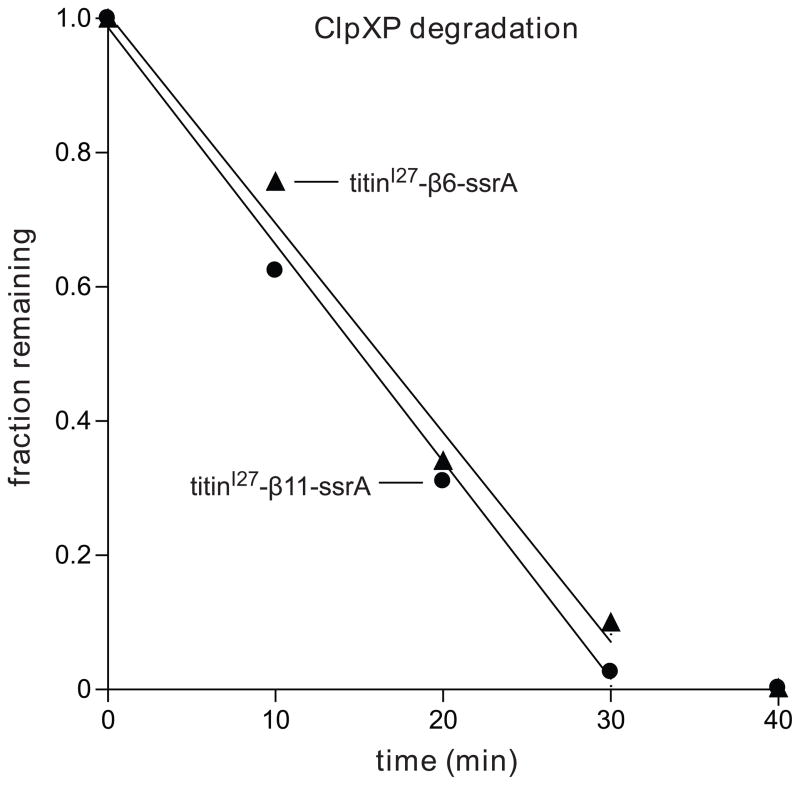
Because the initial step in ClpXP degradation of GFP involves extraction of the terminal strand, the enzyme would then need to pull on the extracted sequence as it attempted to unfold the resulting 10-stranded barrel. As shown above, ClpXP degraded cp6-SFGFP-ssrA, which does not stall, with a ~3-fold higher Vmax than SFGFP-ssrA, which does stall. To test if these differences in degradation rates result from the differential ability of ClpXP to grip the sequences that form β-strands 6 or 11, we cloned these sequences between a folded protein domain (titinI27) and a C-terminal ssrA-tag. Importantly, ClpXP degraded each substrate at a similar rate (Fig. 7), suggesting that it grips the strand-6 and strand-11 sequences almost equally well during unfolding. Thus, the properties of the strand-6 and strand-11 sequences do not appear to account for the slower degradation of SFGFP-ssrA compared to cp6-SFGFP-ssrA or for the stalling differences between these substrates.
Fig. 7.
Sequences corresponding to the 6th β-strand (TLVNRIELKGI) or the 11th β-strand strand (HMVLLEFVTAA) of SFGFP were inserted between the titinI27 protein and the ssrA tag (NCBI accession codes JF951871 and JF951872, respectively). ClpXP (1 μM ClpX6; 2 μM ClpP14) degradation of each substrate (10 μM) was monitored by SDS-PAGE, staining with Coomassie blue and densitometry. Reactions contained 4 mM ATP and an ATP-regeneration system.
ATPase dependence
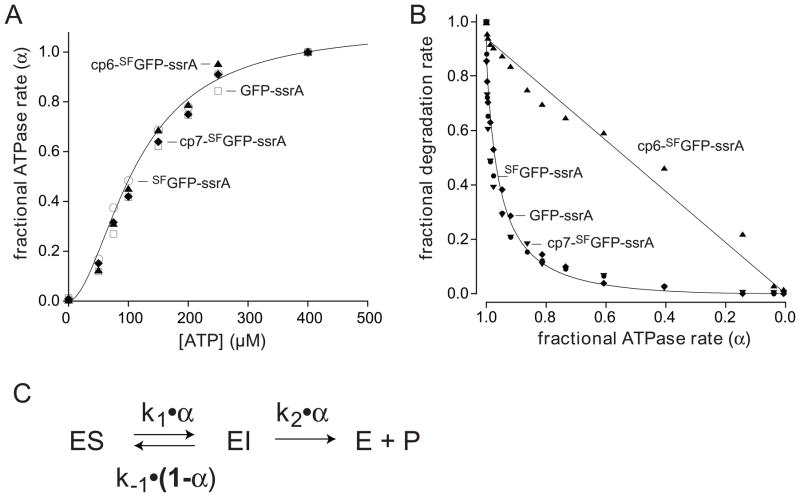
We determined the rates at which ClpXP hydrolyzed different concentrations of ATP in the presence of 10 μM GFP-ssrA, SFGFP-ssrA, cp6-SFGFP-ssrA, or cp7-SFGFP-ssrA. Fig. 8A shows normalized ATPase rates plotted as a function of ATP concentration. For each substrate, the v/Vmax curves were similar and were fit well by the Hill version of the Michaelis-Menten equation, with half-maximal rates at ATP concentrations of 115–135 μM and positively cooperative n-values of 1.8 to 2.2 (Table 1). There were, however, substrate-dependent Vmax differences, with the values for GFP-ssrA, SFGFP-ssrA, and cp7-SFGFP-ssrA being roughly similar (159–203 min−1 enz−1), whereas the value for the non-stalling substrate, cp6-SFGFP-ssrA, was substantially higher (336 min−1 enz−1).
Fig. 8.
(A) Fractional rates of ATP hydrolysis (α = v/Vmax) by ClpXP (1 μM ClpX6; 2 μM ClpP14) at different concentrations of ATP in the presence of 10 μM GFP-ssrA (squares), SFGFP-ssrA (circles), cp6-SFGFP-ssrA (triangles), or cp7-SFGFP-ssrA (diamonds). The solid line is a fit of the SFGFP-ssrA data to α = 1/(1+(KM/[ATP])n), the Hill form of the Michaelis-Menten equation. KM, Vmax, and n values for each protein substrate are listed in Table 1. (B) Fractional degradation rates (v/Vmax) for ClpXP (1 μM ClpX6; 2 μM ClpP14) proteolysis of 10 μM GFP-ssrA (diamonds), SFGFP-ssrA (closed circles), cp6-SFGFP-ssrA (triangles), and cp7-SFGFP-ssrA (downward triangles) are plotted as a function of the fractional rate of ATP hydrolysis (α). Solid lines are fits to the equation ka•α2/(kb•(1−α)+α) for GFP-ssrA and cp6-SFGFP-ssrA, where ka = k1k2/(k1+k2) and kb = k-1/(k1+k2). (C) Single-intermediate model for enzymatic unfolding, where ES represents the complex of ClpXP with intact GFP and EI represents a complex in which the terminal β strand of the substrate has been extracted, leaving a 10-stranded β barrel.
Next we determined rates of degradation for GFP-ssrA, SFGFP-ssrA, cp6-SFGFP-ssrA, or cp7-SFGFP-ssrA over a wide range of ATP concentrations by measuring loss of 467-nm fluorescence. Fig. 8B shows the fractional degradation rate plotted as a function of the fractional ATPase rate (defined as α). Strikingly, degradation of GFP-ssrA, SFGFP-ssrA, and cp7-SFGFP-ssrA, the substrates in which the strand-extracted intermediate is stable, fell off in similar, very steep non-linear fashions. Indeed, a modest initial decline in the ATPase rate from 100 to 90% of maximal resulted in a ~5-fold decrease in the degradation rate of these substrates. By contrast, ClpXP degradation of cp6-SFGFP-ssrA, the substrate in which the strand-extracted intermediate is unstable, decreased in a roughly linear manner with the ATPase rate.
We found that single-intermediate model generally accounted for both the stalling and non-stalling behaviors of the different GFP substrates (Fig. 8C). In this model, the ClpXP-substrate complex (ES) forms a strand-extracted intermediate (EI) with a rate constant of k1•α, and EI is subsequently denatured/degraded with a rate constant of k2•α or refolds to ES with a rate constant of k−1• (1−α). The latter term, disfavors refolding of the strand-extracted intermediate at high ATPase rates, when most ClpXP enzymes are ATP bound. Assuming steady-state for [EI] and substrate saturation, this model predicts that the fractional degradation rate equals k1•k2•α2/(k−1•(1−α)+(k1+k2)•α). Fig. 8B shows fits to this equation for one of the stalling substrates (GFP-ssrA; R > 0.99) and for the non-stalling substrate (cp6-SFGFP-ssrA; R > 0.99). Good fits were also obtained for SFGFP-ssrA and cp7-SFGFP-ssrA (R > 0.98). Although the single-intermediate model recapitulates most of the observed stalling features, it seems likely that more than one unfolding intermediate is populated to some degree, potentially including intermediates which the C-terminal β strand is only partially dislodged or extracted.
Discussion
To explain why degradation of GFP-ssrA ceases at low rates of ATP hydrolysis, Martin et al. proposed that ClpXP extracts the ssrA-tagged terminal strand from the GFP β barrel but the intermediate refolds before further unfolding and degradation can occur.7 Our results strongly support this stepwise model for ClpXP unfolding of GFP, which leads to futile cycles of strand extraction and refolding at low concentrations of ATP. For example, we designed a split variant in which the ssrA-tagged 11th strand was non-covalently bound to the remaining GFP structure and found that ClpXP extracted this strand without denaturing the rest of the protein. Moreover, the rate of strand extraction in this experiment was fast enough to account for the rate of GFP proteolysis, as expected for an on-pathway step in the degradation reaction. Notably, when ClpXP extracted the 11th and 10th strands of another split GFP variant, the remaining portions of GFP did denature. Thus, GFP lacking its C-terminal 11th strand is reasonably stable but subsequent extraction of the adjacent 10th strand results in unfolding. GFP-ssrA lacking its C-terminal strand loses 400-nm fluorescence, which depends upon Glu222 in strand 11, but maintains normal 467-nm fluorescence. Importantly, we found that ClpXP produces GFP species with a reduced ratio of 400/467 fluorescence both under low-ATP stalling conditions and high-ATP degradation conditions, as predicted if a strand-extracted intermediate is populated under these conditions.
We designed a circularly permuted variant of superfolder GFP (cp7-SFGFP-ssrA), which should also form a stable 10-stranded intermediate after extraction of its C-terminal strand.29 Like non-permuted SFGFP-ssrA, this variant was resistant to ClpXP degradation at low ATP-hydrolysis rates. By contrast, another circularly permuted variant (cp6-SFGFP-ssrA) unfolded following extraction of its C-terminal strand and did not stall ClpXP. Thus, the ability to resist ClpXP degradation at low ATP concentrations correlates with the ability of the substrate to form a stable unfolding intermediate, even though stalling did not correlate with the global stabilities of different substrates. At saturating ATP, ClpXP degraded stalling GFP variants more slowly than the non-stalling variant. Thus, the presence of a stable unfolding intermediate appears to slow normal degradation. We note, however, that Vmax for ClpXP degradation of SFGFP-ssrA was less than half the value of the other stalling substrates (Table 1), suggesting that changes in the rates of strand extraction and/or in the rates of unfolding of the 10-stranded barrel also play roles in determining the overall rates of degradation of these proteins.
ClpXP appears to unfold proteins using a power-stroke mechanism.15–17 Specifically, each cycle of ATP hydrolysis by ClpX is thought to result in an attempt to translocate a segment of the substrate polypeptide, thereby pulling the native protein against the narrow axial channel and creating a transient unfolding force. For stable substrates, like the titinI27 domain, hundreds of cycles of ATP hydrolysis can be required before denaturation becomes statistically probable.15 This result suggests that a power stroke must coincide with a stochastic decrease in protein stability to successfully extract the terminal structural element of the substrate. For titinI27, this initial ClpXP-mediated unfolding event appears to cause global denaturation.7 Studies of ClpXP unfolding of a multidomain filamin substrate assayed by optical-trapping nanometry also support this model.16 For example, in different single-molecule experiments, the dwell time before unfolding of a specific filamin domain varied from a few to more than 100 s, with the latter time being sufficient to hydrolyze several hundred ATP molecules. However, once unfolding of a filamin domain commenced, highly cooperative denaturation was typically complete in less than 1 ms. Subsequent ATPase cycles then resulted in translocation of the unfolded protein in steps of 5–8 amino acids at an average rate of ~30 residues s−1.
Our current view of the mechanism by which ClpXP unfolds GFP-ssrA begins with repeated enzymatic tugging on the 11th or C-terminal strand that is attached to the ssrA tag. During this process, enzymatic pulling will occasionally partially or completely dislodge the terminal strand, leaving a native 10-stranded barrel. Multiple cycles of ATP hydrolysis are then required to finish translocation of the extracted strand and preceding turn (~18 residues) and to unfold the remaining 10-stranded structure to allow degradation. The time required for completion of these events will increase as the ATPase rate decreases. Refolding of the extracted strand before unfolding of the 10-stranded barrel would restore the original substrate, necessitating renewed attempts to begin denaturation of the 11-stranded barrel. Strand refolding probably requires slipping of the substrate from the grip of ClpXP and is therefore more likely to occur at low ATPase rates when a higher fraction of enzymes are in an ATP-free state. Indeed, good fits of the observed ATP dependence of GFP degradation by ClpXP required a strand-refolding rate proportional to 1 minus the fractional ATP-hydrolysis rate.
The protein-unfolding activities of different AAA+ proteases display considerable variation.31 For example, the HslUV and FtsH proteases fail to degrade GFP proteins with suitable C-terminal recognition tags, whereas Lon degrades such substrates about 200-fold more slowly than ClpXP or ClpAP.32–36 For the proteases that degrade these GFP variants poorly, it is presently unclear if these AAA+ enzymes fail to dislodge the C-terminal strand or if they fail in a subsequent step in unfolding and degradation. Our results suggest that this question could be resolved by assaying 400-nm and 467-nm fluorescence of appropriate GFP variants during unfolding attempts by Lon, HslUV, and FtsH. Moreover, preliminary experiments suggest that appropriately tagged versions of some of our circularly permuted GFP proteins will be useful model substrates for Lon and HslUV, allowing convenient fluorescence-based assays of the unfolding and degradation activities of these AAA+ proteases. We note that ClpXP extraction of the terminal elements of split proteins also provides a powerful new tool with which to investigate the sequence determinants of protein structure and function. GFP-fusion proteins are commonly used to study protein localization and turnover in vivo, but interpretations can be complicated if partial degradation generates free GFP. This problem could potentially be overcome by fusing proteins to a circularly permutated GFP that can be completely degraded.
Materials and Methods
Protein Expression, Purification, and Cleavage
E. coli ClpX and E. coli ClpP were purified as described.14,37 A His6-tagged variant of E. coli SspB was purified by Ni2+-NTA chromatography and S200 size-exclusion chromatography.38 GFP or titinI27 substrates were expressed in E. coli strain X90, which had been transformed with appropriate overproducing plasmids. Cells were grown at 37 °C to OD600 = 0.8, protein expression from the T7 promoter was induced by addition of 1 mM IPTG, and growth was continued at room temperature for 3.5 h, before the cells were harvested and lysed.
The coding sequence for SFGFP was obtained from the Registry of Standard Biological Parts (BBa I746916). SFGFP variants were cloned into pCOLADuet-1 with an N-terminal H6 tag (MGSHHHHHH) and a C-terminal ssrA tag (AANDENYALAA). Table 1 lists the different SFGFP or GFP variants used for these studies and NCBI accession numbers for their amino-acid sequences. Most variants were constructed in the super-folder GFP sequence background (SFGFP).25 Circularly permuted GFP variants are designated with a cp# prefix, where # represents the C-terminal β strand of the permuted structure, and were cloned with a GGTGGS sequence connecting the residues corresponding to wild-type N terminus and C terminus.39 In some GFP variants, a GGTEGSLVPRGSGESGGS sequence for thrombin cleavage was inserted into the loop between two β strands to allow production of split proteins. These variants have names like SFGFP-10/11-ssrA, where 10/11 indicates insertion of the cleavage site between strands 10 and 11. The gene and amino-acid sequences of the substrates used in this work have been deposited in the databases of the National Center for Biotechnology Information; accession codes are listed in Table 1 or in figure legends.
All GFP variants were purified by Ni2+-NTA affinity (Qiagen) and S200 size-exclusion chromatography, and were stored in PD buffer (50 mM HEPES [pH 7.5], 200 mM KCl, 5 mM MgCl2, 10% glycerol). Cleavage of appropriate substrates with thrombin (GE Healthcare; 4 units/mg substrate) was performed in 20 mM Tris-HCl (pH 7.5), 150 mM NaCl, 2.5 mM CaCl2 for 2 h at 37 °C.
Substrates containing the human titinI27 domain, linker regions, and a C-terminal ssrA-tag were cloned in pACYC, purified by Ni2+-NTA affinity, and stored in PD buffer.
Biochemical Assays
Degradation and unfolding assays were performed at 30 °C in PD buffer and were monitored by SDS-PAGE, and/or by loss of fluorescence emission at 511 nm after excitation at 400 nm or at 467 nm. Degradation reactions contained ssrA-tagged substrates, E. coli ClpX6, E. coli ClpP14, and an ATP-regeneration system (16 mM creatine phosphate, 6 μg/mL creatine phosphokinase). Some degradation reactions also contained E. coli SspB, which helps deliver ssrA-tagged substrates for ClpXP degradation.40 Rates of ATP hydrolysis were determined at 30 °C in PD buffer using an assay in which production of ADP is coupled to enzymatic oxidation of NADH.41–42 Equilibrium and kinetic stability assays were performed at 30 °C in PD buffer supplemented with different concentrations of GuHCl and were monitored by changes in 467-nm fluorescence.
Research highlights.
ClpX extracts the C-terminal β-strand of GFP-ssrA first, leaving a 10-stranded barrel
The strand-extracted intermediate is populated during ClpX degradation
Refolding of the extracted strand is favored at low ATP, causing degradation to stall
Low-ATP stalling is not observed for one circularly permuted GFP variant
The strand-extracted intermediate is unstable for this non-stalling substrate
Acknowledgments
We thank Peter Chien, Santiago Lima, and Randall Mauldin for helpful discussions. Supported by NIH grant AI-15706. T.A.B. is an employee of the Howard Hughes Medical Insititute.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Baker TA, Sauer RT. ATP-dependent proteases: recognition logic and operating principles. Trends Biochem Sci. 2006;31:647–653. doi: 10.1016/j.tibs.2006.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Palombella VJ, Rando OJ, Goldberg AL, Maniatis T. The ubiquitin-proteasome pathway is required for processing the NF-κB1 precursor protein and the activation of NF-κB. Cell. 1994;78:773–785. doi: 10.1016/s0092-8674(94)90482-0. [DOI] [PubMed] [Google Scholar]
- 3.Levitskaya J, Sharipo A, Leonchiks A, Ciechanover A, Masucci MG. Inhibition of ubiquitin/proteasome-dependent protein degradation by the Gly-Ala repeat domain of the Epstein-Barr virus nuclear antigen 1. Proc Natl Acad Sci USA. 1997;94:12616–12621. doi: 10.1073/pnas.94.23.12616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lee C, Schwartz MP, Prakash S, Iwakura M, Matouschek A. ATP-dependent proteases degrade their substrates by processively unraveling them from the degradation signal. Mol Cell. 2001;7:627–637. doi: 10.1016/s1097-2765(01)00209-x. [DOI] [PubMed] [Google Scholar]
- 5.Kenniston JA, Baker TA, Sauer RT. Partitioning between unfolding and release of native domains during ClpXP degradation determines substrate selectivity and partial processing. Proc Natl Acad Sci USA. 2005;102:1390–1395. doi: 10.1073/pnas.0409634102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tian L, Holmgren RA, Matouschek A. A conserved processing mechanism regulates the activity of transcription factors Cubitus interruptus and NF-kappaB. Nat Struct Mol Biol. 2005;12:1045–1053. doi: 10.1038/nsmb1018. [DOI] [PubMed] [Google Scholar]
- 7.Martin A, Baker TA, Sauer RT. Protein unfolding by a AAA+ protease: critical dependence on ATP-hydrolysis rates and energy landscapes. Nat Struct Mol Biol. 2008;15:139–145. doi: 10.1038/nsmb.1380. [DOI] [PubMed] [Google Scholar]
- 8.Wojtkowiak D, Georgopoulos C, Zylicz M. Isolation and characterization of ClpX, a new ATP-dependent specificity component of the Clp protease of Escherichia coli. J Biol Chem. 1993;268:22609–22617. [PubMed] [Google Scholar]
- 9.Gottesman S, Clark WP, de Crecy-Lagard V, Maurizi MR. ClpX, an alternative subunit for the ATP-dependent Clp protease of Escherichia coli Sequence and in vivo activities. J Biol Chem. 1993;268:22618–22626. [PubMed] [Google Scholar]
- 10.Gottesman S, Roche E, Zhou YN, Sauer RT. The ClpXP and ClpAP proteases degrade proteins with C-terminal peptide tails added by the SsrA tagging system. Genes Dev. 1998;12:1338–1347. doi: 10.1101/gad.12.9.1338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Siddiqui SM, Sauer RT, Baker TA. Role of the protein-processing pore of ClpX, an AAA+ ATPase, in recognition and engagement of specific protein substrates. Genes Dev. 2004;18:369–374. doi: 10.1101/gad.1170304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Martin A, Baker TA, Sauer RT. Pore loops of the AAA+ ClpX machine grip substrates to drive translocation and unfolding. Nat Struct Mol Biol. 2008;15:1147–1151. doi: 10.1038/nsmb.1503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Martin A, Baker TA, Sauer RT. Diverse pore loops of the AAA+ ClpX machine mediate unassisted and adaptor-dependent recognition of ssrA-tagged substrates. Mol Cell. 2008;29:441–450. doi: 10.1016/j.molcel.2008.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kim YI, Burton RE, Burton BM, Sauer RT, Baker TA. Dynamics of substrate denaturation and translocation by the ClpXP degradation machine. Mol Cell. 2000;5:639–648. doi: 10.1016/s1097-2765(00)80243-9. [DOI] [PubMed] [Google Scholar]
- 15.Kenniston JA, Baker TA, Fernandez JM, Sauer RT. Linkage between ATP consumption and mechanical unfolding during the protein processing reactions of an AAA+ degradation machine. Cell. 2003;114:511–520. doi: 10.1016/s0092-8674(03)00612-3. [DOI] [PubMed] [Google Scholar]
- 16.Aubin-Tam ME, Olivares AO, Sauer RT, Baker TA, Lang MJ. Single-molecule protein unfolding and translocation by an ATP-fueled proteolytic machine. Cell. 2011;145:256–267. doi: 10.1016/j.cell.2011.03.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Maillard RA, Chistol G, Sen M, Righini M, Tan J, Kaiser CM, Hodges C, Martin A, Bustamante C. ClpX(P) generates mechanical force to unfold and translocate its protein substrates. Cell. 2011;145:459–469. doi: 10.1016/j.cell.2011.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Singh SK, Grimaud R, Hoskins JR, Wickner S, Maurizi MR. Unfolding and internalization of proteins by the ATP-dependent proteases ClpXP and ClpAP. Proc Natl Acad Sci USA. 2000;97:8898–8903. doi: 10.1073/pnas.97.16.8898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Heim R, Prasher DC, Tsien RY. Wavelength mutations and posttranslational autoxidation of green fluorescent protein. Proc Natl Acad Sci USA. 1994;91:12501–12504. doi: 10.1073/pnas.91.26.12501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cubitt AB, Heim R, Adams SR, Boyd AE, Gross LA, Tsien RY. Understanding, improving and using green fluorescent proteins. Trends Biochem Sci. 1995;20:448–455. doi: 10.1016/s0968-0004(00)89099-4. [DOI] [PubMed] [Google Scholar]
- 21.Ormö M, Cubitt AB, Kallio K, Gross LA, Tsien RY, Remington SJ. Crystal structure of the Aequorea victoria green fluorescent protein. Science. 2006;273:1392–1395. doi: 10.1126/science.273.5280.1392. [DOI] [PubMed] [Google Scholar]
- 22.Chattoraj M, King BA, Bublitz GU, Boxer SG. Ultra-fast excited state dynamics in green fluorescent protein: multiple states and proton transfer. Proc Natl Acad Sci USA. 1996;93:8362–8367. doi: 10.1073/pnas.93.16.8362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kent KP, Childs W, Boxer SG. Deconstructing green fluorescent protein. J Am Chem Soc. 2008;130:9664–9665. doi: 10.1021/ja803782x. [DOI] [PubMed] [Google Scholar]
- 24.Stoner-Ma D, Melief EH, Nappa J, Ronayne KL, Tonge PJ, Meech SR. Proton relay reaction in green fluorescent protein (GFP): Polarization-resolved ultrafast vibrational spectroscopy of isotopically edited GFP. J Phys Chem B. 2006;110:22009–22018. doi: 10.1021/jp065326u. [DOI] [PubMed] [Google Scholar]
- 25.Pédelacq JD, Cabantous S, Tran T, Terwilliger TC, Waldo GS. Engineering and characterization of a superfolder green fluorescent protein. Nat Biotechnol. 2006;24:79–88. doi: 10.1038/nbt1172. [DOI] [PubMed] [Google Scholar]
- 26.Cabantous S, Terwilliger TC, Waldo GS. Protein tagging and detection with engineered self-assembling fragments of green fluorescent protein. Nat Biotechnol. 2005;223:102–107. doi: 10.1038/nbt1044. [DOI] [PubMed] [Google Scholar]
- 27.Reid BG, Flynn GC. Chromophore formation in green fluorescent protein. Biochemistry. 1997;36:6786–6791. doi: 10.1021/bi970281w. [DOI] [PubMed] [Google Scholar]
- 28.Stoner-Ma D, Jaye AA, Ronayne KL, Nappa J, Meech SR, Tonge PJ. An alternate proton acceptor for excited-state proton transfer in green fluorescent protein; rewiring GFP. J Am Chem Soc. 2008;130:1227–1235. doi: 10.1021/ja0754507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Huang YM, Bystroff C. Complementation and reconstitution of fluorescence from circularly permuted and truncated green fluorescent protein. Biochemistry. 2009;48:929–940. doi: 10.1021/bi802027g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kenniston JA, Burton RE, Siddiqui SM, Baker TA, Sauer RT. Effects of local protein stability and the geometric position of the substrate degradation tag on the efficiency of ClpXP denaturation and degradation. J Struct Biol. 2004;146:130–140. doi: 10.1016/j.jsb.2003.10.023. [DOI] [PubMed] [Google Scholar]
- 31.Koodathingal P, Jaffe NE, Kraut DA, Prakash S, Fishbain S, Herman C, Matouschek A. ATP-dependent proteases differ substantially in their ability to unfold globular proteins. J Biol Chem. 2009;284:18674–18684. doi: 10.1074/jbc.M900783200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Flynn JM, Levchenko I, Seidel M, Wickner SH, Sauer RT, Baker TA. Overlapping recognition determinants within the ssrA degradation tag allow modulation of proteolysis. Proc Natl Acad Sci USA. 2001;11:10584–10589. doi: 10.1073/pnas.191375298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Herman C, Prakash S, Lu CZ, Matouschek A, Gross CA. Lack of a robust unfoldase activity confers a unique level of substrate specificity to the universal AAA protease FtsH. Mol Cell. 2003;11:659–669. doi: 10.1016/s1097-2765(03)00068-6. [DOI] [PubMed] [Google Scholar]
- 34.Kwon AR, Trame CB, McKay DB. Kinetics of protein substrate degradation by HslUV. J Struct Biol. 2004;146:141–147. doi: 10.1016/j.jsb.2003.11.003. [DOI] [PubMed] [Google Scholar]
- 35.Choy JS, Aung LL, Karzai AW. Lon protease degrades transfer-messenger RNA-tagged proteins. J Bacteriol. 2007;189:6564–6571. doi: 10.1128/JB.00860-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gur E, Sauer RT. Recognition of misfolded proteins by Lon, a AAA+ protease. Genes Dev. 2008;22:2267–2277. doi: 10.1101/gad.1670908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Neher SB, Sauer RT, Baker TA. Distinct peptide signals in the UmuD and UmuD′ subunits of UmuD/D′ mediate tethering and substrate processing by the ClpXP protease. Proc Natl Acad Sci USA. 2003;100:13219–13224. doi: 10.1073/pnas.2235804100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bolon DN, Wah DA, Hersch GL, Baker TA, Sauer RT. Bivalent tethering of SspB to ClpXP is required for efficient substrate delivery: a protein-design study. Mol Cell. 2004;13:443–449. doi: 10.1016/s1097-2765(04)00027-9. [DOI] [PubMed] [Google Scholar]
- 39.Baird GS, Zacharias DA, Tsien RY. Circular permutation and receptor insertion within green fluorescent proteins. Proc Natl Acad Sci USA. 1999;96:11241–11246. doi: 10.1073/pnas.96.20.11241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Levchenko I, Seidel M, Sauer RT, Baker TA. A specificity-enhancing factor for the ClpXP degradation machine. Science. 2000;289:2354–2356. doi: 10.1126/science.289.5488.2354. [DOI] [PubMed] [Google Scholar]
- 41.Nørby JG. Coupled assay of Na+,K+-ATPase activity. Methods Enzymol. 1988;156:116–119. doi: 10.1016/0076-6879(88)56014-7. [DOI] [PubMed] [Google Scholar]
- 42.Burton RE, Siddiqui SM, Kim YI, Baker TA, Sauer RT. Effects of protein stability and structure on substrate processing by the ClpXP unfolding and degradation machine. EMBO J. 2001;20:3092–3100. doi: 10.1093/emboj/20.12.3092. [DOI] [PMC free article] [PubMed] [Google Scholar]