Abstract
High levels of viremia and chemokines and cytokines underlie the progression of severe dengue disease. Dengue virus (DENV) preferentially infects peripheral blood monocytes, which secrete elevated levels of immunomediators in patients with severe disease. Further, DENV nonstructural proteins (NS) are capable of modifying intracellular signaling, including interferon inhibition. We demonstrate that peak secretions of immunomediators such as IL-6, IL-8, IP-10, TNFα or IFNγ in DENV-infected monocytes correlate with maximum virus production and NS4B and NS5 are primarily responsible for the induction of immunomediators. Furthermore, we demonstrate that sequential NS4AB processing initiated by the viral protease NS2B3(pro) and via the intermediate 2KNS4B significantly enhances immunomediator induction. While the 2K-signal peptide is not essential for immunomediator induction, it plays a synergistic role with NS4B. These data suggest that NS4B maturation is important during innate immune signaling in DENV-infected monocytes. Given similar NS4B topologies and polyprotein processing across flaviviruses, NS4B may be an attractive target for developing Flavivirus-wide therapeutic interventions.
Keywords: Dengue virus, DENV; dengue hemorrhagic fever; chemokines and cytokines; nonstructural protein NS5, NS4B; 2K-signal sequence; THP-1 monocytes
Introduction
Dengue virus (DENV) causes considerable risk to human health worldwide infecting an estimated 50–100 million people annually and causing explosive outbreaks with infection rates as high as 80–90% among individuals previously unexposed to the virus (World Health Organization, 1997). The incidence of dengue diseases has dramatically increased during the past two decades as the result of an expanding geographical distribution of the Aedes mosquito vector and increased human travel (Gubler, 2002; Gubler, 2006; World Health Organization, 1997). Over 90% of DENV infections are asymptomatic or result in self-limiting dengue fever (DF) cases that resolve without complications (Guzman and Isturiz, 2010; Guzman and Kouri, 2002). However, a subset of infected patients progresses to severe dengue hemorrhagic fever (DHF) and dengue shock syndrome (DSS), resulting in over a half-million hospitalizations each year worldwide (Gubler, 1998; World Health Organization, 2009). No definitive mechanisms explain the progression of DHF/DSS, which can be defined in part as bleeding and increased plasma leakage into the pleural cavities and peripheral tissue without morphological damage to the capillary endothelium (Chang, Harn, and Nimmannitya, 1990; Srikiatkhachorn et al., 2007). Clinical studies indicate that patients who progress to severe disease demonstrate elevated viremia (Libraty et al., 2002; Murgue et al., 2000; Vaughn et al., 2000) and high levels of interleukin (IL)-6, IL-8 and tumor necrosis factor alpha (TNFα) in the bloodstream (Hober et al., 1993; Huang et al., 2000; Juffrie et al., 2001; Nguyen et al., 2004; Priyadarshini et al., 2010; Raghupathy et al., 1998). Similarly, peripheral blood monocytes from patients with DHF/DSS display elevated DENV antigen and increased expression of activation markers and production of immunomediators (Durbin et al., 2008; Halstead, 1989; Kou et al., 2008), implicating monocytes as important cells during infection and severe disease pathogenesis. Moreover, DENV-infected primary monocytes secrete DHF/DSS-associated immunomediators (Bosch et al., 2002; Chen and Wang, 2002).
DENV belongs to the family Flaviviridae and consists of four genetically distinct serotypes having a positive-sense, single-stranded RNA genome of approximately 11 kilobases (kb) in length. The RNA encodes for a polyprotein precursor that is co- and post-translationally processed into three structural (C, PrM and E) and seven nonstructural proteins (NS1, NS2A, NS2B, NS3, NS4A, NS4B and NS5) (Bell et al., 1985; Chambers et al., 1990). The NS are responsible for various enzymatic activities during replication, including the NS5 RNA-dependent RNA polymerase (RdRP) and methyltransferase activity required for viral RNA capping (Chung et al., 2010; Dong et al., 2010; Geiss et al., 2009; Selisko et al., 2010; Zou et al., 2011), the NS3 helicase and the NS2B3 protease (NS2B3pro) (Cahour, Falgout, and Lai, 1992; Falgout et al., 1991; Padmanabhan et al., 2006). Several NS, such as NS3, NS4B and NS5, interact as part of the viral replication complex facilitating transcription and translation of the viral genome (Harris et al., 2006; Khromykh et al., 1999; Khromykh, Sedlak, and Westaway, 1999; Mackenzie, Kenney, and Westaway, 2007; Roosendaal et al., 2006; Westaway et al., 1997a; Westaway et al., 1997b). Further, accumulating evidence suggests that intrinsic DENV genetic characteristics within NS4B and NS5 are associated with severe disease outcomes (Leitmeyer et al., 1999).
DENV NS5 induces IL-8 transcription and protein secretion in human embryonic kidney cells, (Medin, Fitzgerald, and Rothman, 2005) and inhibits the interferon alpha (IFNα) response through binding and degradation of STAT2 (Ashour et al., 2009; Mazzon et al., 2009). Also, NS4B strongly inhibits the IFN transduction cascade by interfering with STAT1 phosphorylation (Munoz-Jordan et al., 2003); and processing of NS4AB by viral and host proteases is required to initiate an IFN-antagonistic function (Munoz-Jordan et al., 2005). Nonstructural protein-induced subversion of the host IFN response and induction of immunomediators may simultaneously promote DENV survival while increasing the risk of severe disease outcomes. Based on the aforementioned data, to further understand the role of NS in dengue immunopathogenesis, we hypothesized that NS5 and maturation of NS4B expressed in monocytes would induce DHF-associated immunomediators. In this report, we demonstrate that both NS5 and NS4B induce immunomediators and that NS4B maturation via cleavage of the NS4AB polypeptide, in a 2KNS4B-dependent manner, significantly enhanced immunomediator production in monocytes.
Results
Elevated secretion of immunomediators from DENV-infected THP-1 monocytes corresponds with peak viral titers and copy numbers
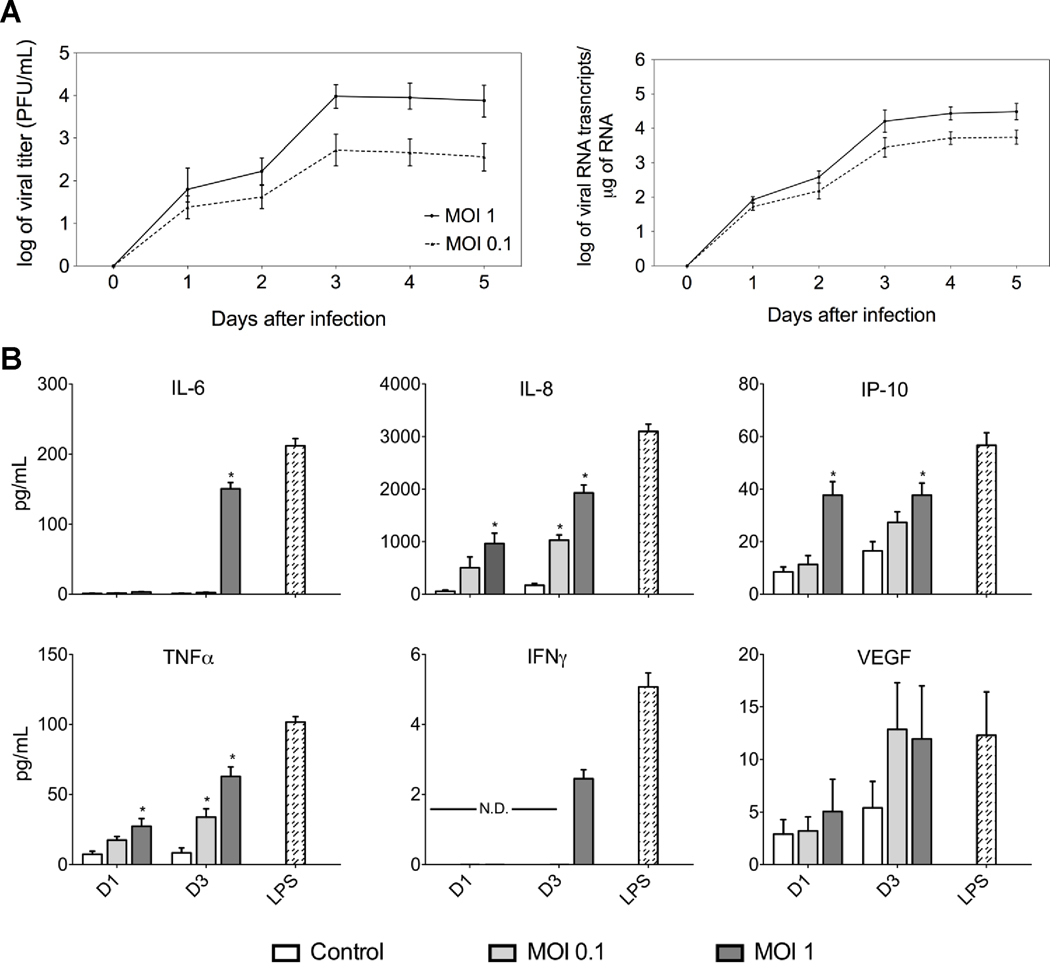
To establish whether DENV-infected monocytes secrete DHF-associated immunomediators, we infected THP-1 cells with DENV-2 New Guinea (NGC) strain and collected cells and culture supernatants each day for five consecutive days. Plaque assay and quantitative real-time polymerase chain reaction (qRT-PCR) data demonstrated that peak viral titers and copy numbers for both MOI-0.1 and -1 occurred on day 3 after infection; however, infection with MOI-1 resulted in approximately log 1.4 higher titer and log 0.5 higher viral RNA transcripts than infection with an MOI-0.1 on day 3 after infection (Fig. 1A). We determined by qRT-PCR that the peak induction of IL-8 and TNFα transcripts occurred at day 3 after infection (Supplemental Fig. 1); therefore, we assayed the THP-1 supernatants for immunomediators at days 1 and 3 after infection. The Luminex® multiplex data corresponded with transcript data in that the THP-1 cells infected with an MOI-1 induced peak secretion of IL-6, IL-8, interferon gamma-induced protein 10 (IP-10) and TNFα at day 3 after infection (Fig. 1B). Levels of IL-6, IL-8, IP-10, and TNFα peaked at 52, 2,000, 40 and 65 pg/mL, respectively. Also slight overall increases were observed in VEGF and IFNγ levels at day 3 after infection. THP-1 cells incubated with lipopolysaccharide (LPS) significantly produced all of the immunomediators tested both at the transcript and protein levels (Supplemental Fig. 1 and Fig. 1B) whereas THP-1 cells mock-infected with UV-treated virus produced low expression levels similar to those observed in untreated THP-1 cells (Fig. 1B). Overall, these results demonstrated that DENV-2 NGC-infected THP-1 cells produced DHF-associated immunomediators and that maximum transcripts expression and protein secretion levels corresponded with peak viral titers and copy numbers. These data are consistent with other in-vitro data demonstrating that infected THP-1 cells secrete elevated chemokines and cytokines, which corresponds with increased viral titers (Chareonsirisuthigul, Kalayanarooj, and Ubol, 2007; Hober et al., 1996; Ubol et al., 2008).
Fig. 1.
Kinetics of chemokines and cytokines secretion in DENV-infected THP-1 cells. THP-1 cells were infected with DENV-2 NGC strain at MOI-0.1 and -1 and cells and supernatants were collected every 24 h after infection for five consecutive days. (A) Viral titers and RNA copy numbers were determined by plaque assay and qRT-PCR, respectively, on samples collected each day after infection (MOI-1, solid line; MOI-0.1, dotted line). (B) Chemokine and cytokine levels in mock- and DENV-infected THP-1 cell culture supernatants collected at days 1 and 3 after infection (mock-infected, white bars; DENV-infected MOI-0.1, light gray bars; DENV-infected MOI-1, dark gray bars; and LPS treatment, striped bars). Data points and bars represent the mean ± SD of three independent experiments conducted in duplicate. (*) indicates statistical significance at p < 0.05 as compared to the same day controls; N.D., not detected.
NS4B or NS5 expressed in THP-1 cells induces IL-6, IL-8 and IP-10
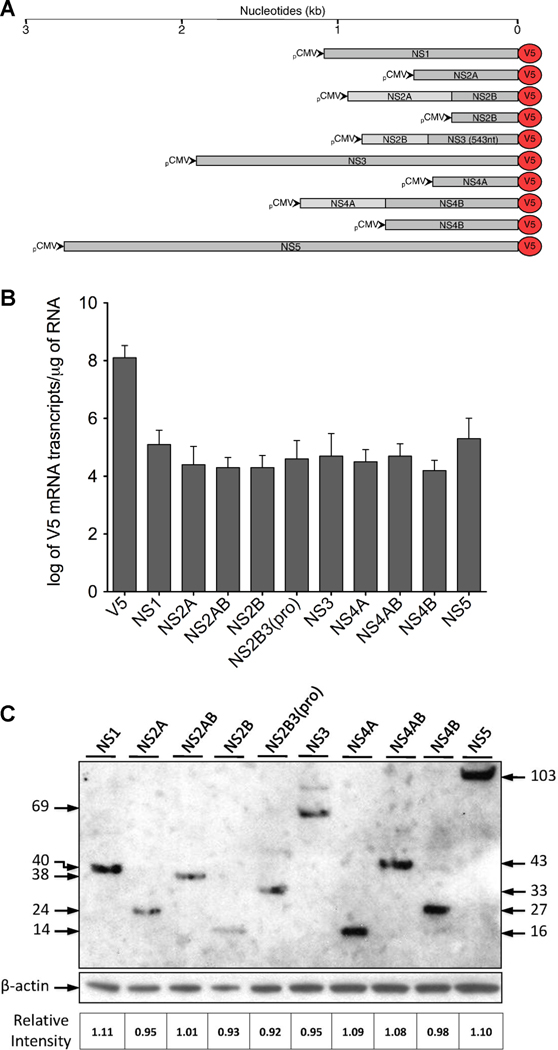
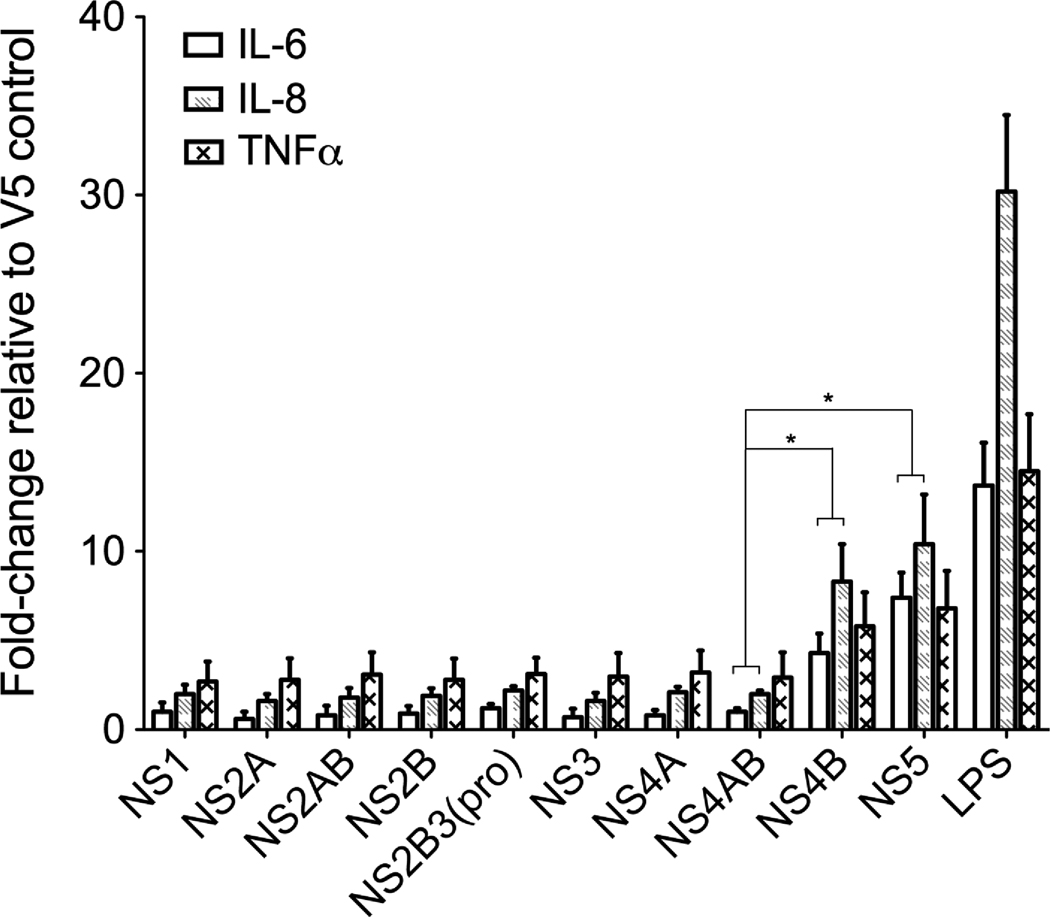
To evaluate the role of DENV NS on the observed induction of immunomediators, we constructed plasmids (Fig. 2A) with a commercially available V5 expression vector using primers designed from the wild-type NGC genome (Table 1). Following the manufacturer’s optimization instructions for the Neon™ system, we determined that maximum expression levels of the V5 control plasmid or NS5-V5 plasmid and THP-1 cell viability (approximately 70% for all plasmids) occurred at 40 h after transfection (Supplemental Fig. 2). Therefore, successful expression of each DENV plasmid was confirmed at 40 h after transfection by immunofluorescence assay (data not shown), qRT-PCR and western blot (Fig. 2B–C). We conducted qRT-PCR using primers designed to detect plasmid expressing V5 mRNA (Table 2) and demonstrated similar mRNA expression levels, wherein approximately log 4.25 copies of transcripts were expressed by all DENV-V5 fusion plasmids with no significant difference between the minimum (NS4B) and maximum (NS5) expression levels (p=0.32) (Fig. 2B). Also, we conducted western blot and demonstrated similar relative intensities (RIs), calculated as described in the materials and methods section, for all DENV proteins, wherein NS1 reached a maximum RI of 1.11 and NS2B3(pro) reached a minimum RI of 0.92 (Fig. 2C). Having established similar transcript and protein expression levels for all DENV plasmids at 40 h after electroporation, we screened transfected cells for immunomediator induction using qRT-PCR. We demonstrated that NS4B significantly increased IL-6 transcripts approximately 4-fold and IL-8 almost 9-fold relative to NS4AB (Fig. 3). Moreover, NS5 significantly increased IL-6 transcripts approximately 8-fold and IL-8 over 11-fold relative to NS4AB (Fig. 3). However, relative to NS4AB expression NS4B (p=0.07) or NS5 (p=0.09) did not significantly increase TNFα transcripts levels (Fig. 3).
Fig. 2.
Expression of nonstructural DENV-V5 fusion protein constructs in THP-1 cells. THP-1 cells were transfected with each DENV nonstructural gene plasmid and cell lysates were collected at 40 h after transfection for expression analysis by qRT-PCR and western blot. (A) Constructs and approximate nucleotide number (kb) of cloned viral genes. Each gene was constructed using a V5 reporter vector having the CMV promoter for expression in mammalian cells. (B) V5 copy numbers were determined for each expressing plasmid using qRT-PCR and reported as log of V5 mRNA transcripts per µg of RNA. A standard curve was prepared by serially diluting known amount of V5 transcripts. (C) Western blot analysis of DENV-V5 proteins expression. Twenty µL (approximately 50 µg) of each cell lysate was loaded and electrophoresed on SDS-PAGE followed by immunoblotting with V5 antibody. Protein loading amounts were confirmed using β-actin and a relative intensity (RI) was calculated by dividing the absolute intensity of the DENV-V5 protein band by the absolute intensity of its corresponding β-actin band. The cell lysate loaded in each well is specified at the top and its corresponding molecular mass (kDa) is depicted at the left or right. Bars represent the mean ± SD of three independent experiments conducted in duplicate. No significant differences (p < 0.05) were found across transcripts.
Table 1.
Primers employed for the construction of DENV gene plasmids.
| Construct and Primer Namesa |
Nucletide Positionb | Primer Sequence (5' - 3')c | Tm (°C) | Amplicon Size (bp) |
PCR Cycling Conditions |
|---|---|---|---|---|---|
| NS1 | |||||
| NS1-Forward (F) NS1-Reverse (R) |
2422–2443 3477-3449 |
accaccatgGATAGTGGTTGCGTTGTGAGCT GGCTGTGACCAAGGAGTTGACCAAATTCT |
65.9 62.3 |
1056 | 95°C 5 min 1 cycle; 95°C 45 s, 57°C 45 s, 72°C 1 min, 30 cycles; 72°C 7 min 1 cycle |
| NS2A | |||||
| NS2A-F NS2A-R |
3478–3499 4131-4102 |
accaccatgGGACATGGGCAGATTGACAACT CCTTTTCTTGTTGGTTCTTGAAAGGGTTGT |
66.1 59.6 |
654 | 95°C 5 min 1 cycle; 95°C 30 s, 55°C 30 s, 72°C 45 s, 30 cycles; 72°C 7 min 1 cycle |
| NS2B | |||||
| NS2B-F NS2B-R |
4132–4149 4521–4995 |
accaccatggctAGCTGGCCACTAAATGAG CCGTTGTTTCTTCACTTCCCACAGGTA |
64.9 60.5 |
390 | 95°C 5 min 1 cycle; 95°C 30 s, 55°C 30 s, 72°C 45 s, 30 cycles; 72°C 7 min 1 cycle |
| NS2AB | |||||
| NS2A-F NS2B-R |
3478–3499 4521–4995 |
accaccatgGGACATGGGCAGATTGACAACT CCGTTGTTTCTTCACTTCCCACAGGTA |
66.1 60.5 |
1044 | 95°C 5 min 1 cycle; 95°C 45 s, 55°C 45 s, 72°C 1 min, 30 cycles; 72°C 7 min 1 cycle |
| NS2B3(pro) | |||||
| NS2B-F NS3pro-R |
4132–4149 5064-5035 |
accaccatggctAGCTGGCCACTAAATGAG GTCATCTTCGATCTCTGGATTGTCTTCAAT |
64.9 58.1 |
933 | 95°C 5 min 1 cycle; 95°C 45 s, 53°C 45 s, 72°C 1 min, 30 cycles; 72°C 7 min 1 cycle |
| NS3 | |||||
| NS3-F NS3-R |
4522–4543 6375-6348 |
accaccatgGCTGGAGTATTGTGGGATGTCC CTTTCTTCCAGCTGCAAACTCCTTGAAT |
66.0 59.5 |
1854 | 95°C 5 min 1 cycle; 95°C 45 s, 55°C 45 s, 72°C 1 min, 30 cycles; 72°C 7 min 1 cycle |
| NS4A | |||||
| NS4A-F NS4A-R |
6376–6393 6825-6800 |
accaccatggctTCCCTGACCCTGAACCTA TGCCATGGTTGCGGCCACCACTGTGA |
64.1 68.6 |
450 | 95°C 5 min 1 cycle; 95°C 30 s, 59°C 30 s, 72°C 45 s, 30 cycles; 72°C 7 min 1 cycle |
| NS4A(-2K) | |||||
| NS4A-F NS4A(-2K)-R |
6376–6393 6756-6731 |
accaccatggctTCCCTGACCCTGAACCTA TCTCTGTTTTTCTGGTTCTGGAATAA |
64.1 59.3 |
381 | 95°C 5 min 1 cycle; 95°C 30 s, 59°C 30 s, 72°C 45 s, 30 cycles; 72°C 7 min 1 cycle |
| NS4B | |||||
| NS4B-F NS4B-R |
6826–6846 7569-7540 |
accaccatggctAACGAGATGGGTTTCCTGGAA CCTTCTCGTGTTGGTTGTGTTCTTCATGAT |
66.4 60.5 |
744 | 95°C 5 min 1 cycle; 95°C 45 s, 55°C 45 s, 72°C 1 min, 30 cycles; 72°C 7 min 1 cycle |
| NS4AB | |||||
| NS4A-F NS4B-R NS4B-GFP-R |
6376–6393 7569-7540 7569-7540 |
accaccatggctTCCCTGACCCTGAACCTA CCTTCTCGTGTTGGTTGTGTTCTTCATGAT gCCTTCTCGTGTTGGTTGTGTTCTTCATGAT |
64.1 60.5 60.5 |
1194 | 95°C 5 min 1 cycle; 95°C 45 s, 55°C 45 s, 72°C 1 min, 30 cycles; 72°C 7 min 1 cycle |
| 2KNS4B | |||||
| 2KNS4B-F NS4B-R |
6757–6777 7569-7540 |
accaccatggctACACCCCAAGATAACCAATTG CCTTCTCGTGTTGGTTGTGTTCTTCATGAT |
64.4 60.5 |
813 | 95°C 5 min 1 cycle; 95°C 45 s, 55°C 45 s, 72°C 1 min, 30 cycles; 72°C 7 min 1 cycle |
| NS5 | |||||
| NS5-F NS5-R |
7570–7591 10269-10239 |
accaccatgGGAACTGGCAACATAGGAGAGA CCACAGGACTCCTGCCTCTTCCTCTTCTTTT |
65.2 64.0 |
2700 | 95°C 5 min 1 cycle; 95°C 1 min, 59°C 60 s, 72°C 1 min, 30 cycles; 72°C 7 min 1 cycle |
Forward, sense sequence; reverse, antisense sequence
Determined from NCBI accession number M29095 (DENV NGC strain)
Kozac sequence shown as bold lower-case letters, fill-in nucleotides for in-frame translation as lower case letters and DENV sequence as upper case letters
Table 2.
Primers employed for the analysis of DENV and host genes by qRT-PCR.
| Gene and Primer Namesa | Primer Sequence (5' - 3') | Tm (°C) | Amplicon Size (bp) |
|---|---|---|---|
| DENV2 Capsidb | |||
| DN-Forward (F) DN-Reverse (R) |
CAATATGCTGAAACGCGAGAG CATCTATTCAGAATCCCTGCT |
55.8 58.3 |
167 |
| V5 Epitopeb | |||
| V5-F V5-R |
TCTGCAGATATCCAGCACAGT ACCGAGGAGAGGGTTAGGGAT |
55.8 59.2 |
87 |
| GAPDHc | |||
| GAPDH-F GAPDH-R |
AGTTAGCCGCATCTTCTTTTGC CAATACGACCAAATCCGTTGAC |
56.1 55.3 |
100 |
| IL-6c | |||
| IL6-F IL6-R |
CCAGGAGCCCAGCTATGAAC CCCAGGGAGAAGGCAACTG |
57.8 58.2 |
63 |
| IL-8c | |||
| IL8-F IL8-R |
GAACTGAGAGTGATTGAGAGT CTCTTCAAAAACTTCTCCACA |
55.1 56.6 |
133 |
| IP-10c | |||
| IP10-F IP10-R |
CCACGTGTTGAGATCATTGC GCTCCCCTCTGGTTTTAAGG |
54.3 55.4 |
139 |
| TNF-αc | |||
| TNFα-F TNFα-R |
CCCAGGGACCTCTCTCTAATCA GCTACAGGCTTGTCACTCGG |
55.9 58.6 |
79 |
Forward, sense sequence; reverse, antisense sequence
Real-time PCR primers used to determine DENV2 NGC and V5 copy numbers
Real-time RT-PCR primers to amplify immunomediator genes
Fig. 3.
DENV NS4B and NS5 induce IL-6, IL-8, and TNFα transcripts in THP-1 cells. THP-1 cells expressing individual DENV nonstructural genes were harvested at 40 h after transfection and total mRNA was isolated for immunomediator transcript analysis by qRT-PCR. The fold-change induction of IL-6, IL-8, and TNFα transcripts was determined for each DENV gene relative to V5 controls. LPS was used as an positive control and GAPDH was used as the endogenous control. Bars represent the mean ± SD of three independent experiments conducted in duplicate. (*) indicates statistical significance at p < 0.05 as compared to transcript levels induced by the NS4AB-V5 fusion protein.
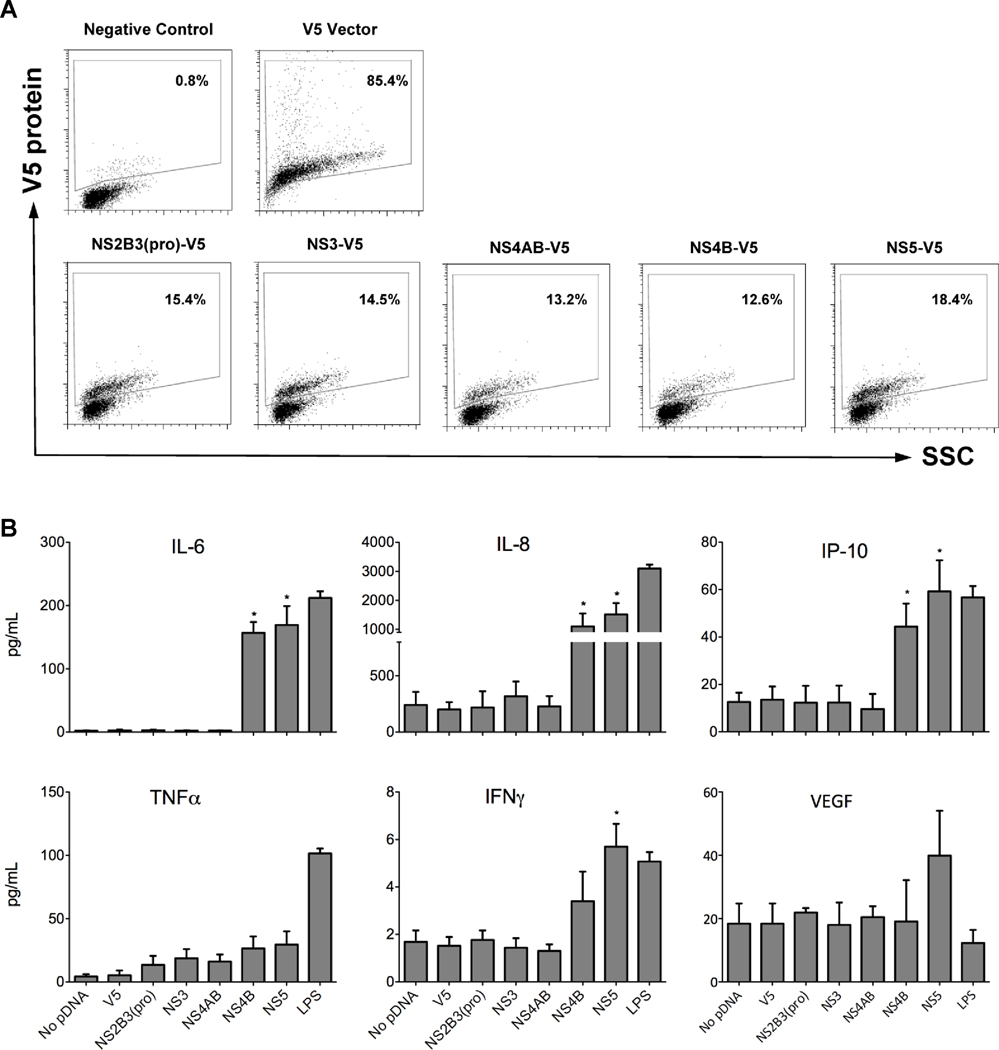
Given that NS4B and NS5 induced DHF-associated immunomediator transcripts, we examined the culture supernatant for secreted immunomediators induced by NS4B, NS5 and the viral replication complex-associated proteins NS2B3(pro), NS3 and NS4AB. As an alternative method to confirm plasmid expression efficiencies, we conducted flow cytometry using a V5 antibody to detect each DENV-V5 fusion protein (Fig. 4A). We observed similar protein levels for all plasmids tested wherein a range of 12–18% of THP-1 cells expressed DENV-V5 fusion proteins (Fig. 4A). Having established similar NS-V5 transcript and protein expression levels by qRT-PCR, western blot and flow cytometry, we quantitated secreted immunomediator levels in the culture supernatants using the Luminex® technology. As expected, NS4B or NS5 significantly induced IL-6, IL-8, IP-10 and IFNγ when compared to cells expressing only the V5 epitope (Fig. 4B). Cells expressing NS4B secreted 57, 1,100 and 48 pg/mL of IL-6, IL-8 and IP-10, respectively, while cells expressing NS5 secreted slightly higher levels of these immunomediators (Fig. 4B). NS4B and NS5 increased TNFα production but the difference was not significant, p=0.11 and 0.08, respectively. However, DENV-infected or LPS-treated cells significantly induced TNFα (Fig. 1B) indicating an alternative route of induction. When we electroporated the THP-1 cells without plasmid DNA (pDNA) or the V5 vector control, we observed chemokine and cytokine expression levels similar to endogenous THP-1 levels (Fig. 4B and data not shown). Cell viability reached approximately 70% at 40 h after treatment, including the no-plasmid transfected and LPS-treated control cells (Supplemental Fig. 2), further indicating that the differences observed were due to the expression of NS4B and NS5.
Fig. 4.
DENV NS4B and NS5 induce DHF-associated immunomediators in THP-1 cells. THP-1 cells were electroporated with DENV NS and after 40 h, cells and supernatants were collected to measure expression efficiencies and immunomediator protein levels, respectively. (A) Percent of THP-1 cells expressing DENV-V5 fusion proteins as determined by flow cytometry. Cells were fixed, permeabilized and V5-expression proteins were detected using an anti-V5 primary antibody and Alexa Fluor® 594 secondary antibody. (B) Chemokine and cytokine levels (pg/mL) in the supernatants of THP-1 cells expressing NS at 40 h after electroporation and as determined by the Luminex® multiplex technology. Bars represent the mean ± SD of three independent experiments conducted in duplicate and (*) indicates statistical significance at p < 0.05 as compared to vector controls.
Maturation of NS4B via cleavage of NS4AB enhances IL-6 and IL-8 production
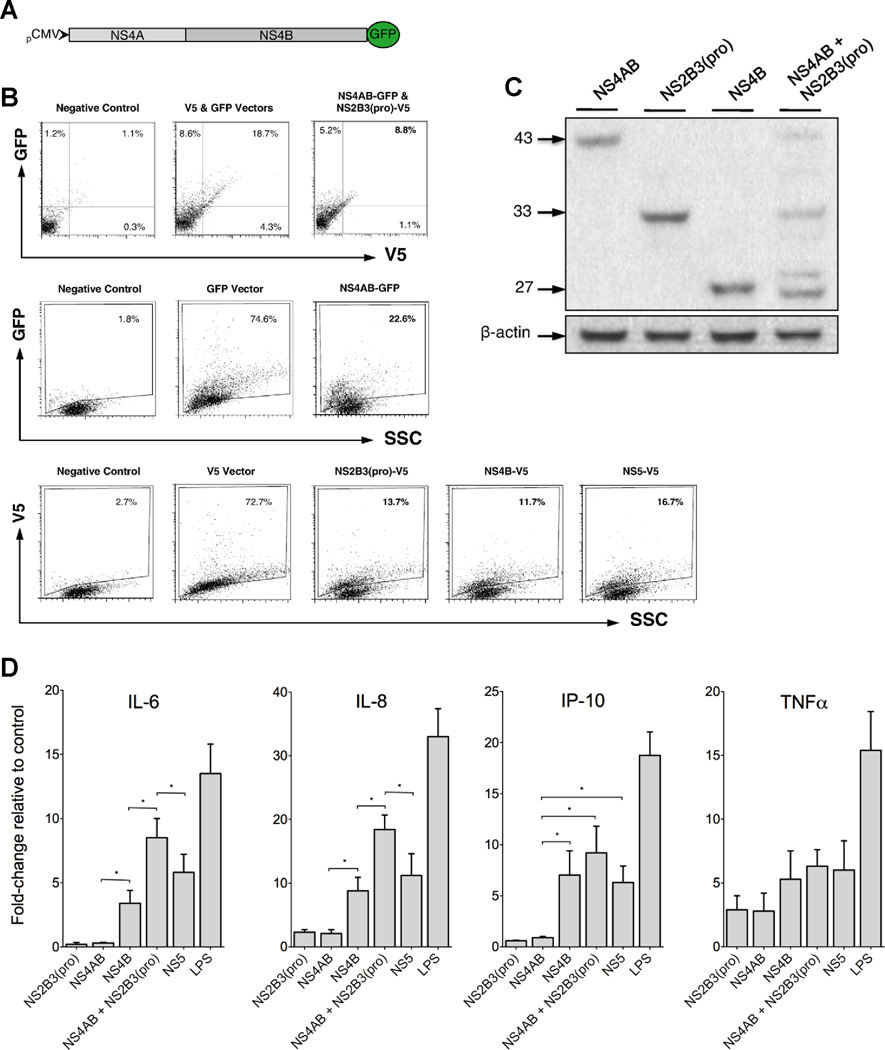
During replication, the DENV protease NS2B3(pro) interacts with the NS4AB polyprotein presumably as part of the viral replication complex. Previous work has demonstrated sequential processing of NS4AB wherein NS2B3(pro) cleaves NS4AB, releasing NS4A from NS4B (Cahour, Falgout, and Lai, 1992; Miller, Sparacio, and Bartenschlager, 2006). We proposed that expressing both NS4AB and NS2B3(pro) in THP-1 cells would best mimic natural processing and maturation of NS4B and possibly affect the immunomediator induction pattern observed during the expression of individual viral proteins. To detect THP-1 cells expressing both NS4AB and NS2B3(pro), we constructed the NS4AB-GFP fusion protein plasmid (Fig. 5A) for co-expression with the NS2B3(pro)-V5 fusion protein. We demonstrated by flow cytometry that approximately 9% of electroporated THP-1 cells co-expressed NS4AB-GFP and NS2B3(pro)-V5, 23% expressed NS4AB-GFP and 12, 14 and 17% expressed NS4B, NS2B3(pro) and NS5, respectively, 40 h after electroporation (Fig. 5B). To confirm the flow cytometry data and examine functional cleavage events, we conducted western blot using cells co-expressing NS4AB and the viral protease NS2B3(pro). As expected, we observed that the co-expression resulted in the cleavage of NS4AB which produced four distinct bands: NS4AB, NS2B3(pro), 2KNS4B and NS4B (Fig. 5C). Co-expression of NS4AB and NS2B3(pro) significantly enhanced the induction of IL-6 and IL-8 transcripts when compared to NS4B or NS5 alone, approximately doubling the relative fold change of IL-6 transcripts from 4- to 8-fold and IL-8 transcripts from 9- to 18-fold (Fig. 5D). Given that NS4B co-localizes with NS3 and NS5 during IFN antagonistic function and as part of the viral replication complex (Mazzon et al., 2008; Miller, Sparacio, and Bartenschlager, 2006; Umareddy et al., 2006), we tested the induction potential of NS4B with NS3 or NS5 and demonstrated that the induction potential was unaltered relative to NS4B or NS5 alone for IL-6 or IL-8 (data not shown), further supporting our results that enhancement occurs primarily during NS4B maturation and polyprotein cleavage events. Interestingly, the cleavage event did not significantly enhance the induction of IP-10, suggesting that multiple pathways may be responsible for inducing immunomediators. Although this result is unexpected, signal transduction crosstalk is regulated in a dynamic manner and may differ under homeostatic and pathologic conditions making it difficult to determine the exact mechanisms responsible for induction outcomes (Hu et al., 2007). However, taken together these results suggest that NS4AB expressed with NS2B3(pro) initiates sequential processing of NS4AB and maturation of NS4B (Miller et al., 2007; Miller, Sparacio, and Bartenschlager, 2006), thereby enhancing the induction of immunomediators.
Fig. 5.
Maturation of NS4B via processing of the polyprotein, NS4AB, enhances the production of IL-6 and IL-8. (A and B) The NS4AB gene was cloned into a GFP reporter vector and co-expressed with NS2B3(pro)-V5 fusion protein in THP-1 cells for detection by flow cytometry. The expression efficiencies were measured at 40 h after transfection and expressed as percent of THP-1 cells expressing DENV-V5-GFP, V5 or GFP fusion proteins. (C) Western blot analysis of THP-1 cells expressing DENV-V5 fusion proteins NS 4AB, 2B3(pro), 4B and both 4AB and 2B3(pro). β-actin was used as the endogenous control and estimated molecular weight markers (kDa) are shown on the left. (D) Induction of immunomediator transcripts in THP-1 cells relative to vector controls as determined by qRT-PCR. Bars represent mean ± SD of six experimental samples and (*) indicates statistical significance at p < 0.05.
Expression of the 2K-signal peptide with NS4B is sufficient to enhance IL-6 and IL-8 induction
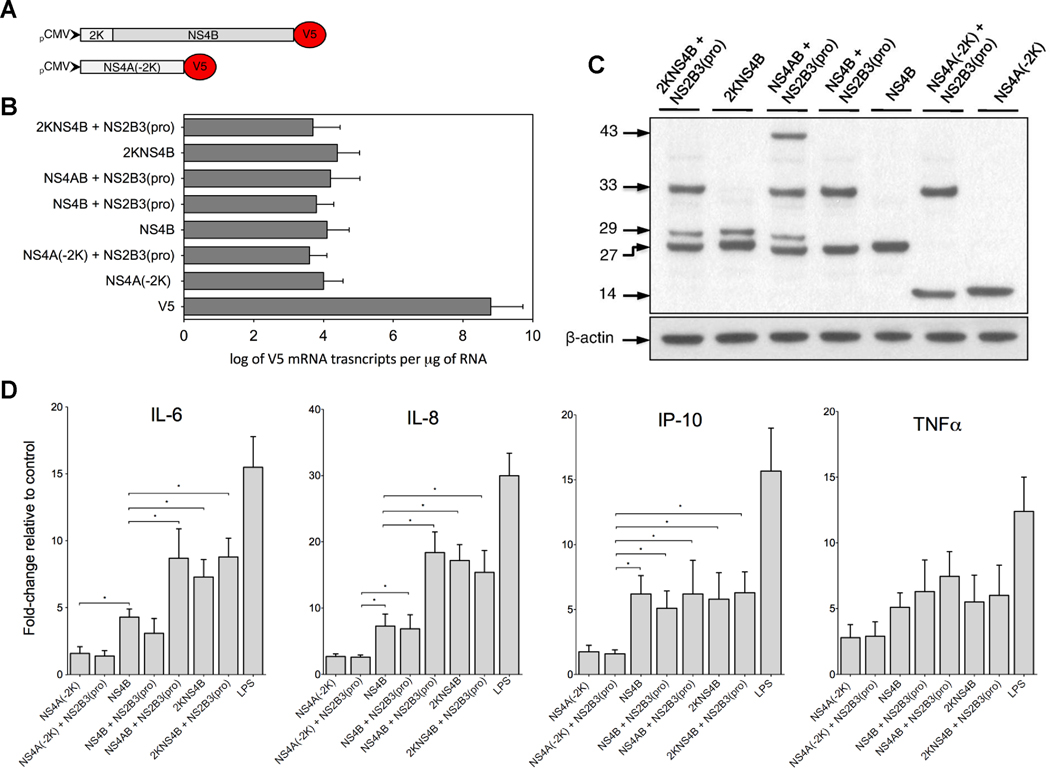
The DENV 2K-signal sequence is a 17-amino acid peptide linking NS4A with NS4B and has little known function. Having observed the cleavage product 2KNS4B in the co-transfected cells, we questioned whether the 2K-signal sequence was responsible for the observed enhanced induction of immunomediator transcripts. To address this question, we constructed the plasmids 2KNS4B and NS4A without 2K (-2K) (NS4A-2K) (Fig. 6A) and expressed them alone or in combination with other NS, particularly NS2B3(pro) due to its intrinsic enzymatic properties and involvement in the replication complex. By using qRT-PCR and western blot, we demonstrated the production of approximately log 4 copies of V5 mRNA transcripts by each plasmid or combination of plasmids (Fig. 6B) and cleavage of 2KNS4B, respectively (Fig. 6C). Importantly, 2KNS4B expression and cleavage, presumably by host proteases, significantly enhanced the induction of IL-6 and IL-8 transcripts (Fig. 6D). Expression of NS4A(-2K) with or without NS2B3(pro) did not induce transcripts of any of the immunomediators tested while NS4B expressing with NS2B3(pro) showed slightly reduced induction potential, suggesting that the viral protease may interfere with NS4B during the induction process (Fig. 6D). Overall, these data suggest that the induction of DHF-associated chemokines and cytokines by NS4B is enhanced when NS4AB is cleaved by NS2B3(pro), specifically during 2KNS4B processing by host signalases and possibly during NS4B localization into the membrane structures (Miller et al., 2007; Miller, Sparacio, and Bartenschlager, 2006; Roosendaal et al., 2006; Umareddy et al., 2006; Westaway et al., 1997a).
Fig. 6.
NS4B with the 2K-signal sequence is sufficient to enhance the induction of immunomediators. (A) The DENV genes 2KNS4B and NS4A without the 2K-signal peptide, NS4A(-2K), were cloned in the V5 reporter vector. (B) Log of V5 mRNA transcripts in THP-1 cells expressing nonstructural plasmids at 40 h after transfection as determined by qRT-PCR. (C) Western blot analysis of THP-1 cells expressing DENV-V5 fusion proteins NS 2B3(pro) and 2K4B, 2K4B, 4AB and 2B3(pro), 2B3(pro) and 4B, 4B, 2B3(pro) and 4A(-2K), and 4A(-2K). Estimated molecular weights (kDa) are depicted on the left and β-actin was detected as the endogenous control. (D) Immunomediators mRNA fold-changes as compared to corresponding V5 controls; GAPDH was used as the endogenous control gene. Bars represent mean ± SD of six independent experimental samples and (*) indicates statistical significance at p < 0.05.
Discussion
It is well established that elevated levels of immunomediators detected in DHF/DSS patients can adversely alter endothelial cell integrity leading to plasma leakage and unresolved vascular permeability (Basu and Chaturvedi, 2008; Green and Rothman, 2006; Leong et al., 2007; Srikiatkhachorn, 2009). However, mechanisms underlying severe DENV immunopathogenesis remain elusive. The lack of an animal model to study dengue immunopathogenesis has lead to extensive clinical and in-vitro studies suggesting that both host and viral factors influence elevated circulating immunomediators and severe disease outcomes (Kurane and Takasaki, 2001; Pang, Cardosa, and Guzman, 2007). DENV is capable of modulating intracellular signaling, such as the IFN response, or host inflammatory responses such as activation of the transcription factor NF-κB (Mogensen and Paludan, 2001a; Mogensen and Paludan, 2001b). DHF/DSS is presumed to occur when the virus outcompetes host antiviral responses and establishes infection while initiating a robust inflammatory response, which is critical for the progression of severe dengue disease. Although other investigators have examined DENV initiated IFN inhibition, very little is known regarding the mechanism by which the DENV proteins initiate DHF/DSS-associated immunomediators. In this report, we demonstrate that maximum production of immunomediators from DENV-infected THP-1 cells corresponds with peak levels of secreted infectious virus and cellular viral RNA. Moreover, using a plasmid construct system that consistently expresses similar levels of viral gene transcripts and viral-V5 fusion proteins, we demonstrate that DENV NS4B and NS5 are potent inducers of DHF/DSS-associated immunomediators in monocytes and that maturation of NS4B via host protease cleavage of 2KNS4B is a critical step during the observed induction.
Peak levels of chemokines and cytokines in DENV-infected THP-1 cells correspond with maximum virus yield
We demonstrate that DENV-infected THP-1 monocytes induce transcription and secretion of immunomediators such as IL-6, IL-8, IP-10, TNFα and IFNγ and that peak levels of these chemokine and cytokines correspond with an increased generation of infectious virus and viral RNA copy numbers. These data are consistent with previous reports demonstrating that secreted immunomediator levels directly correspond with the amount of virus production in THP-1 cells (Chareonsirisuthigul, Kalayanarooj, and Ubol, 2007; Ubol et al., 2008). Other investigators have demonstrated no infectious virus production from DENV-infected THP-1 cells using low passage clinical isolates and in the absence of enhancing antibodies (Diamond et al., 2000). This inconsistency may be due to differences in virus passage, as high-passage virus isolates in-vitro may acquire dominant mutations that exhibit aberrant phenotypic characteristics not found in low-passage isolates (Rico-Hesse et al., 1998). Our data suggest that immunomediator induction occurs during virus replication when viral RNA copies are elevated. Moreover, others have demonstrated that the antiviral drug ribavirin inhibits DENV replication and IL-6 and IL-8 production in a dose dependent manner (Huang et al., 2000). Regardless, it remains difficult to pinpoint the precise immunomediator initiation step due to the dynamic characteristics of virus replication. It is possible that immunomediator induction corresponds with the initial translation of input positive-strand viral RNA, negative-strand viral RNA synthesis, viral protein production or packaging, and/or secretion of virions. Functional studies demonstrate that the expression of all NS in THP-1 cells induces the activation of NFκB, the ubiquitous transcription factor involved in the rapid production of many chemokines and cytokines (Hershkovitz et al., 2008). Similarly, expression of NS or NS5 alone in human embryonic kidney cells induces IL-8 transcription and protein secretion (Medin, Fitzgerald, and Rothman, 2005; Pryor et al., 2007), further suggesting that NS effectively modulate intracellular signaling and initiate immunomediator induction.
NS5 induces DHF-associated immunomediators in THP-1 monocytes
We demonstrate that NS5 induces high levels of IL-8 as well as IL-6, IP-10 and IFNγ transcripts and protein secretion in THP-1 cells. Moreover, it appears that NS5 triggers a rapid and sustained production of immunomediators as demonstrated by increased transcript and protein secretion at 24 and 72 h after DENV infection, and 20 and 40 h after transfection with NS5 constructs, respectively. NS5 has a potent induction potential as demonstrated by consistent and equivalent expression levels of all tested viral gene transcripts and V5 fusion proteins. Although we observed that NS5 localized to the nucleus and the cytoplasm at 20 and 40 h after transfection, most NS5 localized to the nucleus (data not shown) consistent with other reports (Pryor et al., 2007; Rawlinson et al., 2009).
The largest and most highly conserved protein among the flaviviruses, NS5 serves as the RNA-dependent RNA polymerase (RdRP) and methyltransferase (Chung et al., 2010; Dong et al., 2010; Egloff et al., 2007; Harris et al., 2006; Zhou et al., 2007; Zou et al., 2011). Other investigators have reported that DENV NS5 induces IL-8 via the activation of the CAAT/enhancer binding protein (c/EBP) and NFκB (Medin, Fitzgerald, and Rothman, 2005) as it shuttles between nuclear and cytoplasmic compartments (Pryor et al., 2007; Rawlinson et al., 2009). Upon positive-sense RNA translation and polyprotein processing in the cytoplasm, soluble NS5 is thought to undergo phosphorylation allowing the host proteins importin-α/β to bind at two distinct nuclear localization sites (NLS) and import NS5 into the nucleus (Pryor et al., 2007). While hyperphosphorylated NS5 localizes to the nucleus, its dephosphorylation elicits a conformational change, allowing exportin-1 to bind and translocate NS5 to the cytoplasm where it interacts with NS3 as part of the viral replication complex (Kapoor et al., 1995; Mackenzie, Kenney, and Westaway, 2007; Rawlinson et al., 2009). Although the function of nuclear NS5 is unclear, greater nuclear accumulation of NS5 results in decreased IL-8 production and vice versa (Pryor et al., 2007; Rawlinson et al., 2009), suggesting that the observed induction of immunomediators occurs when NS5 is localized in the cytoplasm. Moreover, because the majority of NS5 localizes to the nucleus and only a relatively small amount of NS5 is required for virus replication in the cytoplasm (Grun and Brinton, 1987; Uchil and Satchidanandam, 2003), it is probable that very low levels of cytoplasmic NS5 is required to initiate immunomediator induction. Although the induction mechanisms by NS5 have not been defined, it is possible that NS5 triggers c/EBP and NFκB signaling events during NS5/importin interactions in the cytoplasm. However, this theory remains to be tested. Recent studies demonstrate that NS5 interacts with NS4B in the cytoplasm, and inhibits IFN signaling by binding and preventing STAT2 phosphorylation (Ashour et al., 2009; Mazzon et al., 2009; Mazzon et al., 2008), further suggesting that viral/host protein interactions are important for modulating host signaling.
NS4B induces DHF-associated immunomediators in THP-1 monocytes
In screening the induction potential of DENV NS in monocytes, we demonstrate for the first time that NS4B induces the secretion of IL-6, IL-8, IP-10 and IFNγ. The potent NS4B induction pattern is a surprise given its highly hydrophobic nature and intimate integration within the ER membrane. Yet, we demonstrate that NS4B has an early and sustained induction potential as suggestive of its ability to induce IL-8 at 20 and 40 h after transfection. A highly hydrophobic protein, NS4B consists of three endoplasmic reticulum (ER) membrane-spanning segments and may serve as an anchor for the viral replication complex (Miller et al., 2007; Miller, Sparacio, and Bartenschlager, 2006; Welsch et al., 2007). NS4B can act as a potent IFN antagonist via interference and degradation of cellular STAT1 (Munoz-Jordan et al., 2003) which is synchronized with NS4B maturation via NS4AB cleavage by the viral protease, NS2B3(pro) (Munoz-Jordan, 2010; Munoz-Jordan et al., 2005). Interestingly, IFN-β-mediated inhibition of IL-8 expression requires the presence of STAT1 and STAT2 (Laver, Nozell, and Benveniste, 2008). Given that NS4B maturation inhibits IFN signaling via STAT1 degradation, we hypothesized that NS4B maturation via NS4AB proteolytic modification(s), including host signalase cleavage of 2KNS4B, would lead to increased levels of IL-8 and possibly other immunomediators. As such, we tested the induction potential of the processing events involved in NS4B maturation.
Maturation of NS4B via processing of 2KNS4B enhances the induction of immunomediators
During polyprotein processing, NS4AB is cleaved by the viral protease NS2B3(pro), releasing NS4A from NS4B having the 2K-signal peptide (2KNS4B) before a host signalase cleaves the 2KNS4B junction in the lumen of the ER to release mature NS4B and allowing for its integration into the ER membrane (Cahour, Falgout, and Lai, 1992; Lin et al., 1993; Miller, Sparacio, and Bartenschlager, 2006). To mimic natural NS4B maturation via polyprotein processing, we co-expressed NS4AB with NS2B3(pro) and observed that polyprotein cleavage events initiated a more potent induction of IL-6, IL-8 and IP-10 transcripts than NS4B or NS5 alone. The enhancement appears to be due to NS4AB processing or maturation and localization of NS4B in the ER, not increased levels of NS expression as demonstrated by similar expression levels of viral protein and gene transcripts. Moreover, our data suggest that the 2K-signal peptide is not required for NS4B induction, yet post-translational proteolytic cleavage events including host signalase cleavage of the 2K-peptide from NS4B is primarily responsible for the enhancement.
The 2K-signal peptide appears to play an important role during a variety of intracellular alterations, including anchoring of NS4B to the ER membrane (Lin et al., 1993). Proteolytic removal of the 2K-peptide from NS4A during DENV replication induces membrane alterations that may harbor the viral replication complex (Miller et al., 2007) and the West Nile virus (WNV) 2K-peptide is important for cytoplasmic rearrangements, foci development and Golgi trafficking (Roosendaal et al., 2006). Moreover, IFN inhibition by NS4B appears to require the presence of the 2K-signal peptide for at least three flaviviruses, DENV, WNV and yellow fever virus (YFV) (Munoz-Jordan et al., 2005). Interestingly, the initial 125 amino acid residues of DENV 2KNS4B, predicted to be located in the cytoplasm between the first and second transmembrane domains, are required for IFN antagonism (Munoz-Jordan et al., 2005), while the YFV 2K-peptide may be important for the translocation of cleaved NS4B to the lumen side of the ER (Chambers, McCourt, and Rice, 1989).
Given that IFN-β-mediated inhibition of IL-8 production requires the presence of STAT1 and STAT2 (Laver, Nozell, and Benveniste, 2008), and NS5 or processing of 2KNS4B by host proteases is synchronized with IFN inhibition via STAT1 and STAT2 degradation, it will be interesting to determine whether both IFN inhibition and immunomediator induction occur in the presence of mature NS4B as intracellular STAT1 and STAT2 diminish. Overall, we demonstrate that the viral protease has no intrinsic induction potential alone, yet it cleaves NS4AB and generates NS4A(-2K) and 2KNS4B, which is then cleaved by a host protease, perhaps allowing NS4B to reorient across the ER and initiate IFN inhibition and immunomediator induction.
Conclusions
Effective host defenses against DENV infection include a robust IFN response and a rapid production of chemokines and cytokines important for immune signaling. Importantly, DENV and other flaviviruses are capable of inhibiting host IFN signaling to establish productive infections and concurrently promote an overproduction of immunomediators associated with severe disease. In this report, we describe DENV-associated immunomediator induction patterns in the monocytic cell line, THP-1. We have demonstrated that THP-1 cells sustain excellent viability after electroporation, are permissive to DENV infection, and thus are an ideal model to examine early events after infection or expression of DENV NS proteins. We also demonstrated that NS4B maturation events, specifically cleavage of 2KNS4B by host proteases, induce significantly higher levels of immunomediators than NS4B or NS5 alone. Therefore, it appears that maturation of NS4B is important for both IFN inhibition and immunomediator induction. Given the shared phenotype of IFN antagonism across the virus family, it is quite possible that IFN inhibition and immunomediator induction patterns induced during NS4B maturation are similar for other flaviviruses. Even though Flavivirus-wide NS4B sequences show divergent and sometimes negligible genotypic resemblance (Lundin et al., 2003; Qu, McMullan, and Rice, 2001), predicted flavivirus NS4B structural topologies, including the multiple transmembrane and cytoplasmic regions, are strikingly similar as well as NS4AB cleavage events. Reduced flavivirus protein expression or polyprotein processing during infection may dampen virus-induced IFN inhibition and the overproduction of potentially deleterious immunomediators, minimizing the risk of severe disease. As such, NS4B may very well be an attractive target for the development of Flavivirus-wide therapeutic interventions.
Materials and Methods
Virus and cell culture
DENV-2 New Guinea C (NGC) strain was obtained from Dr. Duane Gubler at the University of Hawaii at Manoa. A virus stock was produced by passaging virus twice in C6/36 cells. Given the extensive use of the THP-1 cell line by others examining DENV pathogenesis and due to their permissibility to DEVN infection (Chareonsirisuthigul, Kalayanarooj, and Ubol, 2007; Chen et al., 2009; Hershkovitz et al., 2008; Hober et al., 1996; Ubol et al., 2008), we chose to work with THP-1 cells in this study. The THP-1 cells were obtained from American Type Culture Collection (ATCC, Rockwell, MD) and were cultured in RPMI-1640 (ATCC) supplemented with 1% penicillin/streptomycin, 10% heat-inactivated fetal bovine serum (FBS) (Gibco Labs, Grand Island, NY) and 2-mercaptoethanol to a final concentration of 0.05 mM; cells were incubated at 37°C in a 5% CO2 atmosphere. The Vero cells (monkey kidney epithelial cells) (ATCC) were maintained in M199 and supplemented with 1% penicillin/streptomycin and 10% FBS.
Infection of THP-1 cells
For infection experiments, 1 × 106 THP-1 cells were infected with DENV at MOI-0.1 or -1. After 1.5 h at 37°C and 5% CO2, the cells were washed and further cultured with fresh growth media. UV-inactivated DENV was generated using previously published protocols (Anderson et al., 1997; Verma et al., 2009). Briefly, DENV was diluted in 500 µL PBS in a 35-mm culture plate and exposed to UV radiation using a Stratalinker 2400 device (Stratagene) for 10 min. Inactivation of virus infectivity was verified by plaque assay using Vero cells. Mock-infected control THP-1 cells were infected with UV-inactivated DENV and for positive controls, THP-1 cells were incubated with 1 µg/mL of LPS for 1 hour, washed and incubated with fresh growth media until collection at 24 h. Every 24 h, cells and supernatants were collected while remaining cells were replenished with fresh growth media. Infectious virus released from infected cells was confirmed by plaque assay on Vero cells, as described below.
Plaque assay
To determine the amount of infectious virus released from DENV-infected THP-1 cells, plaque assay was conducted using Vero cell monolayers as described previously (Lambeth et al., 2005). Briefly, 2.5 × 105 Vero cells per well were seeded in 6-well culture plates (Corning, Lowell, MA) and incubated for 2 to 3 days until confluent. Supernatants from DENV-infected THP-1 cells were serially diluted using 10-fold dilutions in Dulbecco’s Modified Eagles Medium (DMEM) (Gibco Labs) supplemented with 10% FBS and 100 µl of each dilution was added to each well of the Vero cells followed by incubation at 37°C and 5% CO2 for 1 hour with rocking every 15 minutes. Three mL of primary nutrient agar containing 1% SeaKem® LE agarose (Lonza, Walkersville, MD) was added to each well and the plates were incubated for 5 days. Three mL of secondary nutrient agar (primary agar containing 1% neutral red) was added to each well and the plates were incubated for an additional two days before counting plaques and calculating viral titers (Lambeth et al., 2005). Titers were expressed as plaque forming units (PFU) per mL.
Construction of plasmids
We employed standard molecular biology techniques to clone the DENV genes (Sambrook and Russell, 2001). Each primer listed in Table 1 was designed from the DENV-2 NGC reference genome, NCBI accession number M29095. All forward primers contain the Kozac sequence and ATG start codon (Table 1). The PCR step was conducted using AmpliTaq Gold® DNA polymerase (Applied Biosystems, Carlsbad, CA) with either the first-strand cDNA template (iScript® cDNA synthesis kit, BioRad Inc., Hercules, CA) synthesized from the wild-type DENV-2 NGC RNA extracted from stock viral supernatant using the QIAamp viral RNA mini kit (Qiagen, Hilden, Germany) or the DENV-2 NGC DNA infectious clone (kindly provided by Barry Falgout, Food and Drug Administration) (Polo et al., 1997). Following the manufacturer’s protocol, PCR products were cloned into either the pcDNA3.1/V5-His-TOPO® or the pcDNA3.1/CT-GFP-TOPO® vector (Invitrogen, Carlsbad, CA) having the CMV promoter for mammalian expression and detection. Proper gene orientation and identity were confirmed by DNA sequencing (Greenwood Molecular Biology Facility, University of Hawaii) and sequence analysis was conducted using Sequencer® 4.10.1 (Gene Codes Corporation, Ann Arbor, MI). Selected plasmids were isolated using Cesium Chloride ethidium bromide equilibrium centrifugation as previously described (Sambrook and Russell, 2001).
Transfections
In order to express the DENV-V5 fusion proteins, we transfected each construct into THP-1 cells using the Neon™ Transfection System (Invitrogen) after performing a series of optimization protocols as specified by the manufacturer. Briefly, 5 × 105 cells were electroporated with 1 µg of plasmid DNA using 1,250 volts and 40 milliseconds for 1 pulse. Cells were directly added to growth media without penicillin/streptomycin and immediately incubated at 37°C and 5% CO2. The supernatants and cells were harvested at 40 h after electroporation and used for the assays described below.
Flow cytometry
Intracellular expression of DENV-V5 or -GFP fusion proteins was detected at 40 h after transfection using a Guava EasyCyte flow cytometer (Guava, Hayward, CA). Cells expressing DENV-V5 proteins were washed, fixed and permeabilized (Fix & Perm reagents, Invitrogen) for intracellular labeling with a mouse monoclonal V5 antibody (Invitrogen) diluted 1:500 in PBS and a secondary antibody against mouse IgG coupled with Alexa Fluor® 594 antibody (Invitrogen). Similarly, cells expressing DENV-GFP were washed, fixed and resuspended in PBS for flow cytometry. Data were analyzed using FlowJo software version 4.3 and expressed as percent cells expressing DENV protein.
Quantitative real-time RT-PCR (qRT-PCR)
Total cellular RNA was extracted from DENV-infected-, pDNA-transfected- and control -THP-1 cells harvested at various time points using the RNeasy® Plus kit with RNase-Free DNase (Qiagen) as per manufacturer protocol. cDNA was synthesized using 1 µg of RNA using the BioRad iScript® kit in a 20 µl reaction volume. BioRad iCycler iQ™ Multicolor Real-Time PCR Detection System was employed to conduct qRT-PCR for quantitation of DENV and V5 copy numbers using Bio-Rad iQ™ SYBR® Green Supermix, 2 µl of 1:3 diluted cDNA, and 10 pmol each of forward and reverse primers (Table 2) in a final reaction volume of 20 µl. Thermal cycling reactions for both DENV and V5 amplifications were initiated with a denaturing step of 4 minutes at 95°C, followed by 40 cycles of 95°C (30 sec) and 55°C (30 sec for DENV and 15 sec for V5). A standard curve was developed from 10-fold serial dilutions of linear DENV or V5 gene having known concentrations to quantitate the dynamic range of detection of 101 to 108 copies per µg of RNA. Host cellular gene changes relative to the glyceraldehyde-3-phosphate dehydrogenase (GAPDH) housekeeping gene were determined as previously described (Verma et al., 2006). Primers used to measure cellular gene changes are listed in Table 2.
Cytokine quantitation
IL-6, IL-8, IP-10, TNFα, VEGF and IFNγ levels were measured in the supernatants of DENV-infected and pDNA transfected THP-1 cells using a Milliplex human cytokine and chemokine 6-plex immunoassay kit (Millipore Corp., Billerica, MA) together with the Luminex® 100™ System (Luminex, Austin, TX) to determine mean fluorescent intensities (MFI) as recommended by the manufacturer. Protein concentrations were calculated from MFI data using 10-fold serially diluted standards and Bead View analysis software version 1.0.4 (Millipore). The minimum detectable concentrations were 0.4 pg/mL for IL-6, 0.3 pg/mL for IL-8, 1.3 pg/mL for IP-10, 0.2 pg/mL for TNFα, 10.1 pg/mL for VEGF and 0.4 pg/mL for IFNγ.
Western blot
Total cellular protein extracts were prepared from THP-1 cells at 40 h after electroporation with DENV-V5 fusion plasmids. Cells were washed once with cold PBS and extracted with 200 µL of M-PER mammalian protein extraction buffer (Thermo Scientific, Rockford, IL) or NP-40 detergent buffer containing EDTA-free complete protease inhibitor cocktail (Roche, Indianapolis, IN). Either 20 µL or 50 µg of total protein was fractionated on a 4–12% gradient SDS polyacrylamide gel using the Mini-Protean II (Bio-Rad, Hercules, CA) and then transferred onto a 0.2 µm nitrocellulose filter (Bio-Rad Laboratories) as previously described (Verma et al., 2006). Nonspecific binding sites were blocked using 5% FBS in 1× Tris buffered saline with 0.1% Tween and membranes were incubated at 4°C overnight with primary V5 or β-actin antibodies (dilution 1:1,000) followed by incubation with secondary antibodies conjugated to HRP (dilution 1:5,000) at room temperature for 1 h. Protein was detected with enhanced chemiluminescence (Amersham ECL, GE Healthcare Limited, Buckinghamshire, UK) using Amersham ECL Hyperfilm (Kodak, Rochester, NY). To determine the relative intensity (RI) of protein bands, the absolute intensity of the DENV-V5 protein band was divided by the absolute intensity of its corresponding β-actin band. Absolute intensities were calculated using Photoshop by multiplying the given pixel value and mean intensity of selected bands as previously described (Luhtala and Parker, 2009).
Cell viability assay
To determine the cell viability, 1 × 106 THP-1 cells were transfected with plasmid DNA or incubated with 1 µg/mL of LPS for 1 h, washed and incubated with fresh growth media until collection at 24 h. Transfected cells were collected at 40 h for measurement of cell viability. Cell viability was measured using the CellTiter96 AQueous One Solution Cell Proliferation Assay (Promega, Madison, WI) kit as per the manufacturers’ protocol (Verma et al., 2008).
Statistical Analysis
Statistical tests, including paired and unpaired Student’s t-tests, were conducted for qRT-PCR and cytokine and chemokine immunoassay using GraphPad InStat version 5.0 (GraphPad software, San Diego, CA). Values were expressed as mean ± SD of three independent observations. P values of <0.05 were considered significant.
Supplementary Material
Induction of immunomediator transcripts in DENV-infected THP-1 cells. Cells were infected at MOI-0.1, MOI-1 or incubated with LPS for 1 hour and collected each day for four consecutive days. qRT-PCR was conducted to determine A) IL-8 and B) TNFα mRNA fold-changes as compared to same day controls. GAPDH was used as the endogenous control gene. Bars represent the mean ± SD of four samples and (*) indicates statistical significance at p < 0.05 compared to same day levels induced by MOI-0.1.
Cell viability of THP-1 cells transfected with plasmid DNA or treated with LPS for 1 h. THP-1 cells were grown to confluence and 1 × 106 cells were treated as indicated in the graph. Viability of treated cells was determined by cell proliferation assay. Results are presented as percent viable cells. Data are expressed as mean ± SD of two independent experiments performed in duplicate.
Acknowledgements
We are grateful to Dr. Barry Falgout of the United States Food and Drug Administration for providing the pRS424 DENV-2 NGC plasmid DNA used as a template to clone NS3 and NS5. We thank the staff and students of the Retrovirology Research Laboratory and the Department of Tropical Medicine, Medical Microbiology and Pharmacology for technical assistance. This work was submitted by JFK to the University of Hawaii at Manoa as part of his doctoral thesis project.
This work was supported in part by institutional funds and grants from the Department of Defense (W8IXWH0720073), and Centers of Biomedical Research Excellence (P20RR018727) and Research Centers in Minority Institutions Program (G12RR003061), National Center for Research Resources, National Institutes of Health. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
The authors declare no competing interests.
References
- Anderson R, Wang S, Osiowy C, Issekutz AC. Activation of endothelial cells via antibody-enhanced dengue virus infection of peripheral blood monocytes. J Virol. 1997;71(6):4226–4232. doi: 10.1128/jvi.71.6.4226-4232.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashour J, Laurent-Rolle M, Shi PY, Garcia-Sastre A. NS5 of dengue virus mediates STAT2 binding and degradation. J Virol. 2009;83(11):5408–5418. doi: 10.1128/JVI.02188-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basu A, Chaturvedi UC. Vascular endothelium: the battlefield of dengue viruses. FEMS Immunol Med Microbiol. 2008;53(3):287–299. doi: 10.1111/j.1574-695X.2008.00420.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bell JR, Kinney RM, Trent DW, Lenches EM, Dalgarno L, Strauss JH. Amino-terminal amino acid sequences of structural proteins of three flaviviruses. Virology. 1985;143(1):224–229. doi: 10.1016/0042-6822(85)90110-2. [DOI] [PubMed] [Google Scholar]
- Bosch I, Xhaja K, Estevez L, Raines G, Melichar H, Warke RV, Fournier MV, Ennis FA, Rothman AL. Increased production of interleukin-8 in primary human monocytes and in human epithelial and endothelial cell lines after dengue virus challenge. J Virol. 2002;76(11):5588–5597. doi: 10.1128/JVI.76.11.5588-5597.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cahour A, Falgout B, Lai CJ. Cleavage of the dengue virus polyprotein at the NS3/NS4A and NS4B/NS5 junctions is mediated by viral protease NS2B-NS3, whereas NS4A/NS4B may be processed by a cellular protease. J Virol. 1992;66(3):1535–1542. doi: 10.1128/jvi.66.3.1535-1542.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chambers TJ, Hahn CS, Galler R, Rice CM. Flavivirus genome organization, expression, and replication. Annu Rev Microbiol. 1990;44:649–688. doi: 10.1146/annurev.mi.44.100190.003245. [DOI] [PubMed] [Google Scholar]
- Chambers TJ, McCourt DW, Rice CM. Yellow fever virus proteins NS2A, NS2B, and NS4B: identification and partial N-terminal amino acid sequence analysis. Virology. 1989;169(1):100–109. doi: 10.1016/0042-6822(89)90045-7. [DOI] [PubMed] [Google Scholar]
- Chang CS, Harn MR, Nimmannitya S. Clinical observation of 15 Thai children with dengue hemorrhagic fever. Gaoxiong Yi Xue Ke Xue Za Zhi. 1990;6(3):131–136. [PubMed] [Google Scholar]
- Chareonsirisuthigul T, Kalayanarooj S, Ubol S. Dengue virus (DENV) antibody-dependent enhancement of infection upregulates the production of anti-inflammatory cytokines, but suppresses anti-DENV free radical and pro-inflammatory cytokine production, in THP-1 cells. J Gen Virol. 2007;88(Pt 2):365–375. doi: 10.1099/vir.0.82537-0. [DOI] [PubMed] [Google Scholar]
- Chen LC, Shyu HW, Lin HM, Lei HY, Lin YS, Liu HS, Yeh TM. Dengue virus induces thrombomodulin expression in human endothelial cells and monocytes in vitroDengue virus induces thrombomodulin expression in human endothelial cells and monocytes in vitro. J Infect. 2009;58(5):368–374. doi: 10.1016/j.jinf.2009.02.018. [DOI] [PubMed] [Google Scholar]
- Chen YC, Wang SY. Activation of terminally differentiated human monocytes/macrophages by dengue virus: productive infection, hierarchical production of innate cytokines and chemokines, and the synergistic effect of lipopolysaccharide. J Virol. 2002;76(19):9877–9887. doi: 10.1128/JVI.76.19.9877-9887.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung KY, Dong H, Chao AT, Shi PY, Lescar J, Lim SP. Higher catalytic efficiency of N-7-methylation is responsible for processive N-7 and 2'-O methyltransferase activity in dengue virus. Virology. 2010;402(1):52–60. doi: 10.1016/j.virol.2010.03.011. [DOI] [PubMed] [Google Scholar]
- Diamond MS, Edgil D, Roberts TG, Lu B, Harris E. Infection of human cells by dengue virus is modulated by different cell types and viral strains. J Virol. 2000;74(17):7814–7823. doi: 10.1128/jvi.74.17.7814-7823.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong H, Chang DC, Xie X, Toh YX, Chung KY, Zou G, Lescar J, Lim SP, Shi PY. Biochemical and genetic characterization of dengue virus methyltransferase. Virology. 2010;405(2):568–578. doi: 10.1016/j.virol.2010.06.039. [DOI] [PubMed] [Google Scholar]
- Durbin AP, Vargas MJ, Wanionek K, Hammond SN, Gordon A, Rocha C, Balmaseda A, Harris E. Phenotyping of peripheral blood mononuclear cells during acute dengue illness demonstrates infection and increased activation of monocytes in severe cases compared to classic dengue fever. Virology. 2008;376(2):429–435. doi: 10.1016/j.virol.2008.03.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egloff MP, Decroly E, Malet H, Selisko B, Benarroch D, Ferron F, Canard B. Structural and functional analysis of methylation and 5'-RNA sequence requirements of short capped RNAs by the methyltransferase domain of dengue virus NS5. J Mol Biol. 2007;372(3):723–736. doi: 10.1016/j.jmb.2007.07.005. [DOI] [PubMed] [Google Scholar]
- Falgout B, Pethel M, Zhang YM, Lai CJ. Both nonstructural proteins NS2B and NS3 are required for the proteolytic processing of dengue virus nonstructural proteins. J Virol. 1991;65(5):2467–2475. doi: 10.1128/jvi.65.5.2467-2475.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geiss BJ, Thompson AA, Andrews AJ, Sons RL, Gari HH, Keenan SM, Peersen OB. Analysis of flavivirus NS5 methyltransferase cap binding. J Mol Biol. 2009;385(5):1643–1654. doi: 10.1016/j.jmb.2008.11.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green S, Rothman A. Immunopathological mechanisms in dengue and dengue hemorrhagic fever. Curr Opin Infect Dis. 2006;19(5):429–436. doi: 10.1097/01.qco.0000244047.31135.fa. [DOI] [PubMed] [Google Scholar]
- Grun JB, Brinton MA. Dissociation of NS5 from cell fractions containing West Nile virus-specific polymerase activity. J Virol. 1987;61(11):3641–3644. doi: 10.1128/jvi.61.11.3641-3644.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gubler DJ. Dengue and dengue hemorrhagic fever. Clin Microbiol Rev. 1998;11(3):480–496. doi: 10.1128/cmr.11.3.480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gubler DJ. Epidemic dengue/dengue hemorrhagic fever as a public health, social and economic problem in the 21st century. Trends Microbiol. 2002;10(2):100–103. doi: 10.1016/s0966-842x(01)02288-0. [DOI] [PubMed] [Google Scholar]
- Gubler DJ. Dengue/dengue haemorrhagic fever: history and current status. Novartis Found Symp. 2006;277:3–16. doi: 10.1002/0470058005.ch2. discussion 16-22, 71-3, 251-3. [DOI] [PubMed] [Google Scholar]
- Guzman A, Isturiz RE. Update on the global spread of dengue. Int J Antimicrob Agents. 2010 doi: 10.1016/j.ijantimicag.2010.06.018. [DOI] [PubMed] [Google Scholar]
- Guzman MG, Kouri G. Dengue: an update. Lancet Infect Dis. 2002;2(1):33–42. doi: 10.1016/s1473-3099(01)00171-2. [DOI] [PubMed] [Google Scholar]
- Halstead SB. Antibody, macrophages, dengue virus infection, shock, and hemorrhage: a pathogenetic cascade. Rev Infect Dis. 1989;11 Suppl 4:S830–S839. doi: 10.1093/clinids/11.supplement_4.s830. [DOI] [PubMed] [Google Scholar]
- Harris E, Holden KL, Edgil D, Polacek C, Clyde K. Molecular biology of flaviviruses. Novartis Found Symp. 2006;277:23–39. discussion 40, 71-3, 251-3. [PubMed] [Google Scholar]
- Hershkovitz O, Zilka A, Bar-Ilan A, Abutbul S, Davidson A, Mazzon M, Kummerer BM, Monsoengo A, Jacobs M, Porgador A. Dengue virus replicon expressing the nonstructural proteins suffices to enhance membrane expression of HLA class I and inhibit lysis by human NK cells. J Virol. 2008;82(15):7666–7676. doi: 10.1128/JVI.02274-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hober D, Poli L, Roblin B, Gestas P, Chungue E, Granic G, Imbert P, Pecarere JL, Vergez-Pascal R, Wattre P, et al. Serum levels of tumor necrosis factor-alpha (TNF-alpha), interleukin-6 (IL-6), and interleukin-1 beta (IL-1 beta) in dengue-infected patients. Am J Trop Med Hyg. 1993;48(3):324–331. doi: 10.4269/ajtmh.1993.48.324. [DOI] [PubMed] [Google Scholar]
- Hober D, Shen L, Benyoucef S, De Groote D, Deubel V, Wattre P. Enhanced TNF alpha production by monocytic-like cells exposed to dengue virus antigens. Immunol Lett. 1996;53(2–3):115–120. doi: 10.1016/s0165-2478(96)02620-x. [DOI] [PubMed] [Google Scholar]
- Hu X, Chen J, Wang L, Ivashkiv LB. Crosstalk among Jak-STAT, Toll-like receptor, and ITAM-dependent pathways in macrophage activation. J Leukoc Biol. 2007;82(2):237–243. doi: 10.1189/jlb.1206763. [DOI] [PubMed] [Google Scholar]
- Huang YH, Lei HY, Liu HS, Lin YS, Liu CC, Yeh TM. Dengue virus infects human endothelial cells and induces IL-6 and IL-8 production. Am J Trop Med Hyg. 2000;63(1–2):71–75. doi: 10.4269/ajtmh.2000.63.71. [DOI] [PubMed] [Google Scholar]
- Juffrie M, Meer GM, Hack CE, Haasnoot K, Sutaryo, Veerman AJ, Thijs LG. Inflammatory mediators in dengue virus infection in children: interleukin-6 and its relation to C-reactive protein and secretory phospholipase A2. Am J Trop Med Hyg. 2001;65(1):70–75. doi: 10.4269/ajtmh.2001.65.70. [DOI] [PubMed] [Google Scholar]
- Kapoor M, Zhang L, Ramachandra M, Kusukawa J, Ebner KE, Padmanabhan R. Association between NS3 and NS5 proteins of dengue virus type 2 in the putative RNA replicase is linked to differential phosphorylation of NS5. J Biol Chem. 1995;270(32):19100–19106. doi: 10.1074/jbc.270.32.19100. [DOI] [PubMed] [Google Scholar]
- Khromykh AA, Sedlak PL, Guyatt KJ, Hall RA, Westaway EG. Efficient trans-complementation of the flavivirus kunjin NS5 protein but not of the NS1 protein requires its coexpression with other components of the viral replicase. J Virol. 1999;73(12):10272–10280. doi: 10.1128/jvi.73.12.10272-10280.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khromykh AA, Sedlak PL, Westaway EG. trans-Complementation analysis of the flavivirus Kunjin ns5 gene reveals an essential role for translation of its N-terminal half in RNA replication. J Virol. 1999;73(11):9247–9255. doi: 10.1128/jvi.73.11.9247-9255.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kou Z, Quinn M, Chen H, Rodrigo WW, Rose RC, Schlesinger JJ, Jin X. Monocytes, but not T or B cells, are the principal target cells for dengue virus (DV) infection among human peripheral blood mononuclear cells. J Med Virol. 2008;80(1):134–146. doi: 10.1002/jmv.21051. [DOI] [PubMed] [Google Scholar]
- Kurane I, Takasaki T. Dengue fever and dengue haemorrhagic fever: challenges of controlling an enemy still at large. Rev Med Virol. 2001;11(5):301–311. doi: 10.1002/rmv.324. [DOI] [PubMed] [Google Scholar]
- Lambeth CR, White LJ, Johnston RE, de Silva AM. Flow cytometry-based assay for titrating dengue virus. J Clin Microbiol. 2005;43(7):3267–3272. doi: 10.1128/JCM.43.7.3267-3272.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laver T, Nozell SE, Benveniste EN. IFN-beta-mediated inhibition of IL-8 expression requires the ISGF3 components Stat1, Stat2, and IRF-9. J Interferon Cytokine Res. 2008;28(1):13–23. doi: 10.1089/jir.2007.0062. [DOI] [PubMed] [Google Scholar]
- Leitmeyer KC, Vaughn DW, Watts DM, Salas R, Villalobos I, de C, Ramos C, Rico-Hesse R. Dengue virus structural differences that correlate with pathogenesis. J Virol. 1999;73(6):4738–4747. doi: 10.1128/jvi.73.6.4738-4747.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leong AS, Wong KT, Leong TY, Tan PH, Wannakrairot P. The pathology of dengue hemorrhagic fever. Semin Diagn Pathol. 2007;24(4):227–236. doi: 10.1053/j.semdp.2007.07.002. [DOI] [PubMed] [Google Scholar]
- Libraty DH, Endy TP, Houng HS, Green S, Kalayanarooj S, Suntayakorn S, Chansiriwongs W, Vaughn DW, Nisalak A, Ennis FA, Rothman AL. Differing influences of virus burden and immune activation on disease severity in secondary dengue-3 virus infections. J Infect Dis. 2002;185(9):1213–1221. doi: 10.1086/340365. [DOI] [PubMed] [Google Scholar]
- Lin C, Amberg SM, Chambers TJ, Rice CM. Cleavage at a novel site in the NS4A region by the yellow fever virus NS2B-3 proteinase is a prerequisite for processing at the downstream 4A/4B signalase site. J Virol. 1993;67(4):2327–2335. doi: 10.1128/jvi.67.4.2327-2335.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luhtala N, Parker R. LSM1 over-expression in Saccharomyces cerevisiae depletes U6 snRNA levels. Nucleic Acids Res. 2009;37(16):5529–5536. doi: 10.1093/nar/gkp572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lundin M, Monne M, Widell A, Von Heijne G, Persson MA. Topology of the membrane-associated hepatitis C virus protein NS4B. J Virol. 2003;77(9):5428–5438. doi: 10.1128/JVI.77.9.5428-5438.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackenzie JM, Kenney MT, Westaway EG. West Nile virus strain Kunjin NS5 polymerase is a phosphoprotein localized at the cytoplasmic site of viral RNA synthesis. J Gen Virol. 2007;88(Pt 4):1163–1168. doi: 10.1099/vir.0.82552-0. [DOI] [PubMed] [Google Scholar]
- Mazzon M, Jones M, Davidson A, Chain B, Jacobs M. Dengue virus NS5 inhibits interferon-alpha signaling by blocking signal transducer and activator of transcription 2 phosphorylation. J Infect Dis. 2009;200(8):1261–1270. doi: 10.1086/605847. [DOI] [PubMed] [Google Scholar]
- Mazzon M, Jones M, Strang A, Davidson A, Chain B, Jacobs M. Dengue virus NS5 inhibits IFNa signalling and together with NS4B reduces cellular STAT2 levels. J Inf. 2008;57:431. [Google Scholar]
- Medin CL, Fitzgerald KA, Rothman AL. Dengue virus nonstructural protein NS5 induces interleukin-8 transcription and secretion. J Virol. 2005;79(17):11053–11061. doi: 10.1128/JVI.79.17.11053-11061.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller S, Kastner S, Krijnse-Locker J, Buhler S, Bartenschlager R. The non-structural protein 4A of dengue virus is an integral membrane protein inducing membrane alterations in a 2K-regulated manner. J Biol Chem. 2007;282(12):8873–8882. doi: 10.1074/jbc.M609919200. [DOI] [PubMed] [Google Scholar]
- Miller S, Sparacio S, Bartenschlager R. Subcellular localization and membrane topology of the Dengue virus type 2 Non-structural protein 4B. J Biol Chem. 2006;281(13):8854–8863. doi: 10.1074/jbc.M512697200. [DOI] [PubMed] [Google Scholar]
- Mogensen TH, Paludan SR. Molecular pathways in virus-induced cytokine production. Microbiol Mol Biol Rev. 2001a;65(1):131–150. doi: 10.1128/MMBR.65.1.131-150.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mogensen TH, Paludan SR. Virus-cell interactions: impact on cytokine production, immune evasion and tumor growth. Eur Cytokine Netw. 2001b;12(3):382–390. [PubMed] [Google Scholar]
- Munoz-Jordan JL. Subversion of interferon by dengue virus. Curr Top Microbiol Immunol. 2010;338:35–44. doi: 10.1007/978-3-642-02215-9_3. [DOI] [PubMed] [Google Scholar]
- Munoz-Jordan JL, Laurent-Rolle M, Ashour J, Martinez-Sobrido L, Ashok M, Lipkin WI, Garcia-Sastre A. Inhibition of alpha/beta interferon signaling by the NS4B protein of flaviviruses. J Virol. 2005;79(13):8004–8013. doi: 10.1128/JVI.79.13.8004-8013.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munoz-Jordan JL, Sanchez-Burgos GG, Laurent-Rolle M, Garcia-Sastre A. Inhibition of interferon signaling by dengue virus. Proc Natl Acad Sci U S A. 2003;100(24):14333–14338. doi: 10.1073/pnas.2335168100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murgue B, Roche C, Chungue E, Deparis X. Prospective study of the duration and magnitude of viraemia in children hospitalised during the 1996–1997 dengue-2 outbreak in French Polynesia. J Med Virol. 2000;60(4):432–438. doi: 10.1002/(sici)1096-9071(200004)60:4<432::aid-jmv11>3.0.co;2-7. [DOI] [PubMed] [Google Scholar]
- Nguyen TH, Lei HY, Nguyen TL, Lin YS, Huang KJ, Le BL, Lin CF, Yeh TM, Do QH, Vu TQ, Chen LC, Huang JH, Lam TM, Liu CC, Halstead SB. Dengue hemorrhagic fever in infants: a study of clinical and cytokine profiles. J Infect Dis. 2004;189(2):221–232. doi: 10.1086/380762. [DOI] [PubMed] [Google Scholar]
- Padmanabhan R, Mueller N, Reichert E, Yon C, Teramoto T, Kono Y, Takhampunya R, Ubol S, Pattabiraman N, Falgout B, Ganesh VK, Murthy K. Multiple enzyme activities of flavivirus proteins. Novartis Found Symp. 2006;277:74–84. doi: 10.1002/0470058005.ch6. discussion 84-6, 251-3. [DOI] [PubMed] [Google Scholar]
- Pang T, Cardosa MJ, Guzman MG. Of cascades and perfect storms: the immunopathogenesis of dengue haemorrhagic fever-dengue shock syndrome (DHF/DSS) Immunol Cell Biol. 2007;85(1):43–45. doi: 10.1038/sj.icb.7100008. [DOI] [PubMed] [Google Scholar]
- Polo S, Ketner G, Levis R, Falgout B. Infectious RNA transcripts from full-length dengue virus type 2 cDNA clones made in yeast. J Virol. 1997;71(7):5366–5374. doi: 10.1128/jvi.71.7.5366-5374.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Priyadarshini D, Gadia RR, Tripathy A, Gurukumar KR, Bhagat A, Patwardhan S, Mokashi N, Vaidya D, Shah PS, Cecilia D. Clinical findings and pro-inflammatory cytokines in dengue patients in Western India: a facility-based study. PLoS ONE. 2010;5(1):e8709. doi: 10.1371/journal.pone.0008709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pryor MJ, Rawlinson SM, Butcher RE, Barton CL, Waterhouse TA, Vasudevan SG, Bardin PG, Wright PJ, Jans DA, Davidson AD. Nuclear localization of dengue virus nonstructural protein 5 through its importin alpha/beta-recognized nuclear localization sequences is integral to viral infection. Traffic. 2007;8(7):795–807. doi: 10.1111/j.1600-0854.2007.00579.x. [DOI] [PubMed] [Google Scholar]
- Qu L, McMullan LK, Rice CM. Isolation and characterization of noncytopathic pestivirus mutants reveals a role for nonstructural protein NS4B in viral cytopathogenicity. J Virol. 2001;75(22):10651–10662. doi: 10.1128/JVI.75.22.10651-10662.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raghupathy R, Chaturvedi UC, Al-Sayer H, Elbishbishi EA, Agarwal R, Nagar R, Kapoor S, Misra A, Mathur A, Nusrat H, Azizieh F, Khan MA, Mustafa AS. Elevated levels of IL-8 in dengue hemorrhagic fever. J Med Virol. 1998;56(3):280–285. doi: 10.1002/(sici)1096-9071(199811)56:3<280::aid-jmv18>3.0.co;2-i. [DOI] [PubMed] [Google Scholar]
- Rawlinson SM, Pryor MJ, Wright PJ, Jans DA. CRM1-mediated nuclear export of dengue virus RNA polymerase NS5 modulates interleukin-8 induction and virus production. J Biol Chem. 2009;284(23):15589–15597. doi: 10.1074/jbc.M808271200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rico-Hesse R, Harrison LM, Nisalak A, Vaughn DW, Kalayanarooj S, Green S, Rothman AL, Ennis FA. Molecular evolution of dengue type 2 virus in Thailand. Am J Trop Med Hyg. 1998;58(1):96–101. doi: 10.4269/ajtmh.1998.58.96. [DOI] [PubMed] [Google Scholar]
- Roosendaal J, Westaway EG, Khromykh A, Mackenzie JM. Regulated cleavages at the West Nile virus NS4A-2K-NS4B junctions play a major role in rearranging cytoplasmic membranes and Golgi trafficking of the NS4A protein. J Virol. 2006;80(9):4623–4632. doi: 10.1128/JVI.80.9.4623-4632.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook J, Russell DW. Molecular Cloning: A Laboratory Manual. Third ed. Cold Spring Harbor Laboratory Press; 2001. [Google Scholar]
- Selisko B, Peyrane FF, Canard B, Alvarez K, Decroly E. Biochemical characterization of the (nucleoside-2'O)-methyltransferase activity of dengue virus protein NS5 using purified capped RNA oligonucleotides (7Me)GpppAC(n) and GpppAC(n) J Gen Virol. 2010;91(Pt 1):112–121. doi: 10.1099/vir.0.015511-0. [DOI] [PubMed] [Google Scholar]
- Srikiatkhachorn A. Plasma leakage in dengue haemorrhagic fever. Thromb Haemost. 2009;102(6):1042–1049. doi: 10.1160/TH09-03-0208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srikiatkhachorn A, Krautrachue A, Ratanaprakarn W, Wongtapradit L, Nithipanya N, Kalayanarooj S, Nisalak A, Thomas SJ, Gibbons RV, Mammen MP, Jr, Libraty DH, Ennis FA, Rothman AL, Green S. Natural history of plasma leakage in dengue hemorrhagic fever: a serial ultrasonographic study. Pediatr Infect Dis J. 2007;26(4):283–290. doi: 10.1097/01.inf.0000258612.26743.10. discussion 291-2. [DOI] [PubMed] [Google Scholar]
- Ubol S, Chareonsirisuthigul T, Kasisith J, Klungthong C. Clinical isolates of dengue virus with distinctive susceptibility to nitric oxide radical induce differential gene responses in THP-1 cells. Virology. 2008;376(2):290–296. doi: 10.1016/j.virol.2008.03.030. [DOI] [PubMed] [Google Scholar]
- Uchil PD, Satchidanandam V. Characterization of RNA synthesis, replication mechanism, and in vitro RNA-dependent RNA polymerase activity of Japanese encephalitis virus. Virology. 2003;307(2):358–371. doi: 10.1016/s0042-6822(02)00130-7. [DOI] [PubMed] [Google Scholar]
- Umareddy I, Chao A, Sampath A, Gu F, Vasudevan SG. Dengue virus NS4B interacts with NS3 and dissociates it from single-stranded RNA. J Gen Virol. 2006;87(Pt 9):2605–2614. doi: 10.1099/vir.0.81844-0. [DOI] [PubMed] [Google Scholar]
- Vaughn DW, Green S, Kalayanarooj S, Innis BL, Nimmannitya S, Suntayakorn S, Endy TP, Raengsakulrach B, Rothman AL, Ennis FA, Nisalak A. Dengue viremia titer, antibody response pattern, and virus serotype correlate with disease severity. J Infect Dis. 2000;181(1):2–9. doi: 10.1086/315215. [DOI] [PubMed] [Google Scholar]
- Verma S, Lo Y, Chapagain M, Lum S, Kumar M, Gurjav U, Luo H, Nakatsuka A, Nerurkar VR. West Nile virus infection modulates human brain microvascular endothelial cells tight junction proteins and cell adhesion molecules: Transmigration across the in vitro blood-brain barrier. Virology. 2009;385(2):425–433. doi: 10.1016/j.virol.2008.11.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verma S, Molina Y, Lo YY, Cropp B, Nakano C, Yanagihara R, Nerurkar VR. In vitro effects of selenium deficiency on West Nile virus replication and cytopathogenicity. Virol J. 2008;5:66. doi: 10.1186/1743-422X-5-66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verma S, Ziegler K, Ananthula P, Co JK, Frisque RJ, Yanagihara R, Nerurkar VR. JC virus induces altered patterns of cellular gene expression: interferon-inducible genes as major transcriptional targets. Virology. 2006;345(2):457–467. doi: 10.1016/j.virol.2005.10.012. [DOI] [PubMed] [Google Scholar]
- Welsch C, Albrecht M, Maydt J, Herrmann E, Welker MW, Sarrazin C, Scheidig A, Lengauer T, Zeuzem S. Structural and functional comparison of the non-structural protein 4B in flaviviridae. J Mol Graph Model. 2007;26(2):546–557. doi: 10.1016/j.jmgm.2007.03.012. [DOI] [PubMed] [Google Scholar]
- Westaway EG, Khromykh AA, Kenney MT, Mackenzie JM, Jones MK. Proteins C and NS4B of the flavivirus Kunjin translocate independently into the nucleus. Virology. 1997a;234(1):31–41. doi: 10.1006/viro.1997.8629. [DOI] [PubMed] [Google Scholar]
- Westaway EG, Mackenzie JM, Kenney MT, Jones MK, Khromykh AA. Ultrastructure of Kunjin virus-infected cells: colocalization of NS1 and NS3 with double-stranded RNA, and of NS2B with NS3, in virus-induced membrane structures. J Virol. 1997b;71(9):6650–6661. doi: 10.1128/jvi.71.9.6650-6661.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- World Health Organization. Geneva: WHO; 1997. Dengue haemorrhagic fever: diagnosis, treatment prevention and control. [Google Scholar]
- World Health Organization. Geneva: WHO Media Centre; 2009. Dengue and dengue haemorrhagic fever. Fact sheet #117 ed. [Google Scholar]
- Zhou Y, Ray D, Zhao Y, Dong H, Ren S, Li Z, Guo Y, Bernard KA, Shi PY, Li H. Structure and function of flavivirus NS5 methyltransferase. J Virol. 2007;81(8):3891–3903. doi: 10.1128/JVI.02704-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou G, Chen YL, Dong H, Lim CC, Yap LJ, Yau YH, Shochat SG, Lescar J, Shi PY. Functional analysis of two cavities in flavivirus NS5 polymerase. J Biol Chem. 2011 doi: 10.1074/jbc.M110.214189. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Induction of immunomediator transcripts in DENV-infected THP-1 cells. Cells were infected at MOI-0.1, MOI-1 or incubated with LPS for 1 hour and collected each day for four consecutive days. qRT-PCR was conducted to determine A) IL-8 and B) TNFα mRNA fold-changes as compared to same day controls. GAPDH was used as the endogenous control gene. Bars represent the mean ± SD of four samples and (*) indicates statistical significance at p < 0.05 compared to same day levels induced by MOI-0.1.
Cell viability of THP-1 cells transfected with plasmid DNA or treated with LPS for 1 h. THP-1 cells were grown to confluence and 1 × 106 cells were treated as indicated in the graph. Viability of treated cells was determined by cell proliferation assay. Results are presented as percent viable cells. Data are expressed as mean ± SD of two independent experiments performed in duplicate.