Abstract
Histone deacetylase HDA1, the prototype for the class II mammalian deacetylases, is likely the catalytic subunit of the HDA1-containing complex that is involved in TUP1-specific repression and global deacetylation in yeast. Although the class I RPD3-like enzymatic complexes have been well characterized, little is known about the identity and interactions of the factors that associate to form the HDA1 complex. In this paper, we identify related HDA2 and HDA3 proteins that are found in the HDA1 complex and show that HDA1 interacts with itself and with the HDA2-HDA3 subcomplex to form a likely tetramer. These interactions are necessary for catalytic activity because mutations in any of the three components disrupt activity both in vitro and in vivo. In this respect the HDA1 complex differs from yeast RPD3, which has components such as SIN3 that are not essential for activity in vitro, and yeast HOS3, which has intrinsic in vitro activity as a homodimer in the absence of other subunits.
Histone deacetylases can be recruited to promoter elements to deacetylate adjacent nucleosomes and repress transcription in a background of global acetylation and deacetylation (1–3). The deacetylases have been grouped into two classes as determined by sequence similarity (4). Class I histone deacetylases are similar to RPD3 and include yeast HOS1 and HOS2 (5) and mammalian HDAC1, HDAC2, and HDAC3 (1). These proteins are considered to be putative catalytic subunits because HDAC1 interacts with trapoxin, a competitive inhibitor of the deacetylase (6). In addition, all of these proteins are homologous to HOS3, which has intrinsic catalytic activity as a homodimer when expressed in Escherichia coli in the absence of other deacetylase components (7). The human class I deacetylase complex has been well characterized. It contains HDAC1/HDAC2, RbAp46/48 (which may be involved in histone recognition), SIN3 (which interacts with proteins such as Mad that recruit the deacetylase complex to DNA), and proteins of unknown function such as SAP30 and SAP18 (1). Most of these proteins are conserved between yeast and humans (1). Interestingly, SIN3 is not essential for HDAC1 deacetylase activity in vitro but is essential in vivo (8, 9), presumably due to its role in targeting RPD3 to promoters (1, 10, 11).
Class II deacetylases are similar to yeast HDA1, a putative catalytic subunit, and include mammalian HDAC4, HDAC5, and HDAC6. However, little is known about the protein complexes that include these deacetylases (4). Yeast HDA1 was first identified as a component of the HDA complex, referred to here as the HDA1 complex (12). When the HDA1 complex was purified to near homogeneity it was found to contain at least four peptides: p75/HDA1, p73/HDA2, p72/HDA2, and p71/HDA3. p73 and p72 are thought to represent posttranslationally modified variants of HDA2 because their sequences were found to be identical for each peptide analyzed (12). Because HDA1 and HDA3 were shown to coimmunoprecipitate and disruption of HDA3 caused the loss of deacetylase activity from the chromatographic profile HDA3 is required for HDA1 activity in vitro. Recently, we reported the role of HDA1 in TUP1 repression (13). TUP1 is a repressor in yeast that affects pathways involved in mating, DNA repair, oxygen and glucose utilization, and osmotic stress (14–17). We have shown that TUP1 represses gene activity in vivo in part through its utilization of HDA1 to deacetylate histones H3 and H2B. This deacetylation occurs at a localized region containing the TATA element adjacent to the TUP1 recruitment site at ENA1 and STE6 (13).
In this work we have identified the encoded sequences of HDA2 and HDA3 and have shown that HDA2 coimmunoprecipitates with HDA1 and HDA3. We also have used immunoprecipitation, genetic disruptions, and direct binding glutathione S-transferase (GST)-fusion experiments to determine the order in which HDA1, HDA2, and HDA3 are associated with each other within the deacetylase complex. Finally, we asked whether HDA2 and HDA3 are required for the activity of the HDA1 deacetylase both in vitro and in vivo. The results of these biochemical and genetic experiments are presented.
Materials and Methods
Yeast Strains.
Please refer to the strain list in Table 1. The hda1∷TRP1 mutant allele was generated by transforming pWJ118 (13) digested with SpeI into YDS2. The hda2∷TRP1 mutant allele was generated by digesting pWJ202.1 with SpeI/SacI to transform YDS2, resulting in WJY103. The hda3∷TRP1 mutant allele was generated through digestion of pWJ302 with SacI/KpnI to transform YDS2, resulting in WJY104. The hda2∷KAN, hda3∷TRP1 double mutant strain WJY105 was obtained by transforming SpeI-digested pWJ204.1 into WJY104. To generate the HDA1∷MYC∷URA3 mutant allele, the plasmid pHDA1–4 was digested with PflmI, and then transformed into yeast strains YDS2, WJY103, WJY104, and WJY105, yielding the WJY200, WJY203, WJY204, and WJY205 strains, respectively. The HDA2∷MYC∷URA3 and the HDA3∷MYC∷URA3 mutant alleles were generated similarly, with pHDA2–4(HindIII digested) and pHDA3–4(BglII digested). The integration or deletion of these genes was confirmed by PCR.
Table 1.
Yeast strains used in this study
| Strain | Relevant genotype | Source |
|---|---|---|
| YDS21U | MATa, ade2-1, can1-100, his3-Δ200, leu2-3,112, trp1, URA3∷TelVR | Ref. 5 |
| WJY127 | Same as YDS21U except for hda1∷TRP1 | Ref. 13 |
| WJY128 | Same as YDS21U except for hda2∷TRP1 | This study |
| WJY129 | Same as YDS21U except for hda3∷TRP1 | This study |
| YDS2 | MAT a, ade2-1, can1-100, his3-Δ200, leu2-3,112, trp1, ura3-52 | Ref. 5 |
| WJY110 | Same as YDS2 except for hda1∷TRP1 | This study |
| WJY103 | Same as YDS2 except for hda2∷TRP1 | This study |
| WJY104 | Same as YDS2 except for hda3∷TRP1 | This study |
| WJY105 | Same as YDS2 except for hda2∷TRP1, hda3∷KAN | This study |
| WJY200 | Same as YDS2 except for HDA1∷MYC18∷URA3 | This study |
| WJY201 | Same as YDS2 except for HDA2∷MYC18∷URA3 | This study |
| WJY202 | Same as YDS2 except for HDA3∷MYC18∷URA3 | This study |
| WJY203 | Same as WJY103 except for HDA1∷MYC18∷URA3 | This study |
| WJY204 | Same as WJY104 except for HDA1∷MYC18∷URA3 | This study |
| WJY205 | Same as WJY105 except for HDA1∷MYC18∷URA3 | This study |
| WJY211 | Same as WJY201 except for hda1∷TRP1 | This study |
Plasmid Construction.
pWJ202.1 was made through the replacement of the AseI fragment (encoding HDA2 amino acid residues 122–478) of plasmid pSKH2 with the 1.2-kb TRP1 gene, whereas the plasmid pSKH2 was made by inserting the 2.5-kb SpeI/NheI PCR fragment containing HDA2 gene and flanking sequence into pBluescript KS(+) (Stratagene). pWJ204.1 was made through the replacement of the AseI fragment of pSKH2 with 2.5-kb KanMX4 gene pFA6a-KanMX4 (18). pWJ302 was constructed by replacing the EcoRV/HindIII fragment (encoding HDA3 amino acid residues 54–607) of the plasmid pWJ301 with the TRP1 gene. pWJ301 was constructed by inserting the 3.5-kb EcoRI/SacI PCR fragment containing the HDA3 gene sequence into pBluescript KS(+). pHDA1–4 was made by inserting the MYC-18 gene (SpeI fragment) (19) in-frame before the TAA of HDA1 coding sequence in plasmid pHDA1–3. The SpeI site in pHDA1–3 before the TAA of the HDA1 gene was made through mutagenesis in plasmid pHDA1–2. pHDA1–2 was constructed by subcloning the HDA1 gene (SacI/XhoI fragment containing HDA1 C-terminal coding sequence and 3′ sequence) into pBluescript KS(+). pHDA2–4 and pHDA3–4 were constructed in the same way as pHDA1–4. pWJ153 was constructed by inserting the PCR fragment of HDA1 coding sequence into the BamHI/SalI site pGEX-4T-1 to express GST-HDA1. The primers used were: 5′-GACGGATCCATGGATTCTGTAATGGT-3′ and 5′-AGCGTCGACTCATTCTTCATCACTCC-3′. pWJ155 was constructed by inserting the PCR fragment of the HDA3 coding sequence into the BamHI/SalI site pGEX-4T-1 to express GST-HDA3. The primers used were: 5′-GACGGATCCATGGATTTACTACGCAT-3′ and 5′-AGCGTCGACTTATACATTTCCTGGTTT-3′. pGST-HDA2(164–254) used for raising α-HDA2 antibody was constructed as follows. The HDA2 sequence was generated by PCR using the following oligonucletides: 5′- CGTGGATCCAGAACCAAGAGACTTTCCGG-3′ and 5′-GATGAATTCCAGCCAATAGGTATTGATCGTG-3′, and the PCR product was digested with BamHI and EcoRI and ligated into the pGEX-2T (Amersham Pharmacia).
Protein Binding Assays.
GST and GST fusion proteins were expressed and purified in E. coli BL21(DE3) according to the procedures provided by the manufacturer (Amersham Pharmacia). HDA1, HDA2, and HDA3 were translated in vitro with plasmids pWJ402, pWJ403, pWJ406 (13), and the TNT in vitro translation system, according to the procedures provided by the manufacturer (Promega). Protein binding assays were performed according to ref. 20. One hundred percent of the eluate and 10% of the input materials were resolved by SDS/PAGE and visualized by fluorography.
Immunoprecipitation and Histone Deacetylase Activity Assay.
The rabbit α-HDA2 antibody was generated against a GST-fusion protein between amino acid residues 164–254 of the HDA2 protein by using standard procedures. Immunoprecipitation was performed according to ref. 21. The α-HDA1 (12) antibody (1:1,000) or α-myc antibody (1:20) coupled to Sepharose 4B-beads (Babco, Richmond, CA) were added to the whole-cell lysates for immunoprecipitation. The dilutions of antibodies for Western blots were: α-HDA1, 1:3,000; α-HDA2, 1:1,000; α-HDA3, 1:3,000; and α-myc, 1:1,000. Deacetylase activities were assayed with half of the immunoprecipitated pellet. The assay was carried out by using 3H-labeled HeLa histones as in ref. 12.
Molecular Weight Determinations.
Calculations to determine molecular weights were determined as in ref. 22. M = 6 πηNas/(1 − vbarρ) with M = molecular weight of the complex; π = 3.142; η = the viscosity of the medium (0.01009 × 10−8 g/cm × sec); N = Avogadro's number (6.02 × 1023); a = Stokes radius (60.3 × 10−8 cm); s = the sedimentation coefficient (9.7 S); vbar = partial specific volume (0.722); and ρ = the density of a 12.5% (average) sucrose solution (1.047 g/ml).
Chromatin Immunoprecipitation (Chr-IP) and PCR Analysis.
Yeast strains were grown to OD600 of 1.0 in yeast extract/peptone/dextrose, and Chr-IPs were performed with the addition of 5 μl antibody specific for each acetyl-lysine to 50 μl whole-cell extract from 8 × 107 cells according to ref. 9. Antibodies to specific histone acetylation sites are as described (13). PCR was performed with 1/100 of the immunoprecipitated DNA that was used as template. 32P-α-dATP was added to the PCR (1 μCi/12.5 μl) for quantitation by PhosphorImager and imagequant software (Molecular Dynamics,). The TEL band was used as a loading control as described (9). The primer pairs used to amplify the ENA1, DAL80, and TEL promoter were as described (13). The primer pairs used to amplify GAL10 were: 5′-GGCGGCTTCTAATCCGTACTTCAA-3′ and 5′-ACCAATGTATCCAGCACCACCTGT-3′. The primer pairs used to amplify the HO gene were: 5′-CATGATGAAGCGTTCTAAACGCAC-3′ and 5′-TAGCCGTGACGTTTGCGATGTCTT-3′.
Comparison of HDA2 and HDA3 Protein Similarity.
The similarity between HDA2 and HDA3 was determined by using the blast program of the Saccharomyces Genome Database. The alignment between HDA2 and HDA3 was generated by using the Smith–Waterman algorithm in the Saccharomyces Genome Database.
Results and Discussion
Identification of HDA2 and HDA3.
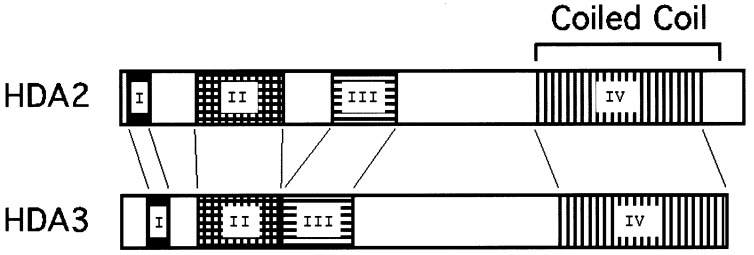
It was previously shown that proteins with apparent relative molecular masses 75, 73, 72, and 71 kDa copurify with histone deacetylase activity in the HDA deacetylase complex (12). p75 was identified as HDA1, and four other proteins (RPD3, HOS1, HOS2, and HOS3) were shown to be highly homologous to HDA1 (5). p73 and p72 were designated as HDA2 and p71 as HDA3. Peptide sequence analysis of HDA2 and HDA3 and their comparison to the yeast database identifies HDA2 (GenBank accession no. U28374) as encoding 674 aa (77 kDa) and HDA3 (GenBank accession no. U25842) as encoding 655 aa (75.4 kDa). For reference, HDA1 encodes 706 aa (80 kDa). The alignment of HDA2 and HDA3 proteins shows ≈21% identity and 48.6% similarity over a region of 668 aa. HDA2 and HDA3 contain four homologous domains (Fig. 1). Domain IV of HDA2 (residues 449–630) and HDA3 (residues 475–650) shows features of coiled-coil structure (Fig. 1). Neither of these proteins have obvious homology with histone deacetylases or with other proteins in either prokaryotic or eukaryotic databases, and they appear to be novel proteins. Therefore, although the coiled-coil sequence homology suggests that HDA2 and HDA3 participate in protein–protein interactions in the deacetylase complex, they are not likely to be catalytic subunits.
Figure 1.
HDA2 and HDA3 have sequence similarity in four domains including one with potential coiled coil structure. Domain I: HDA2 (residues 12–33), HDA3 (residues 31–52), identity 31%, similarity 52%; domain II: HDA2 (residues 84–177), HDA3 (residues 84–169), identity 36%, similarity 54%; domain III: HDA2 (residues 238–299), HDA3 (residues 180–246), identity 25%, similarity 46%; domain IV: HDA2 (residues 449–630), HDA3 (residues 475–650), identity 23%, similarity 47%.
HDA2 Coimmunoprecipitates with HDA1 and HDA3.
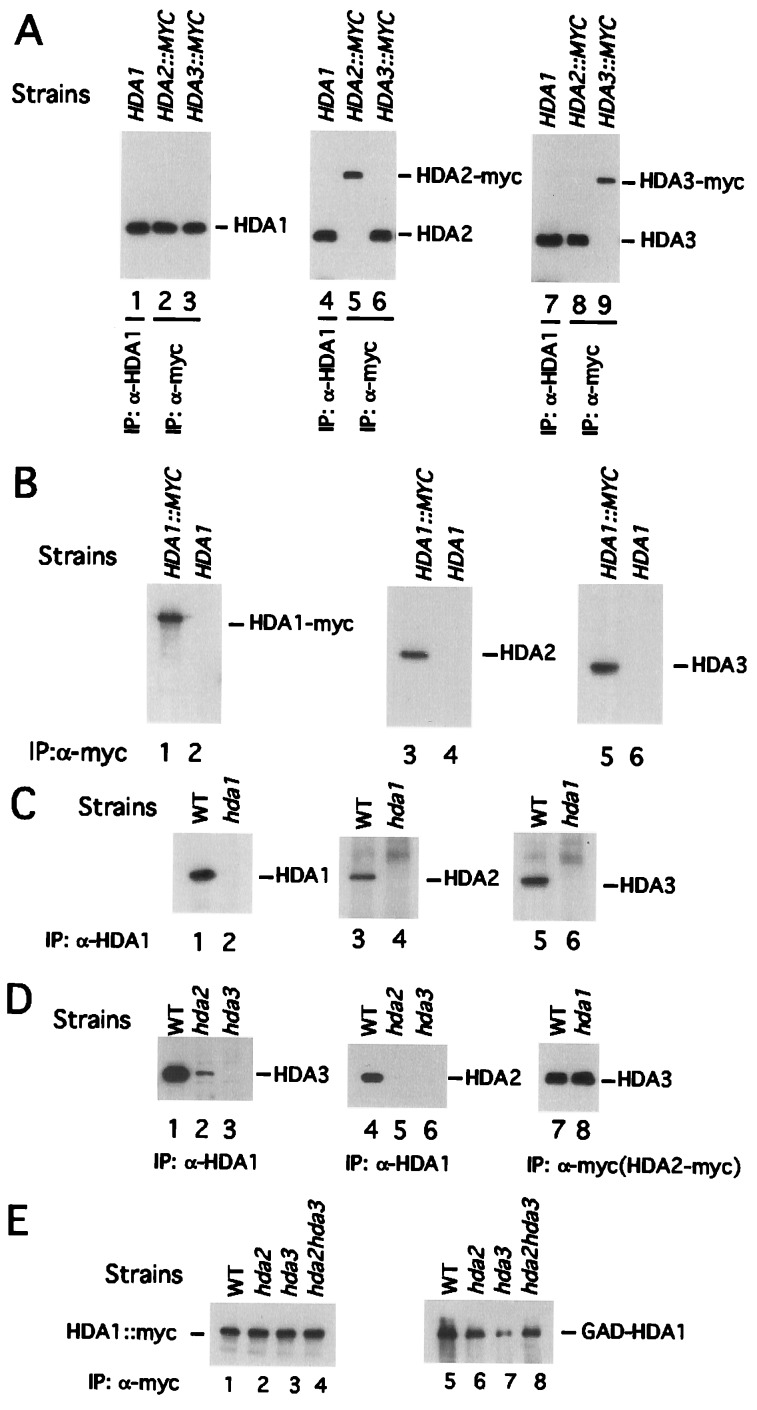
HDA2 and HDA3 cochromatograph with HDA1 (12). HDA1 and HDA3 also coimmunoprecipitate, indicating that these two proteins are in the same macromolecular complex (12). To determine whether HDA2 is a component of the deacetylase complex, HDA2 and HDA3 genes were fused to 18 copies of the sequence coding for the myc epitope at the 3′ end of their coding regions. Then they were integrated into the natural loci to replace the HDA2 and HDA3 genes, resulting in strains WJY201 and WJY202, respectively. Whole-cell extracts were prepared from the wild-type (WT) YDS2 cells (which lack myc-tagged protein), WJY201 and WJY202. α-HDA1 antibody and α-myc antibody then were used to immunoprecipitate HDA1, HDA2-myc, and HDA3-myc proteins from these extracts. The immunoprecipitated materials were subjected to SDS/PAGE and Western blotting in which α-HDA1, α-HDA2, and α-HDA3 antibodies were used to detect the presence of HDA1, HDA2-myc, and HDA3-myc proteins (Fig. 2A). We found that in strain YDS2 α-HDA1 antibody immunoprecipitates HDA1 (Fig. 2A, lane 1), HDA2 (Fig. 2A, lane 4), and HDA3 (Fig. 2A, lane 7). Using WJY201, the HDA2-myc containing strain, we found that α-myc antibody immunoprecipitates HDA2-myc (Fig. 2A, lane 5), HDA1 (Fig. 2A, lane 2), and HDA3 (Fig. 2A, lane 8). Finally, in WJY202 the HDA3-myc containing strain, we found that α-myc antibody immunoprecipitates HDA3-myc (Fig. 2A, lane 9), HDA1 (Fig. 2A, lane 3), and HDA2 (Fig. 2A, lane 6). To rule out the possibility of nonspecific interactions between α-myc antibody and untagged proteins of the HDA1 complex, α-myc antibody was incubated with the cell lysate from YDS2 (which lacks myc-tagged protein). The precipitate was probed separately with α-HDA1, α-HDA2, and α-HDA3 antibody (Fig. 2B). It is evident that the α-myc antibody did not pull down either untagged HDA1 (Fig. 2B, lane 2), HDA2 (Fig. 2B, lane 4), or HDA3 (Fig. 2B, lane 6). Also, to guard against nonspecific interactions between α-HDA1 antibody and HDA2 or HDA3, immunoprecipitates from the hda1Δ strain (WJY110) were probed with α-HDA2 and HDA3 antibodies (Fig. 2C). α-HDA1 antibody does not immunoprecipitate either HDA2 (Fig. 2C, lane 4) or HDA3 (Fig. 2C, lane 6) from the hda1Δ mutant strain. Therefore, we conclude that HDA1, HDA2, and HDA3 are subunits of the same macromolecular complex.
Figure 2.
The interactions among the subunits of the HDA complex HDA1, HDA2, and HDA3. (A) The HDA complex coimmunoprecipitates with any single subunit of the complex. In lanes 1, 4, and 7, the whole-cell extracts were prepared from YDS2 (containing the WT HDA1 gene). In lanes 2, 5, and 8, strain WJY201 (containing HDA2∷MYC gene) was used. In lanes 3, 6, and 9, strain WJY202 (containing HDA3∷MYC gene) was used. The antibodies used for immunoprecipitation are shown. Blots were probed with α-HDA1 (lanes 1–3), α-HDA2 (lanes 4–6), and α-HDA3 (lanes 7–9). (B) The α-myc antibody does not recognize HDA1, HDA2, or HDA3 lacking a myc tag. In lanes 1, 3, and 5, strain WJY200 (HDA1∷myc) was used. In lanes 2, 4, and 6, YDS2 (containing WT HDA1, HDA2, and HDA3 with no myc tag) was used. Blots were probed with α-HDA1 (lanes 1 and 2), α-HDA2 (lanes 3 and 4), and α-HDA3 (lanes 5 and 6). (C) The α-HDA1 antibody does not recognize HDA2 or HDA3. In lanes 1, 3, and 5, strain YDS2 was used. In lanes 2, 4, and 6, the hda1Δ strain WJY110 was used. Blots were probed with α-HDA1 (lanes 1 and 2), α-HDA2 (lanes 3 and 4), and α-HDA3 (lanes 5 and 6). (D) The interaction between HDA2 and HDA3 is independent of HDA1. In lanes 1–6, strains YDS2 (WT, lanes 1 and 4), WJY103 (hda2Δ, lanes 2 and 5) and WJY104 (hda3Δ, lanes 3 and 6) were used. In lanes 7 and 8, the strains WJY201 (WT, lane 7) and WJY211 (hda1Δ, lane 8) were used, where the natural HDA2 gene has been replaced by HDA2∷MYC. The resulting blots were probed with α-HDA3 (lanes 1–3, 7, and 8) and α-HDA2 (lanes 4–6). (E) HDA1-HDA1 interact. HDA1 was tagged with myc18 in WT (WJY200), hda2Δ (WJY203), hda3Δ (WJY204), and hda2Δhda3Δ (WJY205). The plasmid expressing GAL4AD-HDA1 (pWJ104) was transformed into these strains. α-Myc antibody was used to pull down HDA1-myc from cell extracts of these strains, and α-myc antibody (lanes 1–4) and α-GAL4 AD antibody (lanes 5–8) were used to detect the presence of HDA1-myc and GAL4-AD-HDA1. IP, immunoprecipitation.
HDA1 Coimmunoprecipitates with HDA2-HDA3 Subcomplex.
To determine the interactions that occur in vivo between HDA1, HDA2, and HDA3 we examined the coimmunoprecipitation of each of these components in genetic backgrounds lacking individual HDA genes. Whole-cell extracts were prepared from WT (YDS2), hda2Δ (WJY103), and hda3Δ (WJY104) cells. α-HDA1 antibody was used to immunoprecipitate the HDA1 complex from these extracts, which subsequently were probed in Western blots with α-HDA3 and α-HDA2 antibodies. First, we asked whether interaction between HDA1 and HDA3 depends on HDA2. When HDA2 was deleted, α-HDA1 antibody immunoprecipitated only ≈4% of HDA3 (Fig. 2D, lane 2) as compared with the level seen in the WT strain containing HDA2 (Fig. 2D, lane 1) and determined by densitometry. This is in part due to the instability of HDA3 in the hda2Δ mutant strain (data not shown). Therefore, HDA2 influences but is not essential for the interaction between HDA1 and HDA3. Second, we asked whether HDA3 mediates an interaction between HDA1 and HDA2. We found in hda3Δ cells that no HDA2 can be detected in the precipitates pulled down by α-HDA1 (Fig. 2D, lane 6). As a control, we have shown that the amount of HDA2 is not affected by the deletion of HDA3 (data not shown). These data indicate that HDA3 is essential for the HDA1-HDA2 interaction. Finally, to determine whether HDA1 is required for interaction between HDA2 and HDA3, extracts were prepared from WT (WJY201) and hda1Δ (WJY211) cells and α-myc antibody was used to pull down HDA2-myc. The presence of HDA3 was detected with α-HDA3 antibody. We found that α-myc immunoprecipitates HDA3 as well in the hda1Δ strain as in the HDA1 WT strain (Fig. 2D, lanes 7 and 8). Therefore the association of HDA2 with HDA3 is independent of HDA1. These data argue that HDA1 interacts with a subcomplex containing HDA2-HDA3 largely through HDA3.
HDA1-HDA1 Coimmunoprecipitate.
It has been shown that a mammalian histone deacetylase complex contains HDAC1 and HDAC2, arguing for the presence of two similar, but nonidentical, RPD3-like catalytic subunits in the same complex (1). Moreover, it was shown that HOS3 has intrinsic catalytic activity as a homodimer when expressed in E. coli or yeast (7). To test whether HDA1 interacts as a homodimer in the HDA1 complex, two differently tagged HDA1 proteins were expressed simultaneously. One is tagged at its C terminus with myc18; the other at its N terminus with GAL4-AD (GAD). The plasmid expressing GAD-HDA1 was transformed into cells (WJY200) containing the HDA1-MYC18 gene integrated at the natural HDA1 locus. We then asked whether immunoprecipitated HDA1-myc also pulls down GAD-HDA1. Using α-myc and α-GAD antibodies to detect the presence of these proteins in Western blots we found (Fig. 2E) that α-myc antibody immunoprecipitates HDA1-myc and GAD-HDA1 (Fig. 2E, lanes 1 and 5). Therefore, HDA1-HDA1 interact directly or indirectly in the same macromolecular complex.
To determine whether HDA2 or HDA3 mediate HDA1-HDA1 interactions we examined the coimmunoprecipitation of HDA1-myc18 and GAD-HDA1 in hda2Δ (WJY203), hda3Δ (WJY204), and hda2Δhda3Δ (WJY205) mutant strains. As is shown in Fig. 2E lanes 2–4 and 6–8, the deletion of HDA2 and HDA3 does not abolish the interaction between HDA1-myc and GAD-HDA1. The tagging of different HDA1 molecules with GAD and myc 18 in the same strain does not interfere with interactions among HDA1, HDA2 and HDA3 in the WT (WJY200) strain (data not shown). We also examined whether there were interactions between HDA2 and HDA2 or between HDA3 and HDA3 using the same approach and found no such self-interactions (data not shown). We conclude that HDA1-HDA1 interacts in a manner that is independent of HDA2 and HDA3.
Direct Interactions Between HDA1-HDA1, HDA2-HDA3, and HDA1-HDA3.
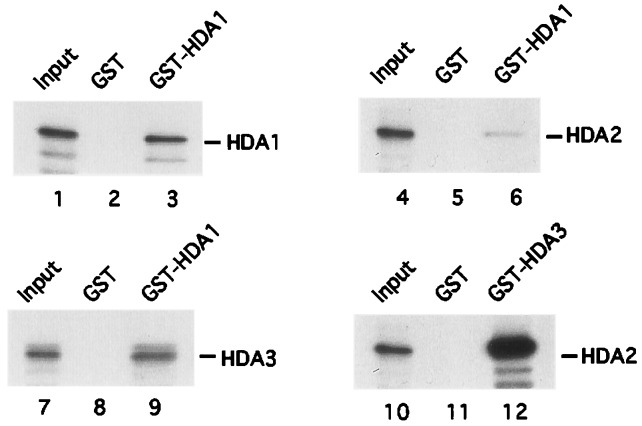
To determine the direct protein–protein interactions between HDA1, HDA2, and HDA3, GST-HDA1 and GST-HDA3 were purified from E. coli, and 35S-labeled HDA1, HDA2, and HDA3 were synthesized by using an in vitro translation system as described in Materials and Methods. First, we asked whether there is a direct interaction between HDA1 and HDA1 and between HDA1 and HDA3. It was found that GST-HDA1 can pull down both HDA1 and HDA3 labeled with 35S (Fig. 3, lanes 3 and 9) whereas the GST protein alone cannot do so (Fig. 3, lanes 2 and 8). Compared with input (Fig. 3, lanes 1 and 7), ≈10% of total HDA1 and HDA3 bound to the GST-HDA1 protein. In contrast, less than 1% 35S-labeled HDA2 binds to GST-HDA1 (Fig. 3, lanes 4 and 6). Next, we tested the interaction between HDA2 and HDA3 by incubating 35S-labeled HDA2 with GST-HDA3. It was found that GST-HDA3 precipitates ≈60% of 35S-labeled HDA2, whereas GST alone cannot retain HDA2 (Fig. 3, lanes 10 and 12). These direct binding experiments demonstrate that HDA1 interacts directly with HDA1 and HDA3. They also indicate that HDA2 interacts most strongly with HDA3. The weakest interaction observed is between HDA1 and HDA2. These data support those of the coimunoprecipitation experiments and argue that an HDA1-HDA1 subcomplex interacts directly with an HDA2-HDA3 subcomplex, and that this interaction occurs most strongly through HDA3.
Figure 3.
Direct interactions between HDA1, HDA2, and HDA3. 35S-Methionine-labeled HDA1, HDA2, and HDA3 were incubated with GST (lanes 2, 5, and 8) or GST-HDA1 (lanes 3, 6, and 9) and immobilized on glutathione-Sepharose beads. 35S-Methionine-labeled HDA2 was incubated with GST (lane 11) and GST-HDA3 (lane 12). The interactions were analyzed according to Materials and Methods.
By following HDA activity enzymatically as described (12) with known protein standards and probing fractions by Western blot we estimate from sucrose density gradient centrifugation an S value of 9.7 S for the HDA1 complex (data not shown). Using Superdex 200 chromatography the Stokes radius of the HDA1 complex was estimated to be 60.3 × 10−8 cm (data not shown). These values allow a calculation of the molecular mass of the HDA1 complex at 299.2 kDa as described (Materials and Methods). The encoded molecular masses of HDA1, HDA2, and HDA3 are 80, 77, and 75.4 kDa, respectively. Therefore we estimate the expected molecular mass of a tetrameric complex containing two molecules of HDA1 to be 312.5 kDa. Because this size correlates well with the molecular mass determined above and we do not find other proteins in the HDA1 complex (12), it is suggested that the HDA1 complex contains a protein tetramer. Interestingly, HOS3 assembles to form a homodimer in yeast and E. coli and it is as a homodimer that it has intrinsic activity (7). Also, HDAC1 and HDAC2 deacetylases have been found in the same HDAC complex in mammalian cells (1). Thus all deacetylase complexes characterized to date, including HDA1, contain at least two putative catalytic deacetylase molecules. Because each nucleosome octamer contains two molecules of each of the four core histone proteins the dimeric HDA1 catalytic component may be required for the deacetylation of both molecules.
HDA2 Is Important for the in Vitro Activity of the HDA1 Complex.
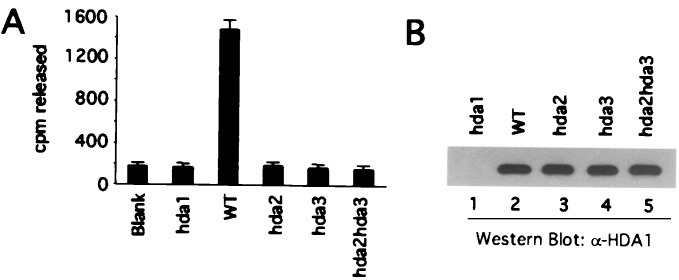
To determine the extent to which HDA2 and HDA3 influence activity of the HDA1 deacetylase, α-HDA1 antibody was used to immunoprecipitate histone deacetylase activity from whole-cell extracts of WT (YDS2), hda1Δ (WJY110), hda2Δ (WJY103), hda3Δ (WJY104), and hda2Δhda3Δ (WJY105) mutant cells. Deacetylase activity was assayed as described (12) using 3H-labeled HeLa core histones as substrate. As shown in Fig. 4A, the deacetylase activity of the WT extract is decreased to background levels when HDA1 is deleted (hda1Δ). hda2Δ, hda3Δ, and hda2Δhda3Δ all cause a similar decrease in deacetylase activity. Moreover, the amounts of HDA1 immunoprecipitated from each of the above strains (except for the hda1Δ strain) are similar as determined by Western blotting (Fig. 4B). Therefore, both HDA2 and HDA3 are required for HDA1 activity in vitro.
Figure 4.
HDA2 and HDA3 are required for the in vitro deacetylase activity of the HDA1 complex. α-HDA1 antibody was used to immunoprecipitate histone deacetylase activity from whole-cell extracts. Yeast strains YDS2 (WT), WJY110 (hda1Δ), WJY103 (hda2Δ), WJY104 (hda3Δ), and WJY105(hda2Δhda3Δ) were used. Half of each precipitate was used to assay the enzymatic activity (A) and half to determine the amount of the HDA1 protein by Western blot (B).
HDA1, HDA2, and HDA3 Are Required for H3/H2B-Specific Deacetylation of the ENA1 Promoter in Vivo.
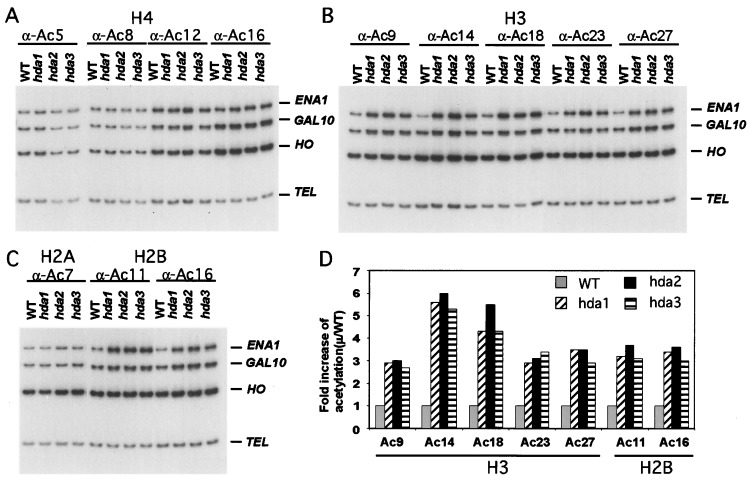
We next asked whether HDA2 and HDA3 are required for the in vivo deacetylation of chromatin. Chr-IP with antibodies directed against individual sites of acetylation in histones has been used to determine the role of HDA1 in deacetylating histones at TUP1-regulated genes such as ENA1. These experiments demonstrate that HDA1 is required specifically for deacetylation of all H3 and H2B sites of acetylation examined but not H4 or H2A sites (13). Therefore, we wanted to determine whether HDA2 and HDA3 are required for similar deacetylation in vivo. Using Chr-IP we examined the acetylation levels of all four core histones at the ENA1 promoter in WT (YDS21U), hda1Δ (WJY127), hda2Δ (WJY128), or hda3Δ (WJY129) strains as described (13). PCR amplification was performed on the promoters of the ENA1, GAL10, and HO genes as well as a subtelomeric region TEL, 500 bp from the end of chromosome VI-R. TEL is used as a loading control because its acetylation state is not affected by HDA1 mutation (9). GAL10 is weakly regulated by TUP1, whereas HO is not. It was found at ENA1 that H3 sites (K9, K14, K18, K23, and K27) and H2B sites (K11 and K16) are hyperacetylated in a similar manner by hda1Δ, hda2Δ, or hda3Δ as compared with the WT control (Fig. 5 B and C; quantitated in Fig. 5D). H4 sites (K5, K8, K12, K16) and H2A site (K7) were relatively unaffected by any of these deletions (Fig. 5 A and C). GAL10 is affected more weakly but in a similar manner by hda1Δ, hda2Δ, or hda3Δ, and HO is apparently unaffected by these deletions (Fig. 5 B and C; quantitations not shown). These data demonstrate that HDA2 and HDA3 are required for the deacetylation of ENA1 (and GAL10) by HDA1.
Figure 5.
HDA2 and HDA3 are required for deacetylation of histones H3 and H2B by HDA1 at the ENA1 promoter. Chr-IP demonstrating the effects of hda1Δ, hda2Δ, and hda3Δ on the acetylation of (A) H4 sites K5, K8, K12, and K16; (B) H3 sites K9, K14, K18, K23, and K27; and (C) H2A site K7 and H2B sites K11 and K16 at the ENA1, GAL10, and HO promoters. Amplification of a 138-bp fragment 0.5 kb from the telomere (Tel) of chromosome VI-R was used as reference to ensure equal loading of samples. Yeast strains used for Chr-IP are WT (YDS2), hda1Δ (WJY127), hda2Δ, (WJY128), and hda3Δ (WJY129). HO was found to be relatively unaffected by HDA1 disruption and was used as a negative control. (D) Quantification of increases in H3 and H2B acetylation in hda1Δ, hda2Δ, and hda3Δ cells relative to WT cells. 32P-α-dATP was added to the PCR mixture, and the PhosphorImager was used to quantitate the intensity of ENA1 PCR bands in the hda1Δ, hda2Δ, and hda3Δ mutants relative to WT after normalizing to the Tel bands.
HDA1, HDA2, and HDA3 Disruptions All Increase Expression of ENA1.
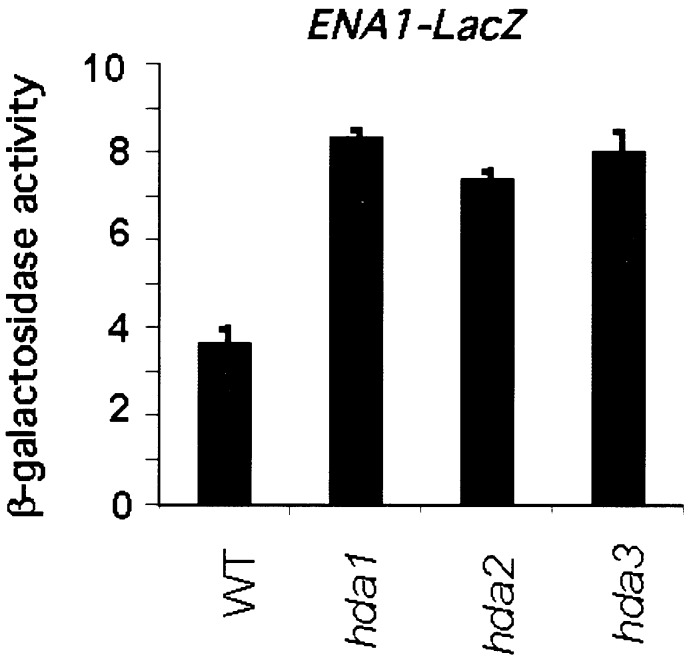
Because it has been shown that HDA1 represses the transcription of ENA1 (13), we wanted to determine the effects of the disruption of HDA2 and HDA3 genes on ENA1 promoter activity. The ENA1-lacZ gene in which the ENA1 promoter was fused to the LacZ reporter gene (23) was integrated into the URA3 locus of WT (YDS2), hda1Δ (WJY110), hda2Δ (WJY103), and hda3Δ (WJY104) yeast. Transcription from the ENA1 promoter was determined by β-galactosidase activity. Fig. 6 shows that the disruptions of HDA1, HDA2, or HDA3 all cause approximate 2-fold increase in ENA1-lacZ transcription. We conclude that HDA2 and HDA3 are repressors of ENA1 in a manner similar to that of HDA1. Moreover, it has been shown that HDA1 disruption increases silencing (telomere position effect, TPE) of URA3 integrated at 2.1 kb from the telomere (5). We find that HDA1, HDA2, and HDA3 disruptions all increase TPE to a similar extent (5.3- to 5.8-fold) (data not shown).
Figure 6.
HDA2 and HDA3 are essential for repression by HDA1. Expression of ENA1-LacZ was derepressed in hda1Δ, hda2Δ, and hda3Δ mutants. Plasmid pFR70i harboring ENA-LacZ was integrated into the URA3 loci of the chromosomes of WT (YDS2), hda1Δ (WJY110), hda2Δ (WJY103), and hda3Δ (WJY104). β-Galactosidase activity is shown in Miller units. Values represent averages for 4–6 transformants. Standard errors were <5%.
Therefore, we conclude that interaction between the HDA1-HDA1 homodimer with the HDA2-HDA3 heterodimer is essential for the catalytic activity of the HDA1 complex in vitro and for all known functions in vivo. Because HDA1 is likely to be the catalytic subunit and interacts directly with the repression domain of TUP1 in vitro (13), it is interesting to speculate on the functions of the other two components. HDA3 also may be involved in TUP1 binding (13); however, these interactions cannot be necessary for the activity of the catalytic subunit in vitro because TUP1 is absent from these reactions. Consequently, it is possible that HDA2 and HDA3 are involved in histone recognition or in altering the structure of the HDA1 dimer in a manner that allows its activity. HDA2 also contains the consensus attachment site at residues 570–585 for phosphopantetheine, the prosthetic group of acyl carrier proteins (documented by Saccharomyces Genome Database/Munich Information Center for Protein Sequences). This raises the intriguing possibility that a noncatalytic component of a deacetylase complex may be required for the capture or transfer of acetyl groups released during histone deacetylation.
Acknowledgments
We are grateful to the members of the Grunstein laboratory for helpful suggestions made throughout the course of this work. We are also especially thankful to Dr. M. Proft for plasmids. This work was supported by National Institutes of Health Grants GM23674 and GM42421 to M.G.
Abbreviations
- GST
glutathione S-transferase
- WT
wild type
- Chr-IP
chromatin-immunoprecipitation
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Knoepfler P S, Eisenman R N. Cell. 1999;99:447–450. doi: 10.1016/s0092-8674(00)81531-7. [DOI] [PubMed] [Google Scholar]
- 2.Ng H H, Bird A. Trends Biochem Sci. 2000;25:121–126. doi: 10.1016/s0968-0004(00)01551-6. [DOI] [PubMed] [Google Scholar]
- 3.Vogelauer M, Wu J, Suka N, Grunstein M. Nature (London) 2000;408:495–498. doi: 10.1038/35044127. [DOI] [PubMed] [Google Scholar]
- 4.Grozinger C M, Hassig C A, Schreiber S L. Proc Natl Acad Sci USA. 1999;96:4868–4873. doi: 10.1073/pnas.96.9.4868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rundlett S E, Carmen A A, Kobayashi R, Bavykin S, Turner B M, Grunstein M. Proc Natl Acad Sci USA. 1996;93:14503–14508. doi: 10.1073/pnas.93.25.14503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Taunton J, Hassig C A, Schreiber S L. Science. 1996;272:408–411. doi: 10.1126/science.272.5260.408. [DOI] [PubMed] [Google Scholar]
- 7.Carmen A A, Griffin P R, Calaycay J R, Rundlett S E, Suka Y, Grunstein M. Proc Natl Acad Sci USA. 1999;96:12356–12361. doi: 10.1073/pnas.96.22.12356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhang Y, Ng H H, Erdjument-Bromage H, Tempst P, Bird A, Reinberg D. Genes Dev. 1999;13:1924–1935. doi: 10.1101/gad.13.15.1924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rundlett S E, Carmen A A, Suka N, Turner B M, Grunstein M. Nature (London) 1998;392:831–835. doi: 10.1038/33952. [DOI] [PubMed] [Google Scholar]
- 10.Kadosh D, Struhl K. Cell. 1997;89:365–371. doi: 10.1016/s0092-8674(00)80217-2. [DOI] [PubMed] [Google Scholar]
- 11.Kadosh D, Struhl K. Mol Cell Biol. 1998;18:5121–51237. doi: 10.1128/mcb.18.9.5121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Carmen A A, Rundlett S E, Grunstein M. J Biol Chem. 1996;271:15837–15844. doi: 10.1074/jbc.271.26.15837. [DOI] [PubMed] [Google Scholar]
- 13.Wu J, Suka N, Carlson M, Grunstein M. Mol Cell. 2001;7:117–126. doi: 10.1016/s1097-2765(01)00160-5. [DOI] [PubMed] [Google Scholar]
- 14.Edmondson D G, Smith M M, Roth S Y. Genes Dev. 1996;10:1247–1259. doi: 10.1101/gad.10.10.1247. [DOI] [PubMed] [Google Scholar]
- 15.Treitel M A, Carlson M. Proc Natl Acad Sci USA. 1995;92:3132–3136. doi: 10.1073/pnas.92.8.3132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Herschbach B M, Arnaud M B, Johnson A D. Nature (London) 1994;370:309–311. doi: 10.1038/370309a0. [DOI] [PubMed] [Google Scholar]
- 17.Huang M, Zhou Z, Elledge S J. Cell. 1998;94:595–605. doi: 10.1016/s0092-8674(00)81601-3. [DOI] [PubMed] [Google Scholar]
- 18.Wach A, Brachat A, Rebischung C, Steiner S, Pokorni K, Hessen S, Philippsen P. Methods Microbiol. 1998;26:67–81. [Google Scholar]
- 19.Cosma M P, Tanaka T, Nasmyth K. Cell. 1999;97:299–311. doi: 10.1016/s0092-8674(00)80740-0. [DOI] [PubMed] [Google Scholar]
- 20.Hecht A, Laroche T, Strahl-Bolsinger S, Gasser S M, Grunstein M. Cell. 1995;80:583–592. doi: 10.1016/0092-8674(95)90512-x. [DOI] [PubMed] [Google Scholar]
- 21.Hecht A, Strahl-Bolsinger S, Grunstein M. Nature (London) 1996;383:92–96. doi: 10.1038/383092a0. [DOI] [PubMed] [Google Scholar]
- 22.Siegel L M, Monty K J. Biochim Biophys Acta. 1966;112:346–362. doi: 10.1016/0926-6585(66)90333-5. [DOI] [PubMed] [Google Scholar]
- 23.Mendoza I, Rubio F, Rodriguez-Navarro A, Pardo J M. J Biol Chem. 1994;269:8792–8796. [PubMed] [Google Scholar]