Abstract
The goal of personalized medicine is to treat each patient with the best drug: optimal therapeutic benefit with minimal side effects. The genomic revolution is rapidly identifying the genetic contribution to the diseased state as well as its contribution to drug efficacy and toxicity. The ability to perform genome-wide studies has led to an overwhelming number of candidate genes and/or their associated variants; however, understanding which are of therapeutic importance is becoming the greatest unmet need in the personalized medicine field. A related issue is the need to improve our methods of identifying and characterizing therapeutic drugs in the context of the complex genomic landscape of the intact body. Drosophila have proven to be a powerful tool for understanding the basic biological mechanisms of human development. This article will review Drosophila as a whole animal tool for gene and drug discovery. We will examine how Drosophila can be used to both sort through the myriad of hits coming from human genome-wide scans and to dramatically improve the early steps in pharmaceutical drug development.
Keywords: cancer, diabetes, Drosophila, heart disease, multigenic
Hippocrates argued that disease was not a punishment from the gods but was the outcome of environmental factors, diet and living habits; therefore, we should use detailed observation as a basis for directing better health. The belief that observation of the patient would lead to optimal individualized therapy required the use of the best tools at hand. Traditional medicine incorporated patient information such as clinical signs, symptoms and personal history. As medical tools advanced, data was incorporated from imaging of the organs affected by the suspected disease to biochemical assays of easily accessible body fluids such as serum and urine.
In the last 100 years this push towards better diagnosis gradually included genetics, leading to further expansion of the medical toolkit. Individualized healthcare included the taking of family history to the cytogenetic analysis of those suffering from rare Mendelian diseases or cancer. Accurate staging of breast cancer relied upon imaging and staging of lumps. Now ErbB receptor status, microarray data or the prediction of risk by BRCA1/2 status is used for progressively more accurate patient prognosis. However, only recently have genetics and genomics taken center stage as perhaps the most promising tool for achieving truly personalized medicine. Recent advances in genomic sciences, medical genetics and human genetics have enabled a more detailed understanding of the role of genetics in human disease. The advances include the determination of the human genome reference sequence, establishment of multisite collaborations that allow the pooling of thousands of human subjects for association studies and the development of systemic whole-genome analysis through the use of arrays (e.g., RNA, chromatin immunoprecipitation [ChIPS], copy number variation). Now, with the next-generation sequencing technologies, we can more clearly define the level of variation among humans and correlate it to the diseased state as demonstrated by the 1000 Genomes Project, The Cancer Genome Atlas and exomic sequencing of rare Mendelian diseases.
The challenge
Genomic approaches continue to lay the groundwork for identifying genes involved in the human disease process, and expectations are high. However, we are beginning to better appreciate that identifying genetic variants is only the first step. At least three additional steps are required. First, we need to determine which mutations are ‘drivers’ of a disease and which are ‘passengers’. This is difficult as genomic studies often identify dozens or hundreds of variants. Second, we need to determine the biological function of these human disease genes, preferably in the context of the whole animal. Third, we need to identify therapeutics that treat the aberrations, again preferably in the context of the whole body.
These steps need to be addressed cheaply and rapidly. In this article, we explore the use of the fruit fly Drosophila as an emerging tool for the next generation ‘Genomics 2.0’. In many ways, the lowly fly is a near-perfect match for genomic studies. We explore recent work by Drosophila researchers to adapt the remarkable tools available in the Drosophila field to the specific needs of genomic studies and personalized medicine. We also consider why the Drosophila world has been slow to embrace genomics and personalized medicine, and why this should change.
Why flies?
To understand the advantages and disadvantages of using Drosophila, let us first consider their most useful features. Flies have a short lifecycle: approximately 10 days depending on the temperature at which they are raised. After embryogenesis, the developing animal develops through three larval molts; larvae are especially useful for drug studies as they are continuously eating with minimal hesitation or regulation. After pupariating and then completing the pupal stage, the emergent adult has many easily scored structures for genetics (sadly, they emerge as picky eaters). Therefore, a major advantage of flies is that they develop quickly with a life-cycle of approximately 9 days. Because they breed rapidly and are readily kept in vials feeding on inexpensive food, flies are also cheap to maintain and experiment with. Table 1 compares some of the common model systems.
Table 1.
Comparing commonly used models.
| Cell culture |
Caenorhabditis elegans (worm) |
Drosophila (fly) |
Danio rerio (zebrafish) |
Musca (mouse) |
|
|---|---|---|---|---|---|
| Lifecycle (days) |
~1 | 3 | 9 | >90 | >50 |
| Epithelia? | No | ± | Yes | Yes | Yes |
| Multigenic (>3) cancer models? |
Yes | No | Yes | No | No |
| Large-scale genetic screening? |
Yes | Yes | Yes | No | No |
| Whole-animal drug screening? |
No | Yes | Yes | Limited | No |
| Approximate cost/month/100 animals |
Low | 12¢ | 20¢ | US$180 | US$2100 |
Drosophila sits in the ‘sweet spot’ of linking genes to therapeutics. It is rapid and inexpensive to work with, but retains important aspects of whole-animal complexity.
Many aspects of these developmental stages have been remarkably well characterized, making Drosophila perhaps the best-understood developing animal in terms of the mechanisms and pathways that direct tissue assembly and maturation. Several basic signaling pathways were first discovered in flies, a point reflected by their unusual names: Hedgehog (larvae cuticle resembles that of a hedgehog), Notch (notched wings) and Wnt (a contraction that includes wingless). The success of Drosophila in identifying key molecular pathways active in development was recognized by the awarding of the Nobel Prize in Medicine in 1995 to Eric Wieschaus, Christiane Nusslein-Volhard, and Edward Lewis. Wieschaus and Nusslein-Volhard used fly genetics to identify every zygotic gene required to assemble the young embryo, a saturation approach that should prove particularly useful as members of the community move towards studying disease mechanisms.
Tools
A key strength of Drosophila is the powerful tools available to manipulate genes. Simply put, the activity of almost any gene in Drosophila can be increased or decreased within nearly any cell or cell type and at any stage of development. This ability to manipulate genes at will represents one of the two most important Drosophila approaches relevant to personalized medicine. The detailed techniques – including the FRT/FLP recombination approach to generate discrete patches of mutant tissue in a wild-type background, the GAL4/upstream activation sequence (UAS) system for mis-expression studies, and the use of inverted repeats to generate RNAi-mediated gene knockdown – have been well reviewed (e.g., [1–3]) and we will introduce the most disease-relevant techniques briefly below. Drosophila has a long history of tool building and with each passing year the ability to generate transgenic Drosophila genes, manipulate gene expression and direct site-specific changes becomes easier, quicker and more powerful.
The genetic modifier screen
Perhaps no tool has proven more powerful in Drosophila than the genetic-modifier screen. If you are wondering how Drosophila can aid your own work, this remarkably powerful technique should likely be your first consideration.
The basis of the genetic modifier screen is that functionally related loci will show a dominant genetic interaction. For example, the EGF receptor (EGFR) acts, in part, through the small GTPase Ras to promote tumorigenesis [4–6]. To model aspects of this process, activated EGFR isoforms were targeted to the Drosophila eye; the result was over-proliferation of the eye epithelium and a ‘rough eye’ in the adult [Levine B, Cagan R, Unpublished Data] [7]. Removing a single genomic copy of Drosophila Ras by itself has no effect on eye development (removing both genomic copies – flies are diploid – is cell and embryonic lethal). But removing a genomic copy of Ras in the presence of activated EGFR suppressed the overproliferation phenotype, yielding a less rough eye. That is, the genomic copy number of Ras is rate-limiting for EGFR activity. Conversely, removing a genomic copy of the pathway inhibitor RasGAP enhanced activated EGFR. In the fly community we refer to Ras and RasGAP as ‘suppressor’ and ‘enhancer’ loci of activated EGFR, respectively, and this data is considered strong evidence for a functional link between these loci. Owing to the availability of off-the-shelf fly stocks containing single mutations that, together, account for most of the predicted loci in the fly genome, screens for functionally linked loci can be readily accomplished. The payoff is identification of truly novel loci that act in your pathway of interest.
Enhancer and suppressor loci are conceptually similar to the modifier/susceptibility loci commonly searched for in personalized medicine studies. Later we will provide disease-related examples to illustrate how genetic modifier screens were used to address specific questions in human therapeutics.
Targeted expression, knockdown
In addition to using genetic tools to test functional links between proteins, advances have made targeted expression or knockdown of genes both easier and more precise. Two major techniques are commonly used to achieve this in stable transgenics, and we briefly outline how they are used.
Targeted transgene expression
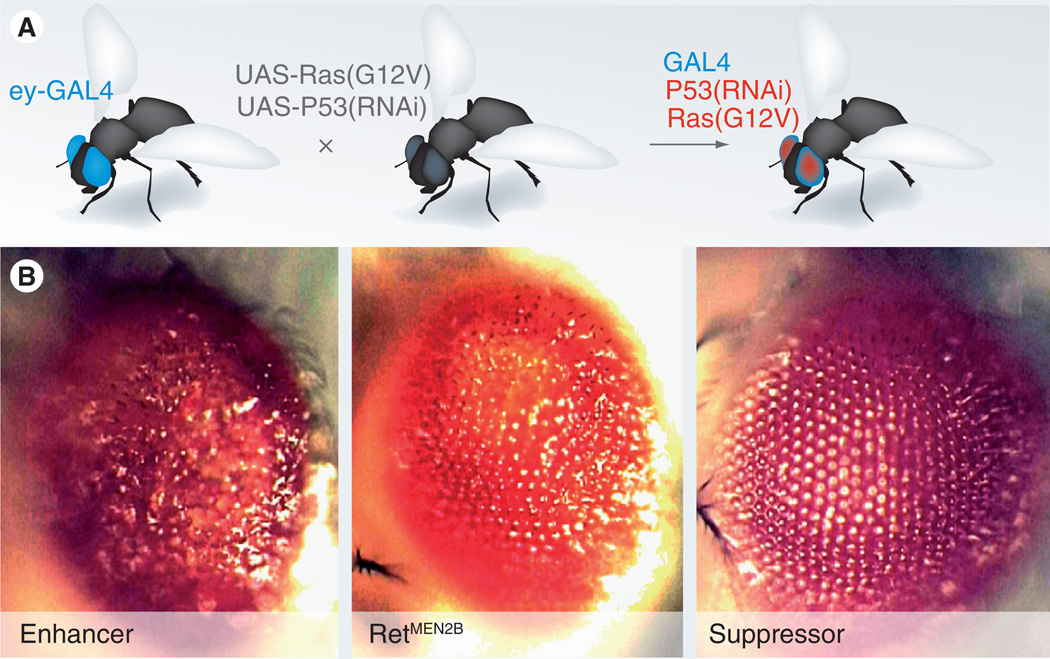
Many diseases are due to the expression of disease-specific isoforms of specific proteins. for example, many solid tumors express the oncogenic Ras isoform RasG12V, leading to activation of the Ras signal transduction pathway and tumorigenesis. Expression of one or more disease genes is readily accomplished in flies: researchers typically use either direct fusion of a useful promoter to the gene of interest or, more commonly, by use of the GAL4/UAS mis-expression system [8]. Figure 1 shows the basic approach, which combines GAL4 fused to a tissue-specific promoter with the transgene of interest (e.g., rasG12V) fused to the GAL4 target sequence UAS. For example, crossing eye-GAL4 flies to uas-rasG12V flies targets oncogenic Ras to the eye; progeny are ready to score in less than 2 weeks. As we discuss below, this approach can provide a useful assay for both genetic screens and for drug discovery/validation.
Figure 1. Tools used to generate disease models.
(A) Crossing a fly line containing eye-targeted GAL4 to a fly line with one or more transgenes results in progeny that express the transgenes in the tissue of interest (e.g., the eye). (B) Targeting expression of the oncogenic isoform RetMEN2B to the eye results in a ‘rough eye’ phenotype, best observed with misplaced outer lenses. Genetic enhancers (left panel) lead to a more extreme phenotype including outgrowths, while genetic suppressors (right panel) bring the eye closer to its normally smooth lens array.
Data taken from [Read R, Cagan R, Unpublished Data].
Targeted knockdown
The Drosophila tool set includes the ability to knock out genes in specific tissues. For disease models, a more common approach has been targeted knockdown of a gene using RNAi; this approach reduces gene activity by destabilizing their transcript. Expression of inverted repeat, dsRNA leads to targeting of homologous sequences for degradation by a Dicer-mediated complex (reviewed in [9]). This approach works particularly well in flies. Currently, three separate collections of flies are available that cover the predicted 20,000 fly transcripts: each of the 20,000 fly lines contains an inverted repeat transgene that targets a single transcript; this transgene is fused with the UAS promoter to allow precise GAL4-mediated targeting of a gene for knockdown (see earlier). For example, knocking down the tumor suppressor P53 in the wing simply requires crossing ‘wing’-GAL4 flies to UAS-P53 flies. The F1 knockdown adults will emerge in less than 2 weeks. The ability to knockdown any predicted transcript with off-the-shelf reagents simplifies the effort, time and cost required.
RNAi: in vivo & cell culture
Many drug and functional screens are more readily accomplished in cell lines. Drosophila cell lines have proven especially useful for this approach, and the Drosophila Genomics Resource Center makes a large number of well-characterized lines available. Although fly cell lines do not take advantage of the ability to screen in a whole-animal setting, they are quicker, cheaper and more quantitative than whole-fly screens. Moreover fly cell lines have unique advantages that have attracted numerous researchers of mammalian systems. They are robust, thrive at room temperature (flies are ectothermic) and, critically, readily take up RNA added to the media. That is, libraries of dsRNA can be screened rapidly through ‘systemic RNAi’ screening, in which single or pools of RNAi constructs are added to the media. The Perrimon laboratory has established an automated facility that permits laboratories to screen a validated dsRNA library, typically against a luciferase readout of the pathway of interest [10]. This approach has been hugely successful in identifying regulators of several pathways including Hedgehog, Wnt, receptor tyrosine kinase/Ras, metabolism, and other pathways [11–15].
Disease models
In studies of development, Drosophila proved useful for its powerful genetics but also its emphasis on examining mechanisms in situ. This whole-animal approach is also invaluable for disease studies. Many of the difficulties translational researchers have had can be traced to the differences between diseased tissue within a whole animal, cell culture models or conceptually related xenograft models. In this section we consider specific examples of how Drosophila can be used to rapidly and inexpensively generate disease models that are useful for studies of both mechanism and therapeutics. Our purpose is not to be comprehensive, but to point to a small number of illustrative examples of how Drosophila can impact specific issues of disease and personalized medicine.
Cancer
Flies provide a natural tool to study the genomics of cancer, as most oncology-related pathways have a long history of study in Drosophila. Indeed, the first tumor suppressor was identified in Drosophila through the pioneering work of Peter Bryant and his colleagues [16,17]. However, only recently has the fly community embraced cancer as a specific field of focus. The number of fly workers studying cancer is rapidly rising. We bring an in situ sensibility to questions of tissue development and homeostasis, and are comfortable considering impact on the animal as a whole. As such, we anticipate flies will have an increasing impact on our understanding of how mutated genes and gene combinations affect both tumor outcome and even drug response.
Multiple endocrine neoplasia type 2
Several recent examples serve to demonstrate the utility of flies in understanding how mutations in oncogenes and tumor suppressors can lead to progressively more aggressive tumors. At the simple end of the mutation spectrum is the ‘one-hit’ solid tumor syndrome multiple endocrine neoplasia type 2 (MEN2). This syndrome is characterized by mutations in the Ret protein, a transmembrane receptor tyrosine kinase. Patients with either spontaneous or inherited oncogenic isoforms of Ret invariably develop medullary thyroid carcinoma (MTC), a potentially aggressive overgrowth of the para-follicular C cells that can lead to metastatic disease if not treated early. MTCs have proven resistant to traditional chemotherapies.
Nearly all patients with MEN2, and many with MTC, have activated isoforms of Ret, suggesting that Ret is both necessary and sufficient for development of MTCs. To develop a whole-animal transgenic model, various oncogenic Ret isoforms were targeted to the developing fly eye epithelium. The result was overproliferation, switches in cell fate, compensatory apoptosis and other aspects of tumorigenesis that suggested this fly MEN2 model could prove useful [18]. The eyes displayed a ‘rough eye’ phenotype that is readily scored with a dissecting microscope, yielding a useful screening tool (Figure 1).
Multigenic cancer models
An important foundation of personalized medicine lies in the ability to recognize and model multigenic disease states. This plays to the strengths of Drosophila, which permit rapid and inexpensive manipulation of multiple genes. In the past decade a small number of Drosophila laboratories have developed multigenic cancer models. Pioneers include the laboratories of Tian Xu, Helena Richardson, Dirk Bohmann and Maria Dominguez; we have also been active in this area. Work with the potent oncogene Ras has demonstrated that it can direct proliferation on its own but that true transformation requires synergy with second loci, a point increasingly embraced in work with other organisms as well. For example, pairing oncogenic Ras isoforms with mutations in the cell polarity protein Scribble, led to strong proliferation and aggressive metastasislike migration by Ras/Scribble cells. Labeling these cells with GFP highlighted their movement to distant sites. More recent work has shown similar synergy between oncogenic Ras and mutations that activate the cytoplasmic tyrosine kinase Src.
In both Drosophila and humans, EGFR and its downstream effectors, Ras and Raf, were also found to synergize with Src kinase activity. The result was both increased proliferation and metastasis-like behavior [19]. This is a particularly clinically relevant pairing: for example, approximately 70% of triple-negative breast tumors show high activity of EGFR plus Src [20–27]. Pairing these not only synergizes with respect to proliferation, but also shows emergent tendencies for transformed cells to undergo metastasis-like migration [28–33]. This migration relied on local interactions between the Ras/Src transformed cells and their neighbors within the epithelium. These increasingly tumor-like phenotypes highlight the potential for flies to screen candidate therapeutics in the context of richer, more complex whole-animal models, and to target aspects such as metastasis, which relies on interactions between tumor cells and their environment.
Work with Drosophila on cancer models has also highlighted another important potential use: the ability to identify candidate modifier loci. An elegant example of this was provided by Dominguez and colleagues [34]. The Drosophila community has developed a number of overexpression tools that permit screening of randomly overexpressed loci [35–37]. Hyperactivation of the Notch signaling pathway has been linked to tumorigenesis owing to its ability to promote excess proliferation [38–43]. Ferres-Marco et al. used an eye overgrowth model to screen and identify two neighboring loci that, when ectopically coexpressed with the Notch ligand-δ, strongly enhanced the Notch-dependent eye overgrowth phenotype (Figure 2) [34]. Pipsqueak and Lola are transcriptional regulators that act by regulating histone modification [44–46]. The authors provide evidence that these two nuclear factors synergize with active Notch pathway signaling to target cell cycle genes including Rb. Similar to work demonstrating synergy between Ras and Scribble or Src, the connection between Notch signaling and epigenetic regulation of the cell cycle points to new targets to explore in cancer.
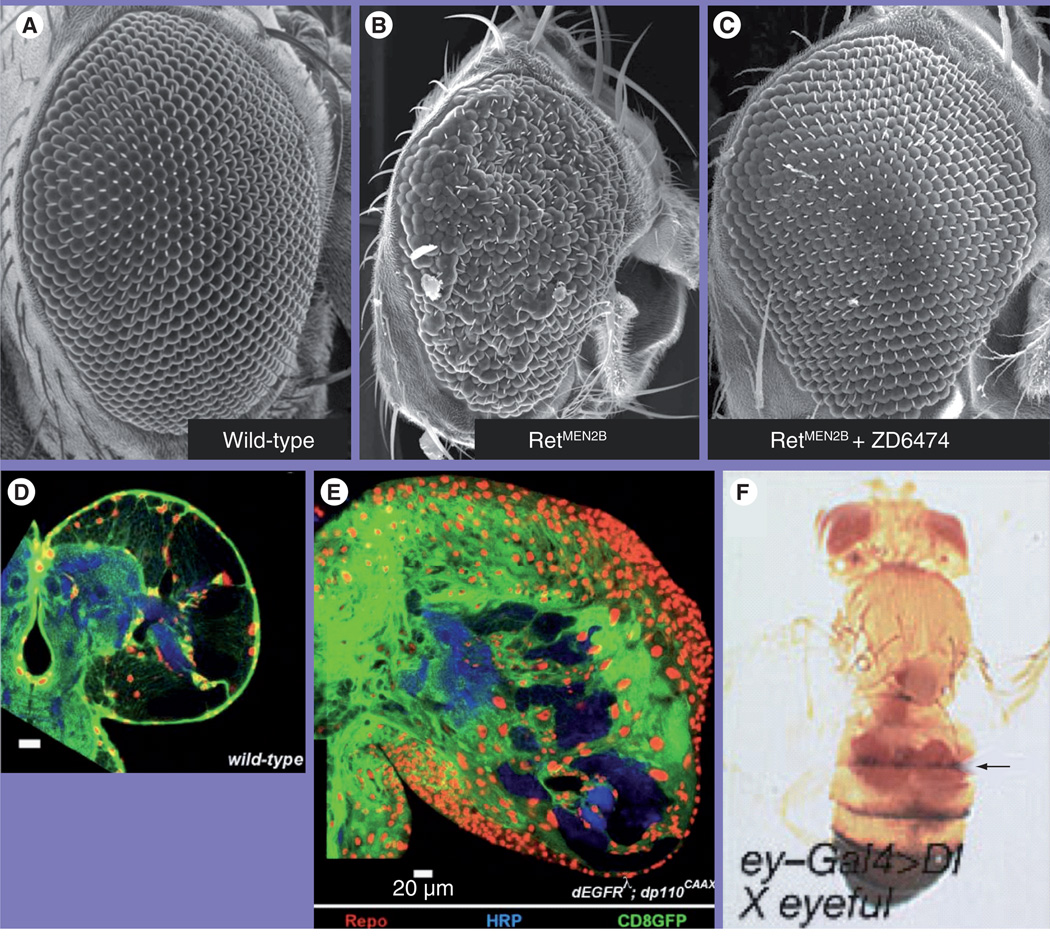
Figure 2. Drosophila cancer models.
Compared to wild-type adult eyes (A), expressing oncogenic Drosophila Ret led to a small ‘rough eye’ phenotype (B). Feeding the compound ZD6474/vandetanib rescued the effects of oncogenic Ret [85] (C). (D–E) Drosophila glioma model based on activated EGF receptor/P110 isoforms targeted to glia. Glial nuclei are labeled with Repo (red), cell bodies with CD8GFP (green); neurons are labeled with HRP (blue) driven by repo-Gal4. These ‘repo>dEGFRλ;dp110CAAX’ brains (E) showed a dramatic increase in glial number and an enlarged brain relative to wild-type (D) [47]. (F) Expressing δ within the developing fly eye led to both enlarged eyes and eye tissue at distant sites (arrow).
HRP: Horseradish peroxidase.
Reproduced with permission from [34].
Glioma
This simple but powerful approach – targeting multiple transgenes with a single driver to establish multigenic cancer models – will be utilized to establish increasingly sophisticated models designed to match the ever-increasingly genomic data. An excellent example of this comes from Read et al., who established and explored a ‘two-hit’ glioma model [47]. Targeting activated isoforms of Drosophila EGFR plus the P110 subunit of PI3K to glia yielded aggressive neoplasias that proved invasive into surrounding nervous system tissue (Figure 2). This transformation was specific to glia; neural tissue did not show similar transformation phenotypes, emphasizing that different cell types respond differently to specific oncogene combinations. Detailed histological and genetic evidence bolstered the view that these flies modeled fundamental aspects of human gliomas. Once in hand, these transgenic flies were then used to explore the downstream signals that mediate these effects, implicating orthologs of Rb, Myc, Elk-1, and so on, as critical regulators. This model should prove useful both for continuing to explore mechanisms and potentially as a first-line screen for candidate therapeutics.
Heart disease
Heart disease remains the number one source of mortality in the developed world, and yet we still only partly understand the mechanisms that lead to progressive heart deterioration. Drosophila does indeed have a heart; they also have structures similar to other organs including the kidneys, lungs and the liver. The pioneers of Drosophila models of heart disease have been Rolf Bodmer and Karen Ocorr, who have taken tools honed in developmental studies to explore specific issues of disease. They have demonstrated progressive heart deterioration in aging adult flies and have shown that this can be reversed by reducing activity of the insulin signaling pathway [48,49]. The structural proteins and channels that regulate human heart function are conserved in Drosophila – several were initially discovered in flies – and likely perform similar functions within a fairly simple, nonchambered tube that moves fly blood (hemolymph) through the body (reviewed in [48,50,51]).
Heart function can be measured in detail and precise M-modes can be generated that demonstrate rhythmicity. With these tools plus high-sensitivity fluorescence imaging [52], the fly heart is improving in its ability to be screened for genes and drugs that compromise or improve function. An excellent example of the utility of flies comes from the study of the KCNQ potassium channel. In humans, mutations in this potassium channel lead to long QT syndrome (LQT) Type 1, a delay in repolarization of the heart that is associated with arrythmia and sudden death. Flies mutated for kcnq survive to adult and, also similar to humans, are susceptible to cardiac failure when the heart is artificially ‘paced’ [53]. Also promising from a personalized medicine standpoint, Drosophila has also conserved the ‘human Ether-a-gogo’ channel (HERG, named after the fly protein due to mutants’ uncontrolled movements). HERG has been linked to drug-induced LQT, a potentially fatal side effect common with many drugs tested in the clinics. Mutation of the fly ortholog led to increased rates of arrythmicity [54], holding the promise that Drosophila could provide a rapid whole-animal assay to detect this serious drug contraindication.
Diabetes
Perhaps the fastest growing disease field in Drosophila research is its use as a tool to explore metabolism-related diseases. Here, we consider work in diabetes, often referred to as an ‘epidemic’ that has tripled in incidence in the USA in just the past 30 years. The utility of Drosophila as a tool for studying diabetes was highlighted by Rulifson, Kim and Nusse [55,56], who developed a model of Type 1 diabetes. To mimic loss of the insulin-secreting β-cells in human patients, they expressed a pro-apoptosis gene in the Drosophila insulin-producing cells (IPCs). Loss of these neuroendocrine cells led to aspects reminiscent of the human disease, including hyperglycemia, hyperinsulinemia, aspects of insulin resistance and altered fat profiles [56]. Further work has demonstrated the presence of cells that function similar to our α-cells; these cells secrete a glucagon-like peptide that is predominant in regulating hemolymph glucose levels [55].
Most diabetes patients suffer from the more common Type 2 diabetes mellitis (T2DM). New Drosophila models of T2DM are being developed that rely on simple high-carbohydrate feeding paradigms [Palankar L, Fink J, Cagan R, Baranski T, Unpublished Data]. These lead to a broad array of effects described in human patients and show both similarities and some differences with Type 1 diabetes. A close examination of heart and kidney function in these high-sugar-fed flies revealed many defects observed in human patients [Na J, Baranski T, Ocorr K, Bodmer R, Cagan R, Unpublished Data]. Genetic screens have indicated a remarkable complexity to the defects imposed by a high-sugar diet, and have highlighted both the complexity of the disease and the need for a powerful genetic system such as Drosophila to sort out the intricacies.
Genomic mapping
Regarding personalized medicine, perhaps the greatest place for future growth in the Drosophila field over the next few years is their potential to provide biological context to the flood of genomic data. Drosophila commonly screen hundreds or even thousands of genes in a study, assessing functional links and generating new pathways based on functional criteria. In this section we cite recent examples in which flies have been used with success to address specific issues that emerged in genomic disease studies.
Susceptibility loci
The potential utility of fly’s genetic tools for identifying susceptibility (modifier) loci is underappreciated, even by fly biologists. Enhancers identified in genetic screens are often excellent candidates to act as modifiers of a primary genetic insult. To explore the potentially utility of Drosophila, we again look to the Drosophila MEN2 model described earlier.
Nearly all patients with MEN2 develop MTC, but only a subset develops additional tumors, such as pheochromocytomas [57–59]. This suggests that susceptibility to these adrenal-based tumors requires differences at secondary loci. A Drosophila genetic-modifier screen – described earlier – identified 140 loci that were functionally linked to the oncogenic activity of Drosophila Ret. We then crossmatched many of these loci to copy number studies in MEN2 tumor tissue and eventually identified two candidate susceptibility loci for the development of pheochromocytomas by MEN2 patients: the cytoplasmic kinase TNIK and the chromatin remodeling complex member CHD3 [18]. These loci are at least biomarkers for susceptibility to pheochromocytomas in these patients; whether they play a similar dominant role in promoting these adrenal tumors is not known.
This work highlights the ability of flies to rapidly identify pathway members that promote disease. Linking these functional genetic screens with high-throughput genomic analysis represents a powerful combination for exploring the subtleties of a disease’s genomic complexities.
Genome-wide association studies
Another underappreciated use of Drosophila genetics is the potential for matching genetic modifiers from Drosophila to SNPs or related genome-wide association study mapping in human patients. Often these approaches identify candidate regions but have difficulty determining, first, the exact gene within a region that actually contributes to the disease, or second, testing whether a candidate gene with a subtle genetic alteration is likely to be functionally linked to the disease. For example, recent efforts to match our Drosophila T2DM models with genome-wide association studies of T2DM patients has yielded interesting preliminary results [Baranski T, Cagan R, Collins F, Unpublished Data]. Drosophila have been able to take several candidate genes within a statistically-defined linkage region and identify a subset that when mutated alter the animal’s response to high-sugar feeding. Incumbent on these studies is the need to demonstrate that flies are predictive of bona fide human susceptibility loci. With this validation, Drosophila may prove a centrally important tool in sorting through the enormous data generated by genomics.
Drug discovery
Drosophila has traditionally been a ‘powerhouse’ as a genetic system. More recently, Drosophila has contributed to the search for compound therapeutics in a small number of other diseases as well. Drosophila holds great promise for drug screening: combining sophisticated multigenic models with the ability to be used in a moderately high-throughput drug screen. This approach may identify compounds with better whole-animal efficacy and reduced whole-animal toxicity. Here, we consider early efforts by the fly community to establish whole-animal screening.
MEN2
Earlier we described MEN2, a cancer syndrome typically triggered by activating mutations in the Ret receptor tyrosine kinase. Santoro and colleagues demonstrated that the anilinoquinazoline-type kinase inhibitor ZD6474/Vandetanib was useful for suppressing oncogenic Ret isoforms in cell culture and xenograft studies [60]. To examine vandetanib in a transgenic model, the drug was fed to the oncogenic Ret fly models: the compound proved effective in suppressing most aspects of oncogenic Ret in situ (Figure 2). The cell culture and subsequent fly work proved predictive: vandetanib has shown clear efficacy in Phase II clinical trials [61] and is completing Phase III trials [Wells S, Pers. Comm.]. This work provides an important validation of the use of Drosophila to predict whole-animal efficacy and toxicity in human patients.
Drosophila still has more to offer MEN2 patients. Most patients that respond to vandetanib show a partial response, and most show significant toxicity at useful doses [61]. The ability of Drosophila to address the full palate of Ret-activated pathways within a whole-animal context promises to find more effective drugs with fewer side effects. Our laboratory has identified new-generation compounds that show increased efficacy with decreased toxicity in fly and human MEN2 cell lines, holding the promise that next-generation drugs will show better therapeutic indices. Furthermore, our increasing knowledge of the genetics of oncogenic Ret will permit a testing of these compounds in the context of multiple genetic backgrounds for more personalized treatment.
Fragile X
We have focused on the use of flies in cancer studies, but the Drosophila community is expanding to other diseases as well. Recently, an elegant approach used flies to identify a candidate therapeutic for Fragile X syndrome [62]. A common form of mental retardation, Fragile X is linked to alterations in the Fragile X mental retardation protein (FMRP), which likely leads to excess glutamate production [63–65]. Mutations in the Drosophila ortholog FMR1 led to lethality if the food of developing animals was supplemented with glutamate. Using an embryo-sorting machine to screen fmr1 mutant embryos against a small chemical library, the authors identified nine compounds that rescued glutamate-dependent lethality in fmr1 flies. Their ‘hits’ were interesting from the standpoint of mechanism: regulators of both GABAergic and muscarinic cholinergic receptors were identified. The latter was especially surprising and pointed to previous unsuspected complexity of the disease. This work highlights the utility of Drosophila to explore neural-based diseases, which can have complex effects through the neural network. Other researchers have reported compound screens against fly models of neuro-degenerative diseases such as triplet-repeat based diseases and Parkinson’s [66–84]. Figure 3 shows an example of a factor identified in a genetic screen that successfully rescued degenerating dopaminergic neurons in a fly model of Parkinson’s as well as the progressive degeneration observed when disease isoforms of ataxin-1 – linked to the triple repeat-class disease spinocerebellar ataxia type 1 (SCA1) – are expressed in the developing eye. Together, this work further demonstrates the utility of Drosophila for whole-animal screening of compounds.
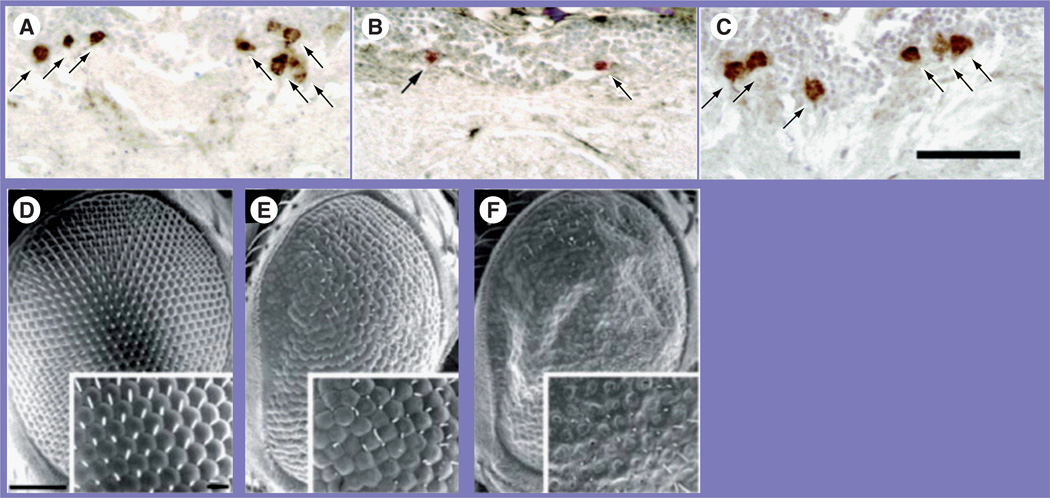
Figure 3. Drosophila neurodegenerative models.
(A–C) Drosophila Parkinson’s model protected by Hsp70 expression. (A) Dopaminergic neurons in dorsomedial neuronal clusters were visible with an antibody to tyrosine hydroxylase (arrows). (B) Overexpressing α-synuclein – linked to human Parkinson’s – led to fewer dopaminergic neurons. (C) A screen [84] identified Hsp70 as protective of dopaminergic neurons when coexpressed with α-synuclein [73]. (C–E) The triplet-repeat disease spinocerebellar ataxia type 1 was modeled in flies. This disease is linked to an expanding glutamine tract within the ataxin-1 protein. (D) Scanning electron micrograph of a control eye. (E) Targeting expression of an ataxin-1 protein containing 30 consecutive glutamine repeats to the developing eye led to a ‘rough eye’ phenotype. (F) Increasing the glutamine repeats to 82 led to a still more severe rough eye phenotype. Reproduced with permission from [73].
Why have Drosophila researchers not contributed more to therapeutics & to personalized medicine?
In this article we have examined the potential for flies as a tool for bringing increased genetic sophistication to issues of therapeutics. This, in turn, is another useful step towards the promise of bringing a similar level of sophistication to therapy in the clinics. Currently, only a few in the Drosophila community have considered the potential of this remarkable insect for exploring issues of therapeutics and of personalized medicine. Most fly biologists are trained as developmental biologists and are not familiar with the different ways the clinical researchers approach problems. In turn, clinical researchers are often not familiar with the power of Drosophila genetics, often emphasizing their differences.
To be sure, Drosophila physiology is considerably different from human physiology. While they have most of the organs we do, the details of how these organs are constructed and the manner in which they function have changed over the past 300 million years. But these issues hold equal weight for development. Yet Drosophila research has perhaps been the single most successful tool for exploring development-related aspects of genetics, morphology and signal transduction. While the details of development differ between flies and mammals, the former was strongly successful in laying out the basic questions, pathways and mechanisms. A remarkable amount of what was learned from flies turned out to be correct for mammals as well, more than was initially anticipated. The ‘master regulators’ of the heart, eye, kidney and so on, proved remarkably well conserved. Even those aspects that were different benefited from asking the right questions derived from fly research.
Future perspective
Our knowledge of complex diseases, such as cancer and diabetes, is at a fairly early stage, mirroring the state of developmental biology two decades ago. We predict that the success of model systems in developmental studies including Drosophila will be repeated for personalized medicine: many assumed differences will disappear as we move from fly knowledge to mammalian exploration. We anticipate that the rapid synergy that emerged in developmental studies will be repeated for disease, and that the fly community further embraces its potential for initiating knowledge in personalized medicine. This will occur because disease studies need to embrace complexity, a strength of Drosophila, and because the science community is increasingly expected to account for issues of disease. This shift in priority is already being felt in the Drosophila community, and we anticipate that multiple disease fields will significantly benefit as a result.
Executive summary.
Challenge of current genomic studies
-
▪
The identification of genetic variants associated with human disease process is only the first step. We need to determine the impact of the genetic variation and the biological function of the gene, as well as identify therapeutics that treat the aberrations, all in the context of the whole body.
Drosophila is a biological system well suited for studies in genomics & personalized medicine
-
▪
Drosophila is inexpensive to maintain in large numbers.
-
▪
Its development has been well characterized and most mechanisms and pathways involved have been conserved throughout the animal kingdom.
There are many powerful molecular tools available to manipulate genes in Drosophila that include the ability to generate transgenic lines, to alter the levels & to direct site-specific changes in gene expression
-
▪These tools include the following:
-
-Genetic modifier screen;
-
-Targeted expression, knockdown;
-
-RNAi, both in vivo and in vitro.
-
-
Many human disease states can be modeled in Drosophila thus allowing mechanistic studies to be performed in the whole animal
-
▪The Drosophila disease models covered are the following:
-
-Cancer – multiple endocrine neoplasia type 2 and multigenic cancers including glioma;
-
-Heart disease – impact by the insulin signaling pathway and long QT syndrome;
-
-Diabetes – Type 1 and 2.
-
-
Functional genomics performed in Drosophila will lend biological context to the myriad of ‘hits’ coming out of the current wave of genomic studies
-
▪
These studies include the characterization of susceptibility loci and genome-wide array studies.
Drosophila holds great promise for drug screening by combining sophisticated multigenic models with the ability to be used in a moderately high-throughput drug screen
-
▪
In preclinical studies, the ZD6474/vandetanib tyrosine kinase inhibitor was shown to be efficacious when fed to a Drosophila transgenic model of MEN2 cancer.
-
▪
Several compounds were identified in a small screen when the compounds rescued a Drosophila transgenic model of Fragile X syndrome.
-
▪
Another screen identified a factor that had a positive impact on a Parkinson’s Drosophila model.
There are still challenges to utilizing Drosophila in a manner that has a greater impact on the field of personalized medicine
-
▪
Drosophila community and clinical researchers need to better understand the different ways they approach their respective fields.
-
▪
Although the physiology of Drosophila and humans are quite different, the molecular details are very well conserved.
Acknowledgments
Ross Cagan is cofounder of Medros, Inc., a company that utilizes Drosophila for disease gene and drug discovery; he retains a financial interest in the company.
Footnotes
Financial & competing interests disclosure
The authors have no other relevant affiliations or financial involvement with any organization or entity with a financial interest in or financial conflict with the subject matter or materials discussed in the manuscript apart from those disclosed.
No writing assistance was utilized in the production of this manuscript.
Bibliography
- 1.Dasgupta R, Perrimon N. Using RNAi to catch Drosophila genes in a web of interactions: insights into cancer research. Oncogene. 2004;23(51):8359–8365. doi: 10.1038/sj.onc.1208028. [DOI] [PubMed] [Google Scholar]
- 2.Venken KJ, Bellen HJ. Transgenesis upgrades for Drosophila melanogaster. Development. 2007;134(20):3571–3584. doi: 10.1242/dev.005686. [DOI] [PubMed] [Google Scholar]
- 3.Venken KJ, Carlson JW, Schulze KL, et al. Versatile P[acman] BAC libraries for transgenesis studies in Drosophila melanogaster. Nat. Methods. 2009;6(6):431–434. doi: 10.1038/nmeth.1331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Casanova ML, Larcher F, Casanova B, et al. A critical role for ras-mediated, epidermal growth factor receptor-dependent angiogenesis in mouse skin. Carcinogenesis Cancer Res. 2002;62(12):3402–3407. [PubMed] [Google Scholar]
- 5.Janes PW, Daly RJ, deFazio A, Sutherland RL. Activation of the Ras signalling pathway in human breast cancer cells overexpressing erbB-2. Oncogene. 1994;9(12):3601–3608. [PubMed] [Google Scholar]
- 6.Martinez-Lacaci I, Kannan S, De Santis M, et al. RAS transformation causes sustained activation of epidermal growth factor receptor and elevation of mitogen-activated protein kinase in human mammary epithelial cells. Int. J. Cancer. 2000;88(1):44–52. doi: 10.1002/1097-0215(20001001)88:1<44::aid-ijc7>3.0.co;2-8. [DOI] [PubMed] [Google Scholar]
- 7.Lesokhin AM, Yu SY, Katz J, Baker NE. Several levels of EGF receptor signaling during photoreceptor specification in wild-type, Ellipse, and null mutant. Drosophila Dev. Biol. 1999;205(1):129–144. doi: 10.1006/dbio.1998.9121. [DOI] [PubMed] [Google Scholar]
- 8.Brand AH, Perrimon N. Targeted gene expression as a means of altering cell fates and generating dominant phenotypes. Development. 1993;118(2):401–415. doi: 10.1242/dev.118.2.401. [DOI] [PubMed] [Google Scholar]
- 9.Kim K, Lee YS, Harris D, Nakahara K, Carthew RW. The RNAi pathway initiated by Dicer-2 in Drosophila. Cold Spring Harb. Symp. Quant. Biol. 2006;71:39–44. doi: 10.1101/sqb.2006.71.008. [DOI] [PubMed] [Google Scholar]
- 10.Mohr S, Bakal C, Perrimon N. Genomic screening with RNAi: results and challenges. Annu. Rev. Biochem. 2010;79:37–64. doi: 10.1146/annurev-biochem-060408-092949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.DasGupta R, Kaykas A, Moon RT, Perrimon N. Functional genomic analysis of the Wnt-wingless signaling pathway. Science. 2005;308(5723):826–833. doi: 10.1126/science.1109374. [DOI] [PubMed] [Google Scholar]
- 12.Lum L, Yao S, Mozer B, et al. Identification of Hedgehog pathway components by RNAi in Drosophila cultured cells. Science. 2003;299(5615):2039–2045. doi: 10.1126/science.1081403. [DOI] [PubMed] [Google Scholar]
- 13.Nybakken K, Vokes SA, Lin TY, McMahon AP, Perrimon N. A genome-wide RNA interference screen in Drosophila melanogaster cells for new components of the Hh signaling pathway. Nat. Genet. 2005;37(12):1323–1332. doi: 10.1038/ng1682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cully M, Genevet A, Warne P, et al. A role for p38 stress-activated protein kinase in regulation of cell growth via TORC1. Mol. Cell. Biol. 30(2):481–495. doi: 10.1128/MCB.00688-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Friedman A, Perrimon N. A functional RNAi screen for regulators of receptor tyrosine kinase and ERK signalling. Nature. 2006;444(7116):230–234. doi: 10.1038/nature05280. [DOI] [PubMed] [Google Scholar]
- 16.Bryant PJ, Levinson P. Intrinsic growth control in the imaginal primordia of Drosophila, and the autonomous action of a lethal mutation causing overgrowth. Dev. Biol. 1985;107(2):355–363. doi: 10.1016/0012-1606(85)90317-3. [DOI] [PubMed] [Google Scholar]
- 17.Woods DF, Bryant PJ. The discs-large tumor suppressor gene of Drosophila encodes a guanylate kinase homolog localized at septate junctions. Cell. 1991;66(3):451–464. doi: 10.1016/0092-8674(81)90009-x. [DOI] [PubMed] [Google Scholar]
- 18.Read RD, Goodfellow PJ, Mardis ER, et al. A Drosophila model of multiple endocrine neoplasia type 2. Genetics. 2005;171(3):1057–1081. doi: 10.1534/genetics.104.038018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Vidal M, Warner S, Read R, Cagan RL. Differing Src signaling levels have distinct outcomes in Drosophila Cancer Res. 2007;67(21):10278–10285. doi: 10.1158/0008-5472.CAN-07-1376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Acharya CR, Hsu DS, Anders CK, et al. Gene expression signatures, clinicopathological features, and individualized therapy in breast cancer. JAMA. 2008;299(13):1574–1587. doi: 10.1001/jama.299.13.1574. [DOI] [PubMed] [Google Scholar]
- 21.Biscardi JS, Maa MC, Tice DA, et al. c-Src-mediated phosphorylation of the epidermal growth factor receptor on Tyr845 and Tyr1101 is associated with modulation of receptor function. J. Biol. Chem. 1999;274(12):8335–8343. doi: 10.1074/jbc.274.12.8335. [DOI] [PubMed] [Google Scholar]
- 22.Bougeret C, Jiang S, Keydar I, Avraham H. Functional analysis of Csk and CHK kinases in breast Cancer Cells. J. Biol. Chem. 2001;276(36):33711–33720. doi: 10.1074/jbc.M104209200. [DOI] [PubMed] [Google Scholar]
- 23.Dimri M, Naramura M, Duan L, et al. Modeling breast cancer-associated c-Src and EGFR overexpression in human MECs: c-Src and EGFR cooperatively promote aberrant three-dimensional acinar structure and invasive behavior. Cancer Res. 2007;67(9):4164–4172. doi: 10.1158/0008-5472.CAN-06-2580. [DOI] [PubMed] [Google Scholar]
- 24.Hitosugi T, Sasaki K, Sato M, Suzuki Y, Umezawa Y. Epidermal growth factor directs sex-specific steroid signaling through Src activation. J. Biol. Chem. 2007;282(14):10697–10706. doi: 10.1074/jbc.M610444200. [DOI] [PubMed] [Google Scholar]
- 25.Ishizawar RC, Miyake T, Parsons SJ. c-Src modulates ErbB2 and ErbB3 heterocomplex formation and function. Oncogene. 2007;26(24):3503–3510. doi: 10.1038/sj.onc.1210138. [DOI] [PubMed] [Google Scholar]
- 26.Moiseeva EP, Heukers R, Manson MM. EGFR and Src are involved in indole-3-carbinol-induced death and cell cycle arrest of human breast cancer cells. Carcinogenesis. 2007;28(2):435–445. doi: 10.1093/carcin/bgl171. [DOI] [PubMed] [Google Scholar]
- 27.Tan M, Li P, Klos KS, et al. ErbB2 promotes Src synthesis and stability: novel mechanisms of Src activation that confer breast cancer metastasis. Cancer Res. 2005;65(5):1858–1867. doi: 10.1158/0008-5472.CAN-04-2353. [DOI] [PubMed] [Google Scholar]
- 28.Long W, Yi P, Amazit L, et al. SRC-3d4 mediates the interaction of EGFR with FAK to promote cell migration. Mol. Cell. 2010;37(3):321–332. doi: 10.1016/j.molcel.2010.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ricono JM, Huang M, Barnes LA, et al. Specific cross-talk between epidermal growth factor receptor and integrin αvβ5 promotes carcinoma cell invasion and metastasis. Cancer Res. 2009;69(4):1383–1391. doi: 10.1158/0008-5472.CAN-08-3612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hiscox S, Morgan L, Green TP, et al. Elevated Src activity promotes cellular invasion and motility in tamoxifen resistant breast cancer cells. Breast Cancer Res. Treat. 2006;97(3):263–274. doi: 10.1007/s10549-005-9120-9. [DOI] [PubMed] [Google Scholar]
- 31.Ishizawar R, Parsons SJ. c-Src and cooperating partners in human cancer. Cancer Cell. 2004;6(3):209–214. doi: 10.1016/j.ccr.2004.09.001. [DOI] [PubMed] [Google Scholar]
- 32.Vadlamudi RK, Sahin AA, Adam L, Wang RA, Kumar R. Heregulin and HER2 signaling selectively activates c-Src phosphorylation at tyrosine 215. FEBS Lett. 2003;543(1–3):76–80. doi: 10.1016/s0014-5793(03)00404-6. [DOI] [PubMed] [Google Scholar]
- 33.Mao W, Irby R, Coppola D, et al. Activation of c-Src by receptor tyrosine kinases in human colon cancer cells with high metastatic potential. Oncogene. 1997;15(25):3083–3090. doi: 10.1038/sj.onc.1201496. [DOI] [PubMed] [Google Scholar]
- 34.Ferres-Marco D, Gutierrez-Garcia I, Vallejo DM, et al. Epigenetic silencers and Notch collaborate to promote malignant tumours by Rb silencing. Nature. 2006;439(7075):430–436. doi: 10.1038/nature04376. [DOI] [PubMed] [Google Scholar]
- 35.Brand AH, Manoukian AS, Perrimon N. Ectopic expression in Drosophila. Methods Cell Biol. 1994;44:635–654. doi: 10.1016/s0091-679x(08)60936-x. [DOI] [PubMed] [Google Scholar]
- 36.Toba G, Ohsako T, Miyata N, et al. The gene search system. A method for efficient detection and rapid molecular identification of genes in Drosophila melanogaster. Genetics. 1999;151(2):725–737. doi: 10.1093/genetics/151.2.725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.McGuire SE, Roman G, Davis RL. Gene expression systems in Drosophila: a synthesis of time and space. Trends Genet. 2004;20(8):384–391. doi: 10.1016/j.tig.2004.06.012. [DOI] [PubMed] [Google Scholar]
- 38.Bolos V, Blanco M, Medina V, et al. Notch signalling in cancer stem cells. Clin. Transl. Oncol. 2009;11(1):11–19. doi: 10.1007/s12094-009-0305-2. [DOI] [PubMed] [Google Scholar]
- 39.Farnie G, Clarke RB. Mammary stem cells and breast cancer – role of Notch signalling. Stem Cell Rev. 2007;3(2):169–175. doi: 10.1007/s12015-007-0023-5. [DOI] [PubMed] [Google Scholar]
- 40.Panelos J, Massi D. Emerging role of Notch signaling in epidermal differentiation and skin cancer. Cancer Biol. Ther. 2009;8(21):1986–1993. doi: 10.4161/cbt.8.21.9921. [DOI] [PubMed] [Google Scholar]
- 41.Moberg KH, Schelble S, Burdick SK, Hariharan IK. Mutations in erupted, the Drosophila ortholog of mammalian tumor susceptibility gene 101, elicit non-cell-autonomous overgrowth. Dev. Cell. 2005;9(5):699–710. doi: 10.1016/j.devcel.2005.09.018. [DOI] [PubMed] [Google Scholar]
- 42.Thompson BJ, Mathieu J, Sung HH, et al. Tumor suppressor properties of the ESCRT-II complex component Vps25 in Drosophila. Dev. Cell. 2005;9(5):711–720. doi: 10.1016/j.devcel.2005.09.020. [DOI] [PubMed] [Google Scholar]
- 43.Brumby AM, Richardson HE. Scribble mutants cooperate with oncogenic Ras or Notch to cause neoplastic overgrowth in Drosophila. EMBO J. 2003;22(21):5769–5779. doi: 10.1093/emboj/cdg548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lehmann M, Siegmund T, Lintermann KG, Korge G. The pipsqueak protein of Drosophila melanogaster binds to GAGA sequences through a novel DNA-binding domain. J. Biol. Chem. 1998;273(43):28504–28509. doi: 10.1074/jbc.273.43.28504. [DOI] [PubMed] [Google Scholar]
- 45.Giniger E, Tietje K, Jan LY, Jan YN. lola encodes a putative transcription factor required for axon growth and guidance in Drosophila. Development. 1994;120(6):1385–1398. doi: 10.1242/dev.120.6.1385. [DOI] [PubMed] [Google Scholar]
- 46.Mishra K, Chopra VS, Srinivasan A, Mishra RK. Trl-GAGA directly interacts with lola like and both are part of the repressive complex of Polycomb group of genes. Mech. Dev. 2003;120(6):681–689. doi: 10.1016/s0925-4773(03)00046-7. [DOI] [PubMed] [Google Scholar]
- 47.Read RD, Cavenee WK, Furnari FB, Thomas JB. A Drosophila model for EGFR-Ras and PI3K-dependent human glioma. PLoS Genet. 2009;5(2):E1000374. doi: 10.1371/journal.pgen.1000374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ocorr K, Perrin L, Lim HY, et al. Genetic control of heart function and aging in Drosophila. Trends Cardiovasc. Med. 2007;17(5):177–182. doi: 10.1016/j.tcm.2007.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wessells RJ, Fitzgerald E, Cypser JR, Tatar M, Bodmer R. Insulin regulation of heart function in aging fruit flies. Nat. Genet. 2004;36(12):1275–1281. doi: 10.1038/ng1476. [DOI] [PubMed] [Google Scholar]
- 50.Bier E, Bodmer R. Drosophila, an emerging model for cardiac disease. Gene. 2004;342(1):1–11. doi: 10.1016/j.gene.2004.07.018. [DOI] [PubMed] [Google Scholar]
- 51.Bodmer R, Venkatesh TV. Heart development in Drosophila and vertebrates: conservation of molecular mechanisms. Dev. Genet. 1998;22(3):181–186. doi: 10.1002/(SICI)1520-6408(1998)22:3<181::AID-DVG1>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- 52.Alayari NN, Vogler G, Taghli-Lamallem O, et al. Fluorescent labeling of Drosophila heart structures. J. Vis. Exp. 2009;32:1423. doi: 10.3791/1423. pii. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ocorr K, Reeves NL, Wessells RJ, et al. KCNQ potassium channel mutations cause cardiac arrhythmias in Drosophila that mimic the effects of aging. Proc. Natl Acad. Sci. USA. 2007;104(10):3943–3948. doi: 10.1073/pnas.0609278104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wessells RJ, Bodmer R. Age-related cardiac deterioration: insights from Drosophila. Front. Biosci. 2007;12:39–48. doi: 10.2741/2047. [DOI] [PubMed] [Google Scholar]
- 55.Kim SO, Avraham S, Jiang S, et al. Differential expression of Csk homologous kinase (CHK) in normal brain and brain tumors. Cancer. 2004;101(5):1018–1027. doi: 10.1002/cncr.20442. [DOI] [PubMed] [Google Scholar]
- 56.Rulifson EJ, Kim SK, Nusse R. Ablation of insulin-producing neurons in flies: growth and diabetic phenotypes. Science. 2002;296(5570):1118–1120. doi: 10.1126/science.1070058. [DOI] [PubMed] [Google Scholar]
- 57.Khosla S, Patel VM, Hay ID, et al. Loss of heterozygosity suggests multiple genetic alterations in pheochromocytomas and medullary thyroid carcinomas. J. Clin. Invest. 1991;87(5):1691–1699. doi: 10.1172/JCI115186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Mulligan LM, Kwok JB, Healey CS, et al. Germ-line mutations of the RET proto-oncogene in multiple endocrine neoplasia type 2A. Nature. 1993;363(6428):458–460. doi: 10.1038/363458a0. [DOI] [PubMed] [Google Scholar]
- 59.Massoll N, Mazzaferri EL. Diagnosis and management of medullary thyroid carcinoma. Clin. Lab. Med. 2004;24(1):49–83. doi: 10.1016/j.cll.2004.01.006. [DOI] [PubMed] [Google Scholar]
- 60.Carlomagno F, Vitagliano D, Guida T, et al. ZD6474, an orally available inhibitor of KDR tyrosine kinase activity, efficiently blocks oncogenic RET kinases. Cancer Res. 2002;62(24):7284–7290. [PubMed] [Google Scholar]
- 61.Wells SA, Jr, Gosnell JE, Gagel RF, et al. Vandetanib for the treatment of patients with locally advanced or metastatic hereditary medullary thyroid cancer. J. Clin. Oncol. 2010;28(5):767–772. doi: 10.1200/JCO.2009.23.6604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Chang S, Bray SM, Li Z, et al. Identification of small molecules rescuing Fragile X syndrome phenotypes in Drosophila. Nat. Chem. Biol. 2008;4(4):256–263. doi: 10.1038/nchembio.78. [DOI] [PubMed] [Google Scholar]
- 63.Hou L, Antion MD, Hu D, et al. Dynamic translational and proteasomal regulation of Fragile X mental retardation protein controls mGluR-dependent long-term depression. Neuron. 2006;51(4):441–454. doi: 10.1016/j.neuron.2006.07.005. [DOI] [PubMed] [Google Scholar]
- 64.Antar LN, Afroz R, Dictenberg JB, Carroll RC, Bassell GJ. Metabotropic glutamate receptor activation regulates Fragile X mental retardation protein and FMR1 mRNA localization differentially in dendrites and at synapses. J. Neurosci. 2004;24(11):2648–2655. doi: 10.1523/JNEUROSCI.0099-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Weiler IJ, Irwin SA, Klintsova AY, et al. Fragile X mental retardation protein is translated near synapses in response to neurotransmitter activation. Proc. Natl Acad. Sci. USA. 1997;94(10):5395–5400. doi: 10.1073/pnas.94.10.5395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Cooper AA, Gitler AD, Cashikar A, et al. A-synuclein blocks ER-Golgi traffic and Rab1 rescues neuron loss in Parkinson’s models. Science. 2006;313(5785):324–328. doi: 10.1126/science.1129462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Clark IE, Dodson MW, Jiang C, et al. Drosophila pink1 is required for mitochondrial function and interacts genetically with parkin. Nature. 2006;441(7097):1162–1166. doi: 10.1038/nature04779. [DOI] [PubMed] [Google Scholar]
- 68.Park J, Lee SB, Lee S, et al. Mitochondrial dysfunction in Drosophila PINK1 mutants is complemented by parkin. Nature. 2006;441(7097):1157–1161. doi: 10.1038/nature04788. [DOI] [PubMed] [Google Scholar]
- 69.Chen L, Feany MB. A-synuclein phosphorylation controls neurotoxicity and inclusion formation in a Drosophila model of Parkinson disease. Nat. Neurosci. 2005;8(5):657–663. doi: 10.1038/nn1443. [DOI] [PubMed] [Google Scholar]
- 70.Hanai S, Kanai M, Ohashi S, et al. Loss of poly(ADP-ribose) glycohydrolase causes progressive neurodegeneration in Drosophila melanogaster. Proc. Natl Acad. Sci. USA. 2004;101(1):82–86. doi: 10.1073/pnas.2237114100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Yang Y, Nishimura I, Imai Y, Takahashi R, Lu B. Parkin suppresses dopaminergic neuron-selective neurotoxicity induced by Pael-R in Drosophila. Neuron. 2003;37(6):911–924. doi: 10.1016/s0896-6273(03)00143-0. [DOI] [PubMed] [Google Scholar]
- 72.Auluck PK, Bonini NM. Pharmacological prevention of Parkinson disease in Drosophila. Nat. Med. 2002;8(11):1185–1186. doi: 10.1038/nm1102-1185. [DOI] [PubMed] [Google Scholar]
- 73.Auluck PK, Chan HY, Trojanowski JQ, Lee VM, Bonini NM. Chaperone suppression of α-synuclein toxicity in a Drosophila model for Parkinson’s disease. Science. 2002;295(5556):865–868. doi: 10.1126/science.1067389. [DOI] [PubMed] [Google Scholar]
- 74.Fernandez-Funez P, Nino-Rosales ML, de Gouyon B, et al. Identification of genes that modify ataxin-1-induced neurodegeneration. Nature. 2000;408(6808):101–106. doi: 10.1038/35040584. [DOI] [PubMed] [Google Scholar]
- 75.Feany MB, Bender WW. A Drosophila model of Parkinson’s disease. Nature. 2000;404(6776):394–398. doi: 10.1038/35006074. [DOI] [PubMed] [Google Scholar]
- 76.Romero E, Cha GH, Verstreken P, et al. Suppression of neurodegeneration and increased neurotransmission caused by expanded full-length huntingtin accumulating in the cytoplasm. Neuron. 2008;57(1):27–40. doi: 10.1016/j.neuron.2007.11.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Sarkar S, Perlstein EO, Imarisio S, et al. Small molecules enhance autophagy and reduce toxicity in Huntington’s disease models. Nat. Chem. Biol. 2007;3(6):331–338. doi: 10.1038/nchembio883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Steffan JS, Agrawal N, Pallos J, et al. SUMO modification of Huntingtin and Huntington’s disease pathology. Science. 2004;304(5667):100–104. doi: 10.1126/science.1092194. [DOI] [PubMed] [Google Scholar]
- 79.Gunawardena S, Her LS, Brusch RG, et al. Disruption of axonal transport by loss of huntingtin or expression of pathogenic polyQ proteins in Drosophila. Neuron. 2003;40(1):25–40. doi: 10.1016/s0896-6273(03)00594-4. [DOI] [PubMed] [Google Scholar]
- 80.Steffan JS, Bodai L, Pallos J, et al. Histone deacetylase inhibitors arrest polyglutamine-dependent neurodegeneration in Drosophila. Nature. 2001;413(6857):739–743. doi: 10.1038/35099568. [DOI] [PubMed] [Google Scholar]
- 81.Kazemi-Esfarjani P, Benzer S. Genetic suppression of polyglutamine toxicity in Drosophila. Science. 2000;287(5459):1837–1840. doi: 10.1126/science.287.5459.1837. [DOI] [PubMed] [Google Scholar]
- 82.Jackson GR, Salecker I, Dong X, et al. Polyglutamine-expanded human huntingtin transgenes induce degeneration of Drosophila photoreceptor neurons. Neuron. 1998;21(3):633–642. doi: 10.1016/s0896-6273(00)80573-5. [DOI] [PubMed] [Google Scholar]
- 83.Chen HK, Fernandez-Funez P, Acevedo SF, et al. Interaction of Akt-phosphorylated ataxin-1 with 14-13-3 mediates neurodegeneration in spinocerebellar ataxia type 1. Cell. 2003;113(4):457–468. doi: 10.1016/s0092-8674(03)00349-0. [DOI] [PubMed] [Google Scholar]
- 84.Warrick JM, Paulson HL, Gray-Board GL, et al. Expanded polyglutamine protein forms nuclear inclusions and causes neural degeneration in Drosophila. Cell. 1998;93(6):939–949. doi: 10.1016/s0092-8674(00)81200-3. [DOI] [PubMed] [Google Scholar]
- 85.Vidal M, Wells S, Ryan A, Cagan R. ZD6474 suppresses oncogenic RET isoforms in a Drosophila model for type 2 multiple endocrine neoplasia syndromes and papillary thyroid carcinoma. Cancer Res. 2005;65(9):3538–3541. doi: 10.1158/0008-5472.CAN-04-4561. [DOI] [PubMed] [Google Scholar]