Abstract
Low density lipoprotein receptor (LDLR) is a major apolipoprotein E (APOE) receptor and thereby is critical to cholesterol homeostasis and, possibly, Alzheimer disease (AD) development. We previously identified a single nucleotide polymorphism (SNP), rs688:C>T, that modulates LDLR exon 12 splicing and is associated with cholesterol levels in pre-menopausal women and with Alzheimer disease in men. To gain additional insights into LDLR splicing regulation, we seek to identify splicing factors that modulate LDLR splicing efficiency. By using an in vitro minigene study, we first found that ectopic expression of SFRS3 (SRp20), SFRS13A (SRp38), SFRS13A-2 (SRp38-2) and RBMX (hnRNP G) robustly decreased LDLR splicing efficiency. While SFRS3 and SFRS13A specifically increased the LDLR transcript lacking exon 11, SFRS13A-2 and RBMX primarily increased the LDLR isoform lacking both exons 11 and 12. When we evaluated the relationship between the expression of these splicing factors and LDLR splicing in human brain and liver specimens, we found that overall SFRS13A expression was significantly associated with LDLR splicing efficiency in vivo. We interpret these results as suggesting that SFRS13A regulates LDLR splicing efficiency and may therefore emerge as a modulator of cholesterol homeostasis.
Keywords: LDLR, splicing, SFRS13A, SRp38, cholesterol
Introduction
Alternative splicing is a major mechanism for regulating gene expression and contributing to protein diversity. Estimates of the proportion of human genes that are alternatively spliced range from 70–80% (Clark, et al., 2007; Johnson, et al., 2003; Kampa, et al., 2004). Alternative splicing is characterized by utilization of different splice sites. Splicing requires that small nuclear ribonucleoproteins (snRNPs) recognize a conserved 5′ splice site, branch point and 3′ splice site to assemble a spliceosome. If these sequences vary from consensus sequence, non-snRNP splicing factors are required to achieve efficient splicing.
The two major non-snRNP splicing factor families are the serine/arginine-rich (SR) protein family and heterogeneous nuclear ribonucleoprotein (hnRNP) family. The SR proteins consist of one or two RNA recognition motifs (RRMs) and a carboxyl-terminal domain enriched with serine/arginine repeats (RS domain). The RRMs determine RNA binding specificity, whereas the RS domain mediates protein-protein interactions (Caceres and Kornblihtt, 2002). Conventionally, SR proteins recognize exonic splicing enhancers (ESEs) and recruit splicing machinery close to the exon/intron boundary and therefore enhance splicing; a minority of SR proteins act to inhibit splicing (reviewed in (Lin and Fu, 2007).
In contrast, hnRNPs were first described as a major group of chromatin-associated RNA-binding proteins (Krecic and Swanson, 1999). These proteins consist of at least one RNA binding motif such as an RRM, an hnRNP K homology (KH) domain or a arginine/glycine-rich (RGG) box as well as auxiliary domains for protein-protein interactions (He and Smith, 2009). HnRNPs recognize ESEs or exonic splicing silencers (ESS) to regulate splicing efficiency.
Changes in DNA sequence that disrupt functional RNA regulatory cis-acting elements can result in aberrant splicing and thereby cause human disease (reviewed in (Cooper, et al., 2009; Tazi, et al., 2009). For example, mutations affecting RNA splicing are the most common cause of neurofibromatosis type 1 (Ars, et al., 2000) while mutations within and around exon 10 of microtubule-associated protein tau that disrupt exon 10 splicing cause frontotemporal dementia with Parkinsonism linked to chromosome 17 (Liu and Gong, 2008). These findings suggest that agents that alter splicing regulation can be a novel means of human disease therapy.
We previously identified a single nucleotide polymorphism (SNP), rs688:C>T, which modulates low-density lipoprotein receptor (LDLR; MIM# 606945) exon 12 splicing efficiency and is associated with cholesterol levels in pre-menopausal women (Zhu, et al., 2007) and with Alzheimer disease (AD) in men (Zou, et al., 2008). We infer from our results that enhancing LDLR exon 12 splicing efficiency will lower cholesterol and reduce AD in relevant populations. However, the regulation of exon 12 splicing is still unclear. Here, we report that SFRS3 (SRp20), SFRS13A (SRp38) and RBMX (hnRNP G) decrease LDLR splicing in vitro. Moreover, SFRS13A expression level is associated with LDLR splicing efficiency in vivo. We interpret these results as suggesting that SFRS13A is a major modulator of LDLR splicing.
Materials and Methods
Cell culture
HepG2 (human hepatocellular carcinoma) cells were grown in Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 10% fetal bovine serum and 1% penicillin/streptomycin at 37°C in a humidified 5% CO2 -95% air atmosphere.
Vectors
LDLR minigenes containing exons 9–14 with rs688T or rs688C in pcDNA3.1 were previously described (Zhu, et al., 2007); the rs688T vector was derived from the rs688C by using site-directed mutagenesis (QuikChange Site-Directed Mutagenesis Kit, Stratagene) according to the manufacturer’s directions. Vectors encoding HNRNPF and HNRNPH, as well as their parent vector pcDNA4, were generous gifts from Dr. Paula Grabowski (University of Pittsburgh) (Han, et al., 2005). Vector encoding HNRNPA2B1 was a kind gift from Dr. Emanuele Buratti (International Centre for Genetic Engineering and Biotechnology, Trieste, IT). Vectors encoding the other splicing factors used here, as well as a pEGFP-C2 negative control vector encoding only EGFP were kindly provided by Dr. Stefan Stamm (University of Kentucky). These vectors, most of which encode the splicing factor as a fusion protein with EGFP, have been previously characterized (Heinrich, et al., 2009; Novoyatleva, et al., 2008; Screaton, et al., 1995).
Screening of splicing factors in vitro
Splicing factor actions on LDLR splicing efficiency were evaluated by co-transfecting vectors encoding each splicing factor with the LDLR exon 9–14 minigene (rs688C or rs688T) into HepG2 cells. Splicing factor effects were considered relative to LDLR minigene co-transfected with either pEGFP or “empty” pcDNA4 (similar results were obtained with each negative control). Vectors were transfected by using FuGene 6 transfection reagent as directed by the manufacturer (Roche Applied Sciences). Briefly, 1.5 × 105 cells were seeded in a 6-well plate in 2 ml of medium without antibiotics one day before performing the transfection. The next day, 1 μg of LDLR minigene and 1 μg of splicing factor construct were mixed with 6 μl of FuGENE reagent in 94 μl of Opti-MEM (Invitrogen) and added to cell culture. Twenty-four hours after transfection, total RNA was isolated (RNeasy Mini Kit, Qiagen) and analyzed for LDLR splicing patterns by reverse transcriptase-PCR (RT PCR) as previously described (Zhu, et al., 2007). One μg of RNA was converted to cDNA (SuperScript III, Invitrogen) and sequences corresponding to LDLR minigene splice products were PCR amplified (Platinum Taq, Invitrogen) with an LDLR exon 10 sense primer 5′ CATCGTGGTGGATCCTGTTC3′ and a vector-specific antisense primer 5′ GGGATAGGCTTACCTTCGAA3′. The PCR profiles consisted of an initial denaturation at 94°C for 4 min, followed by cycles of 94°C for 30 s, 60°C for 30 s, and 72°C for 45 s, and final extension at 72°C for 7 min. The minimal number of PCR cycles necessary to discern products was performed, e.g., 23 cycles. PCR products were separated by polyacrylamide gel electrophoresis (PAGE) and visualized by SYBR-gold fluorescence on a fluorescence imager (Fuji FLA-2000). PCR product identities were determined by gel purification and direct sequencing (Davis Sequencing). The amount of full length and inefficiently spliced LDLR isoforms were quantified by fluorescence intensity. For each sample, fluorescence values were corrected for background and normalized for length differences among amplicons. Sample splicing efficiency was then quantified as the amount of full length (FL) LDLR PCR product containing exons 10–14 divided by the total LDLR PCR product for that sample.
For experiments evaluating the dose-dependence of the splicing factors, 0.01, 0.1, or 1 μg of vectors containing the splicing factor (total amount made up to 1 μg with “negative control” pcDNA4) was co-transfected with 1 μg of LDLR minigene containing rs688C. Twenty-fourhours after transfection, total RNA was collected and splicing efficiency evaluated by RT-PCR as described above.
Western blot
HepG2 cells were co-transfected as described above. Twenty-four hours later, cells were washed with 1 ml of room temperature phosphate buffered saline (PBS) and lysed in 80 μl of RIPA butter (50 mM Tris, pH 8.0, 150 mM NaCl, 1% NP-40, 0.5% deoxycholic acid, 0.1% SDS) containing 1× protease inhibitor cocktail (Roche Applied Science) for 30 minutes on ice with occasional rocking. A cell scraper was used to collect the cell lysate. Lysates were pooled from three wells for each sample and centrifuged at 10,000×g for 10 minutes at 4°C. Fifty μl of protein extract was mixed with 10 μl of 6X SDS sample loading buffer containing β-mercaptoethanol, boiled for 5 minutes and subjected to electrophoresis on a 12% polyacrylamide gel. Proteins were transferred to nitrocellulose membranes (Bio-Rad). The blots were then blocked with 5% non-fat powdered milk in PBS for 1 hour at room temperature and probed overnight with a mouse anti-GFP antibody (1:200 dilution; Cat. No. 11814460001, Roche Applied Science) or with a mouse anti-Actin antibody (1:200 dilution; Cat. No. sc-8432, Santa Cruz Biotechnology) at 4°C. After washing with 0.1% Tween-20 in PBS four times for 5 minutes each, the blots were incubated with peroxidase-conjugated sheep anti-mouse antibody (1:1,000 dilution, Jackson ImmunoResearch) for 1 hour at room temperature. Bound peroxidase was visualized by using a SuperSignal West Pico kit (Pierce) and a molecular imager (ChemiDoc XRS System, Bio-Rad).
Human tissues
Human liver samples were obtained from the Brain and Tissue Bank for Developmental Disorders (Baltimore, MD) and have been previously described (Zhu, et al., 2007). The samples were from deceased individuals with an average age at death for women of 28 ± 9 years (mean ± SD, range of 15–44, n = 15) and for men of 27 ± 10 (range 13–46, n = 20). The average postmortem interval (PMI) for women was 13 ± 5 hours (mean ± SD, range 4–19, n = 15) while the PMI for men was 10 ± 3 hours (range 3–14, n = 20).
Human anterior cingulate brain specimens were generously provided by the Sanders-Brown Alzheimer Disease Center Neuropathology Core and have also been described elsewhere (Zou, et al., 2008). The samples were from deceased individuals with an average age at death for females of 82 ± 7 years (mean ± SD, n = 31) and for males of 82 ± 8 (n = 28). The average postmortem interval (PMI) for females and males was 3.0 ± 0.8 h (mean ± SD, n = 31) and 3.1 ± 0.8 h (n = 28), respectively.
Evaluation of LDLR splicing efficiency in vivo
Total RNA was prepared from the human liver and brain specimens and converted to cDNA in 1μg aliquots with random hexamers and reverse transcriptase (SuperScript III, Invitrogen) (Chomczynski and Sacchi, 1987). The LDLR splicing efficiency in brain specimens was evaluated by RT-PCR as we previously described for the liver specimens (Zhu, et al., 2007). Briefly, an LDLR exon 10 sense primer 5′ CCTGGCCAGCAGCATGCCGTC3′ and an exon 14 antisense primer 5′ CATCGTGGTGGATCCTGTTC3′ were used to PCR-amplify (Platinum Taq, Invitrogen) brain cDNAs corresponding to LDLR exons 10–14, as well as isoforms lacking exons 11 and/or 12. PCR profiles consisted of preincubation at 94°C for 4 min, followed by cycles of 94°C for 30 s, 60°C for 30 s, and 72°C for 45 s, and final extension at 72°C for 7 min. The minimal number of PCR cycles necessary to discern products was performed, e.g., 30 cycles. PCR products were separated by PAGE and quantified as described above.
Evaluation of splicing factor expression in vivo
The expression level of SFRS3, SFRS13A and RBMX in the liver and brain specimens was quantified by real-time quantitative PCR (RT-qPCR). The 20 μl RT-qPCR mixture containing approximately 20 ng of liver or brain cDNA, 1 μM of each primer and 1× SYBR Green PCR Master Mix (Ambion) was subjected to RT-qPCR (MJ Research PTC-200 with Chromo4 detector). PCR profiles consisted of preincubation at 95°C for 10 min, followed by 40 cycles of 95°C for 30 s, 60°C for 30 s, and 72°C for 30 s. Specificity of the reactions was evaluated by showing a single PCR product by gel electrophoresis and by performing a melting curve analysis after the PCR amplification. The copy numbers of PCR product in each sample were determined relative to standard curves that were amplified in parallel and were based upon standardized amounts of purified PCR products. The following mRNAs and primers were evaluated: SFRS3:5′ CGGCTTTGCTTTTGTTGAAT3′ and 5′ TCACCATTCGACAGTTCCAC3′ SFRS13A:5′ ATTTCTACACTCGCCGTCCA3′ and 5′ CCGTCCACAAATCCACTTTC3′ RBMX:5′ GTAGCAGTGGAATGGGAGGA3′ and 5′ CCATCATCTCTTGGGGACAA3′ ribosomal protein L13A (RPL13A): 5′ CATCTCCTTCTCGGCATCA3′ and5′AACCCTGTTGTCAATGCCTC3′ hypoxanthine-guanine phosphoribosyltransferase 1 (HPRT1): 5′GACCAGTCAACAGGGGACAT3′ and 5′AACACTTCGTGGGGTCCTTTTC3′. The geometric mean of RPL13A and HPRT1 was used to normalize splicing factor expression among the samples (Vandesompele, et al., 2002). All RT-qPCR assays were repeated twice.
Statistics
The effect of splicing factor ectopic expression on LDLR mini-gene splicing efficiency was analyzed by using analysis of variance (ANOVA) and a post-hoc Scheffe test (SPSS software, v 17, SPSS Inc., Chicago, IL). The correlation between LDLR splicing efficiency and splicing factor expression was analyzed by using a linear regression model (SPSS).
Results
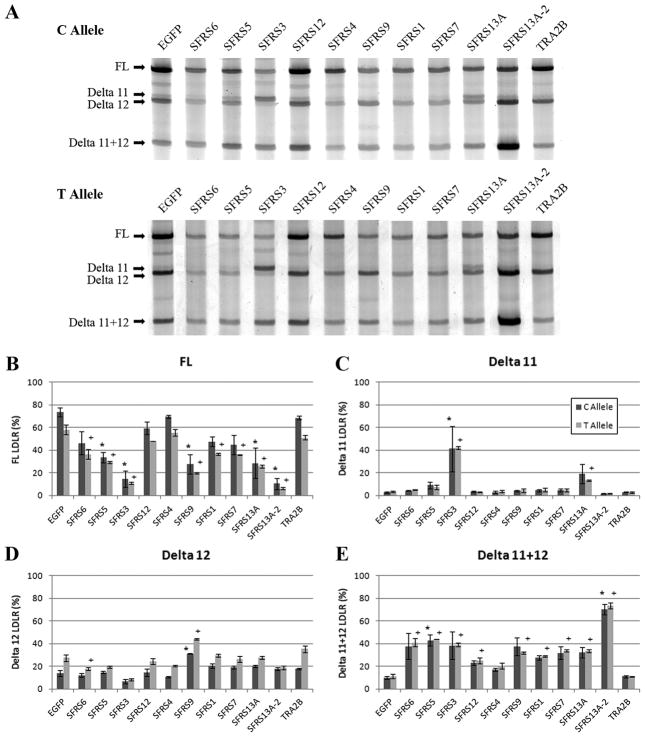
To identify splicing factors that may modulate LDLR splicing efficiency, we quantified the effects of eleven widely studied SR protein family members on splicing of an LDLR exon 9–14 minigene (Zhu, et al., 2007). We co-transfected the LDLR minigenes containing either rs688C or rs688T alleles with vectors encoding candidate SR proteins into HepG2 cells. Splicing efficiency was quantified 24 hours later by RT-PCR. The proportion of FL LDLR transcript was consistently less with the rs688T allele than the rs688C allele (Figure 1), reproducing our earlier observation of a SNP-induced effect on splicing (Zhu, et al., 2007). None of the SR proteins showed a SNP-dependent effect. Rather, many of the SR proteins reduced LDLR minigene splicing efficiency, with SFRS3, SFRS13A, and SFRS13A-2 showing the largest changes (Figure 1B). SFRS3 reduced FL LDLR by specifically increasing an LDLR transcript that skipped exon 11, i.e., Delta 11 LDLR (p < 0.001) (Figure 1C). SFRS13A acted similarly to increase Delta 11 and also increased the LDLR isoform that lacked both exons 11 and 12 (Figure 1C and E). SFRS13A-2 acted primarily by increasing the LDLR isoform that lacked both exons 11 and 12 (Figure 1E). Interestingly, SFRS13A and SFRS13A-2 are alternatively spliced isoforms from the same gene; SFRS13A includes one RNA recognition motif (RRM) and three RS domains while SFRS13A-2 has the same RRM but only one RS domain (Komatsu, et al., 1999). Hence, the increased number of RS domains within SFRS13A relative to SFRS13A-2 appears to mediate differential effects on LDLR splicing.
Figure 1. SR protein family effects on LDLR minigene splicing in vitro.
HepG2 cells were co-transfected with vectors encoding different SR proteins and LDLR rs688C or rs688T in an exon 9–14 minigene. The effects of the SR proteins and rs688 allele on LDLR minigene splicing are shown as representative images (A) and quantitative results (B–E, mean ± SD, n = 3, * and + reflect p < 0.01 when compared to rs688T and rs688C minigenes, respectively, co-transfected with the negative control pEGFP vector). The faint PCR products observed between FL and Delta 11, and between Delta 12 and Delta 11+12 represent non-physiologic LDLR splice variants, i.e., FL LDLR lacking the first 74 bp of exon 14, and a Delta 13 LDLR isoform, respectively.
HnRNPs are also critical splicing regulatory proteins. Therefore, we also screened twelve well-characterized hnRNP family members for their effects on LDLR splicing. We found that RBMX and RBMXL2 showed the largest effects (Figure 2B); each decreased the inclusion of exons 11 and 12 in the final LDLR mRNA product, regardless of which rs688 allele was present (p < 0.001; Figure 2E). Since RBMX has been reported to influence splicing in an RRM-independent fashion (Heinrich, et al., 2009), we further evaluated the effects of a truncated RBMX form that lacks the RRM domain. The result was identical to RBMX (Figure 2B–E); we interpret these results as indicating that RBMX may modulate LDLR splicing by acting as a scaffold protein without binding to LDLR mRNA.
Figure 2. HnRNP family member effects on LDLR minigene splicing in vitro.
HepG2 cells were co-transfected with vectors encoding different hnRNP family members and LDLR rs688C or rs688T-containing minigenes. The effects of the hnRNPs and rs688 allele on LDLR minigene splicing are shown as representative images (A) and quantitative results (B–E, mean ± SD, n = 3, except for HNRNPA1 and A2B1, which reflects mean ± range, n=2, * and + reflect p < 0.01 when compared to rs688T and rs688C minigenes co-transfected with the negative control pEGFP vector).
Overall, our in vitro screening identified SFRS3, SFRS13A, SFRS13A-2, RBMX and RBMXL2 as candidates for modulating LDLR splicing in human tissues. Since the expression of RBMXL2 is restricted to testis (Elliott, et al., 2000), we focused on the first four splicing factors in subsequent studies. To evaluate whether these splicing factors repressed LDLR minigenesplicing in a dose-dependent manner, three doses (0.01, 0.1, and 1 μg) of the vectors encoding the splicing factors were co-transfected with 1 μg of LDLR minigene. Because three of the four splicing factors were encoded as EGFP fusion proteins, we confirmed that expression was indeed dose-dependent by using anti-GFP Western blots (Figure 3A). When we analyzed LDLR minigene splicing by RT-PCR, we found that as the dose of splicing factor increased, the splicing factor effects on LDLR splicing increased as well (Figure 3B–F). SFRS3 and SFRS13A acted mostly by increasing Delta 11, while SFRS13A-2 and RBMX increased Delta 11+12. Overall, these results are consistent with the data shown in Figures 1 and 2. We note that SFRS3 displayed a steeper dose-response curve than the other splicing factors. This may reflect that SFRS3 is more potent than the other factors, or that SFRS3 may act in a cooperative fashion, which has been suggested for other splicing factors previously (Lynch and Maniatis, 1995). In summary, these results confirm that these splicing factors modulate LDLR splicing in a dose-dependent manner.
Figure 3. Splicing factors show dose-dependent effects on LDLR splicing.
The indicated amounts of vectors encoding splicing factors were co-transfected with 1 μg of the vector encoding the rs688C allele LDLR minigene. The total amount of non-LDLR vector in the transfections was held constant at 1 μg by adding “negative control” pcDNA4 vector. The dose-dependent overexpression of SFRS13A, SFRS13A-2 and RBMX was confirmed by Western blots (A). Dose dependent effects on LDLR minigene splicing efficiency are shown as representative images (B) and overall quantitation (C–F, mean ± SD, n = 3).
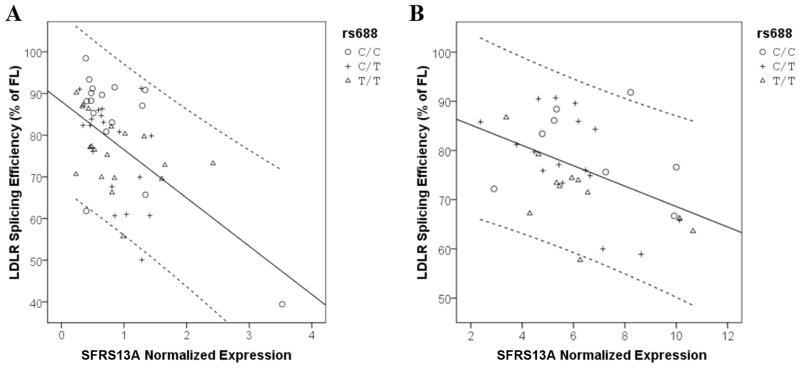
To investigate whether these splicing factors are associated with LDLR splicing efficiency in vivo in a dose-dependent fashion, we quantified SFRS3, SFRS13A, and RBMX expression as well as LDLR splicing in 59 human brain and 35 human liver specimens. Expression of the splicing factors was quantified by RT-qPCR; SFSR13A and SFRS13A-2 were amplified by common primers. LDLR splicing efficiency was evaluated by RT-PCR with primer sequences corresponding to LDLR exon 10 and exon 14. Although others had reported sex-dependent differences in SFRS3 expression and we had seen a sex-dependent difference in LDLR splicing in vivo previously (Zhu, et al., 2007; Zou, et al., 2008), we did not discern any sex-dependent differences in the expression of SFRS3, SFRS13A, or RBMX in brain or liver (Table 1). We proceeded to evaluate the correlation between splicing factor expression and LDLR splicing by using linear regression models that included each of the splicing factors as well as rs688 and sex (Figure 4). We found that SFRS13A expression correlated significantly with LDLR splicing in both the brain (p < 0.001, observed power = 0.999) and liver (p = 0.003, observed power = 0.890). In contrast, SFRS3 and RBMX expression was not significantly associated with LDLR splicing. Interestingly, the inclusion of both SFRS13A expression and rs688 genotype in the model of LDLR splicing tended to reduce the association of rs688 with LDLR splicing, i.e., the association of rs688 with splicing efficiency in the liver remained significant (p = 0.040, observed power = 0.622) while the association of rs688 with splicing in the brain showed a strong trend (p = 0.062, power = 0.544). Consistent with our finding that SFRS13A acted similarly upon the rs688C and rs688T LDLR minigenes, the interaction between SFRS13A expression and rs688 genotype was not significant (p = 0.138). Overall, increased SFRS13A expression is associated significantly with decreased efficiency of LDLR splicing while rs688 genotype is associated significantly with LDLR splicing in the liver and trends with LDLR splicing in the brain.
Table 1.
Splicing factor expression in human brain and liver
| Human Brains
|
p value | Human Livers
|
p value | |||
|---|---|---|---|---|---|---|
| Male (n = 31) | Female (n = 28) | Male (n = 20) | Female (n = 15) | |||
| SFRS3 | 0.82 ± 0.44 | 0.88 ± 0.40 | 0.59 | 3.83 ± 2.18 | 3.55 ± 1.62 | 0.68 |
| SFRS13A | 0.85 ± 0.54 | 0.96 ± 0.63 | 0.47 | 6.15 ± 2.24 | 6.26 ± 2.00 | 0.88 |
| RBMX | 0.48 ± 0.24 | 0.54 ± 0.26 | 0.35 | 2.05 ± 1.30 | 1.80 ± 0.84 | 0.52 |
These results reflect the mean ± SD for the expression of the indicated splicing factors. Splicing factor expression was normalized to the geometric mean of RPL13A and HPRT1 expression as described (Vandesompele, et al., 2002).
Figure 4. SFRS13A and rs688 are associated with LDLR splicing efficiency.
The relationship between LDLR splicing efficiency, SFRS13A expression and rs688 genotype are shown. As expression level of SFRS13A increased, the splicing efficiency of LDLR decreased. (A) In brain, specimens included 16 rs688C/C, 18 rs688C/T, and 19 rs688T/T. The r2 for the model is 0.309. (B) In liver, specimens included 8 rs688C/C, 16 rs688C/T and 15 rs688T/T. The r2 for the model is 0.213. The solid lines represent fit lines and the dashed lines represent 95% confidence intervals.
Discussion
In this study, we identified several splicing factors that influence LDLR minigene splicing in transfected cells. Two of the factors, SFRS3 and SFRS13A, increased Delta 11 LDLR while two other factors, SFRS13A-2 and RBMX, increased Delta 11+12 LDLR. A comparison of splicing factor expression and LDLR splicing efficiency in human brain and liver specimens established that SFRS13A expression correlated with LDLR splicing efficiency. Overall, we interpret our results as suggesting that SFRS13A may modulate LDLR splicing in vivo. Since the association between SFRS13A expression and LDLR splicing is more robust than that between rs688 and LDLR splicing, and rs688 itself is associated with cholesterol and AD (Zhu, et al., 2007; Zou, et al., 2008), we propose that SFRS13A may also emerge as a modulator of cholesterol homeostasis and AD risk.
The critical nature of LDLR to cholesterol homeostasis is well established in that the loss of a single LDLR allele causes familial hypercholesterolemia (Hobbs, et al., 1992). In addition, LDLR is a receptor for apoE in the brain (Cao, et al., 2006; Fryer, et al., 2005). Since alleles of apoE are the major AD genetic risk factors and changes in LDLR expression modify Aβ metabolism (Cao, et al., 2006; Kim, et al., 2009), factors that modulate LDLR expression or splicing represent potential modulators of AD risk. Consistent with this possibility, our previous work demonstrated that rs688:C>T, a SNP within LDLR exon 12, modulates exon 12 splicing, and is associated with cholesterol levels and with AD in at least some case-control series (Zhu, et al., 2007; Zou, et al., 2008). The LDLR isoforms lacking exon 11 and/or 12 are predicted to encode a truncated LDLR protein that contains the ligand binding domain but lacks the transmembrane domain. Since these LDLR isoforms encode non-functional LDLR proteins, further work is necessary to elucidate the regulation of LDLR splicing. In this regard, the rs688 T allele is predicted to neutralize a SFRS5 binding site by ESEfinder (http://rulai.cshl.edu/tools/ESE2/(Cartegni, et al., 2003)) or create an ESS by FAS-ESS (http://genes.mit.edu/fas-ess/(Wang, et al., 2004)). Hence, we hypothesized that ectopic expression of SFRS5 may enhance splicing of the rs688 C allele, or, perhaps, overcome the neutralizing effect of rs688T; however, ectopic expression of SFRS5 decreased the proportion of LDLR that contained exons 11 and 12. We interpret this finding as suggesting that SFRS5 may bind to exons 10 or 13 preferentially to enhance their inclusion at the expense of exons 11 and 12. Indeed, none of the splicing factors that we examined showed a SNP-dependent effect. Hence, the rs688 effect on splicing may be mediated by other splicing factors, changes in RNA secondary structure, or by an unknown mechanism. Since the rs688T allele mainly increases the isoform lacking exon 12, identifying important cis-acting splicing element(s) within exon 12 may help to identify splicing factors that promote exon 12 inclusion.
Several observations support the possibility that SFRS13A is involved in LDLR splicing. First, each of the two SFRS13A isoforms modulated LDLR minigene splicing in vitro. Second, the SFRS13A isoforms acted in a dose-dependent manner to alter LDLR splicing in vitro. Third, SFRS13A overall expression correlated with LDLR splicing efficiency in the brain and liver in vivo. We note that the SFRS13A effect is independent of rs688 allele both in vitro or in vivo, indicating that SFRS13A probably interacts with a cis-element separate from rs688. Although our minigene co-transfection studies evaluated SFRS13A and SFRS13A-2 separately, our efforts to distinguish these SFRS13A isoforms in our RT-qPCR studies were unsuccessful. SFRS13A and SFRS13A-2 share the same first five exons but differ in exon 6; to successfully amplify these cDNAs, our RT-qPCR studies used primers corresponding to exons 2 and 3 and hence amplified both SFRS13A and SFRS13A-2. The proteins encoded by SFRS13A and SFRS13A-2 contain an identical RRM domain and an initial RS domain. However, SFRS13A contains two additional RS domains (Komatsu, et al., 1999). Since RRM domains are responsible for protein-RNA interaction, SFRS13A and SFSR13A-2 may recognize the same RNA sequence but recruit different splicing factors. This may account for the actions of SFRS13A and SFSR13A-2 in the in vitro minigene studies, i.e., SFRS13A and SFRS13A-2 acted similarly to increase Delta 11+12 LDLR while only SFSR13A-2 also increased Delta 11 LDLR. Since we did not detect significant levels of Delta 11 LDLR in our in vivo studies, SFRS13A may be the primary isoform expressed or other factors may suppress Delta 11 LDLR in vivo.
Since we previously found that rs688:C>T modulated LDLR exon 11–12 splicing in a sex-dependent fashion, the positive SFRS3 results in our screening were interesting because SFRS3 expression has been reported to show sex-dependent expression differences. In particular, Antunes-Martins et. al. found that hippocampal SFRS3 mRNA expression was higher in male mice than female mice both at basal state and following its upregulation by memory training, i.e., the spatial version of the Morris water maze and background contextual fear conditioning (Antunes-Martins, et al., 2007). However, we did not detect differences in SFRS3 expression between male and female humans. This may reflect that we compared expression in human anterior cingulate while Antunes-Martins et. al compared murine hippocampus. Characterization of the murine SFRS3 promoter revealed that SFRS3 expression is probably regulated in a cell-specific fashion (Jumaa, et al., 1997). Therefore, it would be interesting to investigate SFRS3 expression in human hippocampus.
Another interesting candidate regarding sex-dependent splicing is RBMX and its paralog, RBMY, which shares 57% sequence homology with RBMX. In humans, RBMX is encoded by the RBMX gene which is located on the X chromosome while RBMY is encoded by RBMY, an RBMX paralog on the Y chromosome (Ma, et al., 1993). Hence, both males and females express RBMX while only males express RBMY, suggesting that RBMY could alter LDLR splicing in a sex-dependent fashion. Here we observed that RBMX and RBMY both decreased LDLR splicing with RBMY having a weaker effect (Figure 2). However, RBMY, like RBMXL2, is only expressed in testis where it is critical for spermatagenesis (Ma, et al., 1993) Therefore, the RBMY modulation of LDLR splicing in vitro may be physiologically relevant only in testis. In contrast, RBMX is ubiquitously expressed. We found that RBMX robustly decreased LDLR minigene splicing in an RRM-independent fashion, indicating that RBMX may interact with other splicing factor(s) and function as a scaffold protein to modulate LDLR splicing. Since RBMX has been reported to interact with TRA2B, (Hofmann and Wirth, 2002; Nasim, et al., 2003), we considered the possibility that TRA2B could mediate RBMX effects However, TRA2B overexpression did not cause a significant change in LDLR minigene splicing (Figure 1), suggesting that RBMX influenced LDLR splicing via other mechanisms in vitro.
In summary, we identified several splicing factors which modulate LDLR splicing in vitro with SFRS3, SFRS13A, SFSR13A-2 and RBMX having the strongest effects. We then found that increased SFRS13A mRNA expression correlated with decreased LDLR splicing efficiency in the human brain and liver. Overall, we interpret our results as suggesting that SFRS13A may be critical in the regulation of LDLR splicing. Since other factors that modulate LDLR splicing, e.g., rs688, have been associated with cholesterol and Alzheimer disease (Zhu, et al., 2007; Zou, et al., 2008), we propose SFRS13A as a candidate for modulating cholesterol homeostasis and AD risk.
Acknowledgments
The authors gratefully acknowledge Drs. Emanuele Buratti, Paula Grabowski and Stefan Stamm for providing vectors encoding splicing factors. Special thanks also to Dr. Stamm for insightful discussions. The authors gratefully acknowledge tissue supplied by the University of Kentucky Alzheimers Disease Center, which is supported by P30AG028383, as well as NIH for their grant support (R01AG026147 and P01AG030128).
References
- Antunes-Martins A, Mizuno K, Irvine EE, Lepicard EM, Giese KP. Sex-dependent up-regulation of two splicing factors, Psf and Srp20, during hippocampal memory formation. Learn Mem. 2007;14(10):693–702. doi: 10.1101/lm.640307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ars E, Serra E, Garcia J, Kruyer H, Gaona A, Lazaro C, Estivill X. Mutations affecting mRNA splicing are the most common molecular defects in patients with neurofibromatosis type 1. Hum Mol Genet. 2000;9(2):237–47. doi: 10.1093/hmg/9.2.237. [DOI] [PubMed] [Google Scholar]
- Caceres JF, Kornblihtt AR. Alternative splicing: multiple control mechanisms and involvement in human disease. Trends Genet. 2002;18(4):186–93. doi: 10.1016/s0168-9525(01)02626-9. [DOI] [PubMed] [Google Scholar]
- Cao D, Fukuchi K, Wan H, Kim H, Li L. Lack of LDL receptor aggravates learning deficits and amyloid deposits in Alzheimer transgenic mice. Neurobiol Aging. 2006;27(11):1632–43. doi: 10.1016/j.neurobiolaging.2005.09.011. [DOI] [PubMed] [Google Scholar]
- Cartegni L, Wang J, Zhu Z, Zhang MQ, Krainer AR. ESEfinder: A web resource to identify exonic splicing enhancers. Nucleic Acids Res. 2003;31(13):3568–71. doi: 10.1093/nar/gkg616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chomczynski P, Sacchi N. Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal Biochem. 1987;162(1):156–9. doi: 10.1006/abio.1987.9999. [DOI] [PubMed] [Google Scholar]
- Clark TA, Schweitzer AC, Chen TX, Staples MK, Lu G, Wang H, Williams A, Blume JE. Discovery of tissue-specific exons using comprehensive human exon microarrays. Genome Biol. 2007;8(4):R64. doi: 10.1186/gb-2007-8-4-r64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper TA, Wan L, Dreyfuss G. RNA and disease. Cell. 2009;136(4):777–93. doi: 10.1016/j.cell.2009.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elliott DJ, Venables JP, Newton CS, Lawson D, Boyle S, Eperon IC, Cooke HJ. An evolutionarily conserved germ cell-specific hnRNP is encoded by a retrotransposed gene. Hum Mol Genet. 2000;9(14):2117–24. doi: 10.1093/hmg/9.14.2117. [DOI] [PubMed] [Google Scholar]
- Fryer JD, Demattos RB, McCormick LM, O’Dell MA, Spinner ML, Bales KR, Paul SM, Sullivan PM, Parsadanian M, Bu G, et al. The low density lipoprotein receptor regulates the level of central nervous system human and murine apolipoprotein E but does not modify amyloid plaque pathology in PDAPP mice. J Biol Chem. 2005;280(27):25754–9. doi: 10.1074/jbc.M502143200. [DOI] [PubMed] [Google Scholar]
- Han K, Yeo G, An P, Burge CB, Grabowski PJ. A combinatorial code for splicing silencing: UAGG and GGGG motifs. PLoS Biol. 2005;3(5):e158. doi: 10.1371/journal.pbio.0030158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He Y, Smith R. Nuclear functions of heterogeneous nuclear ribonucleoproteins A/B. Cell Mol Life Sci. 2009;66(7):1239–56. doi: 10.1007/s00018-008-8532-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heinrich B, Zhang Z, Raitskin O, Hiller M, Benderska N, Hartmann AM, Bracco L, Elliott D, Ben-Ari S, Soreq H, et al. Heterogeneous nuclear ribonucleoprotein G regulates splice site selection by binding to CC(A/C)-rich regions in pre-mRNA. J Biol Chem. 2009;284(21):14303–15. doi: 10.1074/jbc.M901026200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hobbs HH, Brown MS, Goldstein JL. Molecular genetics of the LDL receptor gene in familial hypercholesterolemia. Hum Mutat. 1992;1(6):445–66. doi: 10.1002/humu.1380010602. [DOI] [PubMed] [Google Scholar]
- Hofmann Y, Wirth B. hnRNP-G promotes exon 7 inclusion of survival motor neuron (SMN) via direct interaction with Htra2-beta1. Hum Mol Genet. 2002;11(17):2037–49. doi: 10.1093/hmg/11.17.2037. [DOI] [PubMed] [Google Scholar]
- Johnson JM, Castle J, Garrett-Engele P, Kan Z, Loerch PM, Armour CD, Santos R, Schadt EE, Stoughton R, Shoemaker DD. Genome-wide survey of human alternative pre-mRNA splicing with exon junction microarrays. Science. 2003;302(5653):2141–4. doi: 10.1126/science.1090100. [DOI] [PubMed] [Google Scholar]
- Jumaa H, Guenet JL, Nielsen PJ. Regulated expression and RNA processing of transcripts from the Srp20 splicing factor gene during the cell cycle. Mol Cell Biol. 1997;17(6):3116–24. doi: 10.1128/mcb.17.6.3116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kampa D, Cheng J, Kapranov P, Yamanaka M, Brubaker S, Cawley S, Drenkow J, Piccolboni A, Bekiranov S, Helt G, et al. Novel RNAs identified from an in-depth analysis of the transcriptome of human chromosomes 21 and 22. Genome Res. 2004;14(3):331–42. doi: 10.1101/gr.2094104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J, Castellano JM, Jiang H, Basak JM, Parsadanian M, Pham V, Mason SM, Paul SM, Holtzman DM. Overexpression of Low-Density Lipoprotein Receptor in the Brain Markedly Inhibits Amyloid Deposition and Increases Extracellular Abeta Clearance. Neuron. 2009;64(5):632–644. doi: 10.1016/j.neuron.2009.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komatsu M, Kominami E, Arahata K, Tsukahara T. Cloning and characterization of two neural-salient serine/arginine-rich (NSSR) proteins involved in the regulation of alternative splicing in neurones. Genes Cells. 1999;4(10):593–606. doi: 10.1046/j.1365-2443.1999.00286.x. [DOI] [PubMed] [Google Scholar]
- Krecic AM, Swanson MS. hnRNP complexes: composition, structure, and function. Curr Opin Cell Biol. 1999;11(3):363–71. doi: 10.1016/S0955-0674(99)80051-9. [DOI] [PubMed] [Google Scholar]
- Lin S, Fu XD. SR proteins and related factors in alternative splicing. Adv Exp Med Biol. 2007;623:107–22. doi: 10.1007/978-0-387-77374-2_7. [DOI] [PubMed] [Google Scholar]
- Liu F, Gong CX. Tau exon 10 alternative splicing and tauopathies. Mol Neurodegener. 2008;3:8. doi: 10.1186/1750-1326-3-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch KW, Maniatis T. Synergistic interactions between two distinct elements of a regulated splicing enhancer. Genes Dev. 1995;9(3):284–93. doi: 10.1101/gad.9.3.284. [DOI] [PubMed] [Google Scholar]
- Ma K, Inglis JD, Sharkey A, Bickmore WA, Hill RE, Prosser EJ, Speed RM, Thomson EJ, Jobling M, Taylor K, et al. A Y chromosome gene family with RNA-binding protein homology: candidates for the azoospermia factor AZF controlling human spermatogenesis. Cell. 1993;75(7):1287–95. doi: 10.1016/0092-8674(93)90616-x. [DOI] [PubMed] [Google Scholar]
- Nasim MT, Chernova TK, Chowdhury HM, Yue BG, Eperon IC. HnRNP G and Tra2beta: opposite effects on splicing matched by antagonism in RNA binding. Hum Mol Genet. 2003;12(11):1337–48. doi: 10.1093/hmg/ddg136. [DOI] [PubMed] [Google Scholar]
- Novoyatleva T, Heinrich B, Tang Y, Benderska N, Butchbach ME, Lorson CL, Lorson MA, Ben-Dov C, Fehlbaum P, Bracco L, et al. Protein phosphatase 1 binds to the RNA recognition motif of several splicing factors and regulates alternative pre-mRNA processing. Hum Mol Genet. 2008;17(1):52–70. doi: 10.1093/hmg/ddm284. [DOI] [PubMed] [Google Scholar]
- Screaton GR, Caceres JF, Mayeda A, Bell MV, Plebanski M, Jackson DG, Bell JI, Krainer AR. Identification and characterization of three members of the human SR family of pre-mRNA splicing factors. EMBO J. 1995;14(17):4336–49. doi: 10.1002/j.1460-2075.1995.tb00108.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tazi J, Bakkour N, Stamm S. Alternative splicing and disease. Biochim Biophys Acta. 2009;1792(1):14–26. doi: 10.1016/j.bbadis.2008.09.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vandesompele J, De Preter K, Pattyn F, Poppe B, Van Roy N, De Paepe A, Speleman F. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol. 2002;3(7):RESEARCH0034. doi: 10.1186/gb-2002-3-7-research0034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Rolish ME, Yeo G, Tung V, Mawson M, Burge CB. Systematic identification and analysis of exonic splicing silencers. Cell. 2004;119(6):831–45. doi: 10.1016/j.cell.2004.11.010. [DOI] [PubMed] [Google Scholar]
- Zhu H, Tucker HM, Grear KE, Simpson JF, Manning AK, Cupples LA, Estus S. A common polymorphism decreases low-density lipoprotein receptor exon 12 splicing efficiency and associates with increased cholesterol. Hum Mol Genet. 2007;16(14):1765–72. doi: 10.1093/hmg/ddm124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou F, Gopalraj RK, Lok J, Zhu H, Ling IF, Simpson JF, Tucker HM, Kelly JF, Younkin SG, Dickson DW, et al. Sex-dependent association of a common low-density lipoprotein receptor polymorphism with RNA splicing efficiency in the brain and Alzheimer’s disease. Hum Mol Genet. 2008;17(7):929–35. doi: 10.1093/hmg/ddm365. [DOI] [PMC free article] [PubMed] [Google Scholar]