Abstract
Background
Although global brain structure is highly heritable, there is still variability in the magnitude of genetic influences on the size of specific regions. Yet, little is known about the patterning of those genetic influences, i.e., whether the same genes influence structure throughout the brain or whether there are regionally-specific sets of genes.
Methods
We mapped the heritability of cortical thickness throughout the brain using 3D structural magnetic resonance imaging in 404 middle-aged male twins. To assess the amount of genetic overlap between regions, we then mapped genetic correlations between three selected seed points and all other points comprising the continuous cortical surface.
Results
There was considerable regional variability in the magnitude of genetic influences on cortical thickness. The primary visual (V1) seed point had strong genetic correlations with posterior sensory and motor areas. The anterior temporal seed point had strong genetic correlations with anterior frontal regions, but not with V1. The middle frontal seed point had strong genetic correlations with inferior parietal regions.
Conclusions
These results provide strong evidence of regionally-specific patterns rather than a single, global genetic factor. The patterns are largely consistent with a division between primary and association cortex, as well as broadly-defined patterns of brain gene expression, neuroanatomical connectivity, and brain maturation trajectories, but no single explanation appears to be sufficient. The patterns do not conform to traditionally-defined brain structure boundaries. This approach can serve as a step toward identifying novel phenotypes for genetic association studies of psychiatric disorders, and normal and pathological cognitive aging.
Keywords: heritability, twins, cortical thickness, endophenotypes, MRI, genetic correlation, imaging genetics
Do the same genes influence structural variation throughout the brain, or are there different sets of genes that influence specific regions or lobes? Global brain structure measures are highly heritable (1,2), perhaps suggesting a single, common genetic factor. There is also regional variation in the magnitude of genetic and environmental influences (1,2), but even substantial regional variation in heritability—the proportion of variance explained by genetic factors (3)—is still neutral with regard to common versus independent genetic influences across brain regions. Because the brain functions via coordinated systems, elucidating genetic patterns and interrelationships among brain regions is crucial for understanding both normal and pathological human brain development and brain aging. For example, gray matter loss occurs with normal aging (4,5), but the patterns of loss are different in pathological conditions such as Alzheimer’s disease (6). The patterns of genetic relationships also have important implications for phenotype or endophenotype definition in genetic association studies of psychiatric and neurological disorders.
In addition to studies demonstrating differences in the heritability of specific structures in different brain regions (1,2), two studies have investigated genetic relationships between pre-determined regions of interest (ROIs) (7,8). Such analyses are constrained by the implicit assumption that traditionally-defined structures will map onto genetic determinants of brain anatomy. Compared with studies of pre-defined structures, detailed maps can provide more precise information about patterns of heritability. A continuous map of the heritability of cortical density (proportion of gray matter per voxel) suggested strong genetic influences in frontal regions, including Broca’s area, and also in Wernicke’s areas; heritabilities were also higher in Wernicke’s area than in the homologous right hemisphere region (9).
Although this continuous map approach contributes valuable information, it is still entirely uninformative about genetic relationships between brain regions. To address this issue, it is necessary to examine genetic correlations (rg) between regions (3). The genetic correlation between two variables is defined as their genetic covariance divided by the square root of the product of their genetic variances (3). Essentially, it indicates the amount of genetic overlap. Genetic correlations can be calculated in twin studies because the twin method makes it possible to separate genetic and environmental sources of variance.
Using the cortical surface reconstruction method of Dale and colleagues (10,11), we investigated the heritability of cortical thickness on a continuous basis across the entire cortical surface in a large sample of adult male twins. In the present sample, we have shown that the two components of cortical volume—thickness and surface area—are determined by independent sets of genetic influences (12). Thus, thickness may have advantages over volume measures in genetic studies. In addition to the continuous heritability map, we examined the degree to which the genetic factors that caused variation at selected vertices (seed points) also did so across the entire cortical surface. These seed point analyses indicate genetic correlations of cortical thickness across regions that are not constrained by pre-defined boundaries.
We considered five hypotheses that might account for the observed patterns of genetic correlation: 1) Heritabilities reflect a common set of underlying genetic factors, with little or no anatomical specificity. 2) Different sets of genes or patterns of gene expression are associated with specific circumscribed anatomical regions based on Brodmann-type areas or entire lobes. 3) Different genes cause variation in different types of cortex such as primary versus association cortex or different sensory modalities. 4) Anatomical connectivity or functional systems are the factors that differentiate the genetic correlation patterns. 5) The genetic correlation patterns reflect patterns of brain maturation.
Methods and Materials
Participants
Participants were part of the Vietnam Era Twin Study of Aging (VETSA). An overview of the VETSA project can be found elsewhere (13). A total of 1,237 twins participated in wave 1 of this longitudinal study, and a subset underwent MRIs. This article is based on 474 twins who had analyzable scans to date; the twin analyses included 404 twins: 110 monozygotic (MZ) and 92 dizygotic (DZ) pairs. To date, 56% of the MRI study participants had zygosity determined by 25 microsatellite markers. Consistent with the overall VETSA project, 95% of the original classifications based on questionnaire and blood group information were in agreement with the genotype-based classifications. When differences occurred, genotype-based classifications were used.
Participants live throughout the United States and had the option of traveling to the University of California, San Diego (UCSD) or Boston University for a day-long series of cognitive, health/medical, and psychosocial assessments. MRIs were performed at UCSD or Massachusetts General Hospital (MGH), typically the day after in-lab evaluations. Almost all twins within a pair were assessed at the same site. Potential VETSA MRI participants were screened for standard MR exclusions. Only 6% of those VETSA participants who were invited to undergo MRI declined to participate; 59% were included. The remaining participants were excluded for reasons such as possible metal in the body (7%), claustrophobia (3%), testing conducted in the twins’ hometown (5%), scanner problems (8%), co-twin excluded (9%), and other reasons (3%).
VETSA does not comprise a Veterans Affairs hospital or a patient sample, and the large majority were not exposed to combat. Basic demographic and health characteristics of the VETSA sample are comparable to U.S. census data for similarly aged men (14,15). Mean age of the MRI participants was 55.8 (SD=2.6) years (range: 51–59); the narrow age range maximizes power to examine within-individual change over time. Mean years of education was 13.9 (SD=2.1), and 85.2% were right-handed. Most participants were employed (74.9% full-time, 4.2% part-time), and 11.2% were retired. There were 88.3% non-Hispanic white, 5.3% African-American, 3.4% Hispanic, and 3.0% “other” participants. Self-reported overall health status was as follows: excellent (14.8%); very good (36.5%); good (37.4%); fair (10.4%); and poor (0.9%). These demographic characteristics did not differ from the entire VETSA sample, nor were there significant differences between MZ and DZ twins.
Image Acquisition
Images were acquired on Siemens 1.5 Tesla scanners (241 at UCSD; 233 at MGH) with protocols individualized for each scanner; sagittal T1-weighted MPRAGE sequences were employed with a TI=1000ms, TE=3.31ms, TR=2730ms, flip angle=7 degrees, slice thickness=1.33mm, and voxel size=1.3×1.0×1.3mm. Raw DICOM MRI scans (including two T1-weighted volumes per case) were downloaded to the MGH. These data were reviewed for quality, registered, and averaged to improve signal-to-noise.
Image Processing
The cortical surface was reconstructed to measure thickness at each surface location, or vertex, using a semi-automated approach. Explicit reconstruction of the cortical surface is a complex procedure with a number of subtasks (10,16–18). Intensity variations due to magnetic field inhomogeneities are corrected, a normalized intensity image is created, and the skull is removed from the normalized image. The preliminary segmentation is partitioned using a connected components algorithm, with connectivity not allowed across the established cutting planes. Any interior holes in the components representing white matter are filled, resulting in a single filled volume for each cortical hemisphere. The resulting surface is covered with a polygonal tessellation and smoothed to reduce metric distortions. After construction of the initial surface model, a refinement procedure is applied to obtain a representation of the gray/white boundary. This surface is subsequently deformed outwards to obtain an explicit representation of the pial surface. The thickness at each location on the surface is then calculated based on the distance between the gray/white and pial surface (18), and subsequently smoothed using a Gaussian kernel with 30mm full-width half maximum. Once generated, the cortical surface model is manually reviewed and edited according to standard, objective editing rules. Studies demonstrate a high correlation of automatic and manual measures in vivo and ex vivo (18,19), and good reliability and validity of these image acquisition and processing methods across different sites and platforms (20–24).
Statistical Analysis
In the twin design, the variance of any trait is accounted for by four latent factors: additive genetic influences (A); non-additive genetic influences, often referred to as dominance (D); common or shared environmental influences (C); and non-shared or individual-specific environmental influences (E) (3,25). Because MZ twins share 100% of their genes, they correlate perfectly (r=1.0) with respect to both additive and non-additive genetic influences. DZ twins share on average 50% of their genes, resulting in correlations of 0.50 for additive genetic influences and 0.25 for non-additive genetic influences. Shared environmental influences are defined as any environmental factor that influences both members of a twin pair equally regardless of zygosity; hence they correlate 1.0 across twin pairs. Individual-specific environmental influences are environmental factors that make twins different from one another. Because these are individual-specific factors, they are assumed to be uncorrelated across twins. Because error is assumed to be random (and therefore uncorrelated), measurement error is included in the E term in standard twin models.
At the univariate level, these latent factors are combined into what are referred to as the “ACE” or “ADE” models; due to model under-identification, an ACDE model cannot be tested (3). If cross-twin correlations for MZ twins are larger than those for DZ twins, it suggests that additive genetic influences are present. When the MZ correlation is substantially more than twice the DZ correlation, shared environmental influences are estimated near zero and non-additive genetic influences are assumed to be present (3).
In twin designs, large samples are generally required in order to yield reliable estimates of heritability (26). After generating cortical heritability maps for the entire VETSA MRI sample, we generated maps derived from six randomly selected subsets of 10 MZ and 10 DZ twin pairs. These maps were highly unreliable (results shown in the Supplement, Figure S1). In contrast, maps of the high and low end of the 95% confidence intervals for the entire sample were consistent with the full sample map.
In bivariate analyses, genetic and environmental covariance estimates are used to calculate genetic and environment correlations. Univariate and bivariate analyses were performed using Mx, a maximum-likelihood-based structural equation modeling software package (27). All models were fit against the raw data, with each vertex treated as an independent and continuous variable. In conjunction with Mx, we utilized the Matlab (The MathWorks, Natick, Massachusetts) software environment in order to sequentially perform the analyses. Preliminary univariate analyses revealed that estimates of the common environment were at or near zero, and that ADE models tended to provide the best fits to the data. Therefore, the ADE model was selected to establish the degree of genetic influence on each vertex. Heritability estimates were derived by combining A and D in order to obtain a broad-sense estimate of the total genetic variance. These values were then standardized so that for each vertex, the sum of the broad-sense genetic variance and the nonshared environmental variance would equal 1.0. Although the distinction between additive and non-additive genetic influences on brain structure is of interest, a sample size far larger than the present sample would be needed in order to reliably distinguish between the two. Bivariate (seed point) analyses also utilized ADE models. The genetic correlations represent the standardized broad-sense genetic covariances between vertices derived by adding the additive genetic covariance and the non-additive genetic covariance.
Results
Heritability maps of cortical thickness
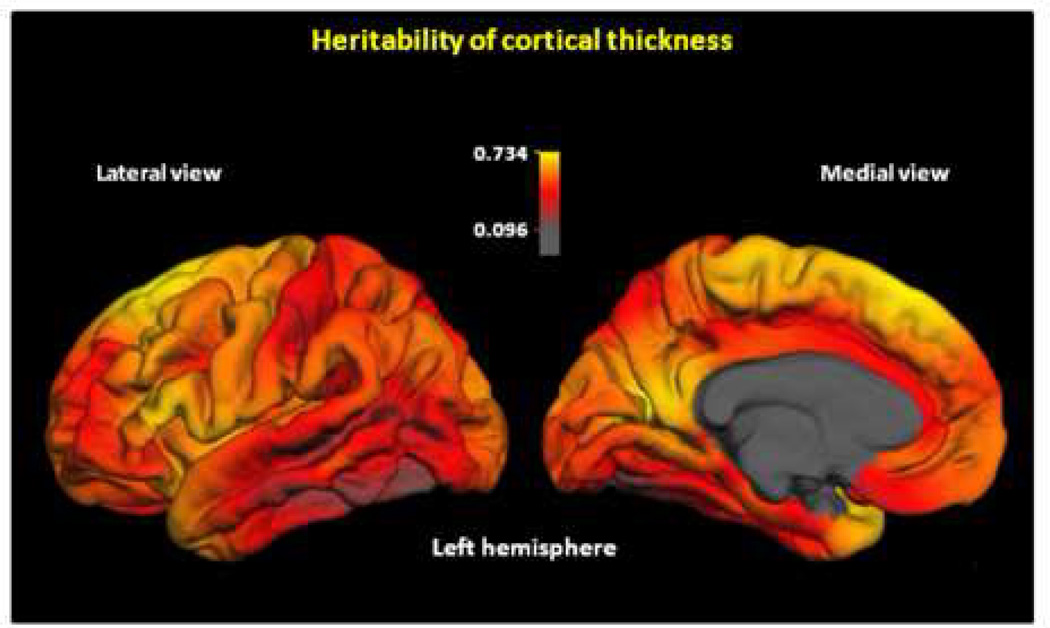
Heritability varied substantially across the cortical surface, from as low as 0.16 to as high as 0.73 (Figure 1; see Figure S2 in the Supplement for other views). The highest heritabilities were observed bilaterally in posterior frontal and anterior medial occipital cortex, and temporal pole in the left hemisphere. The lowest heritability estimates were observed in middle and inferior lateral temporal cortex (especially in the left hemisphere), and anterior prefrontal and orbitofrontal cortex (especially in the right hemisphere).
Figure 1.
Continuous map of heritability of cortical thickness. Color scale indicates heritability values. Heritabilities are based on the total (additive + non-additive) genetic variance. The maps show the variability in the magnitude of genetic influences across the cortex. Maps of the right hemisphere are shown in Figure S2 in the Supplement.
Heritability did not vary simply according to lobar or standard cortical parcellation schemes. For instance, some of the highest as well as some of the lowest heritability estimates were observed within the frontal and temporal lobes. There was relatively high heritability in Broca’s, but somewhat lower heritability in Wernicke’s area. There were no clear differences in heritability between these left hemisphere regions and homologous right hemisphere regions. The middle and inferior left temporal cortices, which are involved in speech perception and language comprehension (28,29), showed some of the lowest heritability estimates.
Seed point analyses: genetic correlation maps
To investigate genetic relationships between regions, we mapped the genetic correlations between cortical thickness at locations on the cortical surface and a specified seed point (3,27). Each seed point shown is in the left hemisphere, but the patterns of genetic correlation to each left hemisphere seed point were very similar to the patterns in the contralateral hemisphere. In addition, when homologous right hemisphere seed points were used, the bilateral patterns were essentially the same.
Primary visual cortex (V1) seed point
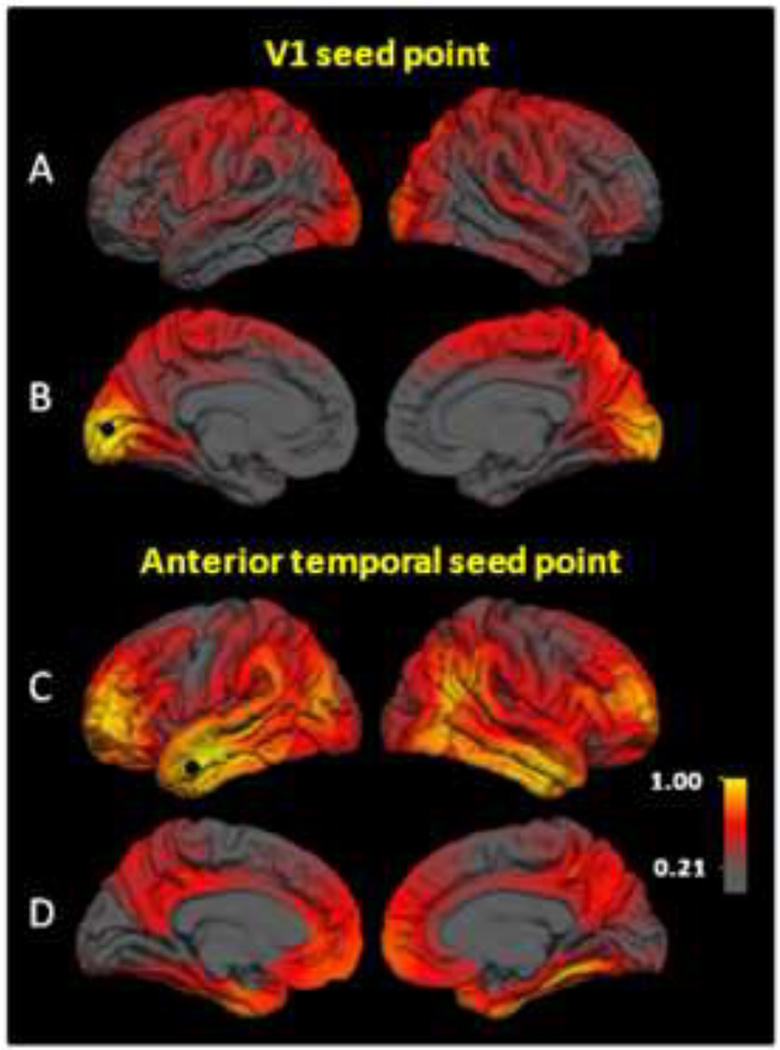
First, we selected a seed point in V1, a well-defined and highly specialized functional area of visual cortex, that can be reliably localized based on sulcal anatomy (17,30). Figures 2a and 2b show genetic correlation maps for the V1 seed point. The highest genetic correlations (rgs=0.8–1.0) were observed bilaterally in medial occipital regions, corresponding to retinotopic visual cortex (31,32). High genetic correlations (rgs≈0.5) were observed with somatosensory, motor, and primary auditory cortex, especially in the right hemisphere. Genetic correlations between the V1 seed point and anterior temporal and anterior frontal regions were essentially zero.
Figure 2.
Maps of genetic correlations with left primary visual cortex (V1) and left anterior temporal gyrus seed points. Color scale indicates the strength of genetic correlations. (a) V1 lateral view. (b) V1 medial view. (c) Anterior temporal lateral view. (d) Anterior temporal medial view. There is a double dissociation with respect to genetic influences; regions with strong genetic associations to the V1 seed point are essentially unrelated genetically to anterior temporal seed point, and vice versa. Strong genetic relationships between anterior frontal and temporal regions (c) correspond to the uncinate fasciculus (37).
Middle temporal gyrus seed point
Second, we chose an anterior middle temporal gyrus seed point (Figures 2c and 2d), an association cortex area that had very low genetic correlations with the V1 seed point. The highest correlations were observed bilaterally with anterior temporal (rgs=0.8–1.0), anterior frontal (rgs=0.8–1.0), and inferior parietal (rgs=0.5–0.95) cortices. There were strikingly low genetic correlations with primary sensory cortex (rgs=0.0–0.2). This pattern was complimentary to the pattern associated with the V1 seed point. There was little overlap in the regions of strong genetic correlation; specific comparisons (Figures 2a vs. 2c; 2b vs. 2d) show that the patterns essentially constitute a double dissociation. That is, regions having high correlations with one seed point are virtually uncorrelated with the other, and vice versa.
Middle frontal gyrus seed point
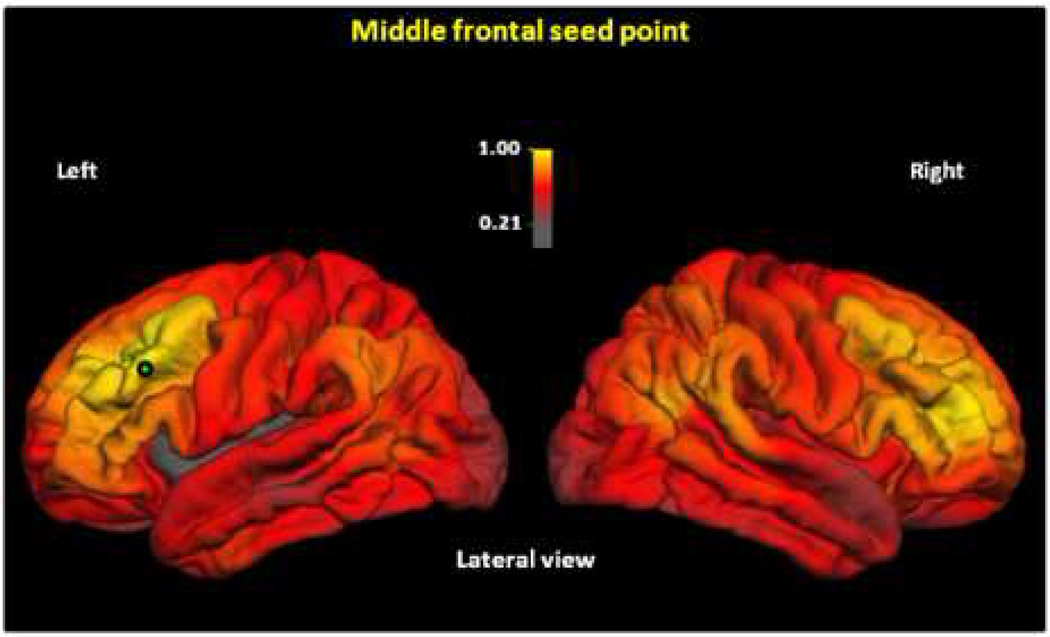
Third, we chose a seed point in another association area, the rostral middle frontal gyrus (Figure 3; see Figure S3 in the Supplement for other views). This analysis showed the highest genetic correlations bilaterally in homologous frontal regions around the seed point (rgs=0.7–1.0), and in the inferior parietal region (including portions of supramarginal and angular gyrus (rgs=0.5–0.9). The lowest genetic correlations were observed in the occipital cortex (rgs=0.3–0.6). Thus, this seed point resulted in genetic correlations with both association cortex and primary sensory and motor cortex, but lower genetic correlations with visual cortex.
Figure 3.
Maps of genetic correlations with left middle frontal gyrus seed point. Color scale indicates strength of genetic correlations. Genetic correlations are calculated as described in Figure 2. The map indicates strong genetic relationships between middle frontal and inferior parietal regions; these correspond to components of the superior longitudinal fasciculus (36). Medial views are shown in Figure S3 in the Supplement.
Discussion
Patterns of heritability
There was substantial variability in the extent of genetic and environmental influences across brain regions, but the pattern does not lend itself to any simple interpretation in terms of cortex type or functional systems. A dissociation between primary and association cortex cannot adequately account for the findings because frontal and temporal association areas are on the opposite extremes of heritability (higher in frontal and lower in temporal), whereas primary visual and somatosensory areas range from moderate to high heritability estimates.
Our results regarding language areas are generally consistent with those of a previous large ROI-based study of children and adolescents (7), but somewhat less consistent with a continuous heritability map of adult twins (9). That is, our results did not indicate substantial left-right differences in heritability or particularly high heritability in Wernicke’s area, although we did find high heritability in Broca’s area. It is perhaps puzzling that our results are more consistent with those in children and adolescents, but heritability estimates based on the adult study may be imprecise given the sample size of only 20 twin pairs (9,26).
Patterns of genetic correlation
We consider five hypotheses that might account for patterns of genetic correlation:
1) Heritabilities reflect a common set of underlying genetic factors, with little or no anatomical specificity
The results cannot be explained by this hypothesis. With all regions showing genetic variation, a global genetic factor cannot explain one seed point having high genetic correlations with some regions while a different seed point has near-zero genetic correlations with the same regions. Rather, these results are consistent with multiple genetic factors with different, but partially overlapping patterns of influence.
2) Different sets of genes or patterns of gene expression are associated with specific circumscribed anatomical regions based on Brodmann-type areas or entire lobes
The idea that the genetic correlation patterns would map to lobar divisions or traditional parcellation schemes was not supported. For example, it can be readily observed in Figure 2c that the regions of highest genetic correlation with inferior frontal cortex include the anterior portion of the left middle temporal gyrus. If we used the rostral middle frontal gyrus ROI from our cortical parcellation based on Desikan et al. (33), the pattern of genetic correlation would have been obscured because the high genetic correlation was restricted to only the anterior portion of that ROI.
The patterns may, however, be consistent with those of gene expression. Animal studies of brain development indicate that there are anterior-posterior gradients of gene expression, although they do not form absolute anterior-posterior boundaries and they do not correspond to anatomically and functionally distinct cortical areas (34,35). Our three different seed point patterns are partially consistent with such expression patterns because the genetic correlation patterns were not completely distinct for anterior and posterior regions. In the context of these smooth, but partially overlapping genetic correlation patterns, it is also noteworthy that V1—the most specialized cortical region in primates with the least overlap of any of the cortical regions (31)—also had the least genetic overlap with other regions in the present study.
3) Different genes cause variation in different types of cortex such as primary versus association cortex or different sensory modalities
The genetic correlations between visual and somatosensory cortex are high, whereas those between visual and association areas are low (based on the V1 and anterior temporal seed points). This pattern is consistent with genetic correlations following the division between primary and association cortex, but not different sensory modalities.
4) Anatomical connectivity or functional systems are the factors that differentiate the genetic correlation patterns
There was partial support for this notion. The high genetic correlations between prefrontal cortex and inferior parietal cortex (supramarginal/angular gyrus; see Figure 3) thickness might be accounted for on the basis of anatomical connectivity because they are at essentially opposite ends of the dorsal component of the superior longitudinal fasciculus, a major inttrahemispheric fiber tract (36). Similarly, the uncinate fasciculus is the major fiber tract connecting anterior temporal (another seed point) with inferior frontal regions (37), and there were high genetic correlations between these regions (see Figure 2b). A genetic association based on functional systems is also possible in that there are extensive prefrontal-parietal connections (38), and functional neuroimaging studies consistently demonstrate combined activation of these regions in tasks involving executive functions and attentional control (39). However, anatomical connectivity does not account for all of the observed patterns. The high genetic correlations between visual cortex and somatosensory cortex are unlikely to be explained in this way because there are no such direct connections between them. Moreover, the high genetic correlations between homologous areas in opposite hemispheres cannot generally be explained by direct connections.
5) The genetic correlation patterns reflect patterns of brain maturation
The genetic correlation patterns are largely consistent with patterns of white and gray matter maturation, which generally proceeds from primary sensory cortices (around the central and calcarine sulci), followed by parietal and mid-frontal regions, and finally, the frontal and temporal poles (40,41). The genetic correlation patterns for the V1 seed point, followed by the middle frontal seed point, followed by the anterior temporal seed point may be viewed as closely paralleling this developmental trajectory.
The genetic correlation patterns in our seed point analyses are also roughly similar to phenotypic correlations observed in in children and adolescents (42). This suggests similar patterns in children and adults, but also that it is largely common genetic influences that drive the phenotypic correlations across brain regions. There is also cross-sectional evidence that the extent of genetic and environmental influences on brain structure may change throughout childhood and adolescence (43). What changes may take place from middle to older age is a question we plan to address in our ongoing follow-up of VETSA participants.
Limitations
It might be argued that the cortical thickness values should be adjusted for average cortical thickness, but doing so would preclude the possibility of testing for a single, common genetic factor. It is also possible that the observed patterns in Figure 1 primarily reflect the degree of measurement error. Greater measurement error would reduce heritability because it would increase non-shared environmental variance (E). However, this explanation seems unlikely because some regions with low heritabilities in Figure 1 were within ROIs that had heritabilities which were average or above average for the 66 cortical parcellation units that we measure (submitted manuscript).
The fact that VETSA participants were not excluded for psychiatric, neurological, or other medical conditions may appear to be a limitation because these are usually considered confounds. However, when we estimate heritabilities, we are estimating the sum total of all genetic influences on a trait. Genes that predispose to certain medical conditions may also predispose to smaller or larger brain structures. For this reason, heritability estimates from samples without exclusions are considered relatively unbiased. Multivariate genetic analyses can examine the extent to which genes that influence medical or psychiatric conditions may overlap with genes that influence brain structure size, but those analyses are beyond the scope of this article.
We do not know how generalizable the results are to women and to other age cohorts. Despite health and demographic similarity to comparably-aged American men, one feature of the sample that does differ from the general population is that 95 (23.5%) participants experienced varying degrees of exposure to combat approximately 35 years prior to the study. The primary concern about this subgroup is usually those individuals who develop post-traumatic stress disorder (PTSD). As of their mid-40s, when diagnostic interviews were conducted, 31 (7.7%) had a lifetime diagnosis of PTSD. This rate is slightly above the 5.0% prevalence for men nationally (44). However, it seems unlikely that this subgroup would substantially alter the results because discordant twin studies demonstrate that cognitive and brain abnormalities are primarily pre-existing risk factors rather than consequences of PTSD (45,46). Also, our primary focus was on the relationships among regions rather than group differences in the thickness of particular regions.
Summary
The results suggest that global brain measures or subregions based on traditional sulcal-based parcellation units may be of limited utility for candidate or genome-wide association studies because different sets of genes influence different subregions. The genetic correlation patterns partially parallel primary versus association divisions of cortex type, anatomical connectivity, the developmental trajectory of early brain maturation, and gene expression patterns found in animal studies. However, none of these provides an entirely adequate explanation. Rather, the observed patterns appear to reflect these processes—and perhaps others—in combination.
Because brain gene expression data cannot easily be collected in adult humans, the twin method provides a valuable alternate approach to studying genetic influences. The information gleaned may lead to more optimal phenotypes for imaging genetic or other genetic association studies. Traditional ROI-based phenotypes may be considered analogous to candidate gene studies, in that they are typically hypothesis-driven. By contrast, our seed-point approach is more similar to the genome-wide association study in that no pre-conceived notion of what constitutes a homogenous region is required and results may emerge that would not be expected a priori. Genetic correlation maps also complement these approaches by providing information about the aggregate patterns of genetic influences on brain development. The three genetic correlation maps in this article serve as heuristic examples, and are not intended to represent an exhaustive assessment of the patterns of genetic influences on cortical thickness. Nevertheless, this approach appears to be promising. It may generate novel parcellation schemes with novel phenotypes that may be better suited than more traditional brain structure phenotypes for use in imaging genetic studies of psychiatric and neurological disorders, or normal and pathological cognitive and brain aging.
Supplementary Material
Acknowledgments
Funded by NIA AG022381, AG018386, AG018384, AG022982. The U.S. Department of Veterans Affairs has provided financial support for the development and maintenance of the Vietnam Era Twin (VET) Registry. Numerous organizations have provided invaluable assistance in the conduct of this study, including: Department of Defense; National Personnel Records Center, National Archives and Records Administration; Internal Revenue Service; National Opinion Research Center; National Research Council, National Academy of Sciences; the Institute for Survey Research, Temple University. Most importantly, the authors gratefully acknowledge the continued cooperation and participation of the members of the VET Registry and their families. Without their contribution this research would not have been possible.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Peper JS, Brouwer RM, Boomsma DI, Kahn RS, Hulshoff Pol HE. Genetic influences on human brain structure: A review of brain imaging studies in twins. Hum Brain Mapp. 2007;28:464–473. doi: 10.1002/hbm.20398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Schmitt JE, Eyler LT, Giedd JN, Kremen WS, Kendler KS, Neale MC. Review of twin and family studies on neuroanatomic phenotypes and typical neurodevelopment. Twin Research and Human Genetics. 2007;10:683–694. doi: 10.1375/twin.10.5.683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Neale MC, Cardon LR. Methodology for genetic studies of twins and families. Dordrecht, The Netherlands: Kluwer Academic Publishers; 1992. [Google Scholar]
- 4.Jernigan TL, Archibald SL, Fennema-Notestine C, Gamst AC, Stout JC, Bonner J, et al. Effects of age on tissues and regions of the cerebrum and cerebellum. Neurobiol Aging. 2001;22:581–594. doi: 10.1016/s0197-4580(01)00217-2. [DOI] [PubMed] [Google Scholar]
- 5.Raz N, Rodrigue KM. Differential aging of the brain: Patterns, cognitive correlates and modifiers. Neurosci Biobehav Rev. 2006;30 doi: 10.1016/j.neubiorev.2006.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Buckner RL. Memory and executive function in aging and AD: Multiple factors that cause decline and reserve factors that compensate. Neuron. 2004;44:195–208. doi: 10.1016/j.neuron.2004.09.006. [DOI] [PubMed] [Google Scholar]
- 7.Schmitt JE, Lenroot RK, Wallace GL, Ordaz S, Taylor KN, Kabani N, et al. Identification of genetically mediated cortical networks: A multivariate study of pediatric twins and siblings. Cereb Cortex. 2008 doi: 10.1093/cercor/bhm211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wright IC, Sham P, Murray RM, Weinberger DR, Bullmore ET. Genetic contributions to regional variability in human brain structure: Methods and preliminary results. Neuroimage. 2002;17:256–271. doi: 10.1006/nimg.2002.1163. [DOI] [PubMed] [Google Scholar]
- 9.Thompson PM, Cannon TD, Narr KL, van Erp T, Poutanen VP, Huttunen M, et al. Genetic influences on brain structure. Nat Neurosci. 2001;4:1253–1258. doi: 10.1038/nn758. [DOI] [PubMed] [Google Scholar]
- 10.Dale AM, Fischl B, Sereno MI. Cortical surface-based analysis. I: Segmentation and surface reconstruction. Neuroimage. 1999;9:179–194. doi: 10.1006/nimg.1998.0395. [DOI] [PubMed] [Google Scholar]
- 11.Rosas HD, Liu AK, Hersch S, Glessner M, Ferrante RJ, Salat DH, et al. Regional and progressive thinning of the cortical ribbon in Huntington's disease. Neurology. 2002;58:695–701. doi: 10.1212/wnl.58.5.695. [DOI] [PubMed] [Google Scholar]
- 12.Panizzon MS, Fennema-Notestine C, Eyler LT, Jernigan TL, Prom-Wormley E, Neale M, et al. Distinct genetic influences on cortical surface area and cortical thickness. Cereb Cortex. 2009 doi: 10.1093/cercor/bhp026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kremen WS, Thompson-Brenner H, Leung YJ, Grant MD, Franz CE, Eisen SA, et al. Genes, environment, and time: The Vietnam Era Twin Study of Aging (VETSA) Twin Research and Human Genetics. 2006;9:1009–1022. doi: 10.1375/183242706779462750. [DOI] [PubMed] [Google Scholar]
- 14.Centers for Disease Control and Prevention Health data for all ages. National Center for Health Statistics; [Google Scholar]
- 15.National Health and Nutrition Examination Survey (NHANES III) (1999–2004): Trends in health and aging. National Center for Health Statistics; [Google Scholar]
- 16.Dale AM, Sereno MI. Improved localization of cortical activity by combining EEG and MEG with MRI cortical surface reconstruction: A linear approach. J Cogn Neurosci. 1993;5:162–176. doi: 10.1162/jocn.1993.5.2.162. [DOI] [PubMed] [Google Scholar]
- 17.Fischl B, Sereno MI, Dale AM. Cortical surface-based analysis. II: Inflation, flattening, and a surface-based coordinate system. Neuroimage. 1999;9:195–207. doi: 10.1006/nimg.1998.0396. [DOI] [PubMed] [Google Scholar]
- 18.Fischl B, Dale AM. Measuring the thickness of the human cerebral cortex from magnetic resonance images. Proceedings of the National Academy of Sciences. 2000;97:11050–11055. doi: 10.1073/pnas.200033797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Walhovd KB, Fjell AM, Reinvang I, Lundervold A, Dale AM, Eilertsen DE, et al. Effects of age on volumes of cortex, white matter and subcortical structures. Neurobiol Aging. 2005;26:1261–1270. doi: 10.1016/j.neurobiolaging.2005.05.020. discussion 1275–1278. [DOI] [PubMed] [Google Scholar]
- 20.Dickerson BC, Fenstermacher E, Salat DH, Wolk DA, Maguire RP, Desikan R, et al. Detection of cortical thickness correlates of cognitive performance: Reliability across MRI scan sessions, scanners, and field strengths. Neuroimage. 2008;39:10–18. doi: 10.1016/j.neuroimage.2007.08.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Han X, Jovicich J, Salat D, van der Kouwe A, Quinn B, Czanner S, et al. Reliability of MRI-derived measurements of human cerebral cortical thickness: The effects of field strength, scanner upgrade and manufacturer. Neuroimage. 2006;32:180–194. doi: 10.1016/j.neuroimage.2006.02.051. [DOI] [PubMed] [Google Scholar]
- 22.Jovicich J, Czanner S, Greve D, Haley E, van der Kouwe A, Gollub R, et al. Reliability in multi-site structural MRI studies: Effects of gradient non-linearity correction on phantom and human data. Neuroimage. 2006;30:436–443. doi: 10.1016/j.neuroimage.2005.09.046. [DOI] [PubMed] [Google Scholar]
- 23.Fennema-Notestine C, Gamst AC, Quinn BT, Pacheco J, Jernigan TL, Thal L, et al. Feasibility of multi-site clinical structural neuroimaging studies of aging using legacy data. Neuroinformatics. 2007;5:235–245. doi: 10.1007/s12021-007-9003-9. [DOI] [PubMed] [Google Scholar]
- 24.Jovicich J, Czanner S, Han X, Salat D, van der Kouwe A, Quinn B, et al. MRI-derived measurements of human subcortical, ventricular and intracranial brain volumes: Reliability effects of scan sessions, acquisition sequences, data analyses, scanner upgrade, scanner vendors and field strengths. Neuroimage. 2009 doi: 10.1016/j.neuroimage.2009.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Eaves LJ, Last KA, Young PA, Martin NG. Model-fitting approaches to the analysis of human behavior. Heredity. 1978;41:249–320. doi: 10.1038/hdy.1978.101. [DOI] [PubMed] [Google Scholar]
- 26.Visscher PM. Power of the classical twin design revisited. Twin Res. 2004;7:505–512. doi: 10.1375/1369052042335250. [DOI] [PubMed] [Google Scholar]
- 27.Neale MC, Boker SM, Xie G, Maes HH. Mx: Statistical Modeling. 6th ed. Richmond, VA: Department of Psychiatry, Medical College of Virginia; 2003. [Google Scholar]
- 28.Nobre AC, Allison T, McCarthy G. Word recognition in the human inferior temporal lobe. Nature. 1994;372:260–263. doi: 10.1038/372260a0. [DOI] [PubMed] [Google Scholar]
- 29.Price C, Thierry G, Griffiths T. Speech-specific auditory processing: Where is it? Trends in Cognitive Science. 2005;9:271–276. doi: 10.1016/j.tics.2005.03.009. [DOI] [PubMed] [Google Scholar]
- 30.Fischl B, Sereno MI, Tootell RBH, Dale AM. High-resolution inter-subject averaging and a coordinate system for the cortical surface. Hum Brain Mapp. 1999;8:272–284. doi: 10.1002/(SICI)1097-0193(1999)8:4<272::AID-HBM10>3.0.CO;2-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sereno MI, Dale AM, Reppas JB, Kwong KK, Belliveau JW, Brady TJ, et al. Borders of multiple visual areas in humans revealed by functional magnetic resonance imaging. Science. 1995;268:889–893. doi: 10.1126/science.7754376. [DOI] [PubMed] [Google Scholar]
- 32.Van Essen DC. Towards a quantitative, probabilistic neuroanatomy of cerebral cortex. Cortex. 2004;40:211–212. doi: 10.1016/s0010-9452(08)70954-7. [DOI] [PubMed] [Google Scholar]
- 33.Desikan RS, Segonne F, Fischl B, Quinn BT, Dickerson BC, Blacker D, et al. An automated labeling system for subdividing the human cerebral cortex on MRI scans into gyral based regions of interest. Neuroimage. 2006;31:968–980. doi: 10.1016/j.neuroimage.2006.01.021. [DOI] [PubMed] [Google Scholar]
- 34.O'Leary DD, Chou SJ, Sahara S. Area patterning of the mammalian cortex. Neuron. 2007;56:252–269. doi: 10.1016/j.neuron.2007.10.010. [DOI] [PubMed] [Google Scholar]
- 35.Kudo LC, Karsten SL, Chen J, Levitt P, Geschwind DH. Genetic analysis of anterior posterior expression gradients in the developing mammalian forebrain. Cereb Cortex. 2007;17:2108–2122. doi: 10.1093/cercor/bhl118. [DOI] [PubMed] [Google Scholar]
- 36.Makris N, Kennedy DN, McInerney S, Sorensen AG, Wang R, Caviness VS, Jr., et al. Segmentation of subcomponents within the superior longitudinal fascicle in humans: A quantitative, in vivo, DT-MRI study. Cereb Cortex. 2005;15:854–869. doi: 10.1093/cercor/bhh186. [DOI] [PubMed] [Google Scholar]
- 37.Ebeling U, von Cramon D. Topography of the uncinate fascicle and adjacent temporal fiber tracts. Acta Neurochirurgica (Wien) 1992;115:143–148. doi: 10.1007/BF01406373. [DOI] [PubMed] [Google Scholar]
- 38.Fuster JM. The prefrontal cortex: Anatomy, physiology, and neuropsychology of the frontal lobe. 2 ed. New York: Raven Press; 1989. [Google Scholar]
- 39.Cabeza R, Nyberg L. Imaging cognition II: An empirical review of 275 PET and fMRI studies. J Cogn Neurosci. 2000;12:1–47. doi: 10.1162/08989290051137585. [DOI] [PubMed] [Google Scholar]
- 40.Yakovlev PI, Lecours AR. The myelogenetic cycles of regional maturation of the brain. In: Mankowski A, editor. Regional development of the brain in early life. Philadelphia: Davis; 1967. pp. 3–69. [Google Scholar]
- 41.Barkovich AJ, Kjos BO, Jackson DE, Jr., Norman D. Normal maturation of the neonatal and infant brain: MR imaging at 1.5 T. Radiology. 1988;166:173–180. doi: 10.1148/radiology.166.1.3336675. [DOI] [PubMed] [Google Scholar]
- 42.Lerch JP, Worsley K, Shaw WP, Greenstein DK, Lenroot RK, Giedd J, et al. Mapping anatomical correlations across cerebral cortex (MACACC) using cortical thickness from MRI. Neuroimage. 2006;31:993–1003. doi: 10.1016/j.neuroimage.2006.01.042. [DOI] [PubMed] [Google Scholar]
- 43.Lenroot RK, Schmitt JE, Ordaz SJ, Wallace GL, Neale MC, Lerch JP, et al. Differences in genetic and environmental influences on the human cerebral cortex associated with development during childhood and adolescence. Hum Brain Mapp. 2007 doi: 10.1002/hbm.20494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kessler RC, Sonnega A, Bromet E, Hughes M, Nelson CB. Posttraumatic stress disorder in the National Comorbidity Survey. Arch Gen Psychiatry. 1995;52:1048–1060. doi: 10.1001/archpsyc.1995.03950240066012. [DOI] [PubMed] [Google Scholar]
- 45.Gilbertson MW, Shenton ME, Ciszewski A, Kasai K, Lasko NB, Orr SP, et al. Smaller hippocampal volume predicts pathological vulnerability to psychological trauma. Nat Neurosci. 2002;5:1242–1247. doi: 10.1038/nn958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kremen WS, Koenen KC, Boake C, Purcell S, Eisen SA, Franz CE, et al. Pretrauma cognitive ability and risk for posttraumatic stress disorder: a twin study. Arch Gen Psychiatry. 2007;64:361–368. doi: 10.1001/archpsyc.64.3.361. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.