Abstract
The transcription factor cAMP-response element binding protein (CREB) mediates the mechanical strain-induced gene expression in the heart. This study investigated which signaling pathways are involved in the straininduced CREB activation using cultured ventricular fibroblasts from adult rat hearts. CREB phosphorylation was analyzed by immunocytochemistry and ELISA. Cyclic mechanical strain (1 Hz and 5% elongation) for 15 min induced CREB phosphorylation in all CREB-positive fibroblasts. Several signaling transduction pathways can contribute to strain-induced CREB activation. The inhibition of PKA, PKC, MEK, p38-MAPK or PI3-kinase partially reduced the strain-induced CREB phosphorylation. Activation of PKA by forskolin or PKC by PMA resulted in a level of CREB phosphorylation comparable to the reduced level of the strain-induced CREB phosphorylation in the presence of PKA or PKC inhibitors. Signaling pathways involving PKC, MEK, p38-MAPK or PI3-kinase seem to converge during strain-induced CREB activation. PKA interacted additively with the investigated signaling pathways. The strain-induced c-Fos expression can be reduced by PKC inhibition but not by PKA inhibition. Our results suggest that the complete strain-induced CREB phosphorylation involves several signaling pathways that have a synergistic effect. The influence on gene expression is dependent on the level and the time of CREB stimulation. These wide-ranging possibilities of CREB activation provide a graduated control system.
Keywords: strain, mechanotransduction, cAMP response element binding protein, c-Fos, fibroblasts.
Introduction
Cardiac ventricular fibroblasts represent the majority of cell numbers in the heart and are subjected to permanent mechanical changes in length and tension that affect cell proliferation, the deposition of the extracellular matrix proteins, and the release of growth and other factors, e.g. cytokines.1,2 Fibroblasts sense mechanical forces via multiple signaling pathways. The mechanotransduction can be classified as initial site with the sensors which are the integrins, the stretch-activated ion channels and the cytoskeleton.3,4 The integrin-mediated transfer of the extracellular matrix movement caused by mechanical strain activates the focal adhesion kinases leading to stimulation of signal cascades.5,6 The stimulation of the stretch-activated ion channels by strain changes the intracellular concentration of Ca2+, Na+ and K+.7 Secondary events include the activation and phosphorylation of membrane associated enzymes.8 Mechanical strain can activate the receptor tyrosine kinases (RTK) leading to the stimulation of mitogenactivated protein kinases and stress-activated protein kinases. Membrane bound enzymes such as G-protein coupled receptors can be activated by mechanical strain leading to an increased level of second messengers, including cAMP level.9 Mechanical stress can cause an induction of reactive oxygen species activating the stress-sensitive p38-MAPK pathway.10,11 The tertiary events include the stimulation of transcription factors regulating gene expression.
Mechanical strain can be considered an extracellular stimulus triggering one of the best characterized stimulus-induced transcription factors, the cAMP response element (CRE)-binding protein (CREB).3 CREB is activated by different signaling pathways leading finally to phosphorylation of a particular protein residue, serine 133 (Ser133). The CREB phosphorylation is required for the CRE-mediated gene expression.12–14 CRE-regions are present in various genes which are important for the cardiovascular system.15 Thus, it is conceivable that changes in the CRE-mediated gene expression contribute to a change in expression of the regulatory proteins.
A previous report presented a differential expression of the CREB family members in the different cell types of the heart.16 It was found that CREB is only expressed in fibroblasts whereas the cAMP response element modulator (CREM) is only expressed in ventricular myocytes. For this reason, investigations into CREB activation could be realized with cardiac fibroblasts as the main cardiac cells of the CREB expression. The aim of this study was to investigate which signaling pathways are responsible for CREB activation by mechanical strain using ventricular fibroblasts of adult rat hearts. We found that several strain-activated signaling pathways contributed to the CREB phosphorylation. The inhibition of several kinases, including PKA, PKC, ERK, p38, Raf-1 kinase, PI3-kinase, MEK, reduced the straininduced CREB phosphorylation. This reduction of the strain-induced CREB phosphorylation was increased by inhibition of two different cascades. These multiple signaling pathways ensure CREB activation during mechanical strain.
Materials and Methods
Animals used in this study were maintained in accordance with the “Guide for the Care and Use of Laboratory Animals” published by the US National Institutes of Health (NIH Publication no. 85–23, revised 1996).
Cell culture and stretch
Fibroblasts were isolated from both ventricles of adult male rat (body weight 292±14 g; n=20) hearts (heart weight 1.14±0.11 g) by means of retrograde perfusion of collagenasecontaining solutions. Details have been previously reported.17 After the perfusion, the cell suspension was centrifuged at 700 rpm for 5 min at room temperature. The cell pellet was resuspended in Dulbecco’s modified Eagle medium (DMEM)/medium 199 (Earle’s salts) at a ratio of 4:1 and 10% fetal calf serum (FCS, Sigma) containing 1% penicillin/streptomycin (Sigma) and 10 µg/mL amikacin (Sigma). Cells were grown to confluency and then passaged once to culture on the Bioflex culture plates coated with collagen I within 24 hrs. Inhibitors were added three hours after the change of the medium. The inhibitors used were 3 µM H89, 1 µM RO-31-8220, 5 µM chelerythrine chloride, 10 µM LY 2940002, 20 µM KN-93, 5 µM SU 6656, 10 µM Raf1 kinase inhibitor I, 10 µM UO 126, 50 µM PD 98059, 2 µM SB 203580 (Calbiochem). For the stretch experiments, the Bioflex culture plates were put in the gasket of the Flexercell Strain Unit (Flexercell; McKeesport, PA, USA) of a tissue incubator (5% CO2, 37°C) for one hour as incubation before the strain. The Flexercell computer system connected the unit with a vacuum pump and controlled the stretch parameters.18 The cells were stretched at an elongation of 5%, frequency of 1 Hz and duration of 15 min. The control groups were handled in the same way but without cyclic deformation.
Immunocytochemistry
After stimulation with strain, 10 µM forskolin (Calbiochem) or 500 nM phorbol-12-myristate-13-acetate (PMA, Calbiochem), cells were fixed in 4% paraformaldehyde for one hour at room temperature. They were then washed three times in 50 mM Tris-HCl, pH 7.4, 150 mM NaCl (TBS) and 0.1% Triton X-100 (TBST) and incubated for one hour in TBST added 2% wt/vol bovine albumin (BSA). Then, cells were incubated with the primary antibodies against CREB (#9192, Cell Signaling) or phospho-CREB (#9191, Cell Signaling) in a dilution of 1:200 overnight at 4°C. After washing, cells were incubated in secondary antibody goat anti-rabbit conjugated with alkaline phosphatase (Sigma) in a dilution 1:50 for four hours. BCIP/NBT (Sigma FastTM) was used as substrate for the alkaline phosphatase. After detection, cells were rinsed and stored in aqua dest. The numbers of CREB as well as phospho-CREB positive nuclei were counted relative to the whole number of cells by ×200 magnification over an area of 0.25 mm2 with the light microscope.
Western blot analysis of c-Fos
For c-Fos analysis, fibroblasts were incubated without inhibitors as control or with 3 µM H89 or 5 µM chelerythrine for 30 min and cyclically mechanically strained (1 Hz, 5% elongation) for one hour. After the strain, the cells were washed with ice-cold PBS and solubilized with sample buffer consisting of 20 mM tris(hydroxymethyl)-aminomethane (TRIS)-HCl, pH 7.5, 150 mM NaCl, 1 mM EDTA, 1 mM EGTA, 1% Triton X-100, 1% vol/vol protease inhibitor cocktail (P8340, Sigma), 1% vol/vol phosphatase inhibitor cocktail I (P2850, Sigma), and 1% vol/vol phosphatase inhibitor cocktail II (P5726, Sigma). The protein content was analyzed with BIORAD protein assay. Proteins (10 µg) were run together on a 10% SDS-polyacrylamide gel. After transfer to nitrocellulose membrane, the membrane was strained with Ponceau Red to control for equal transfer of protein. Membranes were treated with 5% Blotto (5% wt/vol non-fat dry milk in TBS (20 mM Tris-HCl, pH 7.4, 150 mM NaCl) with 0.1% Tween 20 for two hours at room temperature. After washing, the blots were incubated for one hour with a mouse antibody against c-Fos (OP17, Oncogene) in a concentration of 2.5 µg/mL or a mouse antibody against β-actin (A5441, Sigma) in a dilution of 1:1000. Horseradish peroxidase (HRP) conjugated anti-mouse IgG were used as secondary antibody in a dilution of 1:1000. The blots were visualized by using an enhanced chemiluminescence detection system (ECL, Amersham).
Preparation of nuclear extract and analysis of phospho-CREB amount
The ActiveMotif Kit for nuclear extract preparation was used. Fibroblasts were cyclically mechanically strained (1 Hz, 5% elongation) for 15 min without or with inhibitors. After the strain, the cells were washed with ice-cold PBS, scraped off the Bioflex plates and centrifuged at 500 g for 5 min at 4°C. The pellets were lysed according to the instruction manual in a hypotonic puffer. The nuclear fractions were centrifuged at 14,000 g for 10 min at 4°C. Protein concentration was analyzed by BIORAD protein assay.
The amount of phospho-CREB was quantified by using a commercially available ELISA combined with a sensitive and specific assay for transcription factor CREB Trans AM™ pCREB from ActiveMotif. This Kit contains a 96-well plate on which an oligonucleotide with CRE (5′-TGACGTCA-3′)-sequence was immobilized. Samples of 2 µg nuclear fractions were handled according to the instruction manual and the optical density of the samples was measured at 450 nm. The amount of phospho-CREB was related to cms-induced phospho-CREB amount.
Statistical analysis
Data are expressed as mean ±SD. The differences were assessed by one-way analysis of variance combined with the Bonferroni test. A value of P<0.05 was considered to be statistically significant.
Results
cAMP response element binding protein activation by cms
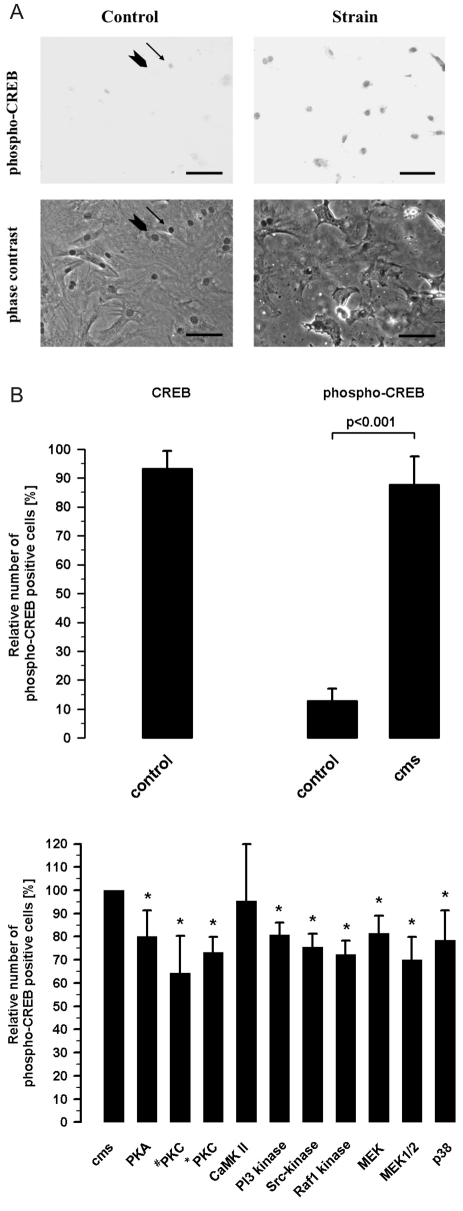
The cultured fibroblasts of adult rat hearts were CREB-positive cells (93.2±6.1%; n=22). In these cultures, a small fraction of 12.9±4.2% were phospho-CREB positive cells (n=10), shown in Figure 1B. Cyclic mechanical strain (cms) increased the number of phospho-CREB positive fibroblasts to 87.8±9.6% (n=10). This result can be achieved by the images of phospho-CREB positive nuclei in fibroblasts without strain as control and under the influence of cms in Figure 1A.
Figure 1.
(A) Qualitative analysis of CREB phosphorylation by cyclical mechanical strain (cms, 5% elongation, 1 Hz, 15 min) in cultured fibroblasts from adult rat hearts in comparison to control without strain by immunocytochemistry. An arrow with head indicates a phospho-CREB positive nucleus and an arrow without head a nucleus without phospho-CREB labeling in the images showing cardiac fibroblasts without cms (control). The comparison between the images in the top line control and strain presents a much higher number of phospho-CREB positive nuclei in the strain image. Scale bar: 50 µm. (B) Quantitative analysis of the number of CREB- and phospho-CREB positive nuclei related to the whole number of cultured fibroblasts from adult rat hearts (n=10). Activation of CREB by cyclical mechanical strain (cms, 5% elongation, 1 Hz, 15 min) analyzed by immunocytochemistry (n=10).
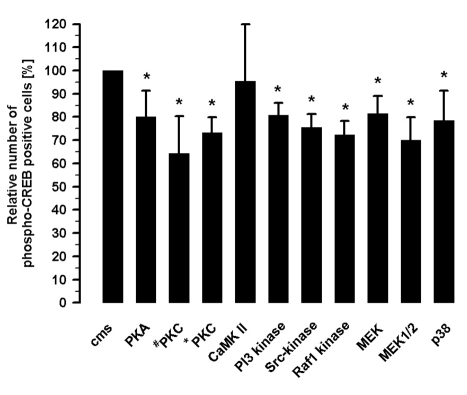
Differential inhibition of CREB phosphorylation suggested the presence of several signaling cascades induced by cyclic mechanical strain (Figure 2). The inhibition of PKA by 3 µM H89 reduced the number of phospho-CREB positive cells to 80.1±11.2% (n=8). The effect of the PKC was investigated with two different inhibitors, 1 µM RO-31-8220 and 5 µM chelerythrine. Addition of RO-31-8220 resulted in 64.3±15.9% and of chelerythrine in 73.3±6.5% phospho-CREB positive cells (n=8) after cms. The number of phospho-CREB positive cells was unchanged by inhibition of CaMK II (20 µM KN-93). Cms can activate tyrosine kinases leading to the CREB phosphorylation. One pathway including the PI3-kinase, inhibiting by 10 µM LY 294002, reduced the number of phospho-CREB positive cells to 80.7±5.4% (n=8). The Src-inhibition by 5 µM SU 6656 resulted in 75.6±5.8% (n=8) phospho-CREB positive cells. A decrease in the phospho-CREB positive cells to 72.3±5.8% (n=8) by Raf1 kinase inhibition (10 µM Raf1-kinaseinhibitor) was registered after cms. The inhibitor PD 98059 (50 µM) which blocked the phosphorylation of MEK resulted in 81.6±7.3% (n=8) phospho-CREB positive cells. The noncompetitive inhibition of MEK (10 µM UO126) revealed 70.1±9.9% phospho-CREB positive cells after cms. The inhibition of p38 MAPK (2 µM SB 203580) caused 78.5±12.8% (n=8) phospho-CREB positive cells under the influence of cms.
Figure 2.
Inhibition of the strain-induced CREB phosphorylation by 3 µM H89 (PKA-inhibition), 1 µM RO-31-8220 (#PKC-inhibition), 5 µM chelerythrine (*PKC-inhibition), 20 µM KN-93 (CaMK II inhibition), 10 µM LY 294002 (PI3-kinase inhibition), 5 µM Su 6656 (Src-kinase inhibition), 10 µM Raf1-kinase inhibitor, 50 µM PD 98059 (MEK- inhibition), 10 µM UO126 (MEK1/2- inhibition), 2 µM SB 203580 (p38-MAPK inhibition) analyzed after 15 min cms by immunocytochemistry (n=8); Mean±SD, *P<0.01 vs.cms.
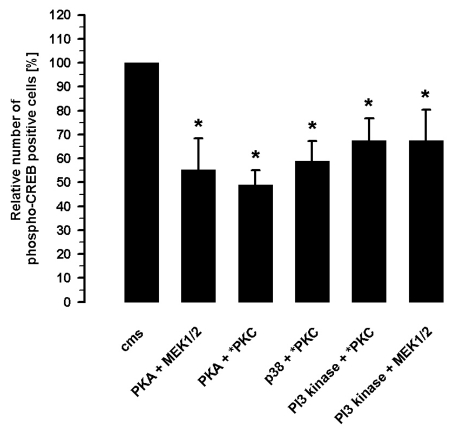
The inhibition of the strain-induced CREB phosphorylation in the presence of two different inhibitors revealed synergistic effects (Figure 3). The inhibition of PKA by 3 µM H89 and MEK by 10 µM UO126 resulted in a reduced number of strain-induced phospho-CREB positive cells to 55.2±13.2% (n=7) presenting an additive effect. Another additive effect on the strain-induced CREB phosphorylation was revealed by the inhibition of PKA (3 µM H89) and PKC (5 µM chelerythrine) which caused 48.9±6.2% (n=7) phospho-CREB positive cells. The inhibition of p38 MAPK (2 µM SB 203580) and PKC (5 µM chelerythrine) showed a subadditive effect on the cms-mediated CREB phosphorylation and reduced the number of phospho-CREB positive cells to 59.0±8.2% (n=7). Another subadditive effect was registered during PKC (5 µM chelerythrine) and PI3-kinase (10 µM LY 294002) inhibition during cms which reduced the number of phospho-CREB positive cells to 67.5±9.2% (n=7). A comparable subadditive effect caused the inhibition of PI3-kinase (10 µM LY 294002) and MEK1/2 (10 µM UO126) during cms resulting in 67.4±12.9% (n=7) phospho-CREB positive cells.
Figure 3.
Inhibition of the strain-induced CREB phosphorylation by the combinations of 3 µM H89 and 10 µM UO126 (PKA and MEK1/2 inhibition) or 3 µM H89 and 5 µM chelerythrine (PKA and PKC inhibition) or 2 µM SB 203580 and 5 µM chelerythrine (p38-MAPK and PKC inhibition) or 10 µM LY 294002 and 5 µM chelerythrine (PI3-kinase and PKC inhibition) or 10 µM LY 294002 and 10 µM UO126 (PI3-kinase and MEK1/2 kinase inhibition) analyzed after 15 min cms by immunocytochemietry (n=7); Mean±SD, *P<0.01 vs. cms.
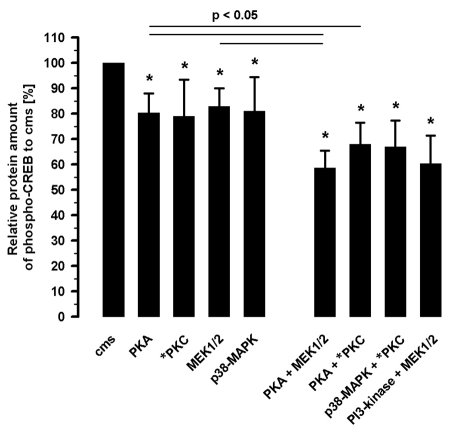
The quantitative analysis of phospho-CREB protein amount in the nuclear fractions showed the same tendency (Figure 4). The extent of the CREB phosphorylation related to cms-induced phospho-CREB amount was reduced by PKA inhibition (3 µM H89) to 80.3±7.7% (n=5), by PKC inhibition (5 µM chelerythrine) to 78.9±14.4% (n=5), by noncompetitve inhibition of MEK (10 µM UO126) to 83.0±7.0% (n=5) and by p38 MAPK (2 µM SB 203589) to 81.1±13.4% (n=5). The inhibition of PKA (3 µM H89) and MEK1/2 (10 µM UO126) reduced the amount of CREB phosphorylation to 58.7±6.7% (n=5) in an additive manner. The PKA (3 µM H89) and PKC (5 µM chelerythrine) inhibition resulted in a reduced amount of phospho-CREB to 68.0±8.3% (n=5). The combination of PKC (5 µM chelerythrine) and p38 MAPK (2 µM SB 203580) inhibition decreased the amount of phospho-CREB to 67.0±10.4% (n=5) a subadditive effect. The inhibition of MEK1/2 (10 µM UO126) and PI3-kinase (10 µM LY 294002) reduced the phospho-CREB amount to 60.4±11.0% (n=5), also a subadditive effect.
Figure 4.
Analysis of straininduced phospho-CREB protein amount inhibited by 3 µM H89 (PKA-inhibition), 5 µM chelerythrine (PKC-inhibition), 10 µM UO126 (MEK1/2- inhibition) and 2 µM SB 203580 (p38-MAPK inhibition), as well as the combination of 3 µM H89 and 10 µM UO126 (PKA and MEK1/2 inhibition) or 3 µM H89 and 5 µM chelerythrine (PKA and PKC inhibition) or 2 µM SB 203580 and 5 µM chelerythrine (p38-MAPK and PKC inhibition) or 10 µM LY 294002 and 10 µM UO126 (PI3-kinase and MEK1/2 kinase inhibition) by ELISA; n=5; Mean±SD, *P<0.05 vs. cms.
cAMP response element binding protein activation by PKA and PKC stimulation
The participation of PKA and PKC in CREB activation was comparable to the parts which the inhibition of PKA or PKC presented by the strain-induced CREB phosphorylation.
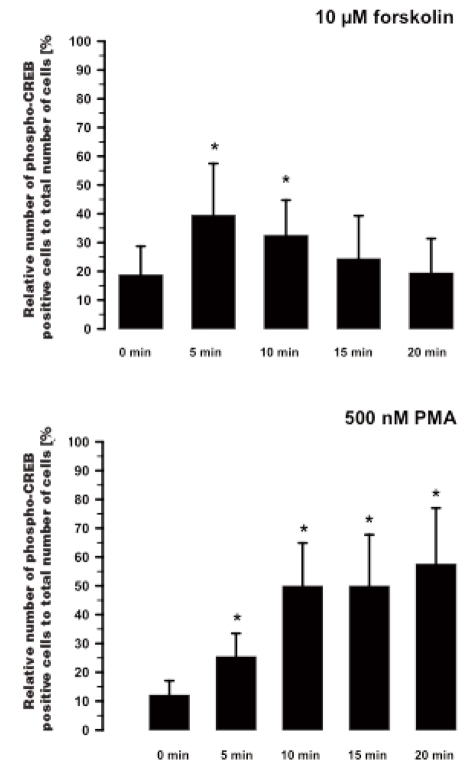
The effect of 10 µM forskolin on the CREB phosphorylation was investigated after four different times (Figure 5). The number of phospho-CREB positive cells was already increased from 18.5±6.3% to 42.9±15.9% (n=8) after 5 min. This enhanced level of phospho-CREB positive cells was still 35.7±8.2% after 10 min. However after 15 min, no forskolin-induced CREB activation was found. Hence, it appears that CREB activation by forskolin is a transient effect.
Figure 5.
Time-dependent CREB activation by 10 µM forskolin (top, n=8) and 500 nM phorbol-12-myristate-13-acetate (PMA, bottom, n=7), analyzed the number of phospho-CREB positive fibroblasts by immunocytochemistry; Mean±SD, *P<0.01 vs. 0 min.
In contrast to the forskolin stimulation, the CREB phosphorylation induced by 500 nM PMA is a sustained effect over time from 10 min to 20 min (Figure 5). The number of phospho-CREB positive cells increased from 12.1±5.0% to 27.6±5.4% after 5 min and to 54.3±10.4% after 10 min (n=6). This enhanced level of phospho-CREB positive cells remained at 53.0±16.8% after 15 min and at 62.8±13.8% after 20 min.
Expression of c-Fos by cms
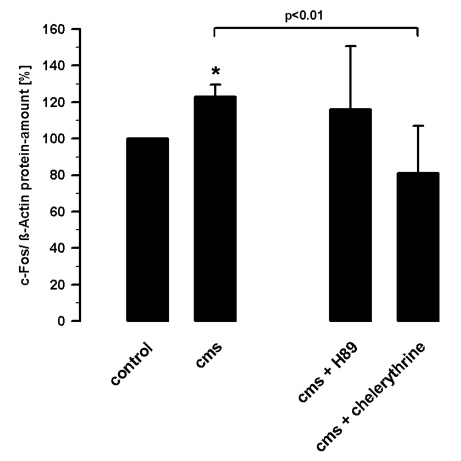
The expression of c-Fos is regulated by CREB. Cms increased the c-Fos protein level by 23±7% (n=4). The effect of cms on the quantity of c-Fos protein was reduced by 42±19% (n=4) in the presence of PKC inhibitor chelerythrine (5 µM). The PKA inhibition with H89 (3 µM) showed no effect on the strain-induced c-Fos expression. The c-Fos expression was measured relative to β-actin by densitometric analysis of Western blots (Figure 6).
Figure 6.
Expression of c-Fos protein by cyclic mechanical strain (5% elongation, 1 Hz, one hour) and in the presence of 3 µM H89 or 5 µM chelerythrine in cultured fibroblasts from adult rat hearts related to β-actin protein analyzed by Western blots (n=4); Mean±SD, * P<0.01 vs. control without strain.
Discussion
This is the first study which investigates in detail the signal transduction from the physical mechanical stimulus of the activation of the transcription factor CREB to the gene expression in cardiac fibroblasts of adult rats. These cultured fibroblasts were identified as pure culture of fibroblasts and were not shown to be myofibroblasts.19 The number of α-smooth muscle actin-positive cells resulted in less than 10% of α-smooth muscle actin-positive fibroblasts. The adult ventricular fibroblasts were also chosen as in vitro model which exclusively expresses CREB as sole member of the CREB family in one of the main cardiac cells.16
The ventricular fibroblasts are subjected to permanent environmental movements in the heart influencing cellular processes and gene expression.3,20 The stimulus of mechanical strain caused CREB phosphorylation in all nuclei of the cultured cardiac fibroblasts. The comparison between the pathway leading to the CREB phosphorylation and the cellular and molecular response to mechanical stress revealed an extensive correspondence of both. The known signaling pathways leading to CREB activation include the cAMP-dependent pathway leading to the PKA stimulation or the activation of receptor tyrosine kinases involved MEK and ERK-1/2.12,13,21 Another signaling pathway activated by tyrosine kinases is the PI3-kinase/Akt pathway mediating the CREB activation.22 Alternative signal cascades involving the p38 and the MAKPAP-2 kinases that are targets of fibroblast growth factor (FGF)- and tumor necrosis factor (TNF)- stimulated pathways result in CREB phosphorylation.23 Thus, the stimulus of mechanical strain can be transformed to activate CREB by multiple signaling pathways.
The inhibition of an individual enzyme of different cascades showed only a partial reduction in the strain-induced CREB phosphorylation. The PKC inhibition contributed the largest share to the strain-induced CREB phosphorylation. This high share could be attributed to interference in the regulation of the MAPK cascade. Different participation of pathways by the CREB activation was recently reported during ischemic pre-conditioning completely involving the p38MAPK pathway and partially involving the ERK1/2 pathway.24
The partial participation of signaling pathways in the CREB activation could be confirmed by the activation of one signaling pathway. The stimulation of cAMP synthesis by forskolin increased the number of phospho-CREB positive cells by approximately 20% and this was not a complete CREB activation. The PKC stimulation caused an enhanced number of phospho-CREB positive cells by approximately 40% and this was again not a complete CREB activation. These results also showed that the participation of PKC to the CREB activation is higher than that of the PKA. The proportion of these different pathways to the CREB phosphorylation is comparable with the different shares of the same pathways by the inhibition of the strain-induced CREB activation.
The expression of c-Fos is regulated by the CRE-region.8,12,13 The strain-induced c-Fos protein level was only reduced by PKC inhibition but not by PKA inhibition. The different participation of the signaling pathways to the straininduced CREB activation is comparable to the effect on the strain-induced c-Fos protein level.
The interaction between different signal transduction pathways was suggested by the inhibition of two individual enzymes of different cascades. The investigated signal cascades had synergistic effects on the CREB activation. Signaling pathways involving PKC, MEK, p38-MAPK or PI3-kinase seem to converge during the strain-induced CREB activation. Additively affecting the CREB activation requires participation of PKA as one of two partners by the strain-induced CREB activation. The reason for the partial participation of several signaling pathways could be to ensure the straininduced CREB activation in any case. Probably, there is a balance between the different signaling pathways to activate the gene expression. It seems dependent on age and environmental conditions such as hypoxia, the signaling pathway of which participates the most in CREB activation.25
CRE-mediated gene expression in the heart has its relevance in the regulation of the proliferation and content of the extracellular matrix (ECM) proteins by the fibroblasts. An influence of an enhanced cAMP level on the collagen synthesis was reported to occur by interaction in CREB with other transcriptional complexes.26 Excessive fibroblast proliferation and increase of ECM proteins induce myocardial stiffness leading to cardiac dysfunction.2 Thus, a changed strain-induced gene expression in the fibroblasts can affect cardiac myocyte function inducing cardiac hypertrophy.8 A role for the transcription factor family CREB/ATF in the physiological and pathological myocardial hypertrophy were reported in cardiac cells.27–29 In summery, several signaling pathways can contribute to strain-induced CREB activation in cardiac fibroblasts. These strain-stimulated signaling pathways seem to converge in part during the CREB activation. An additive interaction of the cascades could be only shown if PKA was involved in the strain-induced CREB activation. The CREB-mediated gene expression seems dependent on the level as well as on the time of the CREB phosphorylation. These wide-ranging possibilities of CREB activation provide a graduated control system and could ensure CREB-mediated gene expression in the heart under several conditions.
References
- 1.Baudino TA, Wayne C, Wayne G, Borg T. Cardiac fibroblasts: friend or foe? Am J Physiol. Heart Circ Physiol. 2006;291:H1015–H1026. doi: 10.1152/ajpheart.00023.2006. [DOI] [PubMed] [Google Scholar]
- 2.Camelliti P, Borg T, Kohl P. Structural and functional characterisation of cardiac fibroblasts. Cardiovasc Res. 2005;65:40–51. doi: 10.1016/j.cardiores.2004.08.020. [DOI] [PubMed] [Google Scholar]
- 3.Bishop J, Lindahl G. Regulation of cardiovascular collagen synthesis by mechanical load. Cardiovasc. Res. 1999;42:27–44. doi: 10.1016/s0008-6363(99)00021-8. [DOI] [PubMed] [Google Scholar]
- 4.MacKenna D, Summerour S, Villarreal F. Role of mechanical factors in modulating cardiac fibroblasts function and extracellular matrix synthesis. Cardiovasc Res. 2000;46:257–63. doi: 10.1016/s0008-6363(00)00030-4. [DOI] [PubMed] [Google Scholar]
- 5.Domingos P, Fonseca P, Nadruz W, Franchini K. Load-induced focal adhesion kinase activation in the myocardium: role of stretch and contractile activity. Am J Physiol Heart Circ Physiol. 2002;282:H556–H564. doi: 10.1152/ajpheart.00534.2001. [DOI] [PubMed] [Google Scholar]
- 6.Ross R, Borg T. Integrins and the myocardium. Circ Res. 2001;88:1112–9. doi: 10.1161/hh1101.091862. [DOI] [PubMed] [Google Scholar]
- 7.Kamkin A, Kiseleva I, Isenberg G. Activation and inactivation of a non-selective cation conductance by local mechanical deformation of acutely isolated cardiac fibroblasts. Cardiovasc Res. 2003;57:793–803. doi: 10.1016/s0008-6363(02)00775-7. [DOI] [PubMed] [Google Scholar]
- 8.Sadoshima J, Izumo S. Mechanical stretch rapidly activates multiple signal transduction pathways in cardiac myocytes: potential involvement of an autocrine/paracrine mechanism. EMBO J. 1993;12:1681–92. doi: 10.1002/j.1460-2075.1993.tb05813.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Meyer C, Alenghat F, Rim P, et al. Mechanical control of cyclic AMP signaling and gene transcription through integrins. Nature Cell Biology. 2000;2:666–8. doi: 10.1038/35023621. [DOI] [PubMed] [Google Scholar]
- 10.Aikawa R, Nagai T, Tanaka M, et al. Reactive oxygen species in mechanical stress-induced cardiac hypertrophy. Biochem. Biophys Res Commun. 2001;289:901–7. doi: 10.1006/bbrc.2001.6068. [DOI] [PubMed] [Google Scholar]
- 11.Pimentel D, Amin J, Xiao L, et al. Reactive oxygen species mediate amplitudedependent hypertrophic and apoptotic responses to mechanical stretch in cardiac myocytes. Circ Res. 2001;89:453–60. doi: 10.1161/hh1701.096615. [DOI] [PubMed] [Google Scholar]
- 12.Lonze Be, Ginty DD. Function and regulation of CREB family transcription factors in the nervous system. Neuron. 2002;35:605–23. doi: 10.1016/s0896-6273(02)00828-0. [DOI] [PubMed] [Google Scholar]
- 13.Mayr B, Montminy M. Transcriptional regulation by the phosphorylation-dependent factor CREB. Mol Cell Biol. 2001;2:599–609. doi: 10.1038/35085068. [DOI] [PubMed] [Google Scholar]
- 14.Shaywitz AJ, Greenberg ME. CREB: a stimulus-induced transcription factor activated by a diverse array of extracellular signals. Annu Rev Biochem. 1999;68:821–61. doi: 10.1146/annurev.biochem.68.1.821. [DOI] [PubMed] [Google Scholar]
- 15.Mueller FU, Neumann J, Schmitz W. Transcriptional regulation by cAMP in the heart. Mol Cell Biochem. 2000;212:11–7. [PubMed] [Google Scholar]
- 16.Husse B, Isenberg G. CREB expression in cardiac fibroblasts and CREB expression in ventricular myocytes. Biochem Biophys Res Commun. 2005;334:1260–5. doi: 10.1016/j.bbrc.2005.06.206. [DOI] [PubMed] [Google Scholar]
- 17.Isenberg G, Klöckner U. Calcium tolerant ventricular myocytes prepared in a KB medium. Pflügers Arch. 1982;395:5–18. doi: 10.1007/BF00584963. [DOI] [PubMed] [Google Scholar]
- 18.Banes A, Link G, Gilbert J, et al. Culturing cells in a mechanically active environment. Am Biotechnol Lab. 1990;8:12–22. [PubMed] [Google Scholar]
- 19.Husse B, Briest W, Homagk L, Isenberg G, Gekle M. Cyclical mechanical stretch modulates expression of collagen I and collagen III by PKC and tyrosine kinase in cardiac fibroblasts. Am J Physiol Regul Comp Physiol. 2007;293:R1898–907. doi: 10.1152/ajpregu.00804.2006. [DOI] [PubMed] [Google Scholar]
- 20.Wang JH, Thampatty BP, Lin JS, Im HJ. Mechanoregulation of gene expression in fibroblasts. Gene. 2007;391:1–15. doi: 10.1016/j.gene.2007.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Du K, Montminy M. CREB is a regulatory target for the protein kinase Akt/PKB. J Biol Chem. 1998;273:32377–9. doi: 10.1074/jbc.273.49.32377. [DOI] [PubMed] [Google Scholar]
- 22.Wiggin G, Soloaga A, Foster J, et al. MSK1 and MSK2 are required for the mitogenand stress-induced phosphorylation of CREB and ATF1 in fibroblasts. Mol Cell Biol. 2002;22:2871–81. doi: 10.1128/MCB.22.8.2871-2881.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tan Y, Rouse J, Zhang A, et al. FGF and stress regulate CREB and ATF-1 via a pathway involving p38 MAP kinase and MAPKAP kinase-2. EMBO J. 996;15:4629–42. [PMC free article] [PubMed] [Google Scholar]
- 24.Nagy N, Shiroto K, Malik G, et al. Ischemic preconditioning involves dual cardio-protective axes with p38MAPK as upstream target. J Mol Cell Cardiol. 2007;42:981–90. doi: 10.1016/j.yjmcc.2007.02.010. [DOI] [PubMed] [Google Scholar]
- 25.Giulio di C, Rapino M, Zingariello M, Antonucci A, Castaldi A. PKCα-mediated CREB activation is oxygen and agedependent in rat myocardial tissue. Histochem Cell Biol. 2007;127:327–33. doi: 10.1007/s00418-006-0245-7. [DOI] [PubMed] [Google Scholar]
- 26.Liu X, Sun S, Hassid A, Ostrom R. cAMP inhibits transforming growth factor-β-stimulated collagen synthesis via inhibition of extracellular signal-regulated kinase 1/2 and Smad signaling in cardiac fibroblasts. Mol Pharmacol. 2006;70:1992–2003. doi: 10.1124/mol.106.028951. [DOI] [PubMed] [Google Scholar]
- 27.Ozgen N, Obreztchikova M, Guo J, et al. Protein kinase D links Gq-coupled receptors to camp response element-binding protein (CREB)-Ser133 phosphorylation in the heart. J Biol Chem. 2008;283:17009–19. doi: 10.1074/jbc.M709851200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Watson P, Reusch J, McCune S, et al. Restoration of CREB function is linked to completion and stabilization of adaptive cardiac hypertrophy in response to exercise. Am J Physiol Heart Circ Physiol. 2007;293:H246–H259. doi: 10.1152/ajpheart.00734.2006. [DOI] [PubMed] [Google Scholar]
- 29.Kehat I, Hasin T, Aronheim A. The role of basic leucine zipper protein-mediated transcription in physiological and pathological myocardial hypertrophy. Ann NY Acad Sci. 2006;1080:97–109. doi: 10.1196/annals.1380.009. [DOI] [PubMed] [Google Scholar]