Abstract
Objective:
By date investigations have indicated the presence of stem cells within the pulp tissue of both temporary and permanent human teeth. In the present study, these stem cells were compared in terms of their growth kinetics and culture requirements.
Materials and Methods:
Stem cells within the pulp of the human third molar (permanent tooth) and the deciduous incisor (temporary tooth) were isolated, culture-expanded and characterized. Then the proliferation potential of the cells was compared using multiple cell growth indices as PDT (Population doubling time), colonogenic activity and growth curve. Furthermore, the cultures of both cells were optimized for maximal proliferation.
Results:
Stem cells of either pulp tissue appeared as fibroblastic cells capable of differentiating into osteoblastic, odontoblastic, adipocytic and chondrocytic cell lineages. In contrast to molar stem cells, those from the incisor tooth expressed neurogenic markers of ßIII Tubulin and Tau. Based on in vitro growth data, the cells from third molar tended to have a lower PDT value (20.79, SD=2.8 versus 25.55, SD=2.9 hours), higher colonogenic activity and better growth curve than those from the deciduous incisor (P<0.05). Both cells exhibited high expansion rate when being plated in a medium with 20% phosphate buffer solution at a density of 100 cells/cm2.
Conclusion:
Given the high proliferation capacity, the stem cells from the human third molar would be an appropriate candidate for use in experimental, preclinical and even clinical setups.
Keywords: Molar, Third; Tooth, Deciduous; Cell Proliferation; Cell Differentiation
INTRODUCTION
By date, multiple dental regions have been reported to contain stem cells. These include the dental pulp of permanent and deciduous teeth [1–2], the periodontal ligament [3], the dental apical papilla [4] and dental follicle [5]. Of dental stem cells, those from the dental pulp are interesting and have drawn relatively considerable attention. The dental pulp stem cells from the permanent third molar were first isolated by Gronthos et al [1] who have described the cells as colonogenic cells capable of producing osteoblastic and chondrocytic cells in vitro and odontoblastic cells in vivo. Following investigations have indicated the broad differentiation capacity of the pulp-derived cells including their ability in giving rise to neural and endothelial cell lineages as well [6–8]. These cells may provide an appropriate source of specialized cells (odontoblast) for pulp regeneration in future regenerative medicine.
Stem cells from deciduous incisor teeth were first investigated by Miura et al [2] who found that an exfoliated human deciduous tooth contains multiopotent stem cells. These stem cells derived from human exfoliated deciduous teeth were called as SHED. The importance of stem cells derived from deciduous teeth is that they are derived from a tissue that is similar in some ways to the umbilical cord. These stem cells would be discarded, if not used. Further investigations have indicated that SHED is originated from the neural crest, hence expressing neural markers. These cells have the ability to differentiate among neural cell lineages in addition to their potential of differentiating along odontoblastic, osteoblastic, chondrocytic and adipocytic cells.
Having the differentiation ability among odontoblastic as well as the other skeletal cell lineages, stem cells with dental origin hold great promise to implement reconstructions for the defects in tooth and skeletal tissue in future regenerative medicine. Several preliminary studies have indeed well indicated the regenerating capacity of these cells using a construct being created with the combination of the cells with appropriate scaffolds [9–13].
Since the dental pulp occurs in small size in the dental chamber, it would be expected that the stem cells could accordingly occur in small number. Therefore, the first step in performing any experimental, preclinical and even clinical investigation is to multiply the cells into sufficient number through cell culture techniques. Given these data, the growth characteristics of the cells would be of critical issue for including them in any procedure. To the best of our knowledge, very little is known regarding the comparative proliferation rate of the stem cells derived from the pulp of permanent and deciduous teeth.
In the present study, stem cells from the pulp of either a permanent (human third molar) or a deciduous (human incisor) tooth were isolated, culture expanded and then characterized. The objectives were to compare the cells in terms of their in vitro growth kinetics and their culture requirements for maximum propagation. A study like this provides information by which investigators will be able to decide on the most appropriate type of stem cells for their experimentations.
MATERIALS AND METHODS
Pulp Tissue and Cell Culture
The teeth were collected at the dental clinic of Shahid Beheshti University under the guidelines approved by the Ethic Committee of Royan Institute. Human third molars were obtained from patients who were young adults aged 20–25 years. Normal human deciduous incisors were collected from 7 to 12-year-old children. At Royan Institute cell culture lab, stem cells were isolated according to the previously-published methods with some modification [1–2].
Briefly, the pulp was removed from the dental chamber and subjected to enzymatic digestion using a solution of 3.0 mg/ml collagenase type I and 4.0 mg/ml dispase (both from Sigma, Germany) for 45 minutes at 37°C. About 3.0 ml Dulbecco’s Modified Eagle Medium (DMEM) (Gibco, Germany) supplemented with 15% Fetal Bovine Serum (FBS) (Gibco, Germany) was added to the digest of pulp tissue and followed by centrifugation at 1200 rpm for 5 minutes. Single-cell suspension was then prepared and the cells were plated at 103 cells/well of 6-well culture plates and incubated in an atmosphere of 5% CO2 and 37°C temperature. After three days, the cultures were washed with Phosphate Buffer Solution (PBS) and provided with a fresh culture medium. Afterwards, the culture medium was changed twice weekly until confluency was achieved. Confluent cultures were passaged at 1:3 ratios till sufficient cells became available for further experiments.
Flow Cytometric Analysis of Cell Surface Epitopes
Flow cytometric analysis was used to characterize the isolated cells with regard to their antigenic phenotype. To prepare the cells for flow cytometry, about 106 cells from passaged-3 cultures were placed in 5 ml tubes, provided with 5 μl of either Propidium Iodide (PI) or Fluorescein Isothiocyanate (FITC)-conjugated antibodies and 5 μl of blocking buffer and followed by incubation at 4°C for 20–25 min in a dark room. The solution was centrifuged at 1200 rpm for 4 minutes, the cells were dispersed in 300–500 μl washing buffer and transferred for analysis by flow cytometry (FACScalibur cytometer equipped with 488 nm argon lasers) apparatus. In this study, IGG2 and IGG1 were taken as isotope control. WinMDI software was used to analyze the flow cytometric results. The following antibodies were used to stain the cells: FITC-conjugated cluster of differentiation 90 (CD90), CD49b, CD31, CD45, CD33, ßIII Tubulin, Tau and PI-conjugated CD44, CD11b, CD34 and CD73 (all purchased from Becton Dickenson, USA).
Multiline-Age Differentiation
Odontoblast differentiation:
To direct differentiation of pulp-derived stem cells towards odontoblast lineages, confluent passaged-3 cell cultures from either group were provided with DMEM supplemented with 0.5 μM vitamin D3 (Sigma, USA), 50 mg/ml ascorbic 2-phosphate (Sigma, USA), 10 nM dexamethazone (Sigma, USA) and 10 mM β glycerol phosphate (Sigma, USA) [14]. After three weeks, odontoblast gene markers and mineralized matrix production were analyzed using reverse transcriptase-polymerase chain reaction (RT-PCR) method and alizarin red staining technique, respectively.
Chondroblast differentiation:
To assay whether or not the isolated cells were able to produce cartilage, a micro mass culture was arranged: about 2.5×105 passaged-3 cells were pelleted under 300 g for five minutes and provided with DMEM supplemented with 10 ng/ml transforming growth factor-ß3 (TGF-ß3, Sigma, Germany), 10 ng/ml bone morphogenetic protein-6 (BMP6, Sigma, Germany), 50mg/ml insulin transferin selenium+ premix (Sigma, Germany), 1.25 mg bovine serum albumin (Sigma, Germany) and 1% FBS [15]. Differentiation culture was kept for about 21 days at the end of which the pellets were subjected to histological processing. The pellets were fixed with 10% formalin, dehydrated in ascending concentrations of ethanol, cleared in xylene, embedded in paraffin, cut into 5-μm-thick sections and finally stained with toluidine blue. In parallel, some pellets were used to extract mRNA in order to further examine cartilage-specific gene expression in the differentiated cells.
Adipocyte differentiation:
Adipogenesis was directed in the confluent cultures of either stem cells (within passage 3) using a differentiation medium consisting of DMEM supplemented with 50 μg/ml ascorbic acid 3-phosphate, 100 nM dexamethazone and 50 μg/ml indomethacine [15]. The cultures were kept in differentiation condition for about 3 weeks and then evaluated for the presence of adipocyte using oil red staining and RT-PCR analysis.
Osteoblast differentiation:
To provide osteogenic conditions, the proliferation medium of the confluent cultures of passaged-3 stem cells from either tooth were removed and substituted with differentiation medium consisting of DMEM supplemented with 50 mg/ml ascorbic 2-phosphate (Sigma, USA), 10 nM dexamethazone (Sigma, USA) and 10 mM β glycerol phosphate (Sigma, USA) [15]. The differentiation cultures maintained for 21 days at the end of which alizarin red staining and RT-PCR analysis were respectively used to detect the production of mineralized bone matrix and the expression of bone-specific genes in cultures.
RT-PCR Analysis
Using RNX™ (-Plus) (RN7713C; CinnaGen Inc., Tehran, Iran), total RNA was extracted from the differentiated pulp stem cells derived from the human third molar and deciduous incisor teeth. To eliminate residual DNA, the RNA samples were treated with 1 U/μl of RNase-free DNaseI (EN0521; Fermentas, Opelstrasse 9, Germany) per 1 μg of RNA in the presence of 40 U/μl of ribonuclease inhibitor (E00311; Fermentas, Germany) and 1×reaction buffer with MgCl2 for 30 min at 37°C. DNaseI was then inactivated by including 3μl of 25 mM EDTA and incubation at 65°C for 10 min. Standard RT reactions were done for 2 μg total RNA using oligo (dt) as a primer and a RevertAid™ First Strand cDNA Synthesis Kit (K1622; Fermentas, Germany) based on the manufacturer’s instructions. For every reaction set, one RNA sample without RevertAid™M-MuLV Reverse Transcriptase (RT-reaction) was used as a negative control in the subsequent PCR. To minimize variation in the RT reaction, all RNA samples from a single experimental setup were reverse transcribed simultaneously. Reaction mixtures for PCR included 20 ng cDNA, 10×PCR buffer (AMS™; CinnaGen Co., Tehran, Iran), 200 mM dNTPs, 5 Pmol of each antisense and sense primer (Table 1), and 1 unit Taq DNA polymerase.
Table 1.
Primers that were used in RT-PCR analysis.
| Gene name | Sense Primer | Anti-Sense Primer |
|---|---|---|
| DMP1 | GCAGAGTGATGACCCAGAG | GCTCGCTTCTGTCATCTTCC |
| DSPP | CCATTCCAGTTCCTCAAAGC | CTGCCCACTTAGAGCCATTC |
| Aggrecan | TCAACAACAATGCCCAAGAC | AGCGACAAGAAGAGGACACC |
| PPAR-Gamma | CTAAAGAGCCTGCGAAAG | TGTCTGTCTCCGTCTTCTTG |
| PPAR- Alpha | TGCTATCATTTGCTGTGGAG | ACTCCGTCTTCTTGATGAT |
| RunX2 | CAAGTAGCAAGGTTCAACGA | CGGTCAGAGAACAAACTAGG |
| Osteocalcin | GGCAGCGAGGTAGTGAAGAG | CAGCAGAGCGACACCCTAGAC |
| GAPDH | CTCATTTCCTGGTATGACACC | CTTCCTCCTGTGCTCTTGCT |
Colonogenic Assays
Colonogenic assay is frequently being performed to estimate the growth potential of MSC-like population in vitro. In the present study, this assay was employed to determine and compare the proliferation capacity of the isolated stem cells from the pulp tissue based on the number and size of the colonies they can produce. For this purpose, passaged-3 cells from either group were plated at 100 cells in 10 cm Petri dish and allowed to grow for 10 days. At the end, the cultures were observed under inverted light microscope to determine the number of colonies. The size of colonies was measured using microscopic objective micrometer.
Calculation of Population Doubling Time
One reliable method to determine the given cell proliferation rate in culture condition is to calculate their population doubling time (PDT) values. Using this method, in the present study the expansion rate of the isolated cells were compared to each other. To determine PDT of the pulp stem cells, passaged-3 cells from either culture were plated at 250 cells/well of 12 well culture plate in a DMEM supplemented with 20% FBS and 100 U/ml penicillin, 100 μg/ml streptomycin. The cultures were incubated till one of them reached confluency, which at this time both cultures were terminated and subjected to hemocytometer count. Using the equation PDT=CT/log N/N0×3.31, PDT was determined [16]. In this equation, CT stands for culture time, N0 for the initiating cell number and N for the harvesting cell number.
Optimizing the Culture Condition
To determine the culture requirements of the isolated cells compared to each other, passaged-3 cells from either culture were plated at varying initiating density in the presence of varying FBS concentration and the cell yield for each condition was recorded. Evaluated cell seeding densities included 10, 50, 100, 200 and 500-cell/cm2 and the used FBS concentrations were as follows: 5%, 10%, 15% and 20% FBS in DMEM, supplemented with 100 U/ml penicillin, 100 mg/ml streptomycin. Using the cell number recorded for each culture condition, fold increase in cell number was determined and in this regard the pulp cells were compared to each other.
Plotting Growth Curve
Stem cells from each group, within passage 3, were cultivated in 12-well culture plate and maintained in the incubator till confluency was achieved. During the culture period some of the culture wells were trypsinized and the cell number was determined on the daily basis using a hemocytometer. The average number for each single day was then calculated and the values were used to plot a growth curve for each stem cell.
Statistical Analysis
In this experimental study, all values are stated as means (standard deviations). The results regarding to colonogenic assay as well as the PDT calculation were analyzed by student t-test and that of culture optimization with the one-way ANOVA. A P-value less than 0.05 was considered as statistically significant.
RESULTS
Cell Culture
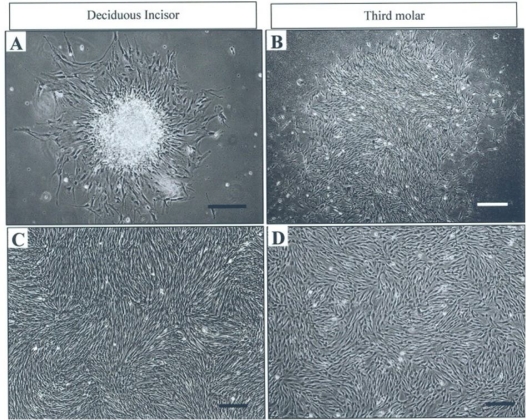
The cultures were observed on the regular basis using an invert light microscope. Single cell suspension from either the third molar or the deciduous incisor was observed to establish the primary cultures with several fibroblastic colonies. In contrast to the third molar culture, most of the colonies produced at the deciduous incisor culture tended to have an aggregate of small clear cell superimposed on the top of the colonies (Fig 1-A,B). The cultures reached confluency in about 10 days. Subcultures from either cell tended to exhibit an accelerated growth, so the cultures reached confluency in shorter time than primary cultures (in about 5–6 days). The fibroblastic morphology of the cells maintained throughout the culture period (Fig 1-C,D). In this regard there was no observed difference between the two cultures.
Fig 1.
Pulp cell cultures from human deciduous incisors and third molar teeth. Primary cultures (A and B), The cells in confluent culture at passage 3 (C and D). Bar=100 μm
Flow Cytometry
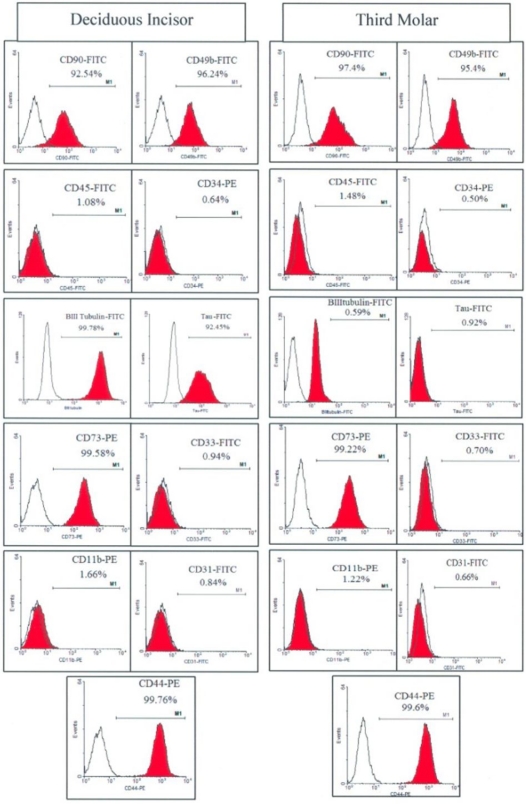
Antigenic profiles of the studied cells exhibited some similarities and some differences (Fig 2). The expression of hematopoietic-endothelial epitopes including, CD 11b, CD 34, CD 31, CD 33, CD 49b and CD 45 were observed in negligible percent of cells from both cultures and the mesenchymal surface epitopes including CD 90, CD 44 and CD 73 were expressed in more than 95% of the cells present at either culture. While stem cells from the third molar pulp did not exhibit the neural cell markers of ßIII Tubulin and Tau, these surface epitopes appeared on more than 90% of the stem cells established from the deciduous incisor pulp.
Fig 2.
Flow cytometric analysis of surface antigens on stem cells derived from human deciduous incisors and third molar teeth.
Multilineage Differentiation
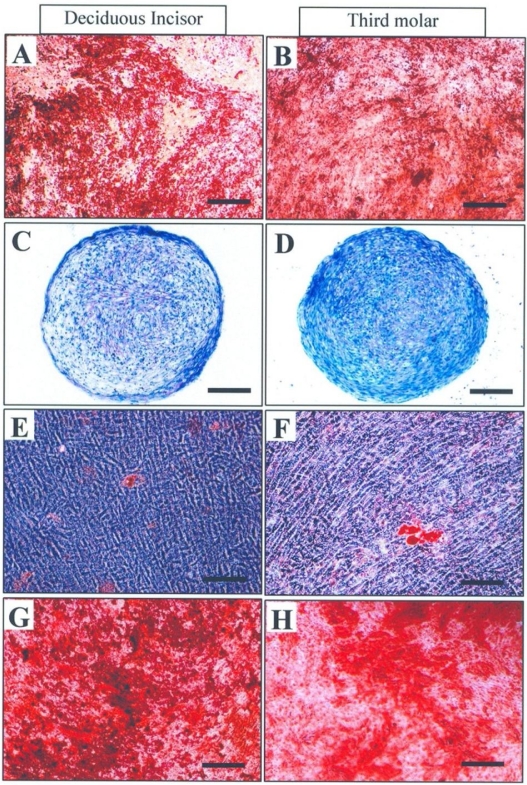
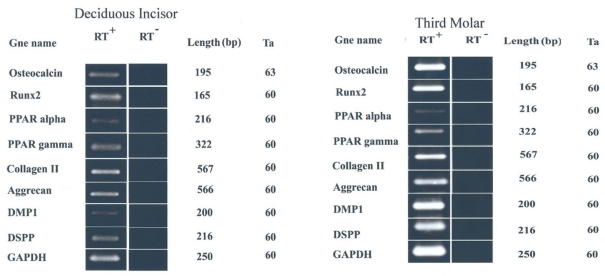
According to our data, the cells from either tooth (third molar or deciduous incisor) succeeded to differentiate among odontoblastic, chondrocytic, adipocytic and osteocytic cell lineages. They were all verified by specific staining as well as RT-PCR analysis (Fig 3). Odontoblastic differentiating cultures were positively stained red by alizarin red which stains the area of mineralized matrix (Fig 3-A,B) and expressed odontoblast-specific genes such as DMP1 (Dentin matrix protein 1) and DSPP (dentin sialophosphoprotein) (Fig 4).
Fig 3.
Differentiation of human dental pulp stem cells: either cell was succeeded to differentiate along odontoblastic (A, B), chondrogenic (C, D), adipocytic (E, F) and osteoblastic (G, H) cell lineages. Bar=100 μm
Fig 4.
RT-PCR analysis of gene expression in the differentiated cells isolated from human deciduous incisors and third molar teeth. Both cells were successfully expressed odontoblastic-, chondrogenic-, adipogenic- and osteogenic-related genes.
In chondrogenic cultures, the cartilage matrix deposited among the cells was detected by toluidine blue staining (Fig 3-C,D). RT-PCR analysis showed further evidence of cartilage differentiation by revealing the expression of cartilage specific genes including aggrecan and collagen II in chondrogenic cultures from both culture groups (Fig 4). Adipogenic differentiation appeared to be sparse; therefore, only a few adipocytes were observed per microscopic field of either culture. Lipid droplets developed in these cells were stained red by oil red (Fig 3-E,F) and the cells itself tended to express adipocyte related genes including PPAR-alpha (Peroxisome proliferators activated-receptor-alpha) and PPAR-gamma (Peroxisome proliferators activated-receptor-gamma) (Fig 4). Osteogenic cultures tended to stain red with alizarin red for mineralized matrix (Fig 3-G,H) and expressed bone specific genes including osteocalcin and Runx2 (Fig 4).
Colonogenic Assay
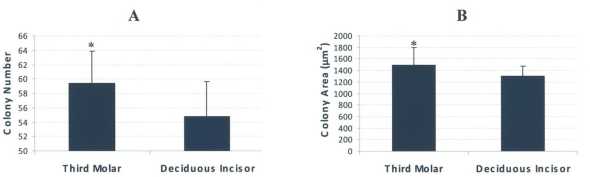
Based on our results, stem cells from the third molar exhibited more colonogenic activity than their counterparts from the deciduous incisor (P<0.05). These cells produced an average of 59.4 colonies (SD=4.5) which was significantly more than the colonies generated at the deciduous incisor (an average of 54.8 colonies, SD=4.8) (Fig 5-A). Regarding the size of the colonies, those of the third molar were significantly (P<0.05) larger than the deciduous incisor (1502.8 μm2, SD=298.8 versus 1307 μm2, SD=166.6) (Fig 5-B).
Fig 5.
Colonogenic assays performed for stem cells derived from human deciduous incisors and third molar teeth. A) The cells from the third molar appeared to create significantly more colonies than their counterpart from deciduous teeth. B) Likewise, the colonies of third molar cells were larger than those of deciduous incisors. * indicates the significant difference of P<0.05.
PDT
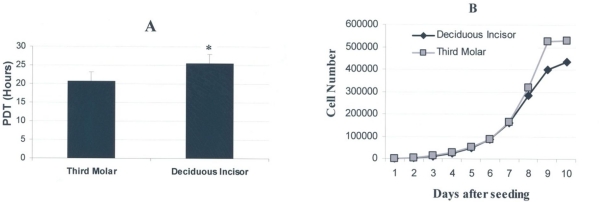
The PDT value for stem cells of the third molar appeared to be significantly lower than that for the deciduous incisor indicating the more rapid rate of proliferation of third molar cells (Fig 6-A). Based on the results, third molar pulp stem cells tended to double their population in the average of 20.79 hours (SD=2.8) while those from the deciduous incisor exhibited a doubling time of 25.55 hours (SD=2.9) (P<0.05). The difference was statistically significant.
Fig 6.
Population doubling time (PDT) and growth curve determined for stem cells derived from human deciduous incisors and third molar teeth. (A) Third molar cells tended to have a shorter PDT value compared to deciduous incisors (P<0.05). * indicates the significant difference of P<0.05. (B) According to the growth curve, third molar cells reached plateau sooner than deciduous incisor cells. Both cells exhibited a lag time of a few days in culture.
Growth Curve
As it can be seen from Fig 6-B the growth curve plotted for either cells indicated that the stem cells from the third molar were the first that reached the plateau phase while at the same time the cells from the deciduous molar were still in the log phase. According to this curve, the lag phase tended to be about 2 days for either culture meaning that both cells started to proliferate two days after being plated. The lag phase is considered to be the adaptation phase to culture conditions.
Culture Optimization
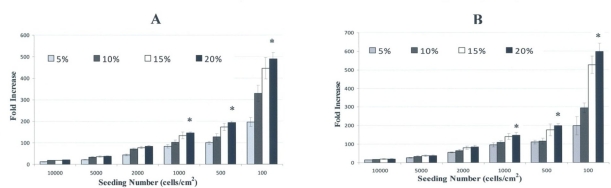
Both cells tended to exhibit a maximum proliferation when being provided with medium supplemented with 20% FBS. Regarding the best cell seeding density, 100 cells/cm2 yields high fold increase in cell number for either cultures (Fig 7).
Fig 7.
Culture optimization for better growth of the stem cells isolated from human deciduous incisors (A) and molar teeth (B). The cells underwent considerable fold increase when are plated at low density in a medium with 20% FBS.
DISCUSSION
To the best of our knowledge, there is very limited information regarding in vitro growth rate and culture needs of the stem cells from human dental pulp. Having the growth characteristics as well as culture requirements of the cells could be of great importance for those who specially are involved in stem cell investigations with the aim of regenerating tissue defects. A study like the current investigation provides some insights on the differences existing between stem cells regarding their basic biological properties in vitro. Using such data, investigators will be able to select the right cells for their experimental, preclinical and clinical researches.
In the present study, stem cells from human teeth which are usually discarded upon extraction were investigated. Since dental pulp stem cells are capable of giving rise to dentin-producing cells (odontoblast) as it was shown in this study, they may be used in regenerative medicine to reconstruct human teeth defects [9–13]. Furthermore, having the ability to differentiate among other tissues including bone and cartilage, pulp derived stem cells would be a great candidate to implement regeneration in skeletal tissue as well. To achieve these goals one necessary step is to culture expand the cells prior to any clinical work. In emergency need for tissue regeneration, quicker expansion of the cells would be desired. In such circumstances cells with rapid expansion rate will be preferable. Based on our results, third molar pulp stem cells tended to have more proliferation capacity than deciduous incisor pulp stem cells and therefore more appropriate for regenerative medicine.
Based on our data stem cells from the pulp of human third molar exhibited a significantly higher expansion rate than those from human deciduous incisor. This data is in contrast with that of Miura et al [2] who reported that SHED proliferate faster with greater PD than the dental pulp stem cells (DPSC). This piece of data has been presented as a part of a figure in Miura et al articles [2] and no further description has been given concerning the condition on which the two cells have been compared. Indeed, the main purpose of Miura et al’s investigation [2] has been the isolation of SHED rather than comparing the two cells. The lack of detailed data however, makes it difficult to interpret the discrepancy existing between our study and Miura et al’s findings [2]. We used the cells within passage 3 in a DMEM medium and allowed them to grow for confluency. In addition, in the current study, multiple methods including colonogenic assays, PDT and plotting growth curve were used to compare the cells. The data obtained by all measuring methods indicated the proliferative superiority of third molar dental pulp stem cells over exfoliated deciduous incisor pulp stem cells.
Owing to the small quantity of dental pulp in the dental chamber, it could be expected that stem cells occur in small number within pulp tissue. On the other hand stem cells are required in large numbers in most experimental, preclinical and clinical setups. This makes expanding of the stem cells an inevitable step prior to their application. Stem cells ex vivo propagation are largely dependent on the presence of FBS in culture medium. According to the former researches, stem cells that are developed in mediums containing FBS would be immunogenic and may transfer bovine pathogens upon transplantation [17]. In spite of the extensive attempts that have been made to substitute bovine serum, less success has been achieved and propagation of stem cells with FBS is still practiced in many laboratories [17–18]. In this study, stem cells from the pulp tissues were investigated in terms of their FBS need for maximum proliferation. According to our results, 20% FBS appeared with maximum cell yield in both cultures. In addition, the 15% FBS tended to have reasonable proliferative effects on both cells, hence the application of 15% FBS rather than 20% for both stem cell ex vivo propagation is suggested. This may help reduce the above-mentioned complications that are associated with FBS use in cell culture.
There are two crucial parameters affecting in vitro cell proliferations; FBS concentrations and initiating cell seeding density. In the present study, the best cell density at culture initiation with maximum cell yield was determined for either cell. The cell proliferation rate is of utmost importance especially in cell therapy strategies where the rapid expansion of cells would be desired. According to our data, both isolated cells possessed the highest fold increase in cell number when the cultures were initiated at the lowest density of 100 cells/cm2. This finding is in agreement with that of Cristofalo et al’s study [19] who reported an inverse correlation between human MSC seeding density and their proliferation.
CONCLUSION
In general, our evaluations indicated rapid proliferation of the molar stem cells compared to deciduous incisor stem cells. In addition, the culture conditions for maximum proliferation of the dental pulp stem cells showed no difference; both culture exhibited maximum cell yield when being plated at low density condition of 100 cells/cm2 and in the presence of 20% FBS concentrations. Such data help investigators decide the appropriate stem cell source for regenerative medicine.
Acknowledgments
The authors wish to thank Royan Institute and Dental Research Center of Shahid Beheshti University of Medical Sciences for their financial support of the present study.
REFERENCES
- 1.Gronthos S, Mankani M, Brahim J, Robey PG, Shi S. Postnatal human dental pulp stem cells (DPSCs) in vitro and in vivo. Proc Natl Acad Sci USA. 2000 Dec 5;97(25):13625–30. doi: 10.1073/pnas.240309797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Miura M, Gronthos S, Zhao M, Lu B, Fisher LW, Robey PG, et al. SHED: stem cells from human exfoliated deciduous teeth. Proc Natl Acad Sci USA. 2003 May 13;100(10):5807–12. doi: 10.1073/pnas.0937635100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Seo BM, Miura M, Gronthos S, Bartold PM, Batouli S, Brahim J, et al. Investigation of multipotent postnatal stem cells from human periodontal ligament. Lancet. 2004 Jul 10–16;364(9429):149–55. doi: 10.1016/S0140-6736(04)16627-0. [DOI] [PubMed] [Google Scholar]
- 4.Sonoyama W, Liu Y, Yamaza T, Tuan RS, Wang S, Shi S, et al. Characterization of the apical papilla and its residing stem cells from human immature permanent teeth: a pilot study. J Endod. 2008 Feb;34(2):166–71. doi: 10.1016/j.joen.2007.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Morsczeck C, Moehl C, Götz W, Heredia A, Schäffer TE, Eckstein N, et al. In vitro differentiation of human dental follicle cells with dexamethasone and insulin. Cell Biol Int. 2005 Jul;29(7):567–75. doi: 10.1016/j.cellbi.2005.03.020. [DOI] [PubMed] [Google Scholar]
- 6.Laino G, Carinci F, Graziano A, d'Aquino R, Lanza V, De Rosa A, et al. In vitro bone production using stem cells derived from human dental pulp. J Craniofac Surg. 2006 May;17(3):511–5. doi: 10.1097/00001665-200605000-00021. [DOI] [PubMed] [Google Scholar]
- 7.d’Aquino R, Graziano A, Sampaolesi M, Laino G, Pirozzi G, De Rosa A, et al. Human postnatal pulp cells co-differentiate into osteoblast and endotheliocytes: a pivotal synergy leading to adult bone tissue formation. Cell Death Differ. 2007 Jun;14(6):1162–71. doi: 10.1038/sj.cdd.4402121. [DOI] [PubMed] [Google Scholar]
- 8.Zhang W, Walboomers XF, Shi S, Fan M, Jansen JA. Multilineage differentiation potential of stem cells derived from human dental pulp after cryopreservation. Tissue Eng. 2006 Oct;12(10):2813–23. doi: 10.1089/ten.2006.12.2813. [DOI] [PubMed] [Google Scholar]
- 9.Huang GT, Sonoyama W, Chen J, Park SH. In vitro characterization of human dental pulp cells: various isolation methods and culturing environments. Cell Tissue Res. 2006 May;324(2):225–36. doi: 10.1007/s00441-005-0117-9. [DOI] [PubMed] [Google Scholar]
- 10.Huang GT, Shagramanova K, Chan SW. Formation of odontoblast-like cells from cultured human dental pulp cells on dentin in vitro. J Endod. 2006 Nov;32(11):1066–73. doi: 10.1016/j.joen.2006.05.009. [DOI] [PubMed] [Google Scholar]
- 11.Huang GT, Sonoyama W, Liu Y, Liu H, Wang S, Shi S. The hidden treasure in apical papilla: the potential role in pulp/dentin regeneration and bioroot engineering. J Endod. 2008 Jun;34(6):645–51. doi: 10.1016/j.joen.2008.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Murray PE, Garcia-Godoy F, Hargreaves KM. Regenerative endodontics: a review of current status and a call for action. J Endod. 2007 Apr;33(4):377–90. doi: 10.1016/j.joen.2006.09.013. [DOI] [PubMed] [Google Scholar]
- 13.Prescott RS, Alsanea R, Fayad MI, Johnson BR, Wenckus CS, Hao J, et al. In vivo generation of dental pulp-like tissue by using dental pulp stem cells, a collagen scaffold, and dentin matrix protein 1 after subcutaneous transplantation in mice. J Endod. 2008 Apr;34(4):421–6. doi: 10.1016/j.joen.2008.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tonomura A, Sumita Y, Ando Y, Iejima D, Kagami H, Honda MJ, et al. Differential inducibility of human and porcine dental pulp-derived cells into odontoblasts. Connect Tissue Res. 2007;48(5):229–38. doi: 10.1080/03008200701507909. [DOI] [PubMed] [Google Scholar]
- 15.Eslaminejad MB, Nikmahzar A, Taghiyar L, Nadri S, Massumi M. Murine mesenchymal stem cells isolated by low density primary culture system. Dev Growth Diff. 2006 Aug;48(6):361–70. doi: 10.1111/j.1440-169X.2006.00874.x. [DOI] [PubMed] [Google Scholar]
- 16.Piera-Velazquez S, Jimenez SA, Stokes D. Increased life span of human osteoarthritic chondrocytes by exogenous expression of telomerase. Arthritis Rheum. 2002 Mar;46(3):683–93. doi: 10.1002/art.10116. [DOI] [PubMed] [Google Scholar]
- 17.Spees JL, Gregory CA, Singh H, Tucker HA, Peister A, Lynch PJ, et al. Internalized antigens must be removed to prepare hypoimmunogenic mesenchymal stem cells for cell and gene therapy. Mol Ther. 2004 May;9(5):747–56. doi: 10.1016/j.ymthe.2004.02.012. [DOI] [PubMed] [Google Scholar]
- 18.Pittenger MF, Mackay AM, Beck SC, Jaiswal RK, Douglas R, Mosca JD, et al. Multilineage potential of adult human mesenchymal stem cells. Science. 1999 Apr 2;284(5411):143–7. doi: 10.1126/science.284.5411.143. [DOI] [PubMed] [Google Scholar]
- 19.Cristofalo VJ, Allen RG, Pignolo RJ, Martin BG, Beck JC. Relationship between donor age and the replicative lifespan of human cells in culture: a reevaluation. Proc Natl Acad Sci USA. 1998 Sep 1;95(18):10614–9. doi: 10.1073/pnas.95.18.10614. [DOI] [PMC free article] [PubMed] [Google Scholar]