Introduction
Previous genomics and transcriptomics studies, including recent RNA-sequencing studies, have now confirmed that only 1–2% of the mammalian genome corresponds to protein-coding genes with the vast majority of the genome does not appear to have protein-coding capacity. Since proteins have been thought to carry out many of the key regulatory functions within cells, it has been puzzling that the number of protein-coding genes does not seem to fluctuate between species with great differences in developmental and physiological complexity.
Interestingly, a number of studies have now shown that protein-coding genes are vastly outnumbered by non-coding RNAs (ncRNAs) especially in eukaryotic genomes [1, 2], and some have estimated that mRNAs only represent 20% of all transcripts [3]. Intriguingly, there seem to be a linear relationship between the complexity of an organism and the number of non-coding RNAs produced [4]. This observation suggests that developmental complexity which is not reflected in the number of protein-coding genes maybe mediated by non-coding RNAs. However, the mechanisms by which non-coding RNAs contribute to this complexity is not completely understood.
One possibility is that non-coding RNAs are slowly acquiring more “power” and essentially are carrying out a “Coup d'état” slowly over evolution? Either by taking on prime responsibility to regulate cellular processes, or alternatively ncRNAs may have been co-opted by proteins to assist in orchestrating the diversity of eukaryotic cellular pathways. Consistent with the latter, almost all known examples of ncRNA function by interfacing with protein coding genes.
In this review we highlight a common theme of large non-coding RNAs forming ribonucleic protein interactions that are known in all kingdoms of life with a specific emphasis on the examples of mammalian ncRNAs that have found protein partners to form ribonucleoprotein complexes resulting a wide range of biological functions (organizational, transcriptional, post-transcriptional, and translational). These ribonucleoprotein complexes have implications in many aspects of biology including genomic imprinting, X inactivation, pluripotency and cancer.
Classic examples of functional ribonucleoprotein complexes
There are many clear examples of non-coding RNAs that function as part of complexes with proteins known as ribonucleoprotein complexes (e.g., ribosome, splicesome, SRP and RNase P). One of the most profound examples of ribonucleoprotein complexes is the ribosome which is required for the translation of messenger (m)RNAs into proteins and is conserved throughout all kingdoms of life. Eukaryotes have 80S ribosomes, each consisting of a small (40S) and large (60S) subunit. Both subunits require RNA and protein components: the 60S large subunit is composed of 5S, 28S, 5.8S ribosomal RNAs and 49 proteins and the 40S subunit has 18S RNA and 33 proteins [5]. The assembly of the ribosome takes place in the nucleolus and requires the coordination of approximately 200 proteins to physically associate with cognate ribosomal RNAs. Although eukaryotes have larger and more complex ribosomes than those present in prokaryotes, nevertheless, ribosomes from all kingdoms of life function according to similar principles. The ribosome represents a great example of a RNA-protein complex that has a clear functional role.
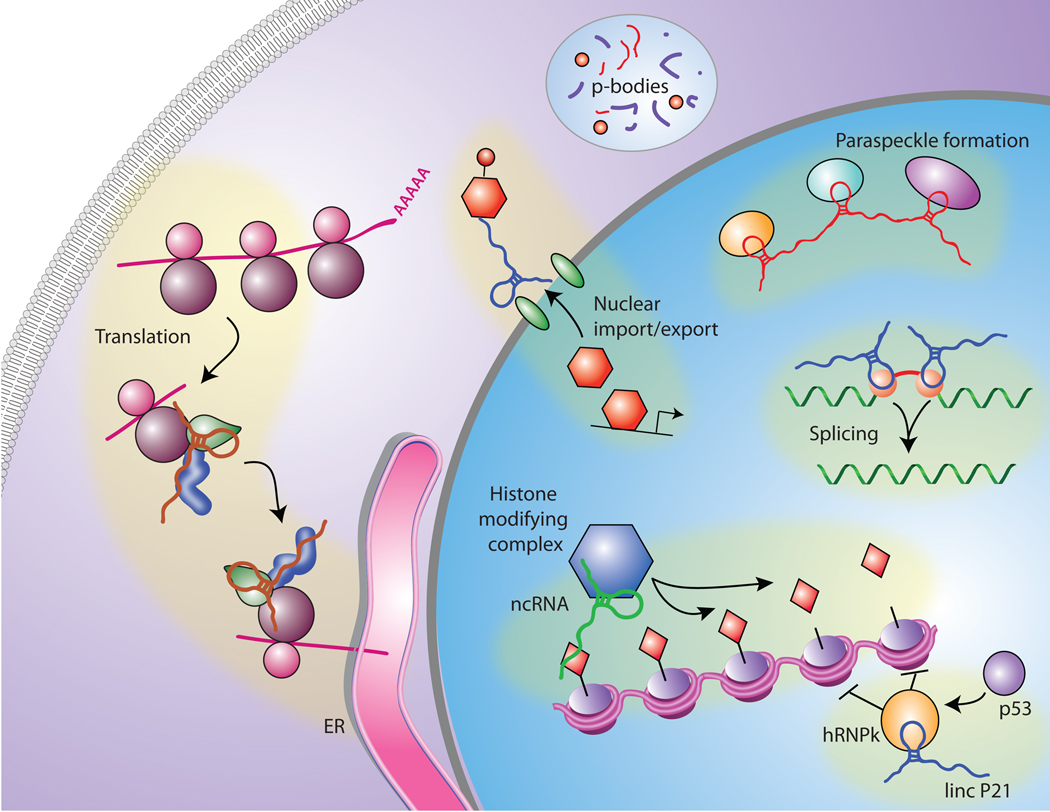
Interestingly, the localization of the ribosome to the endoplasmic reticulum to initiate translation also requires another ribonucleoprotein complex which is referred to as signal recognition particle (SRP) can recognize and bind to the endoplasmic reticulum (ER) signal sequence in a newly synthesized secretory protein and subsequently delivers the protein along with its associated ribosomes to the membrane of the ER to complete the translation of the protein (Figure 1). In prokaryotes and eukaryotes SRP forms a ribonucleic-protein complex with 7SL and 5SL respectively.
Fig 1. Extensive Large Non-coding RNA-protein Interactions in the Mammalian Genome Which are involved in Numerous Biological Processes.
In the nucleus large non-coding RNAs interact with chromatin-modifying complexes (e.g., PRC2, CoREST, LSD1) and are thought to guide these complexes to their site of action and/or serve as scaffolds. Also, large non-coding RNAs play a crucial role in other processes such as splicing as well as nuclear-cytoplasmic shuttling. In the cytoplasm, large non-coding RNAs are also involved in several process including signal recognition particle (SRP) which directs nascent secretory proteins to the endoplasmic reticulum and some large non-coding RNAs have been found in P-bodies.
Another classic example of a functional ribonucleoprotein complex is RNase P [6], this ribonuceloprotein complex is found in all kingdoms of life and it is responsible for the generation of mature 5’ends of tRNAs by cleaving the 5’ leader elements of precursor tRNAs. In bacteria and euckryotes RNase P is composed of a core RNA and 5 and 10 proteins, respectively. In vitro studies have shown that RNase P RNA can function as a ribozyme since the RNA can function without its protein partners [7]. These studies show a remarkable plasticity in RNA-protein interactions throughout evolution.
These examples of ribonucleoprotein complexes (ribosome, RNase P and SRP) demonstrate that RNAs play a central role in the function of RNA-protein complexes in all kingdoms of life and they suggest that other newly discovered non-coding RNAs may function in a similar manner. Although these classic ribonucleoprtein complexes appear to be conserved between prokarotes and eukaryotes, the eukaryotes counterparts tend to be more complex involving more proteins; this is probably due to the fact that the eukaryotic cell has more specialized functions. It is worth noting that a large number of mammalian proteins have RNA binding motifs, and these proteins are involved in a wide range of cellular processes. The significance of many RNAs interaction with proteins is not known and has not been well characterized.
Several technologies have emerged over the years to assay RNA-protein interactions on a large scale with each technology having its advantages and disadvantages. Native RNA co-immunoprecipitation (RIP) which assays RNA-protein interaction without any form of cross-linking has been used to assay RNA interaction with chromatin complexes, Ago proteins and others both in the nucleus and the cytoplasm [8–10]. One caveat to this technology is the potential for RNA molecules to associate with proteins after cell lysis in a non-specific manner [11]. Alternatively, cross-linking by formaldehyde or UV light can be applied prior to assaying RNA-protein interactions to eliminate the potential for non-specific interactions following cell lysis [12–14], however, these methods tend to increase background levels and to be less efficient. Nevertheless, collectively these methodologies have been able to show that RNA-protein interactions are more widely spread than previously thought and these RNP complexes participate in a wide range of biological processes. These observations argue that non-coding RNAs function through interactions with multiprotein complexes – a field that is yet to be fully explored. Thus, paving the way for functional and structural studies to elucidate RNA sequences or secondary structures that maybe important for these interactions.
In the following sections we will discuss emerging ribonucleoprotein complexes of a wide range of non-coding RNAs and their binding protein partners and how these ribonucleoproetin complexes can function in a wide range of cellular processes.
Ribonucleoprotein complexes of large non-coding RNAs and chromatin-modifying complexes
The human body is composed of hundreds of distinct cell types. Each cell must occupy a specific position within the body and perform a specific function. Since all the cells within a multicellular organism contain the same genome, the information that cells utilize to establish their identity is likely to be coded in their Epigenome [15]. The epigenome is comprised of modifications to DNA (i.e., DNA methylation), and modifications to histone proteins at specific amino acid residues (e.g., acetyation, methyaltion, phosphorylation …etc) [16]. Key regulators of the epigenome are chromatin-modifying complexes which can add or remove covalent modifications to chromatin. In contrast to transcription factors, which can recognize and bind to specific DNA sequences, the majority of chromatin-modifying complexes do not have DNA binding capacity. A major gap in our understanding of epigenetic regulation by chromatin-modifying complexes is how these complexes are targeted to specific regions of the genome. Recent studies have recently shown that the Jumanji protein Jarid2 can recruit the polycomb repressive complex (PRC)2 to its target sites in mouse embryonic stem cells (mESC) [17–20], however, jarid2 shows low expression in differentiated cells; and therefore its not clear how PRC2 is targeted to its genomic sites in other cell types. Also, there is a plethora of chromatin-modifying complexes that does not have DNA binding protein partners that direct them to their sites of action.
Several studies have now suggested a potential role for non-coding RNAs in guiding chromatin-modifying complexes to their targets [21]. For example, the large non-coding RNA HOTAIR, which is transcribed from the HOXC locus on chromosome 12, interacts with the chromatin-modifying complex PRC2 and potentially guide this complex to the HOXD region on chromosome 2 to repress the transcription of several HOXD genes in trans [10]. Similarly, Zhao et al found RepA (in the 5’ of Xist) to also interact with PRC2 and affect its localization and activity on the inactive X chromosome in cis [22]. We have recently shown that many large intergenic non-coding RNAs (lincRNAs) associate with several chromatin-modifying complexes including PRC2, SMCX and CoREST [9]. About 20% of expressed lincRNAs associate with components of PRC2 (SUZ12 and EZH2) in several human cell types. Furthermore, siRNA-mediated knockdown of PRC2-associated lincRNAs results in changes in gene expression similar to those observed in PRC2 knockdown suggesting that these lincRNAs function in the same pathway. Collectively, these studies show that large non-coding RNAs association with chromatin modifying complexes is a general phenomenon in mammalian cells; however, future studies are needed to determine if large ncRNAs are guiding chromatin-modifying complexes to genomic loci, to that end one needs to show that depletion of these ncRNAs disrupt the localization of these complexes to their genomic sites, and those large ncRNAs are localized to the same DNA regions.
Similarly several large non-coding RNAs transcribed antisense to protein-coding genes can also interact with chromatin-modifying complexes and affect chromatin landscape. For example, the antisense transcript to the Igf2r gene known as Air is required for the allele-specific silencing of several genes in the mouse placenta functions through direct interaction with the repressive histone methyltransferase G9a [23, 24]. Similarly, the Kcnq1 gene has a nuclear retained antisense transcript, Kcnq1ot1, that associates in a tissue-specific manner with the chromatin complexes G9a and PRC2 and represses several protein-coding genes with a 1 Mb region in cis [25].
A study by Yu et al (2008) found that many tumor suppressor genes have antisense transcripts [26]. For example, p15, a cyclin-dependent kinase inhibitor implicated in leukemia, has an antisense transcript that silences its transcription in cis and in trans by inducing heterochromatin formation without a change in DNA methylation in a Dicer-independent manner. It is possible that these antisense transcripts directly bind and recruit chromatin-modifying complexes to their associated sense transcripts.
The role of non-coding RNAs in chromatin formation has also been observed in plants. A study by liu et al found that targeted 3’ processing of a non-coding antisense transcript to the FLC gene, a major floral repressor gene, results in the recruitment of FLD, a homolog of the human histone demethylase LSD1, which targets H3K4me2 for demethyaltion [27]. This targeting results in the reduced transcription of the FLC sense transcript. Notably, antisense mediated chromatin modifications appear to operate mostly in cis in contrast to intergenic ncRNAs which can operate both in cis and in trans.
What is the role of chromatin associated non-coding RNAs? Although many studies have now shown that large non-coding RNAs form ribonucleic protein interactions with chromatin-modifying complexes in a wide range of cell types and organisms, however, the exact role of these interactions is not well understood at least on a global scale. It is possible that non-coding RNAs play a role in i) facilitating architectural interactions amongst protein coding components of chromatin-modifying complexes; ii) the targeting of these complexes to specific regions of the genome; or iii) the activity of theses complex or all of the above. Future studies are needed to dissect the mechanism of RNA-mediated epigenetic regulation.
RNA-protein interactions in transcriptional regulation
In the previous section we explored how large non-coding RNAs can regulate gene expression via interactions with chromatin-modifying complexes and altering the chromatin at gene promoters to affect their transcriptional output. Alternatively, a non-coding RNA could also form RNA-protein complexes with transcription factors themselves and modulate transcription factor activity. For example, 30 years ago it was demonstrated that a transcription factor (TFIIIA) served a dual role to bind DNA at the internal promoter of the 5S RNA and subsequently forms a ribonucleic protein complex with the 5S RNA [6, 28, 29]. Interestingly, two different regions of the same Zinc Finger binding domain bind DNA and RNA respectively.
Several other recent studies are pointing to a more global role of transcription factors interfacing with RNA to modulate transcriptional regulation. Specifically, the growth arrest-specific (Gas)5 non-coding RNA, which has high expression level in growth-arrested cells due to lack of nutrients. It was recently demonstrated that Gas5 performs this function, in part, by forming a ribonucleoprotein complex with the glucocorticoid receptor (GR) through direct binding to its DNA binding domain (DBD) [30]. This binding of Gas5 RNA to the DBD of GR subsequently modulates the GR transcriptional activity at its downstream target genes. Similarly, it has been demonstrated that a large non-coding RNA termed NRON modulates the nuclear localization of the transcription factor NFAT [31]. Moreover, Martianov et al found a non-coding RNA transcribed from the minor promoter of the human dihydrofolate reductase (DHFR) gene to regulate the transcription of this gene by directly interacting with the major promoter of DHFR and forming a DNA-RNA triplex. Furthermore this non-coding RNA can interact with TFIIB and leads to the dissociation of the pre-initiation complex causing repression of DHFR [32].
Consistent with a global role for RNA interfacing with transcription factors several recent studies have shown that many protein-coding genes have non-coding RNAs transcribed in close proximity to their transcription state site (TSS RNAs) [33]. In addition to TSS RNAs, others have described similar species of ncRNAs that are also transcribed in close proximity to eukaryotic genes which include promoter-associated RNAs (PASR) and transcription-initiation RNAs (tiRNAs) [34, 35]. Although the function of these transcripts is not known, one can speculate that many of these ncRNA maybe playing a regulatory function by interacting with transcription factors and/or chromatin complexes to regulate the transcription of their neighboring genes in a similar manner to the DHFR ncRNA described above. Further studies are needed to elucidate in more detail the nature of these transcripts and if they share a similar regulatory mechanism. Together these, studies point to a potential mechanism of fine-tuning transcription factor functions by interfacing with non-coding RNAs.
Ribonucleoproteins are involved in nuclear organization
The mammalian nucleus is a highly organized structure with specific compartments and subnuclear bodies. Such nuclear compartments are required for various functions, including transcriptional regulation and splicing, and change in response to external stimuli or progression through the cell-cycle. A few studies have shown that large non-coding RNAs alone or through association with protein partners can play a structural role in the formation of some nuclear compartments.
A recently identified subnuclear body, termed paraspeckles, comprises a ribonucleoprotein complex [36]. Paraspeckles are found in a wide range of tissue, primary and transformed cells but not in embryonic stem cells, however they do appear after differentiation. Several proteins have been implicated as core components of paraspeckles including PSF/SFPQ, NONO, and PSPC1 which are members of the Drosophila melanogaster behavior, human splicing family [36]. Subsequently a long non-coding RNA known as NEAT1 has been shown to be required for paraspeckle formation and integrity as siRNA-mediated knockdown of NEAT1 results in loss of paraspeckles [36–38]. More recently, NEAT1 has also been implicated in nuclear export of mRNAs from the nucleus [39].
Another study by Sone et al (2007) found a large non-coding RNA (i.e., Gomafu) to be expressed in a distinct set of neurons. Interestingly, the spliced mature Gomafu RNA is localized to the nucleus and is detected as numerous foci that do not co-localize with any known nuclear domain markers possibly marking a novel nuclear compartment [40]. Collectively, these studies implicate long non-coding RNAs in the formation of specialized compartments within the cell and suggest that other yet to be identified long non-coding RNAs can have structural functions in mammalian cells alone or by binding to protein partners.
Small RNAs based Ribonucleic-protein interactions
Similar to large non-coding RNAs, small RNAs (less than 200 nucleotides in length) also associate with protein partners to perform various functions. In yeast, sense and antisense RNAs transcribed from heterochromatic loci form double-stranded (ds)RNAs that are processed by DICER into small RNAs that interact with Ago proteins as a crucial part of RNA-induced initiation of transcriptional silencing (RITS) complex [41]. RITS is required for the silencing of heterochromatic repeats through the recruitment of chromatin-modifying complexes to these loci [42]. In RNA dependent RNA polymerase I (Rdp1) mutants, RITS fails to localize to the heterochromatic loci and lack siRNAs suggesting that the initial transcription of dsRNA is essential for the generation of siRNAs and the maintenance of heterochromatin [42]. A recent study, however, found a tiny fraction of Dicer-independent small RNAs that are generated through a celluar degradation mechanism which then bind to Ago1 and can recruit chromatin complexes to initiate the addition of specific histone marks (Mozaed paper). The authors suggest that this may be a part of a general surveillance mechanism of the transcriptome.
In mammals, there is also emerging evidence that small RNAs targeted to the promoter region of genes can interact with Ago proteins and results in alteration of chromatin modifications at promoters by the recruitment of chromatin complexes and therefore can affect the transcription of genes. Previously, Janowski and colleagues found that small RNA targeted to the transcription start sites or promoter regions of genes can induce gene silencing through the binding of Ago1 and Ago2 to the DNA (Janowski et al., 2006). A similar study from Kevin Morris’s group found that small RNAs targeted to the promoter region of the UbC gene in human cells result in long term silencing via changes in chromatin modifications in an Ago1-dependent manner (Hawkins et al., 2009). Intriguingly, another study found that small RNAs can also induce gene activation in some cases when targeted to the promoter region of genes (Janowski et al., 2007). A recent study has also found that small RNAs that are transcribed from 5’ end of polycomb target genes in primary T cells and embryonic stem cells, are associated with the chromatin-modifying complex PRC2 [43]. Collectively these studies demonstrate that ribonucleic-protien complexes comprised of small RNAs play key roles in epigenetic regulation from yeast to mammals.
Another major class of functional small RNAs is microRNAs (miRNAs) [44]. MicroRNA biogenesis initiates from primary transcripts (pri-miRNA) that are capped, polyadenylated and processed by Drosha into a short stem-loop structure (pre-miRNA). Pre-miRNAs are then processed by the RNase III-like enzyme Dicer into functional 23 nt mature miRNAs. Each mature miRNA sequence can be complementary to few hundred mRNAs and via this complementarity, a single miRNA can regulate the translation of many proteins and therefore simultaneously influence many cellular processes and pathways [44]. miRNAs also function within a multiprotein complex known as RNA-induced silencing complex (RISC). RISC is composed of several proteins including DICER, the double-stranded RNA binding protein TRBP, and Argonaute 2 [45]. This multiprotein complex becomes a functional ribonucleoprotein when a double-stranded RNA or pre-miRNA and a matching mRNA become incorporated into the complex. Thereafter, the argonaute 2 protein which has the slicing activity within the RISC complex is triggered to destabilize or degrade the mRNA [46]. These studies demonstrate the importance of small RNA-protein interactions in the regulation of translation.
Small ncRNA- protein interactions are also implicated in spermatogenesis. For example, piwi-associated RNAs (piRNAs) are slightly larger than miRNAs and siRNAs, range in size between 26–32 nt have only been found in germ cells thus far and associate with the Piwi family of Argonaut proteins [47–49]. Mice have three piwi proteins: Mili, Miwi, and Miwi2. These proteins are expressed in the various stages of mouse meiosis and interact with piRNAs [50]. Functional studies such as mouse knockouts of these proteins have demonstrated that they are required for spermatogenesis as these mice displayed meiotic defects and sterility [51, 52]. Although all three proteins resulted in spermatogenic arrest, the stage at which the arrest occurs varies between the three proteins suggesting a non-overlapping function (reviewed in Khalil and Wahlestedt, 2008). Only few piRNA clusters have been characterized to date and their functions have been implicated in transposon silencing or guidance of chromatin-modifying complexes [53].
Similar to piRNAs another group of small RNAs have been implicated in germ cell biology. MSY2, a germ-cell-specific member of the Y-box family of DNA-/RNA-binding proteins, have recently been shown to associate with a novel class of small RNAs. MSY2 binds small RNAs (MSY-RNAs) that range in size from 18–36 nt with the majority being 25–31 nt in length, and are expressed in both germ cells and somatic cells [54]. Despite the abundance of piRNAs in the testes which are also similar in size to the immunoprecipitated RNAs with MSY; the authors found only 7% of these MSY-RNA map to known piRNA genes. MSY-RNA map to exons (65%), introns (16%), intergenic regions (12%) and piRNA clusters (7%) with no MSY-RNA matching to rRNA, tRNA or miRNAs. Also, since the majority of MSY-RNAs have a 5’ adenosine, this distinguishes them form piRNAs and miRNAs suggesting that MSY-RNA is a distinct class of small RNAs. Although, the exact function of these MSY-RNA is not known, however, MSY-RNAs are enriched in chromatin in the nucleus, and in the cytoplasm they are detected in both ribonucleoproteins and polysomes, suggesting that MSY-RNA may play a role in regulating genes at the transcriptional or post-transcriptional level. Also, since deletion of Msy2 in mice results in spermatogenic defects this also provide further evidence that MSY-RNAs are likely to be functional. MSY-RNAs and piRNAs may play critical roles during spermatogenesis including regulation of meiotic progression and meiotic recombination with implications for fertility and inherited genetic disorders.
X inactivation and genomic imprinting
One of the most dramatic examples large non-coding based RNPs playing a critical role in chromatin regulation is X chromosome inactivation. Female somatic cells silence one of their two X chromosomes to equalize the dosage of X linked genes with males (XY) [55, 56]. At the center of a complex choreography is a large non-coding RNA known as XIST, specifically, the Repeat A region of Xist physically associates with PRC2 and is required for its localization to the future inactive X chromosome [22]. This ribonucleoprotein complex of Xist and its protein partners confers a transcriptionally repressive environment on most of the inactive X chromosome by modifying histone tails with repressive chromatin marks that likely interfere with transcriptional fidelity.
The interface of large non-coding RNAs and chromatin modifying complexes is becoming a common theme in the maternal or paternal allelic silencing beyond Xist [21]. One hallmark of imprinted regions is that they tend to have a cluster of protein-coding and non-coding RNAs [57]. Moreover, these non-coding RNAs tend to regulate imprinted protein-coding genes in cis through association with chromatin- modifying complexes. The non-coding RNAs Air and Kcnqot1 are two large non-coding RNAs that associate with the chromatin modifying complexes (Air with G9a, and Kcnqot1 with PRC2 and G9a) and target them in cis to neighboring genes to affect chromatin modifications and therefore the transcription of those genes [24, 25]. These studies open up another interesting question: do non-coding RNAs help establish imprints in the germline by guiding chromatin complexes to imprinted loci to establish imprinting marks. These studies are usually hampered by the difficulty of culturing germ cells in vitro and the inability to obtain pure germ cells that are not contaminated by neighboring somatic cells. Nonetheless it is becoming increasingly clear that large non-coding RNAs are key modulators of allelic imprinting.
Potential role of large non-coding RNAs in pluripotency?
Several landmark studies have previously shown that the coordinated effort of several transcription factors is required to maintain or induce pluripotency [58]. These transcription factors occupy the promoters of genes involved in self-renewal, while genes that are involved in cellular differentiation are occupied by repressive complexes such as polycomb proteins [59]. This distribution is not restricted to the promoters of protein-coding genes but also have been observed at promoters of microRNAs [60] and more recently at large non-coding RNAs [61, 62]. These observations suggested a potential role for non-coding RNAs in pluripotency.
For example, Guttman et al identified approximately 100 lincRNA promoters bound by Sox2, Oct4 or Nanog for which direct regulation occurs. A more recent study by Mohamed et al. identified 105 and 335 large noncoding RNA proximal regions bound by Oct4 and Nanog respetively. The authors focused on four conserved long noncoding RNAs that are bound by Oct4 or Nanog. The induction of differentiation of mouse ES cells by retinoic acid affected the levels of these transcripts, similar to Oct4 and Nanog. Also, knockdown of either Oct4 or Nanog resulted in concomitant changes in the levels of these lncRNAs suggesting that these lncRNAs are likely to be direct targets of Oct4 and Nanog. Interestingly, siRNA-mediated knockdown of one of the four lncRNAs (known as Gomafu/Miat) resulted in the downregulation of several pluripotency markers including Oct4 and Nanog. These studies implicate lncRNAs in pluripotency and suggest that they may function in specific pathways that regulate cell fate decisions.
However, the mechanism of long RNAs mediated effects on pluripotency remains a key question. One possibility is that long non-coding RNAs can form ribonucleic-protein complexes with chromatin-modifying complexes to alter the expression of genes that contribute to the maintenance of pluripotency. Consistent with this notion a study found two large non-coding RNA in mouse ES cells to be associated with MLL1 and H3K4me3 [61]. Collectively, these studies point to a key role for large non-coding RNAs interfacing with the core pluripotency transcriptional machinery and chromatin modify complexes
Potential roles for non-coding RNAs and ribonucleoproetins in disease
The key functional roles of long non-coding RNAs has prompted researchers to investigate their contribution to human disease. Numerous studies have now found an association of human disease with mis-regualtion of long non-coding RNAs. However, there has been a great debate whether these long non-coding RNAs are causing the disease or they become mis-regulated as a consequence of disease. Two recent studies point to lincRNAs playing a role in the former [63, 64]. The lincRNA HOTAIR has recently been shown to be sufficient to drive metastasis by interfacing with chromatin-modifying complex PRC2 [63, 64]
Another study demonstrated that a lincRNA termed lincRNA-p21 plays a key role in the p53 tumor suppressor pathway [64]. Specifically, lincRNA-p21 is a direct target of p53 that serves as a global repressor in the p53 pathway. Loss of lincRNA-p21 function resulted in a similar tumor-supressor phenotype to p53. Thus, in contrast to HOTAIR, lincRNA-p21 maybe a “tumor-supressor lincRNA”. The mechanism of lincRNA-p21 based repression of p53 target genes required the ribonucleic-protein association with hnRNP-K. Interestingly, the physcial association between lincRNA-p21 and hnRNP-K was required to modulate the localization of hnRNP-K to genes repressed by p53 via lincRNA-p21. Collectively, HOTAIR and lincRNA-p21 represent a potential new class of “onco-“ and “tumor-suppressor lincRNAs” that are key regulators of cellular transformation by forming ribonucleic-protein interactions.
Long non-coding RNAs have also been shown to be mis-regulated in neurological disorders. Intriguingly, the brain has the second highest level of non-coding RNA expression after the testis. Previously, two independent studies identified two large non-coding RNAs that are transcribed in the opposite direction of FMR1, the gene implicated in fragile X syndrome [65, 66]. The first is a 2.4 kb non-coding RNA known as FMR4 and its transcribed about 200 bp upstream of FMR1. The second is an antisense transcript to FMR1 with multiple splice variants known as ASFMR1. Both FMR4 and ASFMR1 become silenced in fragile X syndrome patients similar to FMR1. However, it is not clear how these non-coding RNAs contribute to the symptoms of fragile X patients. The function of ASFMR1 is not known, however, FMR4 has an anti-apoptotic function and may functions by protecting neuronal cells during development. Also, an antisense non-coding RNA to BACE1, an enzyme that is responsible for cleaving APP, was found to be elevated in Alzheimer’s patients. This non-coding RNA stabilizes BACE1 mRNA in vitro and in vivo which leads to increased BACE1 protein levels and therefore contributes to increased AB-42 accumulation in brain cells [67]. Other lncRNAs have been implicated in cardiac disease, spinocerebeullar ataxia type 8 and many others [57]. These association studies have now set the stage for follow-up functional studies that will reveal how lncRNA contribute to human disease and if they are driving disease state or merely reflecting indirect effects of changes in the transcriptional program of cells.
Discussion
There are extensive RNA-protein interactions in all kingdoms of life. Here we explored their key roles in numerous cellular processes ranging from translation, splicing, nuclear organization and potentially epigenetic regulation of gene expression. A notable commonality amongst the diversity of these regulatory mechanisms is ncRNA bridging diverse protein complexes. The notion of ncRNA as a “molecular scaffold” proposed by Zappula and Cech in the context of telomere replication may also apply to epigenetic regulation of cellular identity [9, 21, 68, 69] and potentially numerous other cellular processes [21]. Thus, RNA-Protein complex diversity could be tantalizing solution to the observation of increasing ncRNA content in parallel with cellular complexity. For example, the multiplicity of lncRNAs could result in an array of novel RNA-protein interactions possibly to fine tune increasingly complex cellular circuitry.
Consistent with this idea is the observation that perturbations of these RNA-Protein interactions can result disease. For example, HOTAIR and lincRNA-p21 serve as modular molecular scaffolds bridging key chromatin modifying complexes that if perturbed can result in cellular transformation [64, 68]. A clear advantage ncRNA as a molecular modulator of protein complex diversity is the inherent ability activate a ncRNA to facilitate a unique ribonucleic-protein complex, with sufficiently less constraints than deriving a protein specific to this function. Collectively the studies reviewed herein, underscore the need for further exploration into the spectrum of RNA-Protein interactions and their importance in disease.
In summary, there is clear evidence across all kingdoms of life that large non-coding RNA-Protein interactions fine-tune a wide range of cellular processes. These ever emerging diversity of RNA-protein interactions are shedding new light into the cell circuitry defining and maintaining cellular complexity.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Carninci P, Kasukawa T, Katayama S, Gough J, Frith MC, Maeda N, et al. The transcriptional landscape of the mammalian genome. Science. 2005;309:1559–1563. doi: 10.1126/science.1112014. [DOI] [PubMed] [Google Scholar]
- 2.Mattick JS, Makunin IV. Non-coding RNA. Hum Mol Genet. 2006;15(Spec No 1):R17–R29. doi: 10.1093/hmg/ddl046. [DOI] [PubMed] [Google Scholar]
- 3.Kapranov P, Cheng J, Dike S, Nix DA, Duttagupta R, Willingham AT, et al. RNA maps reveal new RNA classes and a possible function for pervasive transcription. Science. 2007;316:1484–1488. doi: 10.1126/science.1138341. [DOI] [PubMed] [Google Scholar]
- 4.Taft RJ, Pheasant M, Mattick JS. The relationship between non-protein-coding DNA and eukaryotic complexity. Bioessays. 2007;29:288–299. doi: 10.1002/bies.20544. [DOI] [PubMed] [Google Scholar]
- 5.Holcik M, Pestova TV. Translation mechanism and regulation: old players, new concepts. Meeting on translational control and non-coding RNA. EMBO Rep. 2007;8:639–643. doi: 10.1038/sj.embor.7400988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lundblad EW, Altman S. Inhibition of gene expression by RNase P. N Biotechnol. 2010 doi: 10.1016/j.nbt.2010.03.003. [DOI] [PubMed] [Google Scholar]
- 7.Kikovska E, Svard SG, Kirsebom LA. Eukaryotic RNase P RNA mediates cleavage in the absence of protein. Proc Natl Acad Sci U S A. 2007;104:2062–2067. doi: 10.1073/pnas.0607326104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hendrickson DG, Hogan DJ, McCullough HL, Myers JW, Herschlag D, Ferrell JE, et al. Concordant regulation of translation and mRNA abundance for hundreds of targets of a human microRNA. PLoS Biol. 2009;7 doi: 10.1371/journal.pbio.1000238. e1000238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Khalil AM, Guttman M, Huarte M, Garber M, Raj A, Rivea Morales D, et al. Many human large intergenic noncoding RNAs associate with chromatin-modifying complexes and affect gene expression. Proc Natl Acad Sci U S A. 2009;106:11667–11672. doi: 10.1073/pnas.0904715106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rinn JL, Kertesz M, Wang JK, Squazzo SL, Xu X, Brugmann SA, et al. Functional demarcation of active and silent chromatin domains in human HOX loci by noncoding RNAs. Cell. 2007;129:1311–1323. doi: 10.1016/j.cell.2007.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mili S, Steitz JA. Evidence for reassociation of RNA-binding proteins after cell lysis: implications for the interpretation of immunoprecipitation analyses. RNA. 2004;10:1692–1694. doi: 10.1261/rna.7151404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hafner M, Landthaler M, Burger L, Khorshid M, Hausser J, Berninger P, et al. PAR-CliP--a method to identify transcriptome-wide the binding sites of RNA binding proteins. J Vis Exp. 2010 doi: 10.3791/2034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hafner M, Landthaler M, Burger L, Khorshid M, Hausser J, Berninger P, et al. Transcriptome-wide identification of RNA-binding protein and microRNA target sites by PAR-CLIP. Cell. 2010;141:129–141. doi: 10.1016/j.cell.2010.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Licatalosi DD, Mele A, Fak JJ, Ule J, Kayikci M, Chi SW, et al. HITS-CLIP yields genome-wide insights into brain alternative RNA processing. Nature. 2008;456:464–469. doi: 10.1038/nature07488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bernstein BE, Meissner A, Lander ES. The mammalian epigenome. Cell. 2007;128:669–681. doi: 10.1016/j.cell.2007.01.033. [DOI] [PubMed] [Google Scholar]
- 16.Kouzarides T. Chromatin modifications and their function. Cell. 2007;128:693–705. doi: 10.1016/j.cell.2007.02.005. [DOI] [PubMed] [Google Scholar]
- 17.Li G, Margueron R, Ku M, Chambon P, Bernstein BE, Reinberg D. Jarid2 and PRC2, partners in regulating gene expression. Genes Dev. 2010 doi: 10.1101/gad.1886410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pasini D, Cloos PA, Walfridsson J, Olsson L, Bukowski JP, Johansen JV, et al. JARID2 regulates binding of the Polycomb repressive complex 2 to target genes in ES cells. Nature. 2010 doi: 10.1038/nature08788. [DOI] [PubMed] [Google Scholar]
- 19.Peng JC, Valouev A, Swigut T, Zhang J, Zhao Y, Sidow A, et al. Jarid2/Jumonji coordinates control of PRC2 enzymatic activity and target gene occupancy in pluripotent cells. Cell. 2009;139:1290–1302. doi: 10.1016/j.cell.2009.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shen X, Kim W, Fujiwara Y, Simon MD, Liu Y, Mysliwiec MR, et al. Jumonji modulates polycomb activity and self-renewal versus differentiation of stem cells. Cell. 2009;139:1303–1314. doi: 10.1016/j.cell.2009.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Koziol MJ, Rinn JL. RNA traffic control of chromatin complexes. Curr Opin Genet Dev. 2010;20:142–148. doi: 10.1016/j.gde.2010.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhao J, Sun BK, Erwin JA, Song JJ, Lee JT. Polycomb proteins targeted by a short repeat RNA to the mouse X chromosome. Science. 2008;322:750–756. doi: 10.1126/science.1163045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sleutels F, Zwart R, Barlow DP. The non-coding Air RNA is required for silencing autosomal imprinted genes. Nature. 2002;415:810–813. doi: 10.1038/415810a. [DOI] [PubMed] [Google Scholar]
- 24.Nagano T, Mitchell JA, Sanz LA, Pauler FM, Ferguson-Smith AC, Feil R, et al. The Air noncoding RNA epigenetically silences transcription by targeting G9a to chromatin. Science. 2008;322:1717–1720. doi: 10.1126/science.1163802. [DOI] [PubMed] [Google Scholar]
- 25.Pandey RR, Mondal T, Mohammad F, Enroth S, Redrup L, Komorowski J, et al. Kcnq1ot1 antisense noncoding RNA mediates lineage-specific transcriptional silencing through chromatin-level regulation. Mol Cell. 2008;32:232–246. doi: 10.1016/j.molcel.2008.08.022. [DOI] [PubMed] [Google Scholar]
- 26.Yu W, Gius D, Onyango P, Muldoon-Jacobs K, Karp J, Feinberg AP, et al. Epigenetic silencing of tumour suppressor gene p15 by its antisense RNA. Nature. 2008;451:202–206. doi: 10.1038/nature06468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Liu F, Marquardt S, Lister C, Swiezewski S, Dean C. Targeted 3' processing of antisense transcripts triggers Arabidopsis FLC chromatin silencing. Science. 2010;327:94–97. doi: 10.1126/science.1180278. [DOI] [PubMed] [Google Scholar]
- 28.Engelke DR, Ng SY, Shastry BS, Roeder RG. Specific interaction of a purified transcription factor with an internal control region of 5S RNA genes. Cell. 1980;19:717–728. doi: 10.1016/s0092-8674(80)80048-1. [DOI] [PubMed] [Google Scholar]
- 29.Pelham HR, Brown DD. A specific transcription factor that can bind either the 5S RNA gene or 5S RNA. Proc Natl Acad Sci U S A. 1980;77:4170–4174. doi: 10.1073/pnas.77.7.4170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kino T, Hurt DE, Ichijo T, Nader N, Chrousos GP. Noncoding RNA gas5 is a growth arrest- and starvation-associated repressor of the glucocorticoid receptor. Sci Signal. 2010;3:ra8. doi: 10.1126/scisignal.2000568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Willingham AT, Orth AP, Batalov S, Peters EC, Wen BG, Aza-Blanc P, et al. A strategy for probing the function of noncoding RNAs finds a repressor of NFAT. Science. 2005;309:1570–1573. doi: 10.1126/science.1115901. [DOI] [PubMed] [Google Scholar]
- 32.Martianov I, Ramadass A, Serra Barros A, Chow N, Akoulitchev A. Repression of the human dihydrofolate reductase gene by a non-coding interfering transcript. Nature. 2007;445:666–670. doi: 10.1038/nature05519. [DOI] [PubMed] [Google Scholar]
- 33.Seila AC, Calabrese JM, Levine SS, Yeo GW, Rahl PB, Flynn RA, et al. Divergent transcription from active promoters. Science. 2008;322:1849–1851. doi: 10.1126/science.1162253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Post-transcriptional processing generates a diversity of 5'-modified long and short RNAs. Nature. 2009;457:1028–1032. doi: 10.1038/nature07759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Taft RJ, Glazov EA, Cloonan N, Simons C, Stephen S, Faulkner GJ, et al. Tiny RNAs associated with transcription start sites in animals. Nat Genet. 2009;41:572–578. doi: 10.1038/ng.312. [DOI] [PubMed] [Google Scholar]
- 36.Bond CS, Fox AH. Paraspeckles: nuclear bodies built on long noncoding RNA. J Cell Biol. 2009;186:637–644. doi: 10.1083/jcb.200906113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Clemson CM, Hutchinson JN, Sara SA, Ensminger AW, Fox AH, Chess A, et al. An Architectural Role for a Nuclear Noncoding RNA: NEAT1 RNA Is Essential for the Structure of Paraspeckles. Mol Cell. 2009 doi: 10.1016/j.molcel.2009.01.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sunwoo H, Dinger ME, Wilusz JE, Amaral PP, Mattick JS, Spector DL. MEN varepsilon/beta nuclear-retained non-coding RNAs are up-regulated upon muscle differentiation and are essential components of paraspeckles. Genome Res. 2009;19:347–359. doi: 10.1101/gr.087775.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chen LL, Carmichael GG. Altered nuclear retention of mRNAs containing inverted repeats in human embryonic stem cells: functional role of a nuclear noncoding RNA. Mol Cell. 2009;35:467–478. doi: 10.1016/j.molcel.2009.06.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sone M, Hayashi T, Tarui H, Agata K, Takeichi M, Nakagawa S. The mRNA-like noncoding RNA Gomafu constitutes a novel nuclear domain in a subset of neurons. J Cell Sci. 2007;120:2498–2506. doi: 10.1242/jcs.009357. [DOI] [PubMed] [Google Scholar]
- 41.Noma K, Sugiyama T, Cam H, Verdel A, Zofall M, Jia S, et al. RITS acts in cis to promote RNA interference-mediated transcriptional and post-transcriptional silencing. Nat Genet. 2004;36:1174–1180. doi: 10.1038/ng1452. [DOI] [PubMed] [Google Scholar]
- 42.Sugiyama T, Cam H, Verdel A, Moazed D, Grewal SI. RNA-dependent RNA polymerase is an essential component of a self-enforcing loop coupling heterochromatin assembly to siRNA production. Proc Natl Acad Sci U S A. 2005;102:152–157. doi: 10.1073/pnas.0407641102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kanhere A, Viiri K, Araujo CC, Rasaiyaah J, Bouwman RD, Whyte WA, et al. Short RNAs are transcribed from repressed polycomb target genes and interact with polycomb repressive complex-2. Mol Cell. 2010;38:675–688. doi: 10.1016/j.molcel.2010.03.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 45.Pratt AJ, MacRae IJ. The RNA-induced silencing complex: a versatile gene-silencing machine. J Biol Chem. 2009;284:17897–17901. doi: 10.1074/jbc.R900012200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Liu J, Carmell MA, Rivas FV, Marsden CG, Thomson JM, Song JJ, et al. Argonaute2 is the catalytic engine of mammalian RNAi. Science. 2004;305:1437–1441. doi: 10.1126/science.1102513. [DOI] [PubMed] [Google Scholar]
- 47.Girard A, Sachidanandam R, Hannon GJ, Carmell MA. A germline-specific class of small RNAs binds mammalian Piwi proteins. Nature. 2006;442:199–202. doi: 10.1038/nature04917. [DOI] [PubMed] [Google Scholar]
- 48.Grivna ST, Beyret E, Wang Z, Lin H. A novel class of small RNAs in mouse spermatogenic cells. Genes Dev. 2006;20:1709–1714. doi: 10.1101/gad.1434406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lau NC, Seto AG, Kim J, Kuramochi-Miyagawa S, Nakano T, Bartel DP, et al. Characterization of the piRNA complex from rat testes. Science. 2006;313:363–367. doi: 10.1126/science.1130164. [DOI] [PubMed] [Google Scholar]
- 50.Khalil AM, Wahlestedt C. Epigenetic mechanisms of gene regulation during mammalian spermatogenesis. Epigenetics. 2008;3:21–28. doi: 10.4161/epi.3.1.5555. [DOI] [PubMed] [Google Scholar]
- 51.Carmell MA, Girard A, van de Kant HJ, Bourc'his D, Bestor TH, de Rooij DG, et al. MIWI2 is essential for spermatogenesis and repression of transposons in the mouse male germline. Dev Cell. 2007;12:503–514. doi: 10.1016/j.devcel.2007.03.001. [DOI] [PubMed] [Google Scholar]
- 52.Grivna ST, Pyhtila B, Lin H. MIWI associates with translational machinery and PIWI-interacting RNAs (piRNAs) in regulating spermatogenesis. Proc Natl Acad Sci U S A. 2006;103:13415–13420. doi: 10.1073/pnas.0605506103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Aravin AA, Sachidanandam R, Girard A, Fejes-Toth K, Hannon GJ. Developmentally regulated piRNA clusters implicate MILI in transposon control. Science. 2007;316:744–747. doi: 10.1126/science.1142612. [DOI] [PubMed] [Google Scholar]
- 54.Xu M, Medvedev S, Yang J, Hecht NB. MIWI-independent small RNAs (MSY-RNAs) bind to the RNA-binding protein, MSY2, in male germ cells. Proc Natl Acad Sci U S A. 2009;106:12371–12376. doi: 10.1073/pnas.0903944106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Brown CJ, Ballabio A, Rupert JL, Lafreniere RG, Grompe M, Tonlorenzi R, et al. A gene from the region of the human X inactivation centre is expressed exclusively from the inactive X chromosome. Nature. 1991;349:38–44. doi: 10.1038/349038a0. [DOI] [PubMed] [Google Scholar]
- 56.Avner P, Heard E. X-chromosome inactivation: counting, choice and initiation. Nat Rev Genet. 2001;2:59–67. doi: 10.1038/35047580. [DOI] [PubMed] [Google Scholar]
- 57.Mattick JS. The genetic signatures of noncoding RNAs. PLoS Genet. 2009;5 doi: 10.1371/journal.pgen.1000459. e1000459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Loh YH, Wu Q, Chew JL, Vega VB, Zhang W, Chen X, et al. The Oct4 and Nanog transcription network regulates pluripotency in mouse embryonic stem cells. Nat Genet. 2006;38:431–440. doi: 10.1038/ng1760. [DOI] [PubMed] [Google Scholar]
- 59.Bracken AP, Dietrich N, Pasini D, Hansen KH, Helin K. Genome-wide mapping of Polycomb target genes unravels their roles in cell fate transitions. Genes Dev. 2006;20:1123–1136. doi: 10.1101/gad.381706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Marson A, Levine SS, Cole MF, Frampton GM, Brambrink T, Johnstone S, et al. Connecting microRNA genes to the core transcriptional regulatory circuitry of embryonic stem cells. Cell. 2008;134:521–533. doi: 10.1016/j.cell.2008.07.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Dinger ME, Amaral PP, Mercer TR, Pang KC, Bruce SJ, Gardiner BB, et al. Long noncoding RNAs in mouse embryonic stem cell pluripotency and differentiation. Genome Res. 2008;18:1433–1445. doi: 10.1101/gr.078378.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Guttman M, Amit I, Garber M, French C, Lin MF, Feldser D, et al. Chromatin signature reveals over a thousand highly conserved large non-coding RNAs in mammals. Nature. 2009;458:223–227. doi: 10.1038/nature07672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Gupta RA, Shah N, Wang KC, Kim J, Horlings HM, Wong DJ, et al. Long non-coding RNA HOTAIR reprograms chromatin state to promote cancer metastasis. Nature. 2010;464:1071–1076. doi: 10.1038/nature08975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Huarte M, Guttman M, Feldser D, Garber M, Koziol MJ, Kenzelmann-Broz D, et al. A Large Intergenic Noncoding RNA Induced by p53 Mediates Global Gene Repression in the p53 Response. Cell. 2010 doi: 10.1016/j.cell.2010.06.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Khalil AM, Faghihi MA, Modarresi F, Brothers SP, Wahlestedt C. A novel RNA transcript with antiapoptotic function is silenced in fragile x syndrome. PLoS ONE. 2008;3:e1486. doi: 10.1371/journal.pone.0001486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Ladd PD, Smith LE, Rabaia NA, Moore JM, Georges SA, Hansen RS, et al. An Antisense Transcript Spanning the CGG Repeat Region of FMR1 is Upregulated in Premutation Carriers but Silenced in Full Mutation Individuals. Hum Mol Genet. 2007 doi: 10.1093/hmg/ddm293. [DOI] [PubMed] [Google Scholar]
- 67.Faghihi MA, Modarresi F, Khalil AM, Wood DE, Sahagan BG, Morgan TE, et al. Expression of a noncoding RNA is elevated in Alzheimer's disease and drives rapid feed-forward regulation of beta-secretase. Nat Med. 2008;14:723–730. doi: 10.1038/nm1784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Tsai MC, Manor O, Wan Y, Mosammaparast N, Wang JK, Lan F, et al. Long Noncoding RNA as Modular Scaffold of Histone Modification Complexes. Science. 2010 doi: 10.1126/science.1192002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Zappulla DC, Cech TR. RNA as a flexible scaffold for proteins: yeast telomerase and beyond. Cold Spring Harb Symp Quant Biol. 2006;71:217–224. doi: 10.1101/sqb.2006.71.011. [DOI] [PubMed] [Google Scholar]