Abstract
Although the physiological relevance of retinoids and steroids in vertebrates is very well established, the origin and evolution of the genetic machineries implicated in their metabolic pathways is still very poorly understood. We investigated the evolution of these genetic networks by conducting an exhaustive survey of components of the retinoid and steroid pathways in the genome of the invertebrate chordate amphioxus (Branchiostoma floridae). Due to its phylogenetic position at the base of chordates, amphioxus is a very useful model to identify and study chordate versus vertebrate innovations, both on a morphological and a genomic level. We have characterized more than 220 amphioxus genes evolutionarily related to vertebrate components of the retinoid and steroid pathways and found that, globally, amphioxus has orthologs of most of the vertebrate components of these two pathways, with some very important exceptions. For example, we failed to identify a vertebrate-like machinery for retinoid storage, transport, and delivery in amphioxus and were also unable to characterize components of the adrenal steroid pathway in this invertebrate chordate. The absence of these genes from the amphioxus genome suggests that both an elaboration and a refinement of the retinoid and steroid pathways took place at the base of the vertebrate lineage. In stark contrast, we also identified massive amplifications in some amphioxus gene families, most extensively in the short-chain dehydrogenase/reductase superfamily, which, based on phylogenetic and genomic linkage analyses, were likely the result of duplications specific to the amphioxus lineage. In sum, this detailed characterization of genes implicated in retinoid and steroid signaling in amphioxus allows us not only to reconstruct an outline of these pathways in the ancestral chordate but also to discuss functional innovations in retinoid homeostasis and steroid-dependent regulation in both cephalochordate and vertebrate evolution.
Keywords: Branchiostoma floridae, cephalochordates, gene duplication, metabolic networks, nuclear receptor signaling cascades
Introduction
Acting as intercellular messengers, retinoids and steroids contribute to the coordination of developmental processes and cellular functions in vertebrates. At the signaling level, both signaling systems rely on members of the nuclear hormone receptor family to regulate transcription of target genes, and, classically, the evolution of the retinoid and steroid pathways has been inferred by identification and characterization of nuclear hormone receptors in different animal lineages (Mangelsdorf et al. 1995; Escriva et al. 1997, 2000, 2004; Bertrand et al. 2004; Markov et al. 2008). In contrast, although retinoids and steroids use related machineries and similar strategies to regulate their physiological activity, the evolution of these metabolic pathways is still poorly understood.
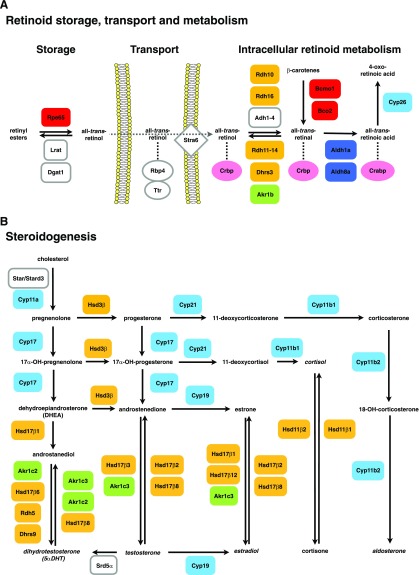
In vertebrates, retinoid metabolism, storage and transport requires no less than seven enzymatic activities, three binding capacities, one cell-surface receptor, and one plasma protein (fig. 1A) (for recent reviews on retinoid metabolism, see Albalat 2009; Theodosiou, et al. 2010). In brief, retinoid metabolism begins when dietary β-carotenes, the main source of retinoids, are cleaved by β,β-carotene-15,15′-oxygenase (Bcmo1; also known as Bco or Bcox), which generates two molecules of retinal. This retinal is subsequently reduced to retinol (vitamin A) by retinaldehyde reductases, a heterogeneous group of enzymes of diverse protein families (Dalfó et al. 2007 and references therein). The retinol is esterified to retinyl esters by lecithin-retinol acyltransferase (Lrat) or acyl-CoA-retinol acyltransferase (Dgat1) enzymes and stored in the liver. Retinyl esters can be mobilized from the liver by retinyl ester hydrolases (including Rpe65) that transform retinyl esters back to retinol. In the blood stream, this retinol is transported by retinol-binding protein 4 (Rbp4) and transthyretin (Ttr) and the subsequent cellular uptake of retinol is mediated by Stra6 (stimulated by retinoic acid protein 6) (Kawaguchi et al. 2007; Theodosiou et al. 2010).
FIG. 1.—
Vertebrate retinoid (A) and steroid (B) metabolism. (A) The main components of retinoid metabolism, storage, and transport in vertebrates are shown. Proteins are boxed, and retinoids are in black. Proteins of the same family are shown in identical colors. Enzymatic reactions are shown with arrows. The dotted arrow indicates retinoid transport and dotted lines highlight retinoid binding to proteins. (B) The main enzymatic reactions of steroidogenesis in vertebrates are indicated. Enzymatic reactions are shown with arrows. Proteins are boxed, and steroids are in black. Proteins of the same family are shown in identical colors.
Within the cell, retinol is oxidized to retinal and, in a second step, to retinoic acid (RA), the main physiologically active retinoid. In this process, retinol oxidation is the rate-limiting step and a key reaction for the physiological control of RA availability. Several dehydrogenases of the medium-chain dehydrogenase/reductase (MDR-Adh) superfamily, including Adh1, Adh3, and Adh4, and of the short-chain dehydrogenase/reductase (SDR) superfamily, such as Rdh16 (also known as Rodh4) and Rdh10, catalyze this rate-limiting reaction in vitro (Gough et al. 1998; Belyaeva, Johnson, et al. 2008). Retinal produced by these enzymes is then transformed to RA through the action of retinaldehyde dehydrogenases (Aldh1a and Aldh8a), which, in close coordination with RA-degrading enzymes of the cytochrome P450 subfamily 26 (Cyp26) class, regulate the spatiotemporal levels of RA (Niederreither et al. 2002; Reijntjes et al. 2005). Inside the cell, retinol, retinal and RA are generally bound to proteins that solubilize and stabilize retinoids in the aqueous environment of the cell. In vertebrates, several cellular retinol-binding proteins (Crbp) and cellular RA binding proteins (Crabp) have been identified (reviewed in Noy 2000), although it is still unclear whether the main physiological task of these binding proteins is to deliver retinoids to or to sequester them from the enzymes.
Similar to retinoid metabolism, steroidogenesis (fig. 1B) is also a complex biochemical pathway that involves numerous cytochrome P450 (Cyp) and hydroxysteroid dehydrogenases (Hsd) enzymes (Payne and Hales 2004). In vertebrates, steroidogenesis is initiated by Cyp11a enzymes that produce pregnenolone from cholesterol transferred from the outer to the inner membrane of mitochondria by Star (for steroidogenic acute regulatory protein, also known as Stard1) or by Stard3 (for Star-related lipid transfer domain containing 3, also known as Mln64) proteins (Clark et al. 1994; Watari et al. 1997; Stocco 2001). Pregnenolone is transformed into progesterone by 3β-hydroxysteroid dehydrogenase/Δ5-Δ4-isomerase (Hsd3β) or to dehydroepiandrosterone (DHEA) by Cyp17. Progesterone can be metabolized to aldosterone, cortisol, or androstenedione through different pathways involving Cyp11b1, Cyp11b2, Cyp17, and Cyp21 enzymes, whereas DHEA can be transformed to androstanediol and to dihydrotestosterone (5αDHT), a potent androgen. 5αDHT can also be synthesized from androstenedione via testosterone by the action of type 1, type 2, and type 3 5α-reductases (Srd5α1, Srd5α2, and Srd5α3) (Russell and Wilson 1994; Uemura et al. 2008). Finally, androstenedione can be transformed to estrone by the aromatase enzyme Cyp19 and further to estradiol by Hsd enzymes of the SDR and aldo-keto reductases (Akr) superfamilies (reviewed in Payne and Hales 2004).
In vertebrates, pairs of Hsd enzymes participate in a prereceptor regulatory system of steroid action through the interconversion of weak to potent hormones (Penning 2003). This prereceptor regulation was first documented for mineralocorticoid receptors and Hsd11β enzymes (Funder et al. 1988; Oppermann et al. 1997). Hsd11β type 1 enzymes convert inactive cortisone into the potent glucocorticoid cortisol, whereas Hsd11β type 2 enzymes catalyze the opposite reaction, hence neutralizing the hormone. Other pairs of activating/deactivating Hsd enzymes are: Hsd17β1 (and Hsd17β12 and Akr1c3) versus Hsd17β2 (and Hsd17β8) enzymes for estrone (weak) to estradiol (potent) conversion (and vice versa), Hsd17β3 (and Akr1c3) and Hsd17β2 (and Hsd17β8) for androstenedione (weak) to testosterone (potent) conversion (and vice versa), and Akr1c2 (and Hsd17β6, Rdh5, and Dhrs9) and Akr1c3 (and Akr1c2 and Hsd17β8) for androstanediol (weak) to 5αDHT (potent) conversion (and vice versa) (Biswas and Russell 1997; Fomitcheva et al. 1998; Penning et al. 2000; Chetyrkin, Belyaeva, et al. 2001; Chetyrkin, Hu, et al. 2001; Huang and Luu-The 2001).
Although the retinoid and steroid metabolic pathways have extensively been studied in vertebrates, the evolutionary origins of these signaling systems are not fully understood. To investigate the evolution of these signaling systems in vertebrates, we have analyzed the genome of the invertebrate chordate amphioxus (Branchiostoma floridae). Phylogenetically positioned at the base of the chordate phylum, amphioxus is the extant invertebrate most closely resembling the last invertebrate ancestor of vertebrates, both in terms of morphology and genome (Schubert, Escriva, et al. 2006; Holland et al. 2008; Putnam et al. 2008). We have identified more than 220 amphioxus genes evolutionarily related to vertebrate components of the retinoid and steroid metabolisms, and our phylogenetic analyses allowed us to reconstruct the retinoid and steroid machineries of the ancestral chordate. Interestingly, we also found compelling evidence for massive expansions of some components of the retinoid and steroid pathways in amphioxus (most distinctively within the SDR superfamily). Based on phylogenetic and genomic linkage data, we hypothesize that these expansions arose by extensive lineage-specific gene duplications, suggesting that the retinoid and steroid pathways in amphioxus are much more complex than initially expected. Taken together, our work demonstrates that functional diversification upon gene duplication in chordates is by no means limited to the vertebrate lineage.
Materials and Methods
Genome Analyses, Gene Identification, and Sequence Alignment
We have carried out exhaustive searches of the amphioxus (B. floridae) genome (versions 1.0 and 2.0) (genome.jgi-psf.org/Brafl1/Brafl1.home.html) for components of retinoid and steroid metabolism. Version 1.0 of the B. floridae genome was preferentially used because it is more inclusive than version 2.0, from which some sequences have been removed (Putnam et al. 2008). Moreover, for the genomic linkage survey of the B. floridae sequences, we needed genome position information, which is unavailable for version 2.0 (Putnam et al. 2008).
As recommended by Kuzniar et al. (2008), we searched the amphioxus genome for orthologous sequences using precomputed sets of homologous genes. In brief, for each component of retinoid and steroid signaling, we downloaded all related sequences from the protein families defined in the Ensembl database (version 52) (www.ensembl.org) (Flicek et al. 2010), to which we added sequences from Uniprot (www.uniprot.org) and from the nonredundant protein database at the National Center for Biotechnology Information (www.ncbi.nlm.nih.gov). Whenever possible, we added other families to the analysis to properly outgroup our phylogenies. Sequences identified in the genome of the cnidarian Nematostella vectensis (version 1.0) (genome.jgi-psf.org/Nemve1/Nemve1.home.html) were also included in the analyses and were used wherever possible to root the phylogenetic trees. Protein alignments were calculated with ClustalW (version 2.0) (Larkin et al. 2007) and Muscle (version 3.6) (Edgar 2004). The results of both alignment tools were merged and subsequently manually refined using SeaView (version 3.2) (Galtier et al. 1996) to exclude redundant and improperly annotated sequences. In contrast to our initial survey of the amphioxus genome, where only human sequences were used for Blast searches (Holland et al. 2008), in the present analysis, all members of a given protein family were used for Blast searches of the complete predicted protein data sets of the amphioxus genome. This comprehensive methodology yielded an inclusive and reproducible set of amphioxus sequences potentially homologous to the different protein families of interest.
All amphioxus sequences obtained by these database searches were retrieved, sorted, and allelic polymorphs were removed using nucleotide and amino acid alignments. In order to identify allelic polymorphs, nucleotide alignments were analyzed by eye and two sequences were considered as alleles, when, with the exception of a few polymorphic changes at third codon positions, large stretches of the compared sequences were identical. Intronic sequences were also aligned, and conservation of intronic nucleotidic sequences stretches were used as additional criterion for the identification of allelic polymorphs. Of the different allelic polymorphs, the sequence with the longest overall alignment with the gene family in question was retained. The sequence identification numbers of all retrieved amphioxus sequences are given in the Supplementary Material online. The genomic position of each amphioxus gene was obtained from version 1.0 of the B. floridae genome (Putnam et al. 2008) and used as a basis for genomic linkage analyses. Automatically annotated genes were manually revised taking into consideration the data available in both version 1.0 and version 2.0 of the B. floridae genome, and errors were corrected to maximize the similarity with known proteins. This additional verification step involving both versions of the B. floridae genome yielded highly consistent results. The treated amphioxus data were subsequently incorporated into the protein family alignments used for the initial Blast analysis of the amphioxus genome. Alignments were obtained using ClustalW (version 2.0) (Larkin et al. 2007) and Muscle (version 3.6) (Edgar 2004). The results of both alignment tools were merged and subsequently subjected to manual refinement. All alignment files are available from the authors.
Phylogenetic Analyses
All sequences retrieved from the amphioxus genome were subjected to phylogenetic analyses in the context of their proper protein families. For each family, phylogenies were calculated with sequences from humans, zebrafish, and amphioxus using N. vectensis sequences as outgroup. The SDR tree was calculated without N. vectensis representatives, and each family hence served as outgroup for the other families, which resulted in a reliable phylogenetic reconstruction of this superfamily. The iLbp tree was also calculated without N. vectensis representatives because we failed to identify iLbp-like sequences in the N. vectensis genome. Phylogenetic tree reconstructions were carried out using both the neighbor joining (NJ) and the maximum likelihood (ML) methods. For both NJ and ML analyses, robustness of the obtained tree topologies was assessed with 1,000 bootstrap replicates. NJ trees were calculated with Phylo_Win (version 2.0) using a Poisson correction and pairwise gap removal (Galtier et al. 1996), whereas ML trees were obtained with PhyML (version 2.4.4) using a Jones, Taylor, and Thornton model, a discrete gamma model with four categories, a gamma shape parameter of two and a ML estimate of the proportion of invariable sites (Guindon and Gascuel 2003). Phylogenetic trees were rooted with NJPlot (version 1.0) (pbil.univ-lyon1.fr/software/njplot.html) (Perrière and Gouy 1996) and formatted using FigTree (version 1.3.1) (tree.bio.ed.ac.uk).
Results
We have carried out an exhaustive search of the amphioxus (B. floridae) genome for components of retinoid and steroid metabolism. To our great surprise, we found evidence for amphioxus-specific duplications in most of the families analyzed. The best example for this tendency, with more than 100 representatives in amphioxus (compared with only 37 in humans), is the SDR superfamily. We will hence first describe our analysis of amphioxus homologs of vertebrate dehydrogenases/reductases with activity against retinol and/or hydroxysteroids, before presenting other elements implicated in retinoid and steroid metabolism.
Identification of Retinol and Hydroxysteroid Dehydrogenases/Reductases in Amphioxus
SDR-Rdh and SDR-Hsd Sequences
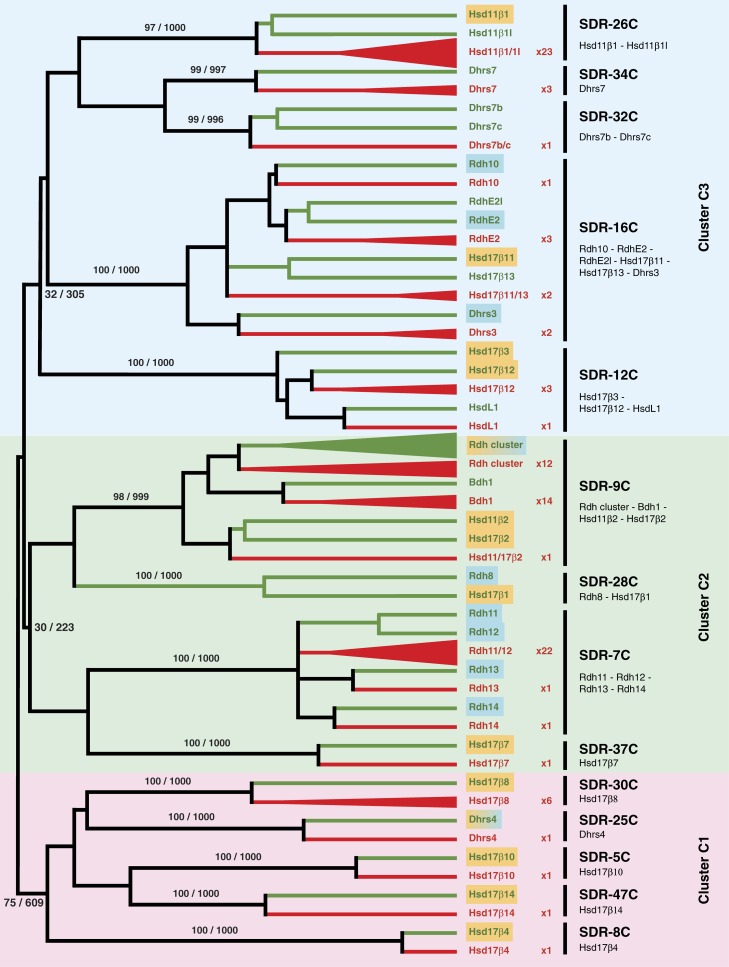
The SDR superfamily includes 73 human members with a substrate spectrum ranging from polyols, retinoids, steroids, and fatty acid derivatives to xenobiotics. We have focused our analysis on those SDR families that include enzymes active against retinoids and steroids (Belyaeva and Kedishvili 2006; Dalfó et al. 2007; Wu et al. 2007). According to the recent reclassification of the SDR superfamily, based on sequence analyses, predicted secondary structure (Bray et al. 2009), and hidden Markov models (Kallberg et al. 2010), we have therefore analyzed the SDR-Rdh and SDR-Hsd enzyme families 5C, 7C, 8C, 9C, 12C, 16C, 25C, 26C, 28C, 30C, 32C, 34C, 37C, and 47C, with a total of 37 members in humans. A survey of the B. floridae genome identified 107 genes encoding SDR enzymes belonging to these families and, hence, with putative functions in amphioxus retinoid or steroid metabolism. Of these 107 amphioxus SDRs, 6 partial sequences were excluded because they were too short for reliable phylogenetic tree reconstruction. Phylogenetic analysis of the retained amphioxus, zebrafish, and human SDR representatives combined with genomic linkage analysis of the amphioxus genes revealed the evolutionary history of the chordate Rdh/Hsd enzymes. These results will be described following the order of the individual SDR families within the overall SDR tree (fig. 2; supplementary fig. S1, Supplementary Material online).
FIG. 2.—
Schematic phylogeny of members of the SDR superfamily with activity against retinoids and/or steroids. The nomenclature of the three major SDR clusters (C1, C2, and C3) as well as of the individual SDR families is based on the recent reclassification of the SDR superfamily (Bray et al. 2009; Kallberg et al. 2010). Bootstrap values (NJ/ML) supporting the three major SDR clusters (C1, C2, and C3) and each of the 14 SDR families are indicated. Retinoid (blue) and steroid (orange) activities of the vertebrate enzymes are indicated, illustrating that both enzymatic activities are commonly found within the SDR superfamily, sometimes even within a single SDR family. Vertebrate branches are in green, whereas amphioxus branches are in red. The number of amphioxus sequences in each branch is indicated.
Family SDR-26C: Hsd11β1-Hsd11β1l Enzymes
Hsd11β type 1 enzymes convert the inactive glucocorticoid cortisone into the potent cortisol, and, in concert with Hsd11β type 2, regulate cortisol levels (Funder et al. 1988). Hsd11β1 enzymes are closely related to the Hsd11β1l proteins (also named Sdr10b or Hsd11β3 in chicken and fish), whose function remains unknown (Baker 2004b). The amphioxus genome contains 23 family SDR-26C genes (fig. 2; supplementary fig. S1, Supplementary Material online; NJ, support value 97 of 100; ML, support value 1,000 of 1,000), most of them are found as tandem duplicates located on four scaffolds of the amphioxus genome (table 1). Our phylogenetic analysis suggests that the amphioxus enzymes are orthologous to both vertebrate Hsd11β1 and Hsd11β1l, and we hence named the sequences Bf_Hsd11β1/1l_1 to 23. The genomic organization together with the tree topology suggest that the multiplicity of family SDR-26C members in amphioxus arose by lineage-specific duplication and further supports the notion that Hsd11β1 and Hsd11β1l diverged early in vertebrate evolution (Baker 2004b).
Table 1.
Genomic Linkage in Amphioxus of Retinoid and Steroid Metabolism Genes
| Superfamily Name | Amphioxus Family Name | Linkage Groups ([Scaffold of the Amphioxus Genome]: Amphioxus Sequence Name) |
| SDR | Hsd11β1/1l | [32]:12,19,21 [43]:13,14,15,16,18,20,22 [45]:23 [66]:2,3,4 [87]:1 [134]:6 [157]:7 [227]:17 [295]:8,9,10,11 [405]:5 |
| Dhrs7 | [34]:1 [42]:3 [46]:2 | |
| Dhrs7b/c | [5] | |
| Rdh10 | [181] | |
| RdhE2 | [1]:1,2,3 | |
| Hsd17β11/13 | [191]:2 [282]:1 | |
| Dhrs3 | [469]:2 [496]:1 | |
| Hsd17β12 | [44]:2,3 [208]:1 | |
| HsdL1 | [34] | |
| Rdh cluster | [34]:10,11 [44]:4 [72]:6 [110]:7 [237]:8,9 [311]:12 [373]:5 [452]:1,2,3 | |
| Bdh1 | [40]:5,7,9,11,14 [79]:1,2,3 [159]:4,6,8,10,12,13 | |
| Hsd11/17β2 | [167] | |
| Rdh11/12 | [7]:5 [8]:13,15,16 [9]:1 [18]:6,9,10,11,12 [52]:7 [74]:2 [87]:8 [180]:14 [189]:3,4 [196]:17,18 [376]:19,20,21,22 | |
| Rdh13 | [18] | |
| Rdh14 | [84] | |
| Hsd17β7 | [214] | |
| Hsd17β8 | [5]:3,4 [69]:2 [190]:5 [238]:1 [723]:6 | |
| Dhrs4 | [390] | |
| Hsd17β10 | [10] | |
| Hsd17β14 | [3] | |
| Hsd17β4 | [193] | |
| Adh | Adh3 | [151] |
| Akr1 | Akr1 | [3]:13 [59]:7,8,9,10,12 [89]:14,16 [229]:2,4,6 [236]:18 [258]:3,5 [281]:15,17 [374]:19 [391]:1 [551]:11 |
| Bco | Bcmo1/Bco2/Rpe65 | [12]:1 [44]:2 [74]:5 [161]:3 [555]:4 |
| Aldh | Aldh2 | [118] |
| Aldh1a | [21]:2 [31]:1,5,6 [155]:3,4 | |
| Aldh8a | [1] | |
| Aldh9a | [152] | |
| iLbp | Crbp | [9] |
| iLbp | [25]:2,4,5,6 [46]:7 [55]:1,8,9 [104]:3 | |
| Fabp1/6 | [12] | |
| NlpC/P60 | Hrasls | [103] |
| Fam84/Hrasls/Lrat | [26]:1,2,4,6,10 [135]:7,12 [265]:3,5,8,9,11 | |
| Dgat | Dgat1 | [76] |
| Soat1/2 | [217] | |
| Lipocalin | Ambp/ApoM/Ptgds/Rbp4 | [11] |
| Apod | [30]:2 [87]:1 [168]:3,4,5 | |
| Ttr/Urah | Ttr/Urah | [60] |
| Stra6 | Stra6l | [4]:1,3 [271]:2 [350]:4 |
| Star | Star/Stard3 | [53] |
| Col4a3bp | [15] | |
| Dlc1/Stard8/13 | [2] | |
| Stard10 | [2] | |
| Stard7 | [338] | |
| Pctp | [166] | |
| Cyp | Cyp27 | [25] |
| Cyp24/27 | [3]:3 [28]:2 [140]:1 | |
| Cyp11/24/27 | [44]:2 [191]:8 [214]:1,3,4,5,6,7 | |
| Cyp3 | [7]:1 [7]:2 | |
| Cyp2 | [44]:4,6,7,10 [61]:1,3 [101]:9 [438]:8 [682]:2 [736]:5 | |
| Cyp17 | [157]:2 [248]:1 | |
| Cyp26 | [164]:1,2,3 | |
| Cyp19 | [9]:1 [484]:2 | |
| Hsd3β | Hsd3β | [1]:3 [7]:1 [31]:2 [33]:5 [37]:6 [68]:4 [89]:7 |
| Sdr42e1 | [5] | |
| Nsdhl | [2] | |
| Srd5α | Srd5α1/2 | [1]:1 [81]:2 |
| Srd5α3 | [107] | |
| Tecr | [41] |
Family SDR-34C: Dhrs7 Enzymes
The human DHRS7 gene was identified from database searches of ESTs from retina and pineal gland (Haeseleer and Palczewski 2000) and, although no biochemical activity has been detected for DHRS7 so far, is expressed in the retina as well as in skeletal muscle and heart. Three amphioxus sequences, Bf_Dhrs7_1 to 3, grouped with the vertebrate Dhrs7 enzymes (fig. 2; supplementary fig. S1, Supplementary Material online; NJ, 99; ML, 997). The tree topology indicates that the three amphioxus Dhrs7 genes duplicated specifically in the cephalochordate lineage.
Family SDR-32C: Dhrs7b-Dhrs7c Enzymes
Human DHRS7b and DHRS7c are predicted genes located on chromosome 17 that encode SDR proteins of unknown function. A single amphioxus sequence, called Bf_Dhrs7b/c, reliably clustered with the vertebrate Dhrs7b–Dhrs7c family (fig. 2; supplementary fig. S1, Supplementary Material online; NJ, 99; ML, 996) indicating the presence of this SDR family in the last common ancestor of amphioxus and vertebrates.
Family SDR-16C: Rdh10–RdhE2–RdhE2l–Hsd17β11–Hsd17β13–Dhrs3 Enzymes
Family SDR-16C includes vertebrate Rdh10, RdhE2 (for epidermal retinaldehyde dehydrogenase 2), RdhE2l (for RdhE2-like), Hsd17β11, Hsd17β13, and Dhrs3 enzymes, which have diverse activities against retinoids and steroids. Vertebrate Rdh10, RdhE2, RdhE2l, and Dhrs3 have been involved in retinoid metabolism during differentiation and development, in pathological states and in the visual cycle. Hsd17β11 has been related to androgen metabolism during steroidogenesis, and the function of Hsd17β13, a hepatic enzyme with a high degree of sequence identity with Hsd17β11, is still unknown. A total of eight amphioxus sequences group with vertebrate family SDR-16C enzymes (fig. 2; supplementary fig. S1, Supplementary Material online; NJ, 100; ML, 1,000). Our phylogenetic analysis shows that one amphioxus sequence, Bf_Rdh10, is similar to Rdh10, three sequences, Bf_RdhE2_1–3, all of which located on the same scaffold (table 1), are related to both RdhE2 and RdhE2l, two sequences, Bf_Hsd17β11/13_1 and 2, are associated with the vertebrate Hsd17β11–Hsd17β13 clade, and two sequences, Bf_Dhrs3_1 and 2, cluster with the vertebrate Dhrs3 enzymes, hence establishing an almost complete repertoire of amphioxus family SDR-16C members. The phylogenetic analysis thus indicates that the diversification of family SDR-16C took place before the cephalochordate–vertebrate split.
Family SDR-12C: Hsd17β3–Hsd17β12–HsdL1 Enzymes
This family includes several enzymes involved in the production of sexual hormones and in the metabolism of fatty acids (Geissler et al. 1994; Huang et al. 2001; Moon and Horton 2003; Luu-The et al. 2006). Vertebrate Hsd17β3 catalyzes the conversion of androstenedione to testosterone, Hsd17β12 transforms estrone into estradiol and participates in the microsomal elongation of fatty acids, and HsdL1 is highly expressed in steroidogenic tissues, and its function has been related to the progression of prostate cancer. Four amphioxus enzymes belong to family SDR-12C (fig. 2; supplementary fig. S1, Supplementary Material online; NJ, 100; ML, 1,000). Three of them, Bf_Hsd17β12_1–3, group with Hsd17β12 and one, Bf_HsdL1, clusters with HsdL1. Of these, Bf_Hsd17β12_2 and Bf_Hsd17β12_3 are located on the same scaffold (table 1). No sequence orthologous to vertebrate Hsd17β3 could be identified in the amphioxus genome.
Family SDR-9C: Rdh cluster–Bdh1–Hsd11β2–Hsd17β2 Enzymes
Family SDR-9C includes three groups of vertebrate enzymes: the Rdh cluster, the Hsd-type 2 group, and the 3-hydroxybutyrate dehydrogenase type 1 (Bdh1) group. Altogether, 27 amphioxus sequences group within this family (fig. 2; supplementary fig. S1, Supplementary Material online; NJ, 98; ML, 999). The vertebrate Rdh cluster includes six human genes, RDH5, RDH16, similar to RDH16, HSD17β6, SDR9C7, and DHRS9. Human RDH5 and RDH16 are involved in visual and general retinol oxidation, respectively (Gough et al. 1998; Yamamoto et al. 1999), although both of them also show some activity toward steroids. In addition, HSD17β6 and DHRS9 are also active against both retinoids and steroids (Biswas and Russell 1997; Chetyrkin, Belyaeva, et al. 2001; Chetyrkin, Hu, et al. 2001; Soref et al. 2001), whereas SDR9C7 does not have a significant activity against steroids and shows only very weak retinaldehyde reductase activity (Kowalik et al. 2009). A total of 12 amphioxus sequences, named Bf_Rdh_1 to 12, group with the vertebrate Rdh cluster (fig. 2; supplementary fig. S1, Supplementary Material online), including two previously characterized retinaldehyde reductases of amphioxus (Bf_Rdh_1 and Bf_Rdh_2) (Dalfó et al. 2001, 2007). The genomic linkage data indicate that at least 7 of the 12 amphioxus Rdh genes cluster together on three distinct scaffolds (table 1). In our phylogenetic tree, vertebrate and amphioxus Rdh sequences group separately, indicating that the extensive expansion of this family in amphioxus and the diversification of the Rdh cluster in vertebrates were independent evolutionary events.
Hsd-type 2 enzymes function as NAD+-dependent oxidases to convert active steroid hormones to their inactive forms. Hsd11β2 oxidizes glucocorticoids, transforming potent cortisol to weak cortisone, whereas Hsd17β2 oxidizes potent sex hormones, such as estradiol and testosterone, into weak estrone and androstenedione, respectively (Wu et al. 1993; Albiston et al. 1994). One amphioxus sequence branches outside a clade that comprises both vertebrate type 2 enzymes (fig. 2; supplementary fig. S1, Supplementary Material online). The amphioxus enzyme orthologous to both vertebrate forms was therefore named Bf_Hsd11/17β2. The tree topology indicates that the 11β- and 17β-forms diverged from an ancestral Hsd-type 2 enzyme duplicated in the lineage leading to extant vertebrates. This divergence in vertebrates of the 11β- and 17β-forms has been supported by the phylogenetic analysis of Hsd-type 2 sequences from sea urchins and acorn worms, which also cluster at the base of the vertebrate Hsd-type 2 clade (Baker 2010a).
We also identified 14 amphioxus sequences that clustered with vertebrate Bdh1 (fig. 2; supplementary fig. S1, Supplementary Material online), a mitochondrial lipid-requiring enzyme that serves to interconvert ketone bodies (Marks et al. 1992). Two of the 14 amphioxus sequences, named Bf_Bdh1_1 and 2, stably group with the vertebrate enzymes, whereas the other 12 sequences, Bf_Bdh1_3 to 14, fall outside this cluster. Interestingly, Bf_Bdh1_1 and 2, together with Bf_Bdh1_3, are linked in the amphioxus genome, with the other 11 amphioxus Bdh1 genes clustering on two different scaffolds (table 1). Together, the tree topology and linkage data suggest an initial duplication of a Bdh1 ancestor predating the cephalochordate–vertebrate split. Although only one of these ancestral Bdh1 duplicates was preserved in vertebrates, both of them were retained and independently expanded in the amphioxus lineage.
Family SDR-28C: Hsd17β1–Rdh8 Enzymes
Vertebrate Hsd17β1 is predominantly expressed in ovaries, breast tissues and placenta, and has been considered as a major determinant of peripheral and gonadal estradiol synthesis (Peltoketo et al. 1988, 1999). Rdh8, also known as photoreceptor retinol dehydrogenase (prRDH), localizes specifically to the outer segments of photoreceptor cells reducing retinal to retinol in the visual cycle (Rattner et al. 2000; Maeda et al. 2005, 2007). Our survey of the amphioxus genome did not reveal any amphioxus sequences clustering with this family. Nevertheless, the identification of several putative Rdh8/Hsd17β1 sequences in cnidarians (Tarrant et al. 2009) and sea urchins (Mindnich and Adamski 2009) suggests that the members of this family might have been lost specifically in the amphioxus lineage.
Family SDR-7C: Rdh11–Rdh12–Rdh13–Rdh14 Enzymes
Vertebrate Rdh11, Rdh12, Rdh13, and Rdh14 are retinaldehyde reductases predominantly involved in retinoid metabolism of the eye (Haeseleer et al. 2002; Kim et al. 2005; Lee et al. 2007; Maeda et al. 2007). In addition, human RDH11 is also involved in the regulation of retinoid homeostasis (Lin et al. 2001; Kedishvili et al. 2002; Belyaeva et al. 2005), RDH12 in retinal reduction of extraocular tissues (Belyaeva et al. 2005), RDH13 in protection against retinal produced from dietary β-carotenes (Belyaeva, Korkina, et al. 2008), and RDH14 in retinal reduction in most human tissues (Belyaeva and Kedishvili 2002). Phylogenetically, Rdh11 and Rdh12 are closely related enzymes that arose from an Rdh11/12 precursor likely during mammalian evolution, whereas Rdh13 and Rdh14 have diverged from the Rdh11/12 ancestor early during evolution. In amphioxus, we found 24 sequences that belonged to the SDR family SDR-7C (fig. 2; supplementary fig. S1, Supplementary Material online; NJ, 100; ML, 1,000). One amphioxus sequence, Bf_Rdh13, is closely related to vertebrate Rdh13 and one, Bf_Rdh14, branches with vertebrate Rdh14, whereas the 22 remaining sequences, named Bf_Rdh11/12_1 to 22, are not clearly associated with any of the vertebrate SDR-7C family members. A total of 20 of the 22 Bf_Rdh11/12 sequences (Bf_Rdh11/12_3 to 22) cluster together in the phylogenetic tree, indicating a lineage-specific amplification of these amphioxus Rdh11/12 sequences. Genomic linkage of 16 of the 22 amphioxus Rdh11/12 genes on five distinct scaffolds (table 1) strongly supports this notion. The internal tree topology does not unambiguously resolve the phylogenetic relationship of the amphioxus and vertebrate enzymes. The phylogenetic analysis nonetheless indicates that the chordate ancestor already had multiple genes encoding SDR-7C family enzymes, including Rdh11/12, Rdh13, and Rdh14, and that some of these ancestral sequences experienced a significant expansion in the amphioxus lineage.
Family SDR-37C: Hsd17β7 Enzymes
Hsd17β7 enzymes show the typical SDR signature at the cofactor-binding motif and at the active site, but their overall sequence identity with other SDR families is low. Hsd17β7, initially described as prolactin receptor–associated protein (PRAP) (Duan et al. 1996), are cholesterogenic enzymes that participate in cholesterol biosynthesis (Mindnich et al. 2004; Shehu et al. 2008). Hsd17β7 has been lost in cholesterol auxotrophic animals that also lost squalene epoxidase (Sqle), a crucial enzyme for cholesterol biosynthesis (Marijanovic et al. 2003). There is a single Hsd17β7 ortholog in the amphioxus genome (fig. 2; supplementary fig. S1, Supplementary Material online; NJ, 100; ML, 1,000). The identification of Bf_Hsd17β7, together with the presence of a putative Sqle enzyme in amphioxus (GenBank accession number XP_002594656), is of evolutionary relevance because these findings suggest that cephalochordates are capable of the de novo synthesis of cholesterol and hence imply that this metabolic pathway was already present in the last common ancestor of all chordates.
Family SDR-30C: Hsd17β8 Enzymes
Hsd17β8 is an oxidative enzyme inactivating estradiol, testosterone, and dihydrotestosterone (Fomitcheva et al. 1998). Whereas in humans and zebrafish, there is only a single Hsd17β8, we found six Hsd17β8 genes encoded in the amphioxus genome, Bf_ Hsd17β8_1 to 6 (fig. 2; supplementary fig. S1, Supplementary Material online; NJ, 100; ML, 1,000). Two of these amphioxus sequences, Bf_ Hsd17β8_3 and 4, are linked in the genome (table 1). Altogether, these data suggest that members of the Hsd17β8 family were already present in the genome of the chordate ancestor and that the amphioxus Hsd17β8 gene complement was expanded by lineage-specific duplication.
Family SDR-25C: Dhrs4 Enzymes
The vertebrate members of this SDR family have been assigned very different enzymatic activities: whereas human DHRS4 has been shown to contribute to the reduction of 3-ketosteroids into 3β-hydroxysteroids (Matsunaga et al. 2008), the mouse Dhrs4 enzyme has been described as a retinaldehyde reductase (Lei et al. 2003). Our analysis identified one amphioxus representative of this SDR family, Bf_Dhrs4 (fig. 2; supplementary fig. S1, Supplementary Material online; NJ, 100; ML, 1,000). Given the presence of an amphioxus representative in this SDR family, the evolutionary origin of Dhrs4 enzymes evidently predated chordate divergence.
Family SDR-5C: Hsd17β10 Enzymes
Vertebrate Hsd17β10 is a multifunctional enzyme that, in addition to a 17β-Hsd activity with sex steroids, acts as 3α-, 7α-, 7β-, 17β-, 20β-, and 21-oxidase of several different substrates (Shafqat et al. 2003). A single amphioxus sequence, named Bf_Hsd17β10, branched at the base of the vertebrate representatives of this SDR family (fig. 2; supplementary fig. S1, Supplementary Material online; NJ, 100; ML, 1,000) suggesting that Hsd17β10 enzymes were already present in the last common ancestor of amphioxus and vertebrates.
Family SDR-47C: Hsd17β14 Enzymes
Hsd17β14 was initially suggested to function in retinol metabolism (Haeseleer and Palczewski 2000), but recent work suggests that the human enzyme acts as a 17β-Hsd with sex steroids in brain, liver, and placenta (Lukacik et al. 2007). In amphioxus, there is a single member of this SDR family, Bf_Hsd17β14 (fig. 2; supplementary fig. S1, Supplementary Material online; NJ, 100; ML, 1,000). Hsd17β14 enzymes were thus very likely already present in the last common ancestor of all chordates suggesting that the evolutionary origin of this SDR family predates the diversification of the chordate lineage.
Family SDR-8C: Hsd17β4 Enzymes
The vertebrate Hsd17β4 enzyme (also called MFE-2, MFP-2, and DBP) is involved in peroxisomal fatty acid β-oxidation and its deficiency causes severe abnormalities in several organs, particularly in the brain (reviewed in Huyghe et al. 2006). In the amphioxus genome, we identified one sequence closely related to vertebrate Hsd17β4. This sequence, Bf_Hsd17β4, marks the base of the vertebrate Hsd17β4 enzymes (fig. 2; supplementary fig. S1, Supplementary Material online; NJ, 100; ML, 1,000) hence indicating that this SDR family might have already been present at the dawn of chordate diversification.
Adh and Akr Sequences in Amphioxus
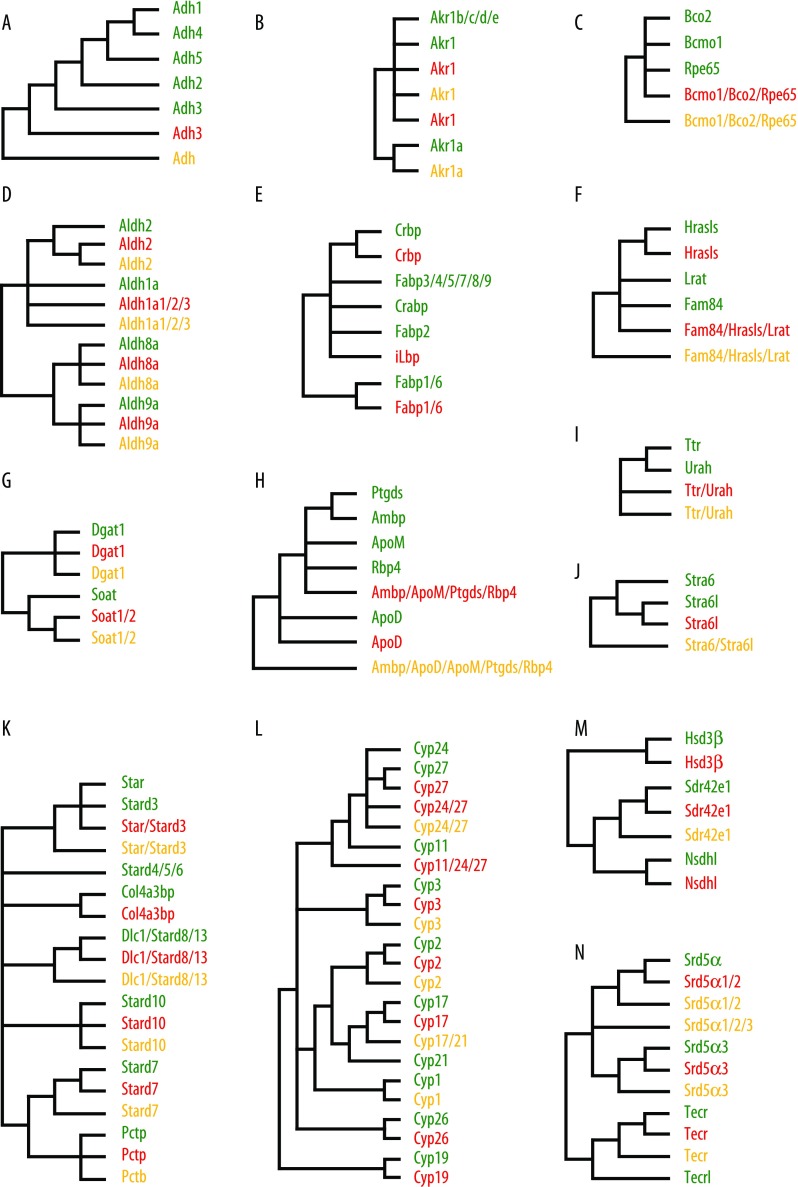
In vertebrates, retinol dehydrogenase activity has also been associated with some members of the Adh family, mainly with the Adh1, Adh3, and Adh4 enzymes. Our survey of the B. floridae genome supports previous work on Adh evolution showing that amphioxus had a single Adh enzyme (Bf_Adh3) that groups with the members of the vertebrate Adh family (fig. 3A, supplementary fig. S2, Supplementary Material online) (Cañestro et al. 2002; reviewed in Gonzàlez-Duarte and Albalat 2005). Biochemically, amphioxus Adh3 is similar to vertebrate Adh3 (Cañestro et al. 2000; Godoy et al. 2006) suggesting that the enzyme activities of Adh1 and Adh4 were vertebrate innovations (Cañestro et al. 2003, 2010).
FIG. 3.—
Diagrammatic tree topologies of the (A) Adh, (B) Akr1, (C) Bco, (D) Aldh, (E) iLbp, (F) NlpC/P60, (G) Dgat, (H) Lipocalin, (I) Ttr/Urah, (J) Stra6, (K) Star, (L) Cyp, (M) Hsd3β, and (N) Srd5α families. Simplified phylogenies of the main components of retinoid and steroid metabolism are shown. Vertebrate branches are in green, amphioxus branches are in red, and branches containing sequences from the cnidarian Nematostella vectensis are in yellow.
Several members of the Akr1 group in the Akr superfamily show activity against retinoids and steroids. Akr1b enzymes that convert glucose to sorbitol can also reduce retinal to retinol (Ruiz et al. 2009), and some Akr1c enzymes can act as Hsd enzymes controlling the interconversion of weak steroid hormones to potent hormones (Dufort et al. 1999). There are 9 Akr1 sequences in humans classified into 5 subgroups (Akr1A to E). The survey of the amphioxus genome revealed 19 Akr1 sequences in amphioxus (Bf_Akr1_1 to 19) and the phylogenetic analysis grouped 18 of these 19 amphioxus sequences into four well-supported clades (fig. 3B, supplementary fig. S3, Supplementary Material online). Genomically linked amphioxus Akr1 genes are found only within a given clade: linkage groups Akr1_2, 4, and 6 and Akr1_3 and 5 branch within the same clade; Akr1_7, 8, 9, 10, and 12 are also grouped in the tree; Akr1_14 and 16 as well as Akr1_15 and 17 together establish one of the four well-supported clades (table 1).
The tree does not efficiently resolve the phylogenetic relationship between the 19 amphioxus Akr1 sequences and their vertebrate orthologs, but given that amphioxus Akr1s within the same clade have the tendency to be linked on the genome, the expansion of Akr1 enzymes in amphioxus was likely independent of the diversification of the vertebrate Akr1 proteins. To further investigate the lineage-specific Akr1 duplicates in amphioxus, we carried out structural analyses of their substrate binding and processing domains. In vertebrate Akr1 enzymes, in addition to the invariant residues Y50 and H113 within the active site, the amino acids at positions 47, 49, 82, 114, 116, 124, 125, 133, 135, 299, and 301 (numbering relative to human AKR1A1) have been associated with substrate binding in the binding pocket (Jez et al. 1997; Penning 1997). In amphioxus Akr1s, the conservation at these positions is highest at residues involved in general substrate recognition and binding (e.g., 100% conservation at positions 47, 50, and 113), is intermediate at residues determining sugar versus steroid specificity (e.g., 63.2% conservation at position 49 and 52.6% conservation at position 114), and is lowest at residues contributing to the definition of steroid specificity (e.g., 26.3% of conservation at position 299 and 15.8% of conservation at position 301). Altogether, this pattern is compatible with the hypothesis that at least some of the amphioxus Akr1s exhibit the capacity to metabolize steroids.
Amphioxus Retinoid Metabolism
Bcmo1, Bco2, and Rpe65 Family
In vertebrates, the Bcmo1 enzyme (also known as Bco) cleaves β-carotene into two molecules of retinal (reviewed in Ross et al. 2000 and Gottesman et al. 2001). Bcmo1 is closely related to Bco2 (for β,β-carotene-9′,10′-oxygenase) and Rpe65 (for retinal pigment epithelium protein of 65 kDa) enzymes. Bco2 catalyzes the asymmetric cleavage of β-carotenes (Kiefer et al. 2001), which has been proposed as an alternative pathway of RA synthesis (Simões-Costa et al. 2008), whereas Rpe65 converts retinyl esters to retinol for photopigment regeneration in the visual cycle (Jin et al. 2005). From an evolutionary perspective, identification of Bcmo1/Bco2/Rpe65 enzymes in urochordates (Takimoto et al. 2006) and Drosophila (von Lintig and Vogt 2000) indicates that the cleavage of β-carotene is an ancient biochemical activity in metazoans.
In total, five amphioxus sequences showed similarity with vertebrate Bcmo1/Bco2/Rpe65 enzymes and grouped with the vertebrate enzymes in our phylogenetic analysis (fig. 3C, supplementary fig. S4, Supplementary Material online; NJ, 94; ML, 993). The amphioxus sequences did not group with any of the three vertebrate enzymes and are not linked in the amphioxus genome (table 1), which is compatible with the notion that Bcmo1, Bco2, and Rpe65 enzymes underwent independent duplication and functional specialization in the cephalochordate and vertebrate lineages.
Aldh1a, Aldh8a, and Cyp26 Enzymes
In vertebrates, retinal is mainly oxidized to RA by three Aldh1a enzymes (Aldh1a1, Aldh1a2, and Aldh1a3) and endogenous RA is degraded to biologically inactive forms by Cyp26 enzymes (Cyp26a, Cyp26b, and Cyp26c). Vertebrate Aldh8a1 (also known as Raldh4 and Aldh12) is also capable of oxidizing retinal (Lin and Napoli 2000; Lin et al. 2003; Liang et al. 2008), although the contribution of Aldh8a1 to RA signaling is not yet fully understood. Previous work has identified six Aldh1a and three Cyp26 in amphioxus, and phylogenetic analyses have suggested that this diversity of Aldh1a and Cyp26 originated by independent duplications in the amphioxus and vertebrate lineages (Cañestro et al. 2006; Marlétaz et al. 2006). Genomic linkage data provide additional evidence for this hypothesis: five of the six amphioxus Aldh1a genes are located on two genome scaffolds and all three amphioxus Cyp26 genes are clustered on a single scaffold (table 1).
We have extended the previous analyses by searching for Aldh8a1 orthologs in amphioxus and identified one amphioxus sequence, Bf_Aldh8a. Phylogenetic analysis showed that Bf_Aldh8a is orthologous to vertebrate Aldh8a1 (fig. 3D, supplementary fig. S5, Supplementary Material online; NJ, 100; ML, 1,000). The identification of Bf_Aldh8a completes the evolutionary analysis in amphioxus of the basic genetic machinery controlling the spatiotemporal levels of endogenous RA (i.e., Aldh1a, Aldh8a, and Cyp26 enzymes) and further strengthens the notion that this machinery was already present in the last common ancestor of all chordates (Cañestro et al. 2006; Marlétaz et al. 2006; Campo-Paysaa et al. 2008; Albalat and Cañestro 2009; Theodosiou et al. 2010).
Cellular Retinoid Binding Proteins: Crbp and Crabp
Intracellular retinoid-binding proteins, such as Crbp and Crabp, bind retinol, retinal and RA to solubilize and stabilize the retinoids in the aqueous environment of the cell. Vertebrate Crbp and Crabp belong to the family of intracellular lipid binding proteins (iLbp), together with three groups of fatty acid--binding proteins (Fabps), namely Fabp1/6, Fabp2, and Fabp3/4/5/7/8/9 (Schaap et al. 2002). In humans, there are 4 Crbp, 2 Crabp, 1 Fabp2, 2 Fabp1/6, and 6 Fabp3/4/5/7/8/9, each one of these proteins being characterized by different ligand binding properties. In amphioxus, we identified 11 iLbp. Although one of these amphioxus iLbp, Bf_Crbp, strongly groups with vertebrate Crbp (fig. 3E, supplementary fig. S6, Supplementary Material online; NJ, 97; ML, 998) and one, Bf_Fabp1/6, groups with vertebrate Fab1/6 (fig. 3E, supplementary fig. S6, Supplementary Material online; NJ, 74; ML, 958), the other nine amphioxus sequences, named Bf_iLbp_1 to 9, are not clearly associated with any vertebrate iLbp group. Because seven of the nine amphioxus iLbp genes are clustered on two genome scaffolds (table 1), it is very likely that these amphioxus genes originated by lineage-specific duplication.
Unlike for Crbp, we were hence unable to identify a clear amphioxus ortholog of vertebrate Crabp. Several iLbp family members with retinoid-binding capacity have been described in protostomes, such as insects and crustaceans (Mansfield et al. 1998; Gu et al. 2002; Folli et al. 2005; Söderhäll et al. 2006), but sequence analyses and 3D structural modeling showed that these protostome iLbps are not orthologous to vertebrate Crbp and Crabp (Gu et al. 2002; Folli et al. 2005; Söderhäll et al. 2006). It is therefore very likely that the capacity to bind retinoids has evolved independently in different iLbp families in the course of bilaterian diversification. Biochemical analyses will hence be required to determine the exact physiological function of each amphioxus member of the iLbp family.
Lrat and Dgat1 Enzymes
The Lrat enzyme catalyzes retinol esterification into retinyl esters, which are accumulated in the vertebrate liver where they can be mobilized to maintain retinoid homeostasis. Lrats hence participate in retinoid storage mechanisms. Lrat enzymes belong to the Lrat-like family of the complex NlpC/P60 superfamily, which unites Lrat with the Hrasl (for Hras-like suppressor, also known as H-rev107-like protein) and Fam84 (for family with sequence similarity 84) subfamilies (Anantharaman and Aravind 2003). We found 13 amphioxus sequences with similarities to vertebrate Lrat-like proteins. Only one of these sequences grouped with one of the three vertebrate subfamilies: Bf_Hrasls, which is weakly associated with the vertebrate Hrasls (fig. 3F, supplementary fig. S7, Supplementary Material online; NJ, 38; ML, 773). The other 12 amphioxus sequences are located on only three scaffolds of the genome (table 1) and cluster together in the phylogenetic tree (fig. 3F, supplementary fig. S7, Supplementary Material online), indicating that they are the result of an extensive lineage-specific duplication. These 12 amphioxus sequences were called Bf_ Fam84/Hrasls/Lrat_1 to 12.
Because phylogenetic analyses did not properly resolve the relationships between amphioxus and vertebrate enzymes, we analyzed the amino acid signatures of the different Lrat-like subfamilies. Lrat-like proteins share two catalytic residues, H60 and C161 (numbers relative to the human LRAT enzyme) (Mondal et al. 2000; Jahng et al. 2003). C161 is the central cysteine of the highly conserved NCE-box of Lrat and Hrasls proteins, whereas in Fam84 members, this cysteine has been replaced by a serine (resulting in a NSE-box). None of the 13 amphioxus sequences exhibits a NSE-box, suggesting that this amino acid replacement in the Fam84 subfamily took place only after the split of the amphioxus and vertebrate lineages. For H60, the conserved amino acid context is HYGIY in Lrat enzymes, HWA/GL/I/VY in Hrasls and HWAV/IF/Y/C in Fam84. For this signature, the 13 amphioxus sequences all show a Hrasls HWA/GL/I /VY context, suggesting that the activity of the amphioxus Lrat-like proteins might be most similar to that of vertebrate Hrasls. In this context, it is interesting to note that the Lrat-like protein EGL-26 of the nematode worm Caenorhabditis elegans has been functionally implicated in lipid metabolism (Estes et al. 2007), a role more similar to that of vertebrate Hrasls (Jin et al. 2007; Nazarenko et al. 2007) than to that of Lrats. Taken together, this suggests that a Hrasls-like activity might be ancestral in this family and that Lrat- and Fam84-like activities arose specifically in the vertebrate lineage after duplication of an ancestral Hrasls-like enzyme.
In addition to Lrat proteins, enzymes involved in the synthesis of triacylglycerol, such as acyl coenzyme A:diacylglycerol acyltransferase (Dgat1), are capable of contributing to retinol esterification by carrying out the acyl coenzyme A:retinol acyltransferase reaction (Ross 1982; O'Byrne et al. 2005; Orland et al. 2005). Although the function of vertebrate Dgat1 in the hepatic control of retinoid homeostasis has been questioned (Batten et al. 2004), in the absence of a clear amphioxus Lrat ortholog, the contribution of Dgat1 enzymes in amphioxus to the esterification of retinol for retinyl ester storage deserves further investigation. Dgat1 enzymes are related to sterol O-acyltransferases (Soat) that catalyze the formation of cholesterol esters from cholesterol. In humans, there are one Dgat1 and two Soat enzymes. We found two sequences similar to vertebrate Dgat1 and Soat enzymes in the amphioxus genome and phylogenetic analysis showed that one of theses sequences, named Bf_Dgat1, groups with vertebrate Dgat1, whereas the other one, called Bf_Soat1/2, branches with vertebrate Soat1 and Soat2 enzymes (fig. 3G, supplementary fig. S8, Supplementary Material online; NJ, 100; ML, 980). Taken together, these data suggest that, in the absence of a clear amphioxus Lrat ortholog, retinol esterification in amphioxus might be catalyzed by Dgat, an enzyme that might have hence played a key role in the biosynthesis of retinyl esters in the ancestral chordate.
Rbp4, Ttr, and Stra6 Proteins
In vertebrates, hepatic retinyl esters can be hydrolyzed back to retinol, which subsequently enters the blood stream in association with Rbp4 and Ttr proteins. Rbp4 belongs to the lipocalin family, a large group of extracellular proteins that bind and transport small hydrophobic molecules. Although the evolutionary origins of Rbp4 remain unclear, another member of this family, apolipoprotein D (ApoD), is considered the most ancient metazoan lipocalin. Vertebrate Rbp4 is evolutionary related to the prostaglandin D2 synthases (Ptgds)-like group that includes Ptgds, alpha-1-microglobulin/bikunin preproprotein (Ambp), and apolipoprotein M (ApoM) (Flower 1996; Ganfornina et al. 2000; Gutiérrez et al. 2000).
In amphioxus, we found six lipocalin-like sequences. A total of five of those, Bf_ApoD_1 to 5, are similar to the vertebrate ApoD proteins, whereas one sequence is more closely related to the vertebrate Ptgds-like group (fig. 3H, supplementary fig. S9, Supplementary Material online; NJ, 72; ML, 761). The amphioxus sequence was named Bf_Ambp/ApoM/Ptgds/Rbp4 to reflect this phylogenetic relationship. Since the exon–intron organization of lipocalin genes contains information about the evolutionary origins of this gene family (Sánchez et al. 2003), we decided to analyze the genomic locus of Bf_Ambp/ApoM/Ptgds/Rbp4. The amphioxus Ambp/ApoM/Ptgds/Rbp4 gene has six coding exons and five introns with a 0,2,1,1,1 pattern of intron phases, which is identical to vertebrate ApoM and Ptgds genes but different from vertebrate Rbp4 genes (Sánchez et al. 2003). It therefore seems that the lipocalin gene of the ancestral chordate was structurally more similar to extant ApoM and Ptgds genes than to Rbp4 genes.
Rbp4 interacts with transthyretin (Ttr) to transport and distribute retinol to peripheral tissues. Vertebrate Ttr proteins are evolutionary related to 5-hydroxyisourate hydrolases (Urah) enzymes (Zanotti et al. 2006). An amphioxus sequence similar to vertebrate Ttr and Urah was identified in our genomic survey. The phylogenetic analysis was unable to resolve, whether the amphioxus sequence is orthologous to either vertebrate Ttr or Urah or to both, and we hence named the sequence Bf_Ttr/Urah (fig. 3I, supplementary fig. S10, Supplementary Material online). Vertebrate Ttr and Urah, however, exhibit different amino acids at key positions for substrate accommodation and catalysis: H11, D49, R51, H102, Y115, and S118 in Urah (numbers refer to the mouse Urah sequence because no human Urah has so far been identified) versus K35, S72, E74, T126, T139, and V142 in Ttr (numbers refer to the human TTR sequence). In the amphioxus Ttr/Urah, the amino acids at these key positions correspond more closely to those of vertebrate Urah, which suggests a 5-hydroxyisourate hydrolase activity of the amphioxus protein. These data therefore support the notion that the hormone transport protein Ttr arose during vertebrate evolution by duplication from an ancestral Urah-like enzyme (Zanotti et al. 2006).
After transport to the target tissue, retinol uptake into the cell is mediated by Stra6, a cell-surface receptor for Rbp4 (Kawaguchi et al. 2007; Theodosiou et al. 2010). Stra6 proteins show similarity with a group of vertebrate proteins of unknown function, called Stra6l (for Stra6-like). We identified four amphioxus sequences with similarity to vertebrate Stra6 in the amphioxus genome, but phylogenetic reconstruction groups the amphioxus sequences with the vertebrate Stra6l proteins (fig. 3J, supplementary fig. S11, Supplementary Material online; NJ, 74; ML, 982). We concluded, therefore, that there is no Stra6 ortholog in amphioxus, suggesting that Stra6 was either lost in cephalochordates or originated specifically in the vertebrate lineage.
In summary, our searches for Rbp4, Ttr, and Stra6 orthologs in the amphioxus genome suggest that amphioxus probably lacks a vertebrate-like system of retinoid transport and uptake. Functional studies are needed to determine whether some of the sequences we have identified in the genome might participate in an alternative system in amphioxus for the mobilization and delivery of retinoids from storage reservoirs and hence whether some kind of retinoid storage, mobilization, and delivery system was active in the ancestral chordate.
Steroidogenesis Enzymes in Amphioxus
Star and Cyp11a
The first reaction in the steroidogenesis pathway is the synthesis of pregnenolone from cholesterol in the inner membrane of mitochondria. In vertebrates, cholesterol is delivered to mitochondria by members of the Star family (Clark et al. 1994; Watari et al. 1997; Stocco 2001), all of which contain a START domain required for intracellular lipid transport, lipid metabolism, and cell signaling processes. Members of the Star family include multiple Stard (for START domain) proteins, Dlc1 (for deleted in liver cancer 1), Col4a3bp (for collagen, type IV, alpha 3 binding protein), and Pctp (for phosphatidylcholine transfer protein). We searched the B. floridae genome for Star family members and found six amphioxus sequences similar to vertebrate Stard proteins that, based on their phylogenetic relationships, were named Bf_Star/Stard3, Bf_Stard7, Bf_Stard10, Bf_Dlc1/Stard8/13, Bf_Col4a3bp, and Bf_Pctp (fig. 3K, supplementary fig. S12, Supplementary Material online). In our phylogenetic analysis, Bf_Star/Stard3 reliably groups with vertebrate Star/Stard3 proteins (fig. 3K, supplementary fig. S12, Supplementary Material online; NJ, 85; ML, 999). This amphioxus sequence is therefore a good candidate to mediate the delivery of cholesterol to the inner mitochondrial membrane of amphioxus.
Once in the mitochondrion, synthesis of pregnenolone from cholesterol is carried out by Cyp11a enzymes, which belong to the mitochondrial clan of the cytochrome P450 enzymes. The mitochondrial Cyp clan includes vertebrate Cyp11, Cyp24, and Cyp27 as well as some invertebrate Cyp enzymes (Nelson 2009). Our survey of the amphioxus genome identified 12 sequences grouping within the mitochondrial Cyp clan (fig. 3L, supplementary fig. S13, Supplementary Material online; NJ, 97; ML, 1,000). The phylogenetic analyses do not resolve the evolutionary relationships between the amphioxus and vertebrate sequences. In total, eight sequences, named Bf_Cyp11/24/27_1 to 8, grouped at the base of the vertebrate mitochondrial Cyp enzymes, three sequences, Bf_Cyp24/27_1 to 3, seem closely related to vertebrate Cyp24 and Cyp27, and one amphioxus sequence, Bf_Cyp27, stably groups with vertebrate Cyp27 (fig. 3L, supplementary fig. S13, Supplementary Material online; NJ, 46; ML, 988). Of the amphioxus mitochondrial Cyp clan members, six of the eight Bf_Cyp11/24/27 genes are genomically linked, clustering on a single genome scaffold (table 1).
Given the absence of crystal structures for mitochondrial Cyp enzymes, only very limited structural information of these proteins is available (Storbeck et al. 2007). We were hence not able to carry out meaningful structural comparisons between the amphioxus and vertebrate sequences to assist with the inference of evolutionary relationships between the different members of this Cyp clan. In conclusion, numerous amphioxus mitochondrial Cyp sequences were identified, but the physiological contribution of these enzymes to the synthesis of pregnenolone from cholesterol remains to be determined.
Hsd3β Enzymes
Pregnenolone is metabolized to progesterone by Hsd3β enzymes. Hsd3β enzymes are unconventional SDR enzymes that belong to the extended category of SDR proteins (Bray et al. 2009). For the sake of clarity, they were not included in the global SDR analysis but have been studied separately. The Hsd3β clade includes three human enzymes, namely HSD3β1, HSD3β2, and HSD3β7, which are evolutionarily related to Nsdhl (for NAD(P)-dependent steroid dehydrogenase-like) and Sdr42e1 (for SDR family 42E, member 1) enzymes (Bray et al. 2009). We identified nine sequences similar to these vertebrate enzymes in the amphioxus genome. Phylogenetic analysis shows that 7 sequences, Bf_Hsd3β_1 to 7, are similar to vertebrate Hsd3β (fig. 3M, supplementary fig. S14, Supplementary Material online; NJ, 100; ML, 1,000), one sequence, Bf_Nsdhl, groups with vertebrate Nsdhl (fig. 3M, supplementary fig. S14, Supplementary Material online; NJ, 100; ML, 1,000), and one sequence, Bf_Sdr42e1, is associated with vertebrate Sdr42e1 (fig. 3M, supplementary fig. S14, Supplementary Material online; NJ, 100; ML, 1,000). Although the seven amphioxus Hsd3β genes do not seem to be linked in the genome (table 1), tree topology nonetheless clearly indicates that the amphioxus genome contains sequences orthologous to vertebrate Hsd3β and that the multiple amphioxus Hsd3βs arose by linage-specific duplication, independent of the Hsd3β diversification of vertebrates.
Cyp17, Cyp19, and Cyp21 Enzymes
In steroidogenesis, vertebrate Cyp17 converts progesterone to 17α-OH-progesterone and to androstenedione, a precursor of active androgens and estrogens. Cyp17 is also capable of transforming pregnenolone to 17α-hydroxypregnenolone and to dehydroepiandrosterone (DHEA). Cyp17 belongs to clan 2 of the cytochrome P450 superfamily, which also includes Cyp1, Cyp2, and Cyp21. We found 12 B. floridae sequences belonging to this Cyp clan 2 (fig. 3L, supplementary fig. S13, Supplementary Material online; NJ, 95; ML, 1,000). Two of these, Bf_Cyp17_1 and 2, are orthologous to vertebrate Cyp17, whereas the other 10, Bf_Cyp2_1 to 10, group with vertebrate Cyp2. Interestingly, our phylogenetic analysis suggests that there is no amphioxus ortholog of vertebrate Cyp21, which is an essential enzyme for the synthesis of corticosteroids in vertebrates.
In vertebrates, androstenedione produced by Cyp17 enzymes is transformed by Cyp19, the so-called aromatase, to estrone. Cyp19 enzymes have also been involved in the synthesis of estradiol from testosterone. In the B. floridae genome, there are two sequences, Bf_Cyp19_1 and 2, that are orthologous to vertebrate Cyp19 (fig. 3L, supplementary fig. S13, Supplementary Material online; NJ, 100; ML, 1,000). These two amphioxus Cyp19 sequences are most likely the result of a lineage-specific duplication.
Srd5α Enzymes
Testosterone can be transformed to 5α-dihydrotestosterone (5αDHT), a potent sex steroid, by the action of type 1, type 2, and type 3 Srd5α enzymes (Russell and Wilson 1994; Uemura et al. 2008). Srd5α enzymes are found in invertebrates and vertebrates and are phylogenetically related to the Tecr enzymes (for trans-2,3-enoyl-CoA reductase, also known as glycoprotein synaptic 2 or Gpsn2) (Markov et al. 2009). We identified two B. floridae sequences, named Bf_Srd5α1/2_1 and 2, that group within the vertebrate Srd5α1/2 clade (fig. 3N, supplementary fig. S15, Supplementary Material online; NJ, 97; ML, 1,000), one sequence, Bf_Srd5α3, that branches with the vertebrate Srd5α3 enzymes (fig. 3N, supplementary fig. S15, Supplementary Material online; NJ, 99; ML, 999), and one sequence, Bf_Tecr, that clusters with the vertebrate Tecr proteins (fig. 3N, supplementary fig. S15, Supplementary Material online; NJ, 100; ML, 1,000). These phylogenetic data support the notion that the amphioxus genome encodes enzymes with Srd5α activity and that some of these amphioxus genes arose by lineage-specific duplication.
Discussion
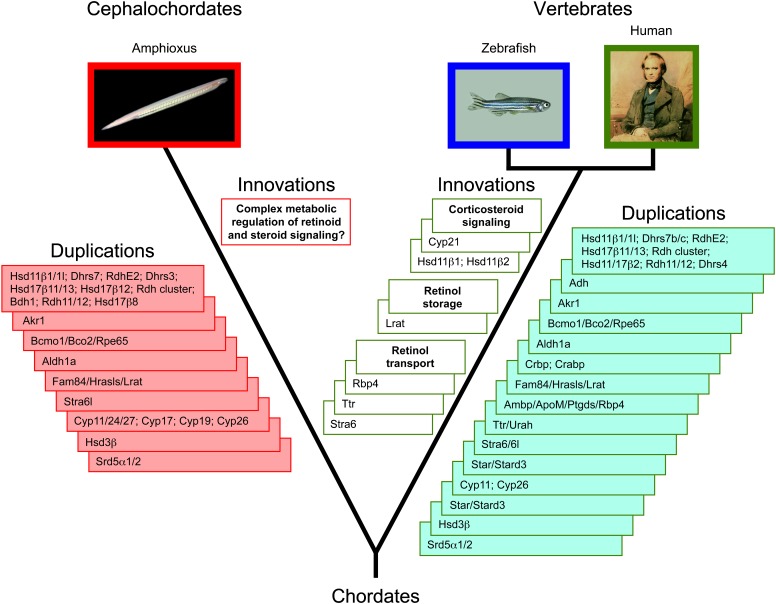
In this study, we have investigated the evolutionary history of the chordate retinoid and steroid signaling systems by carrying out an exhaustive analysis of the amphioxus genome. Our evolutionary inferences are based on the premises that orthologs typically have similar functions (Tatusov et al. 1997; Eisen 1998; Gabaldón and Huynen 2004) and that integrative functional predictions combining phylogenetic information, genomic linkage data, concerted gene gains or losses, and structural analyses are a powerful tool to predict the biological processes an orthologous protein participates in (Gabaldón and Huynen 2004). Using this combinatorial approach, we have thus evaluated the presence of the genetic machinery for retinoid and steroid signaling in amphioxus (fig. 4). We will now explore the implications of our findings on the evolutionary history of retinoid and steroid signaling in chordates, paying special attention to the evolution of new physiological capabilities in the vertebrate lineage.
FIG. 4.—
The evolutionary diversification of the retinoid and steroid genetic machineries in chordates. Gene duplications and functional innovations are shown for both the cephalochordate and the vertebrate lineages. The names of duplicated genes and gene families are indicated for both cephalochordates and vertebrates and several functional innovations resulting from the evolution of novel protein functions are proposed. The “?” highlights that additional studies assessing the biochemical and functional properties of the cephalochordate retinoid and steroid machineries are required to provide experimental support for the hypotheses derived from our evolutionary analyses.
Evolution of Retinoid Signaling
In the vertebrate RA signaling cascade, several enzymes, binding proteins, and transport elements constitute complex machineries that control the physiological levels of RA. Our global analysis of the genome of the cephalochordate amphioxus, the extant species most closely resembling the last invertebrate ancestor of vertebrates (Schubert, Escriva, et al. 2006; Holland et al. 2008; Putnam et al. 2008), gives us the unique opportunity to attempt the reconstruction of the RA pathway in this ancestor and hence to evaluate the roles RA signaling might have played in the evolutionary diversification of vertebrates (fig. 4).
We searched the genome of the amphioxus B. floridae for sequences similar to all vertebrate proteins involved in the retinoid pathway (fig. 1). Phylogenetic, genomic, and structural analyses indicate that amphioxus has 1) SDR-retinol dehydrogenases putatively capable to catalyze retinol oxidation and retinal reduction (Bf_Rdh_1 to 12, Bf_Rdh10, Bf_Rdh11/12_1 to 22, Bf_Rdh13, Bf_Rdh14, Bf_Dhrs3_1 and 2, and Bf_Dhrs4), 2) enzymes for RA synthesis and degradation (Bf_Aldh1a_1 to 6, Bf_Aldh8a and Bf_Cyp26_1 to 3), 3) enzymes likely involved in β-carotene cleavage (Bf_Bcmo1/Bco2/Rpe65_1 to 5), and 4) candidates for lipid binding proteins possibly involved in retinoid stabilization and protection within the cell (Bf_Crbp and Bf_iLbp_1 to 9). Our screening of the amphioxus genome is consistent with the results of previous analyses demonstrating the presence in amphioxus of single copies of RA and retinoid X receptors (RAR and RXR, respectively) (Holland LZ and Holland ND 1996; Cañestro et al. 2001; Escriva et al. 2002; Schubert et al. 2004, 2005; Schubert, Holland, et al. 2006; Koop et al. 2010) and of active retinoid metabolism (Dalfó et al. 2001, 2002). Moreover, the lineage-specific duplications of some key components of the retinoid pathway might actually reflect very intricate control mechanisms for retinoid metabolism throughout the amphioxus life cycle.
In contrast, our searches for Lrat, Rbp4, Ttr, and Stra6 suggest that amphioxus might lack counterparts of these components of vertebrate retinoid signaling. The amphioxus genome contains sequences related to these proteins, but structural analyses indicate that the amphioxus representatives of these families more closely resemble vertebrate proteins with functions that are not directly linked to the retinoid pathway. This concerted absence of the four main components involved in storage, transport, and cellular uptake of retinoids suggests that this system might be a functional novelty of vertebrates. Elaboration of this axis of retinoid metabolism might have enabled vertebrates to improve the control of retinoid homeostasis and to compensate retinoid fluctuations in natural environments.
Evolution of Steroid Signaling
Comparative analyses of steroidogenic enzymes has led to the conclusion that some enzymatic activities arose independently in arthropods and vertebrates and, therefore, that steroidogenesis has been elaborated in parallel in the main bilaterian lineages (Markov et al. 2009). The origin of the enzymatic activities within the chordate phylum, however, remains uncertain, and the survey of steroidogenic enzymes (fig. 1) in amphioxus is crucial to obtain insights into the evolution of the steroid pathway in chordates and vertebrates (fig. 4).
Our analysis reveals that, with some minor and very intriguing exceptions, the amphioxus genome contains most of the genetic machinery for steroid metabolism. The repertoire of steroidogenic enzymes is even significantly larger than previously anticipated (Baker 2004a). Amphioxus has orthologs of vertebrate Cyp17 (Bf_Cyp17_1 and 2), Cyp19 (Bf_Cyp19_1 and 2), Star (Bf_Star/Stard3), Hsd3β (Bf_Hsd3β_1 to 7), Srd5α (Bf_Srd5α1/2_1 and 2 and Bf_Srd5α3) and of mitochondrial Cyp enzymes evolutionarily related to vertebrate Cyp11a (Bf_Cyp11/24/27_1 to 8). Although Cyp19 and Hsd3β activities have been detected in amphioxus gonadal tissues (Callard et al. 1984; Mizuta et al. 2008), and although docking studies predict, for example, that Cyp19 can bind androgens (Callard et al. 2011), the biochemical activity of most of these amphioxus enzymes still requires experimental confirmation. Nevertheless, based on this collection of amphioxus enzymes and supported by the presence of sex steroids in amphioxus (Chang et al. 1985; Fang et al. 2001; Takeda et al. 2003; Mizuta and Kubokawa 2007), we hypothesize that a vertebrate-like sex steroid metabolism exists in cephalochordates. In contrast, amphioxus probably lacks Cyp21, which in vertebrates is an enzyme instrumental for corticosteroid synthesis. The absence of Cyp21 and the fact that C21 hydroxylated steroids could not be detected after incubation of amphioxus tissues with pregnenolone (Mizuta et al. 2008) together suggest that cephalochordates might not be capable of synthesizing corticosteroids and therefore that adrenal steroid hormones were a vertebrate innovation.
In amphioxus, sex steroids, including progesterone, estrone, estradiol and testosterone, have been detected by RIA and immunohistochemical methods (Chang et al. 1985; Fang et al. 2001; Takeda et al. 2003; Mizuta and Kubokawa 2007), and two steroid receptors, ER and SR, have been characterized (Bridgham et al. 2008; Paris et al. 2008; Katsu et al. 2010; Callard et al. 2011). Amphioxus ER is orthologous to vertebrate estrogen receptor (ER) and amphioxus SR is orthologous to the vertebrate steroid receptors (SR), including androgen receptor (AR), glucocorticoid receptor (GR), mineralocorticoid receptor, (MR) and progesterone receptor (PR) (Bridgham et al. 2008; Paris et al. 2008; Schubert et al. 2008; Katsu et al. 2010; Callard et al. 2011). Although amphioxus ER does not activate transcription and seems to be an estrogen insensitive inhibitor of SR activity (Bridgham et al. 2008; Katsu et al. 2010; Callard et al. 2011), amphioxus SR is activated by estrogens and mediates transcriptional activation through ER- and AR-like DNA response elements (Bridgham et al. 2008; Katsu et al. 2010). Although the binding of amphioxus ER and SR to other sex steroids, such as progesterone or testosterone, has not been demonstrated, 3D modeling of amphioxus SR suggests that it may also bind androgen steroids (Baker and Chang 2009). In sum, given the relative complexity of the amphioxus steroidogenic system due to various lineage-specific duplications, the study of the enzymatic activities of the different steroidogenic enzymes in amphioxus, the complete analysis of the binding capacities of the two amphioxus steroid receptors, and the detailed assessment of the physiological roles of steroids in amphioxus are crucial for understanding the elaboration of steroid-dependent signaling not only in cephalochordates but also in vertebrates.
Evolution of Prereceptor Regulation
In the steroid pathway, hydroxysteroid dehydrogenases regulate the steroid response in vertebrates by controlling hormone activation and inactivation, a mechanism known as prereceptor regulation of steroid action (reviewed in Penning 2003). Vertebrate Hsd17β control sex steroids, whereas Hsd11β regulate adrenal hormones. Amphioxus has many Hsd17β enzymes: one amphioxus Hsd11/17β2, six Hsd17β8, one Hsd17β10, and two Hsd17β11/13 are candidates to catalyze oxidation of sex steroids, whereas one Hsd17β7, three Hsd17β12, and one Hsd17β14 might be responsible for steroid reduction. Amphioxus, therefore, has the genetic machinery necessary for enzymatically activating and inactivating sex steroids, pushing back the evolutionary origins for prereceptor control of sex steroid responses to the origin of chordates. In contrast, given the likely absence of the corticosteroidogenic Cyp21 enzyme in amphioxus for synthesis of adrenal steroids, a prereceptor control for corticosteroid action is probably not required in amphioxus. Even though the amphioxus genome contains 23 Hsd11β1/1l-like and 1 Hsd11/17β2-like sequences, evolutionary and functional studies are compatible with these enzymes metabolizing steroids other than corticosteroids (Kusakabe et al. 2003; Baker 2004b, 2010b). The evolution of Hsd11β1 and Hsd11β2 activities against corticosteroids would thus be functional innovations of vertebrates that allowed the elaboration of a prereceptor control of the adrenal hormone response in the vertebrate lineage.
In vertebrates, retinoid action is tightly controlled spatiotemporally by the coordinated activity of enzymes synthesizing (i.e., Aldh1a and Aldh8a) and degrading (i.e., Cyp26) endogenous RA (Niederreither et al. 2002; Reijntjes et al. 2005; Campo-Paysaa et al. 2008; Theodosiou et al. 2010). This metabolic system controlling the physiological levels of retinoids available for receptor occupancy is reminiscent of the prereceptor system for steroid action. Our phylogenetic data indicate that this prereceptor regulation is also functioning in amphioxus and was hence already present in the last common ancestor of amphioxus and vertebrates. Moreover, because in amphioxus the components of retinoid prereceptor regulation have been subjected to lineage-specific duplications (there are six Aldh1a, one Aldh8a, and three Cyp26 enzymes encoded in the amphioxus genome), it is possible that this system has secondarily been elaborated in this lineage, creating a rather complex metabolic regulation of retinoid signaling, which contrasts the simplicity of the retinoid receptor system, which in cephalochordates is based on a single RAR and a unique RXR.
Supplementary Material
Supplementary figures S1–S15 are available at Genome Biology and Evolution online (http://www.gbe.oxfordjournals.org/).
Acknowledgments
The authors would like to thank Cristian Cañestro and Christophe Tiffoche for fruitful discussions and critical reading of the manuscript. We are also grateful to Diego Sánchez for his advice on the evolution of the lipocalin family. This research was supported by funds from ANR (ANR-07-BLAN-0038 and ANR-09-BLAN-0262-02) and CNRS (to M.S.), by CRESCENDO, a European Union Integrated Project of FP6, (to V.L. and M.S.), by a grant from the Ministerio de Ciencia e Innovación (BFU2010-14875) (to R.A.), and by the GDRE-RA Comparative Genomics (to R.A. and F.B.).
References
- Albalat R. The retinoic acid machinery in invertebrates: ancestral elements and vertebrate innovations. Mol Cell Endocrinol. 2009;313:23–35. doi: 10.1016/j.mce.2009.08.029. [DOI] [PubMed] [Google Scholar]
- Albalat R, Cañestro C. Identification of Aldh1a, Cyp26 and RAR orthologs in protostomes pushes back the retinoic acid genetic machinery in evolutionary time to the bilaterian ancestor. Chem Biol Interact. 2009;178:188–196. doi: 10.1016/j.cbi.2008.09.017. [DOI] [PubMed] [Google Scholar]
- Albiston AL, Obeyesekere VR, Smith RE, Krozowski ZS. Cloning and tissue distribution of the human 11 β-hydroxysteroid dehydrogenase type 2 enzyme. Mol Cell Endocrinol. 1994;105:R11–R17. doi: 10.1016/0303-7207(94)90176-7. [DOI] [PubMed] [Google Scholar]
- Anantharaman V, Aravind L. Evolutionary history, structural features and biochemical diversity of the NlpC/P60 superfamily of enzymes. Genome Biol. 2003;4:R11. doi: 10.1186/gb-2003-4-2-r11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker ME. Co-evolution of steroidogenic and steroid-inactivating enzymes and adrenal and sex steroid receptors. Mol Cell Endocrinol. 2004a;215:55–62. doi: 10.1016/j.mce.2003.11.007. [DOI] [PubMed] [Google Scholar]
- Baker ME. Evolutionary analysis of 11β-hydroxysteroid dehydrogenase-type 1, -type 2, -type 3 and 17β-hydroxysteroid dehydrogenase-type 2 in fish. FEBS Lett. 2004b;574:167–170. doi: 10.1016/j.febslet.2004.08.023. [DOI] [PubMed] [Google Scholar]
- Baker ME. 11β-hydroxysteroid dehydrogenase-type 2 evolved from an ancestral 17β-hydroxysteroid dehydrogenase-type 2. Biochem Biophys Res Commun. 2010a;399:215–220. doi: 10.1016/j.bbrc.2010.07.057. [DOI] [PubMed] [Google Scholar]
- Baker ME. Evolution of 11β-hydroxysteroid dehydrogenase-type 1 and 11β-hydroxysteroid dehydrogenase-type 3. FEBS Lett. 2010b;584:2279–2284. doi: 10.1016/j.febslet.2010.03.036. [DOI] [PubMed] [Google Scholar]
- Baker ME, Chang DJ. 3D model of amphioxus steroid receptor complexed with estradiol. Biochem Biophys Res Commun. 2009;386:516–520. doi: 10.1016/j.bbrc.2009.06.079. [DOI] [PubMed] [Google Scholar]
- Batten ML, et al. Lecithin-retinol acyltransferase is essential for accumulation of all-trans-retinyl esters in the eye and in the liver. J Biol Chem. 2004;279:10422–10432. doi: 10.1074/jbc.M312410200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belyaeva OV, Johnson MP, Kedishvili NY. Kinetic analysis of human enzyme RDH10 defines the characteristics of a physiologically relevant retinol dehydrogenase. J Biol Chem. 2008;283:20299–20308. doi: 10.1074/jbc.M800019200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belyaeva OV, Kedishvili NY. Human pancreas protein 2 (PAN2) has a retinal reductase activity and is ubiquitously expressed in human tissues. FEBS Lett. 2002;531:489–493. doi: 10.1016/s0014-5793(02)03588-3. [DOI] [PubMed] [Google Scholar]
- Belyaeva OV, Kedishvili NY. Comparative genomic and phylogenetic analysis of short-chain dehydrogenases/reductases with dual retinol/sterol substrate specificity. Genomics. 2006;88:820–830. doi: 10.1016/j.ygeno.2006.06.004. [DOI] [PubMed] [Google Scholar]
- Belyaeva OV, Korkina OV, Stetsenko AV, Kedishvili NY. Human retinol dehydrogenase 13 (RDH13) is a mitochondrial short-chain dehydrogenase/reductase with a retinaldehyde reductase activity. FEBS J. 2008;275:138–147. doi: 10.1111/j.1742-4658.2007.06184.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belyaeva OV, et al. Biochemical properties of purified human retinol dehydrogenase 12 (RDH12): catalytic efficiency toward retinoids and C9 aldehydes and effects of cellular retinol-binding protein type I (CRBPI) and cellular retinaldehyde-binding protein (CRALBP) on the oxidation and reduction of retinoids. Biochemistry. 2005;44:7035–7047. doi: 10.1021/bi050226k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertrand S, et al. Evolutionary genomics of nuclear receptors: from twenty-five ancestral genes to derived endocrine systems. Mol Biol Evol. 2004;21:1923–1937. doi: 10.1093/molbev/msh200. [DOI] [PubMed] [Google Scholar]
- Biswas MG, Russell DW. Expression cloning and characterization of oxidative 17β- and 3α-hydroxysteroid dehydrogenases from rat and human prostate. J Biol Chem. 1997;272:15959–15966. doi: 10.1074/jbc.272.25.15959. [DOI] [PubMed] [Google Scholar]
- Bray JE, Marsden BD, Oppermann U. The human short-chain dehydrogenase/reductase (SDR) superfamily: a bioinformatics summary. Chem Biol Interact. 2009;178:99–109. doi: 10.1016/j.cbi.2008.10.058. [DOI] [PubMed] [Google Scholar]
- Bridgham JT, Brown JE, Rodríguez-Marí A, Catchen JM, Thornton JW. Evolution of a new function by degenerative mutation in cephalochordate steroid receptors. PLoS Genet. 2008;4:e1000191. doi: 10.1371/journal.pgen.1000191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Callard GV, Pudney JA, Kendall SL, Reinboth R. In vitro conversion of androgen to estrogen in amphioxus gonadal tissues. Gen Comp Endocrinol. 1984;56:53–58. doi: 10.1016/0016-6480(84)90060-1. [DOI] [PubMed] [Google Scholar]
- Callard GV, et al. Evolutionary origins of the estrogen signaling system: insights from amphioxus. J Steroid Biochem Mol Biol. Forthcoming 2011 doi: 10.1016/j.jsbmb.2011.03.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campo-Paysaa F, Marlétaz F, Laudet V, Schubert M. Retinoic acid signaling in development: tissue-specific functions and evolutionary origins. Genesis. 2008;46:640–656. doi: 10.1002/dvg.20444. [DOI] [PubMed] [Google Scholar]
- Cañestro C, Albalat R, Escriva H, Gonzàlez-Duarte R. Endogenous β-galactosidase activity in amphioxus: a useful histochemical marker for the digestive system. Dev Genes Evol. 2001;211:154–156. doi: 10.1007/s004270100137. [DOI] [PubMed] [Google Scholar]
- Cañestro C, Albalat R, Postlethwait JH. Oikopleura dioica alcohol dehydrogenase class 3 provides new insights into the evolution of retinoic acid synthesis in chordates. Zool Sci. 2010;27:128–133. doi: 10.2108/zsj.27.128. [DOI] [PubMed] [Google Scholar]
- Cañestro C, Godoy L, Gonzàlez-Duarte R, Albalat R. Comparative expression analysis of Adh3 during arthropod, urochordate, cephalochordate and vertebrate development challenges its predicted housekeeping role. Evol Dev. 2003;5:157–162. doi: 10.1046/j.1525-142x.2003.03022.x. [DOI] [PubMed] [Google Scholar]
- Cañestro C, Postlethwait JH, Gonzàlez-Duarte R, Albalat R. Is retinoic acid genetic machinery a chordate innovation? Evol Dev. 2006;8:394–406. doi: 10.1111/j.1525-142X.2006.00113.x. [DOI] [PubMed] [Google Scholar]
- Cañestro C, et al. Amphioxus alcohol dehydrogenase is a class 3 form of single type and of structural conservation but with unique developmental expression. Eur J Biochem. 2000;267:6511–6518. doi: 10.1046/j.1432-1327.2000.01711.x. [DOI] [PubMed] [Google Scholar]
- Cañestro C, et al. Ascidian and amphioxus Adh genes correlate functional and molecular features of the ADH family expansion during vertebrate evolution. J Mol Evol. 2002;54:81–89. doi: 10.1007/s00239-001-0020-2. [DOI] [PubMed] [Google Scholar]
- Chang C, Liu Y, Zhu H. Steroid hormones and their functional regulation in amphioxus. In: Lofts B, Holmes WN, editors. Current trends in comparative endocrinology. Hong Kong (China): Hong Kong University Press; 1985. pp. 205–207. [Google Scholar]
- Chetyrkin SV, Belyaeva OV, Gough WH, Kedishvili NY. Characterization of a novel type of human microsomal 3α-hydroxysteroid dehydrogenase: unique tissue distribution and catalytic properties. J Biol Chem. 2001;276:22278–22286. doi: 10.1074/jbc.M102076200. [DOI] [PubMed] [Google Scholar]
- Chetyrkin SV, Hu J, Gough WH, Dumaual N, Kedishvili NY. Further characterization of human microsomal 3α-hydroxysteroid dehydrogenase. Arch Biochem Biophys. 2001;386:1–10. doi: 10.1006/abbi.2000.2203. [DOI] [PubMed] [Google Scholar]
- Clark BJ, Wells J, King SR, Stocco DM. The purification, cloning, and expression of a novel luteinizing hormone-induced mitochondrial protein in MA-10 mouse Leydig tumor cells. Characterization of the steroidogenic acute regulatory protein (StAR) J Biol Chem. 1994;269:28314–28322. [PubMed] [Google Scholar]
- Dalfó D, Albalat R, Molotkov A, Duester G, Gonzàlez-Duarte R. Retinoic acid synthesis in the prevertebrate amphioxus involves retinol oxidation. Dev Genes Evol. 2002;212:388–393. doi: 10.1007/s00427-002-0254-z. [DOI] [PubMed] [Google Scholar]
- Dalfó D, Cañestro C, Albalat R, Gonzàlez-Duarte R. Characterization of a microsomal retinol dehydrogenase gene from amphioxus: retinoid metabolism before vertebrates. Chem Biol Interact. 2001;130–132:359–370. doi: 10.1016/s0009-2797(00)00261-1. [DOI] [PubMed] [Google Scholar]
- Dalfó D, Marqués N, Albalat R. Analysis of the NADH-dependent retinaldehyde reductase activity of amphioxus retinol dehydrogenase enzymes enhances our understanding of the evolution of the retinol dehydrogenase family. FEBS J. 2007;274:3739–3752. doi: 10.1111/j.1742-4658.2007.05904.x. [DOI] [PubMed] [Google Scholar]
- Duan WR, Linzer DI, Gibori G. Cloning and characterization of an ovarian-specific protein that associates with the short form of the prolactin receptor. J Biol Chem. 1996;271:15602–15607. doi: 10.1074/jbc.271.26.15602. [DOI] [PubMed] [Google Scholar]
- Dufort I, Rheault P, Huang XF, Soucy P, Luu-The V. Characteristics of a highly labile human type 5 17β-hydroxysteroid dehydrogenase. Endocrinology. 1999;140:568–574. doi: 10.1210/endo.140.2.6531. [DOI] [PubMed] [Google Scholar]
- Edgar RC. MUSCLE: a multiple sequence alignment method with reduced time and space complexity. BMC Bioinformatics. 2004;5:113. doi: 10.1186/1471-2105-5-113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisen JA. Phylogenomics: improving functional predictions for uncharacterized genes by evolutionary analysis. Genome Res. 1998;8:163–167. doi: 10.1101/gr.8.3.163. [DOI] [PubMed] [Google Scholar]
- Escriva H, Bertrand S, Laudet V. The evolution of the nuclear receptor superfamily. Essays Biochem. 2004;40:11–26. doi: 10.1042/bse0400011. [DOI] [PubMed] [Google Scholar]
- Escriva H, Delaunay F, Laudet V. Ligand binding and nuclear receptor evolution. Bioessays. 2000;22:717–727. doi: 10.1002/1521-1878(200008)22:8<717::AID-BIES5>3.0.CO;2-I. [DOI] [PubMed] [Google Scholar]
- Escriva H, Holland ND, Gronemeyer H, Laudet V, Holland LZ. The retinoic acid signaling pathway regulates anterior/posterior patterning in the nerve cord and pharynx of amphioxus, a chordate lacking neural crest. Development. 2002;129:2905–2916. doi: 10.1242/dev.129.12.2905. [DOI] [PubMed] [Google Scholar]
- Escriva H, et al. Ligand binding was acquired during evolution of nuclear receptors. Proc Natl Acad Sci U S A. 1997;94:6803–6808. doi: 10.1073/pnas.94.13.6803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Estes KA, Kalamegham R, Hanna-Rose W. Membrane localization of the NlpC/P60 family protein EGL-26 correlates with regulation of vulval cell morphogenesis in Caenorhabditis elegans. Dev Biol. 2007;308:196–205. doi: 10.1016/j.ydbio.2007.05.020. [DOI] [PubMed] [Google Scholar]
- Fang YQ, Weng YZ, Hu XX. Distribution of sex steroid hormones and their receptors in the gonads and nervous system of amphioxus (Branchiostoma belcheri) Curr Zool. 2001;47:398–403. [Google Scholar]
- Flicek P, et al. Ensembl’s 10th year. Nucleic Acids Res. 2010;38:D557–D562. doi: 10.1093/nar/gkp972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flower DR. The lipocalin protein family: structure and function. Biochem J. 1996;318:1–14. doi: 10.1042/bj3180001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Folli C, Ramazzina I, Percudani R, Berni R. Ligand-binding specificity of an invertebrate (Manduca sexta) putative cellular retinoic acid binding protein. Biochim Biophys Acta. 2005;1747:229–237. doi: 10.1016/j.bbapap.2004.11.013. [DOI] [PubMed] [Google Scholar]
- Fomitcheva J, Baker ME, Anderson E, Lee GY, Aziz N. Characterization of Ke 6, a new 17β-hydroxysteroid dehydrogenase, and its expression in gonadal tissues. J Biol Chem. 1998;273:22664–22671. doi: 10.1074/jbc.273.35.22664. [DOI] [PubMed] [Google Scholar]
- Funder JW, Pearce PT, Smith R, Smith AI. Mineralocorticoid action: target tissue specificity is enzyme, not receptor, mediated. Science. 1988;242:583–585. doi: 10.1126/science.2845584. [DOI] [PubMed] [Google Scholar]
- Gabaldón T, Huynen MA. Prediction of protein function and pathways in the genome era. Cell Mol Life Sci. 2004;61:930–944. doi: 10.1007/s00018-003-3387-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galtier N, Gouy M, Gautier C. SEAVIEW and PHYLO_WIN: two graphic tools for sequence alignment and molecular phylogeny. Comput Appl Biosci. 1996;12:543–548. doi: 10.1093/bioinformatics/12.6.543. [DOI] [PubMed] [Google Scholar]
- Ganfornina MD, Gutiérrez G, Bastiani M, Sánchez D. A phylogenetic analysis of the lipocalin protein family. Mol Biol Evol. 2000;17:114–126. doi: 10.1093/oxfordjournals.molbev.a026224. [DOI] [PubMed] [Google Scholar]
- Geissler WM, et al. Male pseudohermaphroditism caused by mutations of testicular 17 β-hydroxysteroid dehydrogenase 3. Nat Genet. 1994;7:34–39. doi: 10.1038/ng0594-34. [DOI] [PubMed] [Google Scholar]
- Godoy L, Gonzàlez-Duarte R, Albalat R. S-Nitrosogluthathione reductase activity of amphioxus ADH3: insights into the nitric oxide metabolism. Int J Biol Sci. 2006;2:117–124. doi: 10.7150/ijbs.2.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzàlez-Duarte R, Albalat R. Merging protein, gene and genomic data: the evolution of the MDR-ADH family. Heredity. 2005;95:184–197. doi: 10.1038/sj.hdy.6800723. [DOI] [PubMed] [Google Scholar]
- Gottesman ME, Quadro L, Blaner WS. Studies of vitamin A metabolism in mouse model systems. Bioessays. 2001;23:409–419. doi: 10.1002/bies.1059. [DOI] [PubMed] [Google Scholar]
- Gough WH, VanOoteghem S, Sint T, Kedishvili NY. cDNA cloning and characterization of a new human microsomal NAD+-dependent dehydrogenase that oxidizes all-trans-retinol and 3α-hydroxysteroids. J Biol Chem. 1998;273:19778–19785. doi: 10.1074/jbc.273.31.19778. [DOI] [PubMed] [Google Scholar]
- Gu PL, Gunawardene YI, Chow BC, He JG, Chan SM. Characterization of a novel cellular retinoic acid/retinol binding protein from shrimp: expression of the recombinant protein for immunohistochemical detection and binding assay. Gene. 2002;288:77–84. doi: 10.1016/s0378-1119(02)00430-4. [DOI] [PubMed] [Google Scholar]
- Guindon S, Gascuel O. A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst Biol. 2003;52:696–704. doi: 10.1080/10635150390235520. [DOI] [PubMed] [Google Scholar]
- Gutiérrez G, Ganfornina MD, Sánchez D. Evolution of the lipocalin family as inferred from a protein sequence phylogeny. Biochim Biophys Acta. 2000;1482:35–45. doi: 10.1016/s0167-4838(00)00151-5. [DOI] [PubMed] [Google Scholar]
- Haeseleer F, Palczewski K. Short-chain dehydrogenases/reductases in retina. Methods Enzymol. 2000;316:372–383. doi: 10.1016/s0076-6879(00)16736-9. [DOI] [PubMed] [Google Scholar]
- Haeseleer F, et al. Dual-substrate specificity short chain retinol dehydrogenases from the vertebrate retina. J Biol Chem. 2002;277:45537–45546. doi: 10.1074/jbc.M208882200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holland LZ, Holland ND. Expression of AmphiHox-1 and AmphiPax-1 in amphioxus embryos treated with retinoic acid: insights into evolution and patterning of the chordate nerve cord and pharynx. Development. 1996;122:1829–1838. doi: 10.1242/dev.122.6.1829. [DOI] [PubMed] [Google Scholar]
- Holland LZ, et al. The amphioxus genome illuminates vertebrate 1425 origins and cephalochordate biology. Genome Res. 2008;18:1100–1111. doi: 10.1101/gr.073676.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang XF, Luu-The V. Characterization of the oxidative 3α-hydroxysteroid dehydrogenase activity of human recombinant 11-cis-retinol dehydrogenase. Biochim Biophys Acta. 2001;1547:351–358. doi: 10.1016/s0167-4838(01)00200-x. [DOI] [PubMed] [Google Scholar]
- Huang Y, et al. A novel human hydroxysteroid dehydrogenase like 1 gene (HSDL1) is highly expressed in reproductive tissues. Mol Biol Rep. 2001;28:185–191. doi: 10.1023/a:1015726217890. [DOI] [PubMed] [Google Scholar]
- Huyghe S, Mannaerts GP, Baes M, Van Veldhoven PP. Peroxisomal multifunctional protein-2: the enzyme, the patients and the knockout mouse model. Biochim Biophys Acta. 2006;1761:973–994. doi: 10.1016/j.bbalip.2006.04.006. [DOI] [PubMed] [Google Scholar]
- Jahng WJ, Xue L, Rando RR. Lecithin retinol acyltransferase is a founder member of a novel family of enzymes. Biochemistry. 2003;42:12805–12812. doi: 10.1021/bi035370p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jez JM, Bennett MJ, Schlegel BP, Lewis M, Penning TM. Comparative anatomy of the aldo-keto reductase superfamily. Biochem J. 1997;326:625–636. doi: 10.1042/bj3260625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin M, Li S, Moghrabi WN, Sun H, Travis GH. Rpe65 is the retinoid isomerase in bovine retinal pigment epithelium. Cell. 2005;122:449–459. doi: 10.1016/j.cell.2005.06.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin XH, et al. Discovery and characterization of a Ca2+-independent phosphatidylethanolamine N-acyltransferase generating the anandamide precursor and its congeners. J Biol Chem. 2007;282:3614–3623. doi: 10.1074/jbc.M606369200. [DOI] [PubMed] [Google Scholar]
- Kallberg Y, Oppermann U, Persson B. Classification of the short-chain dehydrogenase/reductase superfamily using hidden Markov models. FEBS J. 2010;277:2375–2386. doi: 10.1111/j.1742-4658.2010.07656.x. [DOI] [PubMed] [Google Scholar]
- Katsu Y, Kubokawa K, Urushitani H, Iguchi T. Estrogen-dependent transactivation of amphioxus steroid hormone receptor via both estrogen and androgen response elements. Endocrinology. 2010;151:639–648. doi: 10.1210/en.2009-0766. [DOI] [PubMed] [Google Scholar]
- Kawaguchi R, et al. A membrane receptor for retinol binding protein mediates cellular uptake of vitamin A. Science. 2007;315:820–825. doi: 10.1126/science.1136244. [DOI] [PubMed] [Google Scholar]
- Kedishvili NY, et al. Evidence that the human gene for prostate short-chain dehydrogenase/reductase (PSDR1) encodes a novel retinal reductase (RalR1) J Biol Chem. 2002;277:28909–28915. doi: 10.1074/jbc.M202588200. [DOI] [PubMed] [Google Scholar]
- Kiefer C, et al. Identification and characterization of a mammalian enzyme catalyzing the asymmetric oxidative cleavage of provitamin A. J Biol Chem. 2001;276:14110–14116. doi: 10.1074/jbc.M011510200. [DOI] [PubMed] [Google Scholar]
- Kim TS, et al. Delayed dark adaptation in 11-cis-retinol dehydrogenase-deficient mice: a role of RDH11 in visual processes in vivo. J Biol Chem. 2005;280:8694–8704. doi: 10.1074/jbc.M413172200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koop D, et al. Retinoic acid signaling targets Hox genes during the amphioxus gastrula stage: insights into early anterior-posterior patterning of the chordate body plan. Dev Biol. 2010;338:98–106. doi: 10.1016/j.ydbio.2009.11.016. [DOI] [PubMed] [Google Scholar]
- Kowalik D, Haller F, Adamski J, Moeller G. In search for function of two human orphan SDR enzymes: hydroxysteroid dehydrogenase like 2 (HSDL2) and short-chain dehydrogenase/reductase-orphan (SDR-O) J Steroid Biochem Mol Biol. 2009;117:117–124. doi: 10.1016/j.jsbmb.2009.08.001. [DOI] [PubMed] [Google Scholar]
- Kusakabe M, Nakamura I, Young G. 11β-hydroxysteroid dehydrogenase complementary deoxyribonucleic acid in rainbow trout: cloning, sites of expression, and seasonal changes in gonads. Endocrinology. 2003;144:2534–2545. doi: 10.1210/en.2002-220446. [DOI] [PubMed] [Google Scholar]
- Kuzniar A, van Ham RC, Pongor S, Leunissen JA. The quest for orthologs: finding the corresponding gene across genomes. Trends Genet. 2008;24:539–551. doi: 10.1016/j.tig.2008.08.009. [DOI] [PubMed] [Google Scholar]
- Larkin MA, et al. Clustal W and Clustal X version 2.0. Bioinformatics. 2007;23:2947–2948. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- Lee SA, Belyaeva OV, Popov IK, Kedishvili NY. Overproduction of bioactive retinoic acid in cells expressing disease-associated mutants of retinol dehydrogenase 12. J Biol Chem. 2007;282:35621–35628. doi: 10.1074/jbc.M706372200. [DOI] [PubMed] [Google Scholar]
- Lei Z, Chen W, Zhang M, Napoli JL. Reduction of all-trans-retinal in the mouse liver peroxisome fraction by the short-chain dehydrogenase/reductase RRD: induction by the PPARα ligand clofibrate. Biochemistry. 2003;42:4190–4196. doi: 10.1021/bi026948i. [DOI] [PubMed] [Google Scholar]
- Liang D, et al. Expressions of Raldh3 and Raldh4 during zebrafish early development. Gene Expr Patterns. 2008;8:248–253. doi: 10.1016/j.gep.2007.12.007. [DOI] [PubMed] [Google Scholar]
- Lin B, et al. Prostate short-chain dehydrogenase reductase 1 (PSDR1): a new member of the short-chain steroid dehydrogenase/reductase family highly expressed in normal and neoplastic prostate epithelium. Cancer Res. 2001;61:1611–1618. [PubMed] [Google Scholar]
- Lin M, Napoli JL. cDNA cloning and expression of a human aldehyde dehydrogenase (ALDH) active with 9-cis-retinal and identification of a rat ortholog, ALDH12. J Biol Chem. 2000;275:40106–40112. doi: 10.1074/jbc.M008027200. [DOI] [PubMed] [Google Scholar]
- Lin M, Zhang M, Abraham M, Smith SM, Napoli JL. Mouse retinal dehydrogenase 4 (RALDH4), molecular cloning, cellular expression, and activity in 9-cis-retinoic acid biosynthesis in intact cells. J Biol Chem. 2003;278:9856–9861. doi: 10.1074/jbc.M211417200. [DOI] [PubMed] [Google Scholar]
- Lukacik P, et al. Structural and biochemical characterization of human orphan DHRS10 reveals a novel cytosolic enzyme with steroid dehydrogenase activity. Biochem J. 2007;402:419–427. doi: 10.1042/BJ20061319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luu-The V, Tremblay P, Labrie F. Characterization of type 12 17β-hydroxysteroid dehydrogenase, an isoform of type 3 17β-hydroxysteroid dehydrogenase responsible for estradiol formation in women. Mol Endocrinol. 2006;20:437–443. doi: 10.1210/me.2005-0058. [DOI] [PubMed] [Google Scholar]
- Maeda A, et al. Role of photoreceptor-specific retinol dehydrogenase in the retinoid cycle in vivo. J Biol Chem. 2005;280:18822–18832. doi: 10.1074/jbc.M501757200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maeda A, et al. Redundant and unique roles of retinol dehydrogenases in the mouse retina. Proc Natl Acad Sci U S A. 2007;104:19565–19570. doi: 10.1073/pnas.0707477104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mangelsdorf DJ, et al. The nuclear receptor superfamily: the second decade. Cell. 1995;83:835–839. doi: 10.1016/0092-8674(95)90199-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mansfield SG, et al. Molecular cloning and characterization of an invertebrate cellular retinoic acid binding protein. Proc Natl Acad Sci U S A. 1998;95:6825–6830. doi: 10.1073/pnas.95.12.6825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marijanovic Z, et al. Closing the gap: identification of human 3-ketosteroid reductase, the last unknown enzyme of mammalian cholesterol biosynthesis. Mol Endocrinol. 2003;17:1715–1725. doi: 10.1210/me.2002-0436. [DOI] [PubMed] [Google Scholar]
- Markov GV, Paris M, Bertrand S, Laudet V. The evolution of the ligand/receptor couple: a long road from comparative endocrinology to comparative genomics. Mol Cell Endocrinol. 2008;293:5–16. doi: 10.1016/j.mce.2008.06.011. [DOI] [PubMed] [Google Scholar]
- Markov GV, et al. Independent elaboration of steroid hormone signaling pathways in metazoans. Proc Natl Acad Sci U S A. 2009;106:11913–11918. doi: 10.1073/pnas.0812138106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marks AR, et al. Molecular cloning and characterization of (R)-3-hydroxybutyrate dehydrogenase from human heart. J Biol Chem. 1992;267:15459–15463. [PubMed] [Google Scholar]
- Marlétaz F, Holland LZ, Laudet V, Schubert M. Retinoic acid signaling and the evolution of chordates. Int J Biol Sci. 2006;2:38–47. doi: 10.7150/ijbs.2.38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsunaga T, et al. Characterization of human DHRS4: an inducible short-chain dehydrogenase/reductase enzyme with 3β-hydroxysteroid dehydrogenase activity. Arch Biochem Biophys. 2008;477:339–347. doi: 10.1016/j.abb.2008.06.002. [DOI] [PubMed] [Google Scholar]
- Mindnich R, Adamski J. Zebrafish 17β-hydroxysteroid dehydrogenases: an evolutionary perspective. Mol Cell Endocrinol. 2009;301:20–26. doi: 10.1016/j.mce.2008.12.002. [DOI] [PubMed] [Google Scholar]
- Mindnich R, Moller G, Adamski J. The role of 17β-hydroxysteroid dehydrogenases. Mol Cell Endocrinol. 2004;218:7–20. doi: 10.1016/j.mce.2003.12.006. [DOI] [PubMed] [Google Scholar]
- Mizuta T, Asahina K, Suzuki M, Kubokawa K. In vitro conversion of sex steroids and expression of sex steroidogenic enzyme genes in amphioxus ovary. J Exp Zool A Ecol Genet Physiol. 2008;309:83–93. doi: 10.1002/jez.438. [DOI] [PubMed] [Google Scholar]
- Mizuta T, Kubokawa K. Presence of sex steroids and cytochrome P450 (CYP) genes in amphioxus. Endocrinology. 2007;148:3554–3565. doi: 10.1210/en.2007-0109. [DOI] [PubMed] [Google Scholar]
- Mondal MS, Ruiz A, Bok D, Rando RR. Lecithin retinol acyltransferase contains cysteine residues essential for catalysis. Biochemistry. 2000;39:5215–5220. doi: 10.1021/bi9929554. [DOI] [PubMed] [Google Scholar]
- Moon YA, Horton JD. Identification of two mammalian reductases involved in the two-carbon fatty acyl elongation cascade. J Biol Chem. 2003;278:7335–7343. doi: 10.1074/jbc.M211684200. [DOI] [PubMed] [Google Scholar]
- Nazarenko I, Schafer R, Sers C. Mechanisms of the HRSL3 tumor suppressor function in ovarian carcinoma cells. J Cell Sci. 2007;120:1393–1404. doi: 10.1242/jcs.000018. [DOI] [PubMed] [Google Scholar]
- Nelson DR. The cytochrome p450 homepage. Hum Genomics. 2009;4:59–65. doi: 10.1186/1479-7364-4-1-59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niederreither K, et al. Genetic evidence that oxidative derivatives of retinoic acid are not involved in retinoid signaling during mouse development. Nat Genet. 2002;31:84–88. doi: 10.1038/ng876. [DOI] [PubMed] [Google Scholar]
- Noy N. Retinoid-binding proteins: mediators of retinoid action. Biochem J. 2000;348:481–495. [PMC free article] [PubMed] [Google Scholar]
- O'Byrne SM, et al. Retinoid absorption and storage is impaired in mice lacking lecithin:retinol acyltransferase (LRAT) J Biol Chem. 2005;280:35647–35657. doi: 10.1074/jbc.M507924200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oppermann UC, Persson B, Jörnvall H. Function, gene organization and protein structures of 11β-hydroxysteroid dehydrogenase isoforms. Eur J Biochem. 1997;249:355–360. doi: 10.1111/j.1432-1033.1997.t01-1-00355.x. [DOI] [PubMed] [Google Scholar]
- Orland MD, et al. Acyl coenzyme A dependent retinol esterification by acyl coenzyme A: diacylglycerol acyltransferase 1. Biochim Biophys Acta. 2005;1737:76–82. doi: 10.1016/j.bbalip.2005.09.003. [DOI] [PubMed] [Google Scholar]
- Paris M, et al. An amphioxus orthologue of the estrogen receptor that does not bind estradiol: insights into estrogen receptor evolution. BMC Evol Biol. 2008;8:219. doi: 10.1186/1471-2148-8-219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Payne AH, Hales DB. Overview of steroidogenic enzymes in the pathway from cholesterol to active steroid hormones. Endocr Rev. 2004;25:947–970. doi: 10.1210/er.2003-0030. [DOI] [PubMed] [Google Scholar]
- Peltoketo H, Isomaa V, Maentausta O, Vihko R. Complete amino acid sequence of human placental 17 β-hydroxysteroid dehydrogenase deduced from cDNA. FEBS Lett. 1988;239:73–77. doi: 10.1016/0014-5793(88)80548-9. [DOI] [PubMed] [Google Scholar]
- Peltoketo H, Luu-The V, Simard J, Adamski J. 17β-hydroxysteroid dehydrogenase (HSD)/17-ketosteroid reductase (KSR) family; nomenclature and main characteristics of the 17HSD/KSR enzymes. J Mol Endocrinol. 1999;23:1–11. doi: 10.1677/jme.0.0230001. [DOI] [PubMed] [Google Scholar]
- Penning TM. Molecular endocrinology of hydroxysteroid dehydrogenases. Endocr Rev. 1997;18:281–305. doi: 10.1210/edrv.18.3.0302. [DOI] [PubMed] [Google Scholar]
- Penning TM. Hydroxysteroid dehydrogenases and pre-receptor regulation of steroid hormone action. Hum Reprod Update. 2003;9:193–205. doi: 10.1093/humupd/dmg022. [DOI] [PubMed] [Google Scholar]
- Penning TM, et al. Human 3α-hydroxysteroid dehydrogenase isoforms (AKR1C1-AKR1C4) of the aldo-keto reductase superfamily: functional plasticity and tissue distribution reveals roles in the inactivation and formation of male and female sex hormones. Biochem J. 2000;351:67–77. doi: 10.1042/0264-6021:3510067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perrière G, Gouy M. WWW-Query: an on-line retrieval system for biological sequence banks. Biochimie. 1996;78:364–369. doi: 10.1016/0300-9084(96)84768-7. [DOI] [PubMed] [Google Scholar]
- Putnam NH, et al. The amphioxus genome and the evolution of the chordate karyotype. Nature. 2008;453:1064–1071. doi: 10.1038/nature06967. [DOI] [PubMed] [Google Scholar]
- Rattner A, Smallwood PM, Nathans J. Identification and characterization of all-trans-retinol dehydrogenase from photoreceptor outer segments, the visual cycle enzyme that reduces all-trans-retinal to all-trans-retinol. J Biol Chem. 2000;275:11034–11043. doi: 10.1074/jbc.275.15.11034. [DOI] [PubMed] [Google Scholar]
- Reijntjes S, Blentic A, Gale E, Maden M. The control of morphogen signalling: regulation of the synthesis and catabolism of retinoic acid in the developing embryo. Dev Biol. 2005;285:224–237. doi: 10.1016/j.ydbio.2005.06.019. [DOI] [PubMed] [Google Scholar]
- Ross AC. Retinol esterification by rat liver microsomes. Evidence for a fatty acyl coenzyme A: retinol acyltransferase. J Biol Chem. 1982;257:2453–2459. [PubMed] [Google Scholar]
- Ross SA, McCaffery PJ, Drager UC, De Luca LM. Retinoids in embryonal development. Physiol Rev. 2000;80:1021–1054. doi: 10.1152/physrev.2000.80.3.1021. [DOI] [PubMed] [Google Scholar]
- Ruiz FX, et al. Aldo-keto reductases from the AKR1B subfamily: retinoid specificity and control of cellular retinoic acid levels. Chem Biol Interact. 2009;178:171–177. doi: 10.1016/j.cbi.2008.10.027. [DOI] [PubMed] [Google Scholar]
- Russell DW, Wilson JD. Steroid 5α-reductase: two genes/two enzymes. Annu Rev Biochem. 1994;63:25–61. doi: 10.1146/annurev.bi.63.070194.000325. [DOI] [PubMed] [Google Scholar]
- Sánchez D, Ganfornina MD, Gutiérrez G, Marín A. Exon-intron structure and evolution of the Lipocalin gene family. Mol Biol Evol. 2003;20:775–783. doi: 10.1093/molbev/msg079. [DOI] [PubMed] [Google Scholar]
- Schaap FG, van der Vusse GJ, Glatz JF. Evolution of the family of intracellular lipid binding proteins in vertebrates. Mol Cell Biochem. 2002;239:69–77. [PubMed] [Google Scholar]
- Schubert M, Escriva H, Xavier-Neto J, Laudet V. Amphioxus and tunicates as evolutionary model systems. Trends Ecol Evol. 2006;21:269–277. doi: 10.1016/j.tree.2006.01.009. [DOI] [PubMed] [Google Scholar]
- Schubert M, Holland ND, Escriva H, Holland LZ, Laudet V. Retinoic acid influences anteroposterior positioning of epidermal sensory neurons and their gene expression in a developing chordate (amphioxus) Proc Natl Acad Sci U S A. 2004;101:10320–10325. doi: 10.1073/pnas.0403216101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schubert M, Holland ND, Laudet V, Holland LZ. A retinoic acid-Hox hierarchy controls both anterior/posterior patterning and neuronal specification in the developing central nervous system of the cephalochordate amphioxus. Dev Biol. 2006;296:190–202. doi: 10.1016/j.ydbio.2006.04.457. [DOI] [PubMed] [Google Scholar]
- Schubert M, et al. Retinoic acid signaling acts via Hox1 to establish the posterior limit of the pharynx in the chordate amphioxus. Development. 2005;132:61–73. doi: 10.1242/dev.01554. [DOI] [PubMed] [Google Scholar]
- Schubert M, et al. Nuclear hormone receptor signaling in amphioxus. Dev Genes Evol. 2008;218:651–665. doi: 10.1007/s00427-008-0251-y. [DOI] [PubMed] [Google Scholar]
- Shafqat N, et al. Expanded substrate screenings of human and Drosophila type 10 17β-hydroxysteroid dehydrogenases (HSDs) reveal multiple specificities in bile acid and steroid hormone metabolism: characterization of multifunctional 3α/7α/7β/17β/20β/21-HSD. Biochem J. 2003;376:49–60. doi: 10.1042/BJ20030877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shehu A, et al. Prolactin receptor-associated protein/17β-hydroxysteroid dehydrogenase type 7 gene (Hsd17β7) plays a crucial role in embryonic development and fetal survival. Mol Endocrinol. 2008;22:2268–2277. doi: 10.1210/me.2008-0165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simões-Costa MS, Azambuja AP, Xavier-Neto J. The search for non-chordate retinoic acid signaling: lessons from chordates. J Exp Zool B Mol Dev Evol. 2008;310:54–72. doi: 10.1002/jez.b.21139. [DOI] [PubMed] [Google Scholar]
- Söderhäll I, et al. Characterization of a hemocyte intracellular fatty acid-binding protein from crayfish (Pacifastacus leniusculus) and shrimp (Penaeus monodon) FEBS J. 2006;273:2902–2912. doi: 10.1111/j.1742-4658.2006.05303.x. [DOI] [PubMed] [Google Scholar]
- Soref CM, et al. Characterization of a novel airway epithelial cell-specific short chain alcohol dehydrogenase/reductase gene whose expression is up-regulated by retinoids and is involved in the metabolism of retinol. J Biol Chem. 2001;276:24194–24202. doi: 10.1074/jbc.M100332200. [DOI] [PubMed] [Google Scholar]
- Stocco DM. StAR protein and the regulation of steroid hormone biosynthesis. Annu Rev Physiol. 2001;63:193–213. doi: 10.1146/annurev.physiol.63.1.193. [DOI] [PubMed] [Google Scholar]
- Storbeck KH, Swart P, Swart AC. Cytochrome P450 side-chain cleavage: insights gained from homology modeling. Mol Cell Endocrinol. 2007;265–266:65–70. doi: 10.1016/j.mce.2006.12.005. [DOI] [PubMed] [Google Scholar]
- Takeda N, Kubokawa K, Matsumoto G. Immunoreactivity for progesterone in the giant Rohde cells of the amphioxus, Branchiostoma belcheri. Gen Comp Endocrinol. 2003;132:379–383. doi: 10.1016/s0016-6480(03)00099-6. [DOI] [PubMed] [Google Scholar]
- Takimoto N, Kusakabe T, Horie T, Miyamoto Y, Tsuda M. Origin of the vertebrate visual cycle: III. Distinct distribution of RPE65 and β-carotene 15,15'-monooxygenase homologues in Ciona intestinalis. Photochem Photobiol. 2006;82:1468–1474. doi: 10.1562/2006-01-14-RA-775. [DOI] [PubMed] [Google Scholar]
- Tarrant AM, et al. Steroid metabolism in cnidarians: insights from Nematostella vectensis. Mol Cell Endocrinol. 2009;301:27–36. doi: 10.1016/j.mce.2008.09.037. [DOI] [PubMed] [Google Scholar]
- Tatusov RL, Koonin EV, Lipman D. A genomic perspective on protein families. Science. 1997;278:631–637. doi: 10.1126/science.278.5338.631. [DOI] [PubMed] [Google Scholar]
- Theodosiou M, Laudet V, Schubert M. From carrot to clinic: an overview of the retinoic acid signaling pathway. Cell Mol Life Sci. 2010;67:1423–1445. doi: 10.1007/s00018-010-0268-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uemura M, et al. Novel 5α-steroid reductase (SRD5A3, type-3) is overexpressed in hormone-refractory prostate cancer. Cancer Sci. 2008;99:81–86. doi: 10.1111/j.1349-7006.2007.00656.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Lintig J, Vogt K. Filling the gap in vitamin A research. Molecular identification of an enzyme cleaving β-carotene to retinal. J Biol Chem. 2000;275:11915–11920. doi: 10.1074/jbc.275.16.11915. [DOI] [PubMed] [Google Scholar]
- Watari H, et al. MLN64 contains a domain with homology to the steroidogenic acute regulatory protein (StAR) that stimulates steroidogenesis. Proc Natl Acad Sci U S A. 1997;94:8462–8467. doi: 10.1073/pnas.94.16.8462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu L, et al. Expression cloning and characterization of human 17β-hydroxysteroid dehydrogenase type 2, a microsomal enzyme possessing 20α-hydroxysteroid dehydrogenase activity. J Biol Chem. 1993;268:12964–12969. [PubMed] [Google Scholar]
- Wu X, Lukacik P, Kavanagh KL, Oppermann U. SDR-type human hydroxysteroid dehydrogenases involved in steroid hormone activation. Mol Cell Endocrinol. 2007;265–266:71–76. doi: 10.1016/j.mce.2006.12.006. [DOI] [PubMed] [Google Scholar]
- Yamamoto H, et al. Mutations in the gene encoding 11-cis retinol dehydrogenase cause delayed dark adaptation and fundus albipunctatus. Nat Genet. 1999;22:188–191. doi: 10.1038/9707. [DOI] [PubMed] [Google Scholar]
- Zanotti G, et al. Structure of zebrafish HIUase: insights into evolution of an enzyme to a hormone transporter. J Mol Biol. 2006;363:1–9. doi: 10.1016/j.jmb.2006.07.079. [DOI] [PubMed] [Google Scholar]