Abstract
Arbuscular mycorrhizal fungi (AMF) represent an ecologically important and evolutionarily intriguing group of symbionts of land plants, currently thought to have propagated clonally for over 500 Myr. AMF produce multinucleate spores and may exchange nuclei through anastomosis, but meiosis has never been observed in this group. A provocative alternative for their successful and long asexual evolutionary history is that these organisms may have cryptic sex, allowing them to recombine alleles and compensate for deleterious mutations. This is partly supported by reports of recombination among some of their natural populations. We explored this hypothesis by searching for some of the primary tools for a sustainable sexual cycle—the genes whose products are required for proper completion of meiotic recombination in yeast—in the genomes of four AMF and compared them with homologs of representative ascomycete, basidiomycete, chytridiomycete, and zygomycete fungi. Our investigation used molecular and bioinformatic tools to identify homologs of 51 meiotic genes, including seven meiosis-specific genes and other “core meiotic genes” conserved in the genomes of the AMF Glomus diaphanum (MUCL 43196), Glomus irregulare (DAOM-197198), Glomus clarum (DAOM 234281), and Glomus cerebriforme (DAOM 227022). Homology of AMF meiosis-specific genes was verified by phylogenetic analyses with representative fungi, animals (Mus, Hydra), and a choanoflagellate (Monosiga). Together, these results indicate that these supposedly ancient asexual fungi may be capable of undergoing a conventional meiosis; a hypothesis that is consistent with previous reports of recombination within and across some of their populations.
Keywords: comparative genomics, meiosis, fungi, arbuscular mycorrhizal fungi, genome evolution, ancient asexuals
Introduction
Meiosis, a hallmark of eukaryotic cells, is necessary for the production of gametes (e.g., spores). It is a major driver of recombination in all eukaryotes, resulting in the shuffling of genomic material between chromosomes. Although predominant throughout eukaryotes (Malik et al. 2008), the advantages versus costs of sexual reproduction and meiosis are still a matter of debate (Ackerman et al. 2010; Archetti 2010). Indeed, although evolutionary theory predicts a rapid extinction of asexual lineages as a consequence of the accumulation of deleterious mutations (Otto and Lenormand 2002; Otto 2009), a number of eukaryotes commonly referred to as “ancient asexuals” (Maynard-Smith 1986) have thrived across diverse ecosystems for millions of years without sex. These ancient asexuals include evolutionarily distant groups such as the bdelloid rotifers, the arbuscular mycorrhizal fungi (AMF) and a number of protist lineages (Maynard-Smith 1986; Haig 1993; Judson and Normark 1996; Gordo and Charlesworth 2000; Normark 2003; Schurko et al. 2009), which are all derived from sexual ancestors (Ramesh et al. 2005).
In the last decade, research on ancient asexuals has provided new insights into how these organisms coped with the absence of observable sexual cycles. First, the majority of these “asexuals” have been found to exhibit genetic recombination consistent with sexual reproduction (Schurko et al. 2009; Heitman 2010). Second, all former putatively asexual taxa whose completely sequenced genomes have been surveyed so far—including a number of medically important pathogens (i.e., Giardia intestinalis, Trichomonas vaginalis, Entamoeba histolytica, several microsporidia, Candida spp., Cryptococcus neoformans, Aspergillus spp., etc.)— have “core meiotic gene” homologs that encode a set of proteins that only function during meiosis in model animals, fungi, and plants studied, and other general DNA repair proteins that are required for proper completion of meiotic recombination in model organisms (Villeneuve and Hillers 2001; Wong et al. 2003; Ramesh et al. 2005; Lee et al. 2008, 2010; Malik et al. 2008), consistent with the hypothesis that the core meiotic gene products would also function in meiotic recombination (or a similar recently derived parasexual process) in these organisms. Orthologs of meiosis-specific genes would be under relaxed functional constraint and accumulate deleterious mutations in asexual organisms and could not be detected by comparative genomic approaches (discussed further by Schurko and Logsdon 2008; Schurko et al. 2009 and references therein), whereas general function DNA repair proteins required for meiotic and mitotic recombination would persist (e.g., Rad51, Mlh1, Pms1, Msh2, Msh6, Rad50, Mre11, etc.). Third, sexual reproduction has recently been identified in other organisms long thought to reproduce only clonally, such as the fungus Aspergillus fumigatus, (Poggeler 2002; Heitman 2006, 2010; O'Gorman et al. 2009), indicating that asexuality should not be assumed based solely on absence of a recognizable sexual stage.
AMF represent one of those ancient asexual lineages whose genomes have yet to be explored for the evidence of sex. These coenocytic and multinucleate fungi represent a group of plant symbionts that play a dramatic role in terrestrial ecosystems and that are thought to date back 500 My (Humphreys et al. 2010). Attempts to explain their extreme longevity in the absence of sex have generally focused on their atypical cellular content, which includes hundreds of nuclei per spore that may, or may not, be genetically divergent (Kuhn et al. 2001; Pawlowska and Taylor 2004; Hijri and Sanders 2005; Pawlowska 2005; Stukenbrock and Rosendahl 2005). These nuclei are thought to be exchanged at a certain rate among different AMF through anastomosis (fusion between hyphae) (Croll et al. 2008; Angelard et al. 2010) and to regularly recombine.
In this study, we explore the potential for meiotic recombination in AMF by searching for the core meiotic genes across the genomes of four AMF species. This “core” of genes, a term first coined by Villeneuve and Hillers (2001) with reference to meiotic recombination machinery of model animal, fungal, and plant systems, and later expanded by phylogenomic analyses to include diverse protists (Ramesh et al. 2005; Malik et al. 2008), encodes 30 proteins that comprise the conserved meiotic recombination machinery of eukaryotes. The present comparative genomic survey allows us to form a hypothetical model for meiosis-like recombination in a cryptic sexual (or parasexual) cycle in AMF, which is consistent with previous reports of recombination in these organisms and may be tested further by future functional studies.
Materials and Methods
Acquisition of a Low Coverage Survey of Glomus diaphanum, Glomus irregulare, Glomus clarum, and Glomus cerebriforme
Low coverage genome surveys of G. diaphanum (MUCL 43196), G. irregulare (DAOM-197198), G. cerebriforme (DAOM 227022), and G. clarum (DAOM 234281) were obtained using the 454 pyrosequencing facility at the Génome Québec Innovation Centre (McGill University, Canada). In all cases, an average of 350 Mb of genome data have been generated using an average read length of 336 bp (median length 368 bp). The assemblies resulted in an average of 46,000 contigs for all species with an average length of 1,010 bp. All AMF sequences identified in this study and their relative accession numbers are shown in table 1 and the supplementary table S1, Supplementary Material online.
Table 1.
List of 51 AMF Genes Directly or Indirectly Involved in the Process of Meiosis
| Gene | Accession Number | CS-Blast Best Hit |
Pfam/NCBI CDD | |||
| Organism | Alignment Score | e Value | % Identity (Identities—Positives) | |||
| DMC1a | FR775428 | Sc | 512 | ×10−146 | 60—74 | PF08423.5 (Rad51); PLN03187 (meiotic recombinase Dmc1) |
| DNL4 | GW_113491.1 | Nc | 340 | 5.00 × 10−94 | 53—73 | PF01068.1 (DNA_ligase_A_M), PF04679.9 (DNA_ligase_A_C) |
| EXO1 | GW_104190.1 | Sm | 125 | 4.00 × 10−30 | 43—74 | — |
| HOP2a | FR775431 | Sc | 174 | 7.00 × 10−45 | 21—45 | PF07106 (TBPIP) |
| HRR25 | HQ262397 | Sm | 259 | 9.00 × 10−70 | 96—97 | PF00069.19 (Pkinase) |
| HTA1 | GW_094651.1 | Nc | 166 | 5.00 × 10−42 | 77—83 | PF00125.18 (Histone) |
| HTA2 | GW_095958.1 | Nc | 130 | 2.00 × 10−31 | 84—88 | PF00125.18 (Histone) |
| MEC1 | FR775435 | Sm | 260 | 6.00 × 10−70 | 46—69 | PF00613.14 (PI3Ka) |
| MLH1 | FR775257 | Nc | 319 | 1.00 × 10−87 | 43—56 | — |
| MLH3 | GW_092048.1 | Sm | 229 | 1.00 × 10−60 | 34—53 | — |
| MND1a | GW_112099.1 | Sc | 212 | 3.00 × 10−56 | 27—46 | PF03962.9 (Mnd1) |
| MRE11 | GW_118657.1 | Sm | 454 | ×10−128 | 47—64 | PF00149.22 (Metallophos) |
| MSC1 | FR822520 | Sm | 148 | 1.00 × 10−36 | 45—59 | — |
| MSC7 | BM027003.1 | Sc | 290 | 6.00 × 10−79 | 39—63 | PF00171.1 (Aldedh) |
| MSH2 | HQ262398 | Sc | 224 | 3.00 × 10−59 | 60—70 | PF00488.15 (MutS_V) |
| MSH4 | FR877640 | Sc | 212 | 9.00 × 10−56 | 35—62 | PF05192.12 (MutS_III), cd03282 (ABC_MSH4_euk) |
| MSH5b | FR775430 | Sc | 231 | 1.00 × 10−80 | 34—51 | PF00488.15 (MutS_V); cd03243 (ABC_MSH5_euk) |
| MSH6 | GW_100645.1 | Sm | 346 | 8.00 × 10−96 | 61—76 | PF01624.14 (MutS_I), PF05188.11 (MutS_II) |
| MUS81 | FR775258 | Sm | 463 | ×10−131 | 38—55 | PF02732.9 (ERCC4) |
| NAM8/MRE2 | FR775259 | Sm | 216 | 1.00 × 10−56 | 61—79 | PF00076.16 (RRM_1) |
| PDS5 | GW_121462.1 | Sm | 257 | 5.00 × 10−69 | 25—42 | — |
| PMS1 | GW_084934.1 | Nc | 137 | 4.00 × 10−33 | 46—72 | PF01119.13 (DNA_mis_repair) |
| RAD1 | FR775256 | Sm | 251 | 2.00 × 10−67 | 53—68 | PF02732.9 (ERCC4) |
| RAD17 | FR822519 | Nc | 253 | 4.00 × 10−68 | 47—60 | PF03215.9 (Rad17) |
| Rad2 | GW_108841.1 | Sm | 206 | 8.00 × 10−54 | 40—57 | — |
| Rad21 | FR877641 | Nc | 198 | 2.00 × 10−51 | 39—55 | — |
| RAD23 | BM959423.1 | Nc | 151 | 1.00 × 10−37 | 43—63 | PF09280.5 (XPC-binding), PF00627.25 (UBA) |
| RAD50 | FR775261 | Sm | 279 | 1.00 × 10−75 | 36—55 | PF02463.13 (SMC_N) |
| RAD51 | FR877639 | Nc | 377 | ×10−105 | 70—83 | PF08423.5 (Rad51) |
| RAD52 | GW_121846.1 | Sc | 130 | 5.00 × 10−31 | 51—68 | PF04098.9 (Rad52_Rad22) |
| RAD54b | GW_109886.1 | Sm | 195 | 8.00 × 10−51 | 39—57 | PF00271.25 (Helicase_C) |
| REC8 | FR775434 | Nc | 146 | 2.00 × 10−36 | 30—51 | PF04825 (Rad21_Rec8) |
| RFA1 | FR838021 | Nc | 278 | 1.00 × 10−75 | 24—43 | PF08646.4 (Rep_fac-A_C) |
| RFA2 | FR838020 | Nc | 198 | 2.00 × 10−51 | 20—32 | — |
| SCC3 | GW_117977.1 | Nc | 299 | 1.00 × 10−81 | 23—41 | — |
| SGS1 | FR775427 | Nc | 189 | 8.00 × 10−49 | 32—50 | PF00271.25 (Helicase_C) |
| SLX1 | FR775433 | Nc | 197 | 4.00 × 10−51 | 35—47 | PF01541.18 (GIY-YIG) |
| SMC1 | GW_121295.1 | Sm | 224 | 5.00 × 10−59 | 66—79 | PF02463.13 (SMC_N) |
| SMC2 | GW_094523.1 | Nc | 286 | 8.00 × 10−78 | 70—83 | PF02463.13 (SMC_N) |
| SMC3 | GW_093795.1 | Nc | 257 | 4.00 × 10−69 | 69—84 | PF02463.13 (SMC_N) |
| SMC4 | GW_088627.1 | Sm | 269 | 1.00 × 10−72 | 66—82 | PF02463.13 (SMC_N) |
| SMC5 | FR775432 | Sm | 300 | 9.00 × 10−82 | 26—50 | — |
| SMC6 | FR775262 | Sm | 293 | 6.00 × 10−80 | 37—61 | PF02463.13 (SMC_N) |
| SPO11 | FR822517 | Nc | 167 | 2.00 × 10−42 | 38—66 | TP6A_N (Type IIB DNA topoisomerase), PLN00060 (meiotic recombination protein SPO11) |
| SRS2a | FR822518 | Sc | 127 | 3.00 × 10−31 | 51—74 | PF00580.15 (UvrD-helicase) |
| TOP1 | FR775263 | Sc | 491 | ×10−139 | 48—63 | PF01028.14 (Topoisom_I) |
| TOP2 | FR775264 | Sm | 535 | ×10−152 | 47—62 | PF00521.1 (DNA_topoisoIV) |
| TOP3 | GW_111366.1 | Nc | 403 | ×10−113 | 57—71 | PF01131.14 (Topoisom_bac) |
| YKU70 | FR775265 | Nc | 276 | 7.00 × 10−75 | 27—43 | PF03730.8 (Ku_C) |
| YKU80 | GW_117534.1 | Nc | 304 | 3.00 × 10−83 | 37—55 | PF02735.10 (Ku) |
| Zip4/Spo22 | FR822521 | Nc | 151 | 2.00 × 10−37 | 31—47 | — |
Note.—Pfam families and domains were retrieved from the Sanger Pfam utility server (2), for e values less than or equal to 1 × 10−05 (Finn et al. 2008).
Best CS-BLAST hit information (source organism, score, e value, and % identity) of each Glomus meiotic protein against a database containing the Saccharomyces cerevisiae S288c (Sc), Neurospora crassa OR74A (Nc), and Sordaria macrospora (Sm) genomes or against the Sc genome when no ortholog is present in the two other ascomycetes.
Alignments were performed using CS-BLAST with two iterations. Meiosis-specific genes are highlighted in gray cells. Each Glomus spp. protein best hit information corresponds to the expected fungal ortholog, except for Rad54, for which corresponding hits represent the greatest identity percentage and the greatest hit length.
In Silico Identification of AMF Meiosis Genes
A total number of 87 genes known to be required for proper meiotic recombination in Saccharomyces cerevisiae have been searched across the genomes of G. irregulare (DAOM 181602) and G. diaphanum (MUCL 43196) using three independent, yet highly complementary approaches (table 1; supplementary table S1, Supplementary Material online). This list of genes is based on previous genomic surveys of fungi and other eukaryotes (Malik et al. 2008; Burns et al. 2010; Nowrousian et al. 2010), which we expanded here to include representatives from those extant fungal phyla with publicly available complete or near-completely sequenced genomes in gene depositories, that is, the Basidiomycota C. neoformans, Ustilago maydis, and Coprinus cinereus; the Chytridiomycota Batrachochytrium dendrobatidis and Allomyces macrogynus; and the Zygomycota Rhizopus oryzae and Phycomyces blakesleeanus.
Putative meiotic genes were initially searched using reciprocal TBlastX and TBlastN (Altschul et al. 1997) searches of publicly available expressed sequence tags from G. irregulare deposited in GenBank (National Center for Biotechnology Information [NCBI]). This preliminary analysis allowed the identification of several AMF transcripts with unambiguous homology to meiosis genes from a number of more distantly related fungal taxa. The remaining genes were from a low coverage genome survey of G. diaphanum, G. clarum, and G. cerebriforme using reciprocal BlastX, TBlastX, and TBlastN procedures and by using a polymerase chain reaction (PCR) approach based on degenerate primers. Overall, the combination of these bioinformatics and molecular approaches allowed the identification of 51 meiosis-related genes in AMF (table 1). Only the best hits, exhibiting a cut off e value lower than 1 × 10−05, were retained in Blast searches. Homology was also asserted using sequence context-specific profiles for homology searches (CS-BLAST; table 1) (Biegert and Soding 2009) for all meiosis-related genes. CS-BLAST searches were performed as follows: Glomus spp. proteins were aligned using CS-BLAST with two iterations, against all S. cerevisiae S288c, Neurospora crassa OR74A, and Sordaria macrospora proteins or against S. cerevisiae when no ortholog was present in N. crassa or S. macrospora, S. cerevisiae, and N. crassa proteins were obtained from the NCBI and S. macrospora proteins were downloaded from “The S. macrospora genome site” of the Ruhr-University Bochum ([Nowrousian et al. 2010], http://c4-1-8.serverhosting.rub.de/public/downloads.html). Pfam family and domain information were also retrieved from the Sanger Pfam server (http://pfam.sanger.ac.uk/). HMMER searches failed to identify additional AMF genes that could not be found using traditional Blast procedures. Other publicly accessible genome sequence databases we searched include the Broad Institute's databases for A. macrogynus, R. oryzae, and B. dendrobatidis (http://www.broadinstitute.org/scientific-community/data) and the Joint Genome Institute’s (JGI) databases for P. blakesleeanus, U. maydis, and Monosiga brevicollis (http://genome.jgi-psf.org/).
Polymerase Chain Reaction
The AMF Msh2 and HRR25 homologues were identified by degenerate PCR. Degenerate PCR was performed using combinations of new primers designed on conserved amino acid motifs of proteins encoding Msh2 (Msh2F: 5′-ATH GAR YTI GGN GTI AAR GA-3′; motif IQLGVK and Msh2R: 5′-AAY ATG GGN GGI AAR AGY ACI TA-3′; motif NMGGKST), and HRR25 (Hrr25F 5′-CCI CAR YTI GAR TAY GAR GC-3′; motif PQLEYE and Hrr25R: 5′-GGN GTI GTC ATY TTY TTY TCC AT-3′; motif MEKKMTT). Briefly, PCR reactions were performed using conditions described previously for other AMF genes (Corradi et al. 2004) and DNA extracted from in vitro cultures of G. diaphanum. Successful PCR amplicons were subjected to gel extraction, bacterial cloning, and conventional Sanger DNA sequencing.
Phylogenetic Verification of Glomus spp. Meiosis-Specific Protein-Coding Genes in Gene Families
The identification of meiosis-specific gene homologs in Glomus spp. was subject to further scrutiny by phylogenetic comparison to other representative eukaryotes, to ensure that orthologs (vs. paralogs) of the seven meiosis-specific genes (Rec8, Spo11-1, Dmc1, Hop2, Mnd1, Msh4, and Msh5) were correctly identified. Generally, we compared Glomus spp. sequences with data from representative fungi, animals (Mus musculus, Hydra magnipapillata), and a choanoflagellate (M. brevicollis). Sequences obtained from G. cerebriforme (Msh4, Rad21, Dmc1) and G. clarum (Rad51, Dmc1) were added to the phylogenetic analyses in an effort to improve resolution.
We assembled Glomus spp. meiosis-specific genes and annotated putative open reading frames by using Geneious Pro 5.3.6 (Biomatters Ltd.) with reference to pairwise comparisons made by BlastX of GenBank and to multiple sequence alignments of homologous proteins made with MUSCLE v. 3.7 (Edgar 2004). Where applicable, vector or PCR primer sequences were excluded from the assemblies. Besides Glomus spp., homologs of meiosis-specific proteins were identified by BlastP searches of the nonredundant NCBI database, JGI, and the Broad Institute (see above). Multiple amino acid sequence alignments (MUSCLE v. 3.7 [Edgar 2004]) were inspected and adjusted manually using MacClade 4.08 (Maddison WP and Maddison DR 1989), and only unambiguously aligned amino acid sites were used for phylogenetic analyses.
We used RAxML v. 7.2.8 (Stamatakis 2006) and MrBayes v. 3.12 (Huelsenbeck and Ronquist 2001; Ronquist and Huelsenbeck 2003) for phylogenetic analyses. Amino acid sequence phylogenies were computed using RAxML v. 7.2.8 with the LG model of amino acid substitutions (Le and Gascuel 2008) and 25 γ-distributed substitution rate categories (LG + 25γ) for 1,000 bootstrap replicates. Bootstrap support was estimated from 1,000 replicates using PhyML v3.0 (Guindon and Gascuel 2003) with the LG + I + 8γ model. We ran MrBayes for 106 generations hosted by the CIPRES Science Gateway Portal v. 3.1 at the San Diego Supercomputer Center (Miller et al. 2011), with four incrementally heated Markov chains, a sampling frequency of 103 generations, temperature set at 0.5 and Whelan and Goldman model (WAG) + I + 8γ (Whelan and Goldman 2001). Only the RAxML topologies are shown.
Phylogenetic Analyses of Concatenated DNA Repair Proteins
A fungal phylogeny was inferred using 12 orthologous DNA repair proteins among the meiotic proteins retrieved for all surveyed fungal taxa (highlighted as blue cells; supplementary table S1, Supplementary Material online), as well as in the outgroup species M. brevicollis (Choanoflagellata) and the animals M. musculus and Tetraodon nigroviridis. A multiple sequence alignment was produced using MUSCLE for each protein (Edgar 2004), and divergent or ambiguous positions were removed. Evolutionary models for each protein were determined using ProtTest (Abascal et al. 2005). Several phylogeny inference procedures gave similar trees (data not shown). The alignments were concatenated using Concaterpillar (Leigh et al. 2008). The phylogenetic tree was inferred using PhyML v3.0 (Guindon and Gascuel 2003) and WAG + 4γ with 1,000 bootstrap replicates. A phylogenetic tree was also inferred using MrBayes v3.1.2 (Ronquist and Huelsenbeck 2003), with 107 generations with 4 γ-distributed substitution rate categories and separate substitution models for each protein.
Results and Discussion
In the present study, we identified a total of 51 genes in Glomus spp. that are required for the proper completion of meiosis in S. cerevisiae (fig. 1 and table 1, supplementary table S1, Supplementary Material online). Homologues of S. cerevisiae genes that could not be identified in Glomus spp. were also absent from the representative genomes of other higher fungal groups, including Sordariales among ascomycetes. Overall, among the 87 S. cerevisiae genes we searched, none were missing exclusively from our available AMF sequence data set, suggesting that our in silico and molecular approaches have covered most, if not all, of the available AMF predicted meiotic proteome.
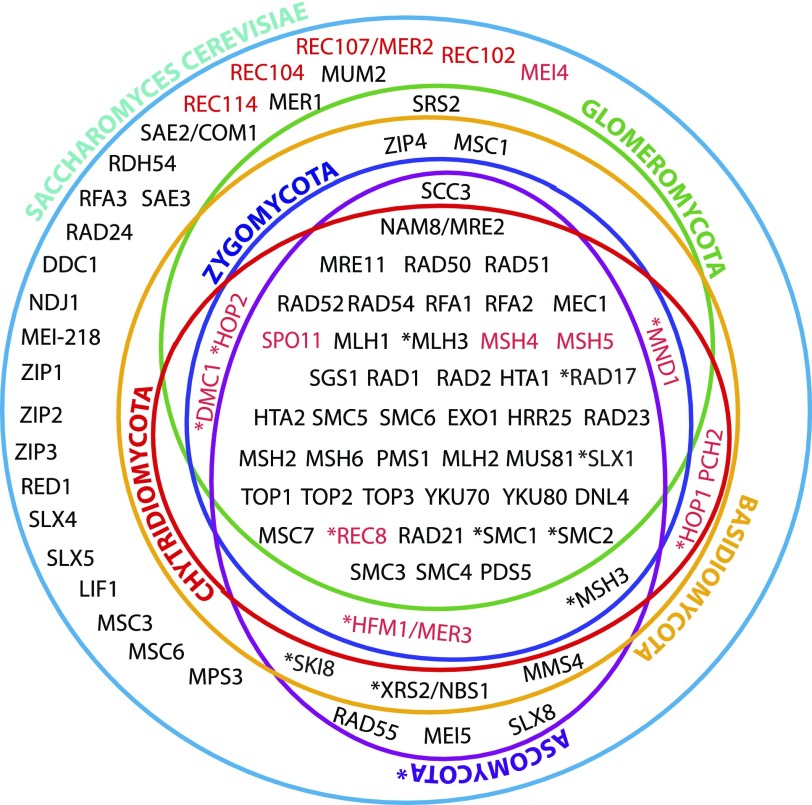
FIG. 1.
Expanded catalog of fungal meiotic genes. Venn diagram showing the presence or absence of the genes known to be directly or indirectly involved in meiotic processes in Saccharomyces cerevisiae (green circle) (Nowrousian et al. 2010). The presence or absence of these genes have been scored in the genomes of fungal relatives, including representative species belonging to the phylum Ascomycota ([Nowrousian et al. 2010], purple circle), Basidiomycota ([Donaldson and Saville 2008; Burns et al. 2010], orange circle), Chytridiomycota (red circle), Zygomycota (dark blue circle), and the AMF Glomeromycota (Green circle), inventoried in detail in supplementary table S1, Supplementary Material online. Meiosis-specific genes are shown in red text. Asterisks represent genes that are sometimes absent in the genome of one or more members of a given phylum. Data included in the purple circle were reported elsewhere (Nowrousian et al. 2010), and we did not repeat the analyses.
Importantly, more than 85% of the core meiotic genes were found to be present in AMF (fig. 2). The only AMF core meiotic genes that could not be detected were homologues of Pch2, Hop1, Mei4, and Mer3; all genes whose loss does not affect the successful completion of meiosis in many fungi (Malik et al. 2008; Kumar et al. 2010). In particular, Pch2, Mei4, Hop1, and Mer3 genes are also absent from the genome of the zygomycete R. oryzae, and Hop1 and Mer3 are absent from known sexual organisms (i.e., N. crassa, Gibberella zeae, and Drosophila melanogaster; Malik et al. [2008]).
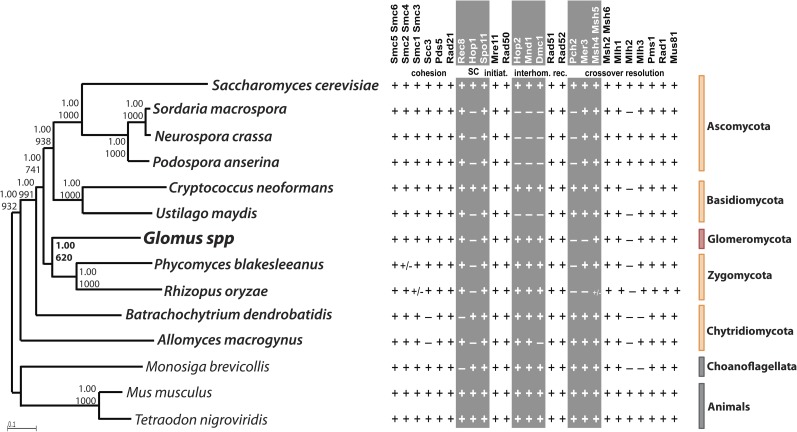
FIG. 2.
Phylogenetic tree of 12 concatenated DNA repair protein sequences (Exo1, Rad1, Mlh1, Mre11, Msh6, Mus81, Smc6, Top1, Top3, Rad23, Rad50, Rad52) (left) and list of the core meiotic genes (right). Left side of the figure: Glomus spp. (G. irregulare and G. diaphanum) are highlighted in red, all surveyed fungi (Ascomycota, Basidiomycota, Chytridiomycota, and Zygomycota) in orange, and outgroup organisms (one choanoflagellate and two animals) in gray (supplementary table S2, Supplementary Material online). Numbers at nodes correspond to Bayesian posterior probabilities (top) and bootstrap supports from 1,000 replicates of maximum likelihood (PhyML) analysis (bottom). Scale bar represents 0.1 amino acid substitutions per site. Right side of the figure: list of the core meiotic proteins (adapted from references San-Segundo and Roeder 1999; Villeneuve and Hillers 2001; Malik et al. 2008; Joshi et al. 2009 and references therein) and their presence (+) and absence (−, i.e., not detected) in the fungal genomes surveyed in this study. +/− denotes the absence of the given genes in some species belonging to that specific phylum. Meiosis-specific proteins are shown in gray columns. A. Ascomycota; B. Basidiomycota; C. Chytridiomycota; Z. Zygomycota; G. Glomeromycota (i.e., AMF). Orthologs of Rad21, Rad51, Pms1, and Mlh and meiosis-specific Spo11-1, Rec8, Hop1, Hop2, Mnd1, and Dmc1 genes of basidiomycetes and B. dendrobatidis were identified with assistance from Arthur Pightling.
Our study indicates that AMF genomes contain genes encoding all the tools necessary for meiotic recombination. In particular, they have genes that encode orthologs of seven meiosis-specific proteins involved in sister-chromatid cohesion (Rec8), double-strand DNA breaks (Spo11-1), interhomolog recombination (Mnd1, Hop2, and Dmc1), and class II crossovers (Msh4 and Msh5) (supplementary figs. S1 and S2, Supplementary Material online). Phylogenetic analyses were used to verify the orthology of these seven meiosis-specific gene homologs in Glomus spp. relative to other fungi, with animals and a choanoflagellate as outgroups (supplementary figs. S1 and S2, Supplementary Material online). These proteins were not selected to trace the species genealogy but are sufficient to determine orthology of all Glomus spp. meiosis-specific genes we identified and their vertical descent (as opposed to them being specifically related to another organism by lateral gene transfer or contaminants in our cultures). In particular, Glomus spp. encode a meiosis-specific Rec8 protein that is distinct from the general Rad21 sister-chromatid cohesion and harbor orthologs of the meiosis-specific transesterase Spo11-1. The meiosis-specific RecA homolog, Dmc1, encoded Glomus spp. is also of fungal origin, as is Rad51, the general eukaryotic recombinase required for homologous recombination. Glomus spp. encode distinct meiosis-specific Mnd1 and Hop2 orthologs; these function with Dmc1 in interhomolog DNA strand exchange during meiosis in model organisms. Glomus spp. are also equipped for mismatch repair with Msh2 and Msh6 proteins and also for meiosis-specific (class II) crossovers that exhibit interference, with Msh4 and Msh5 proteins. Altogether, the presence of these genes in Glomus spp. is compelling evidence for an active, hitherto undetected, meiosis-like program in the life cycle of AMF.
The presence of meiotic recombination proteins in AMF is also supported by other independent signatures of sexuality, namely the presence of many retrotransposons (Ty1-Copia and Ty3-Gypsy; data not shown) (Matic 2001; Wright and Finnegan 2001; Arkhipova 2005; Arkhipova and Meselson 2005; Gollotte et al. 2006) and recombination within their populations (Vandenkoornhuyse et al. 2001; Croll and Sanders 2009; den Bakker et al. 2010). These evolutionary features, combined with the presence of an expanded suite of conserved meiotic recombination genes, are compelling indicators of sexual reproduction in many eukaryotes (Malik et al. 2008; Schurko et al. 2009). Here, we propose a model of meiotic recombination in AMF based on the presence of core meiotic genes (fig. 3).
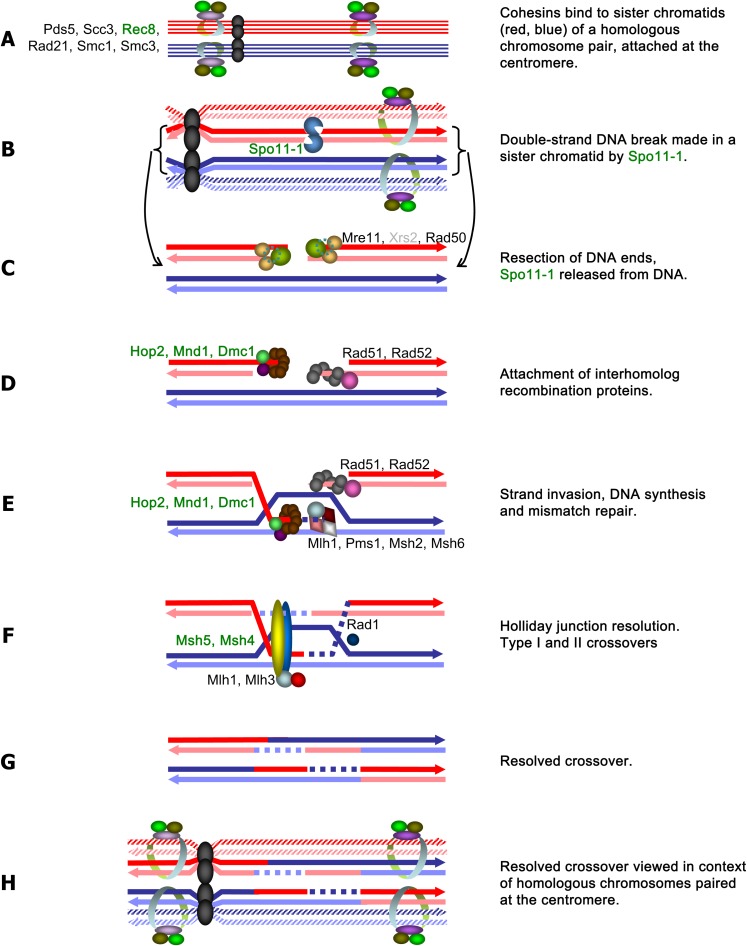
FIG. 3.
Hypothetical model of meiotic recombination in AMF Glomus spp., depicting likely interactions among proteins identified in this study. The names of meiosis-specific proteins are highlighted in green. Exact stoichiometry is not implied. In meiosis I, cohesins bind to sister chromatids (A), after which double-strand DNA breaks occur, with Spo11 and accessory recombination initiation proteins if present (B). Double-strand break repair is initiated (C). Interhomolog recombination and strand exchange proteins are attracted to the double-strand break (accessory proteins not shown) (D). The resulting heteroduplex (E) may be resolved by class II crossovers, which utilize meiosis-specific proteins (F, G) or by gene conversion (proteins not shown) or Class I crossovers (via Mus81), which do not. This model is derived from the general model that was based on details from Saccharomyces cerevisiae, Drosophila melanogaster, Caenorhabditis elegans, and Arabidopsis thaliana, and phylogenomic analyses described in references (Malik et al. 2008) and references within.
We also identified meiosis-specific gene homologs in B. dendrobatidis, a chytridiomycete that lacks any described sexual cycle. Although sex is now known in the other fungi included in our analyses, B. dendrobatidis, the fungal pathogen of amphibians, appears to primarily reproduce asexually (James et al. 2009). Core meiotic genes identified in figure 2 indicate that B. dendrobatidis is also capable of undergoing meiotic recombination.
The acquisition of a large sequence data set allowed us to tackle another interesting aspect of AMF evolution, namely their origin from within the fungal kingdom. In particular, we tested the most recent findings suggesting that these ubiquitous organisms may be more closely related to the Zygomycetes than previously thought (Corradi and Sanders 2006; Lee and Young 2009; Liu et al. 2009). We tested this hypothesis by reconstructing a fungal phylogeny of the DNA repair and recombination proteins encoded by all surveyed taxa (fig. 2), as these are fairly well conserved. The resulting phylogenies were all very similar to those identified recently, showing that AMF cluster with relatively strong support as a sister group of Mucorales (phylum Zygomycota). Obviously, the reduced species sampling in our study does not allow any conclusive evidence about the specific evolutionary origin of AMF within the fungal kingdom. However, this relevant phylogenetic signal, together with a virtually identical set of core meiotic genes between those groups (all genes that are absent in AMF are also absent from R. oryzae), is a highly intriguing relationship that will hopefully bolster future research in this specific area of comparative genomics upon completion of the first AMF genome sequence (Martin et al. 2008).
Recent advances in the field of population genetics have allowed the identification of several events of recombination both within and across several AMF populations (Vandenkoornhuyse et al. 2001; Croll and Sanders 2009; den Bakker et al. 2010). However, conclusions about the origin of such events (i.e., meiotic vs. mitotic recombination) have been systematically shadowed by a lack of evidence for meiosis in these putative ancient asexuals. By providing the first evidence for an expanded and conserved catalog of AMF meiosis-specific genes, the present study fills an important gap in our understanding of the genetics of these ubiquitous ecologically important organisms. In particular, these conclusions open up the exciting perspective that AMF may not be the evolutionary aberration that they have been long held to be and that they may be able to undergo a cryptic sexual cycle. Future studies such as colocalization or genetic disruption are required to demonstrate the conditions in which the meiosis-specific gene homologs we identified in this study encode products functioning in meiosis in Glomus spp. or if they function in a putative parasexual process including interhomolog recombination and crossing over, that is recently derived from a typical meiotic recombination process.
Supplementary Material
Supplementary figures S1 and S2 and tables S1 and S2 are available at Genome Biology and Evolution online (http://www.gbe.oxfordjournals.org/).
Acknowledgments
We would like to thank Arthur Pightling for providing access prior to publication to his analyses of core meiotic genes (in John Logsdon’s laboratory, University of Iowa) and for thoughtful discussion, and we thank Joseph Heitman and Jason Stajich and two anonymous reviewers for their thoughtful comments on a previous version of the manuscript. N.C. and C.H.S. are Scholars of the Canadian Institute for Advanced Research (CIFAR) Program in Integrated Microbial Biodiversity (IMB). S.-B. M. is supported by a CIFAR-IMB Junior Fellowship. This project is supported by grants from the Natural Sciences and Engineering Research Council of Canada (NSERC-Discovery) to N.C., C.H.S., and M.H.
References
- Abascal F, Zardoya R, Posada D. ProtTest: selection of best-fit models of protein evolution. Bioinformatics. 2005;21:2104–2105. doi: 10.1093/bioinformatics/bti263. [DOI] [PubMed] [Google Scholar]
- Ackerman S, Kermany AR, Hickey DA. Finite populations, finite resources, and the evolutionary maintenance of genetic recombination. J Hered. 2010;101:S135–S141. doi: 10.1093/jhered/esq019. [DOI] [PubMed] [Google Scholar]
- Altschul SF, et al. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Angelard C, Colard A, Niculita-Hirzel H, Croll D, Sanders IR. Segregation in a mycorrhizal fungus alters rice growth and symbiosis-specific gene transcription. Curr Biol. 2010;20:1216–1221. doi: 10.1016/j.cub.2010.05.031. [DOI] [PubMed] [Google Scholar]
- Archetti M. Complementation, genetic conflict, and the evolution of sex and recombination. J Hered. 2010;101:S21–S33. doi: 10.1093/jhered/esq009. [DOI] [PubMed] [Google Scholar]
- Arkhipova I, Meselson M. Deleterious transposable elements and the extinction of asexuals. Bioessays. 2005;27:76–85. doi: 10.1002/bies.20159. [DOI] [PubMed] [Google Scholar]
- Arkhipova IR. Mobile genetic elements and sexual reproduction. Cytogenet Genome Res. 2005;110:372–382. doi: 10.1159/000084969. [DOI] [PubMed] [Google Scholar]
- Biegert A, Soding J. Sequence context-specific profiles for homology searching. Proc Natl Acad Sci U S A. 2009;106:3770–3775. doi: 10.1073/pnas.0810767106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burns C, et al. Analysis of the basidiomycete Coprinopsis cinerea reveals conservation of the core meiotic expression program over half a billion years of evolution. PLoS Genet. 2010;6(9):e1001135. doi: 10.1371/journal.pgen.1001135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corradi N, Hijri M, Fumagalli L, Sanders IR. Arbuscular mycorrhizal fungi (Glomeromycota) harbour ancient fungal tubulin genes that resemble those of the chytrids (Chytridiomycota) Fungal Genet Biol. 2004;41:1037–1045. doi: 10.1016/j.fgb.2004.08.005. [DOI] [PubMed] [Google Scholar]
- Corradi N, Sanders IR. Evolution of the P-type II ATPase gene family in the fungi and presence of structural genomic changes among isolates of Glomus intraradices. BMC Evol Biol. 2006;6:21. doi: 10.1186/1471-2148-6-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Croll D, Sanders IR. Recombination in Glomus intraradices, a supposed ancient asexual arbuscular mycorrhizal fungus. BMC Evol Biol. 2009;9:13. doi: 10.1186/1471-2148-9-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Croll D, et al. Nonself vegetative fusion and genetic exchange in the arbuscular mycorrhizal fungus Glomus intraradices. New Phytol. 2009;181:924–937. doi: 10.1111/j.1469-8137.2008.02726.x. [DOI] [PubMed] [Google Scholar]
- den Bakker HC, Vankuren NW, Morton JB, Pawlowska TE. Clonality and recombination in the life history of an asexual arbuscular mycorrhizal fungus. Mol Biol Evol. 2010;27:2474–2486. doi: 10.1093/molbev/msq155. [DOI] [PubMed] [Google Scholar]
- Donaldson ME, Saville BJ. Bioinformatic identification of Ustilago maydis meiosis genes. Fungal Genet Biol. 2008;45:S47–S53. doi: 10.1016/j.fgb.2008.04.012. [DOI] [PubMed] [Google Scholar]
- Edgar RC. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finn RD, et al. The Pfam protein families database. Nucleic Acids Res. 2008;36:281–288. doi: 10.1093/nar/gkm960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gollotte A, et al. Repetitive DNA sequences include retrotransposons in genomes of the Glomeromycota. Genetica. 2006;128:455–469. doi: 10.1007/s10709-006-0019-0. [DOI] [PubMed] [Google Scholar]
- Gordo I, Charlesworth B. The degeneration of asexual haploid populations and the speed of Muller’s ratchet. Genetics. 2000;154:1379–1387. doi: 10.1093/genetics/154.3.1379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guindon S, Gascuel O. A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst Biol. 2003;52:696–704. doi: 10.1080/10635150390235520. [DOI] [PubMed] [Google Scholar]
- Haig D. Alternatives to meiosis: the unusual genetics of red algae, microsporidia, and others. J Theor Biol. 1993;163:15–31. doi: 10.1006/jtbi.1993.1104. [DOI] [PubMed] [Google Scholar]
- Heitman J. Sexual reproduction and the evolution of microbial pathogens. Curr Biol. 2006;16:R711–R725. doi: 10.1016/j.cub.2006.07.064. [DOI] [PubMed] [Google Scholar]
- Heitman J. Evolution of eukaryotic microbial pathogens via covert sexual reproduction. Cell Host Microbe. 2010;8:86–99. doi: 10.1016/j.chom.2010.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hijri M, Sanders IR. Low gene copy number shows that arbuscular mycorrhizal fungi inherit genetically different nuclei. Nature. 2005;433:160–163. doi: 10.1038/nature03069. [DOI] [PubMed] [Google Scholar]
- Huelsenbeck JP, Ronquist F. MRBAYES: Bayesian inference of phylogenetic trees. Bioinformatics. 2001;17:754–755. doi: 10.1093/bioinformatics/17.8.754. [DOI] [PubMed] [Google Scholar]
- Humphreys CP, et al. Mutualistic mycorrhiza-like symbiosis in the most ancient group of land plants. Nat Commun. 2010;1:103. doi: 10.1038/ncomms1105. [DOI] [PubMed] [Google Scholar]
- James TY, et al. Rapid global expansion of the fungal disease chytridiomycosis into declining and healthy amphibian populations. PLoS Pathog. 2009;5:e1000458. doi: 10.1371/journal.ppat.1000458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joshi N, Barot A, Jamison C, Borner GV. Pch2 links chromosome axis remodeling at future crossover sites and crossover distribution during yeast meiosis. PLoS Genet. 2009;5:e1000557. doi: 10.1371/journal.pgen.1000557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Judson OP, Normark BB. Ancient asexual scandals. Trends Ecol Evol. 1996;11:41–46. doi: 10.1016/0169-5347(96)81040-8. [DOI] [PubMed] [Google Scholar]
- Kuhn G, Hijri M, Sanders IR. Evidence for the evolution of multiple genomes in arbuscular mycorrhizal fungi. Nature. 2001;414:745–748. doi: 10.1038/414745a. [DOI] [PubMed] [Google Scholar]
- Kumar R, Bourbon HM, de Massy B. Functional conservation of Mei4 for meiotic DNA double-strand break formation from yeasts to mice. Genes Dev. 2010;24:1266–1280. doi: 10.1101/gad.571710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le SQ, Gascuel O. An improved general amino acid replacement matrix. Mol Biol Evol. 2008;25:1307–1320. doi: 10.1093/molbev/msn067. [DOI] [PubMed] [Google Scholar]
- Lee J, Young JP. The mitochondrial genome sequence of the arbuscular mycorrhizal fungus Glomus intraradices isolate 494 and implications for the phylogenetic placement of Glomus. New Phytol. 2009;183:200–211. doi: 10.1111/j.1469-8137.2009.02834.x. [DOI] [PubMed] [Google Scholar]
- Lee SC, et al. Microsporidia evolved from ancestral sexual fungi. Curr Biol. 2008;18:1675–1679. doi: 10.1016/j.cub.2008.09.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee SC, et al. Evolution of the sex-related locus and genomic features shared in microsporidia and fungi. PLoS One. 2010;5:e10539. doi: 10.1371/journal.pone.0010539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leigh JW, Susko E, Baumgartner M, Roger AJ. Testing congruence in phylogenomic analysis. Syst Biol. 2008;57:104–115. doi: 10.1080/10635150801910436. [DOI] [PubMed] [Google Scholar]
- Liu Y, et al. Phylogenomic analyses predict sistergroup relationship of nucleariids and fungi and paraphyly of zygomycetes with significant support. BMC Evol Biol. 2009;9:272. doi: 10.1186/1471-2148-9-272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maddison WP, Maddison DR. Interactive analysis of phylogeny and character evolution using the computer program MacClade. Folia Primatol. 1989;53:190–202. doi: 10.1159/000156416. [DOI] [PubMed] [Google Scholar]
- Malik S-B, Pightling AW, Stefaniak LM, Schurko AM, Logsdon JM., Jr An expanded inventory of conserved meiotic genes provides evidence for sex in Trichomonas vaginalis. PLoS One. 2008;3:e2879. doi: 10.1371/journal.pone.0002879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin F, et al. The long hard road to a completed Glomus intraradices genome. New Phytol. 2008;180:747–750. doi: 10.1111/j.1469-8137.2008.02671.x. [DOI] [PubMed] [Google Scholar]
- Matic I. Sex and retrotransposons. Trends Microbiol. 2001;9:110. doi: 10.1016/s0966-842x(01)01991-6. [DOI] [PubMed] [Google Scholar]
- Maynard-Smith J. Contemplating life without sex. Nature. 1986;324:300–301. doi: 10.1038/324300a0. [DOI] [PubMed] [Google Scholar]
- Miller MA, Pfeifer W, Schwartz T. Creating the CIPRES Science Gateway for inference of large phylogenetic trees. Proceedings of the Gateway Computing Environments Workshop (GCE); 2010 Nov 14; New Orleans, LA. New Orleans (LA): IEEE. p. 2010:1–8. [Google Scholar]
- Normark BB. Genomic signatures of ancient asexual lineages. Biol J Linn Soc. 2003;71:69–84. [Google Scholar]
- Nowrousian M, et al. De novo assembly of a 40 Mb eukaryotic genome from short sequence reads: Sordaria macrospora, a model organism for fungal morphogenesis. PLoS Genet. 2010;6:e1000891. doi: 10.1371/journal.pgen.1000891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Gorman CM, Fuller HT, Dyer PS. Discovery of a sexual cycle in the opportunistic fungal pathogen Aspergillus fumigatus. Nature. 2009;457:471–474. doi: 10.1038/nature07528. [DOI] [PubMed] [Google Scholar]
- Otto SP. The evolutionary enigma of sex. Am Nat. 2009;174(Suppl 1):S1–S14. doi: 10.1086/599084. [DOI] [PubMed] [Google Scholar]
- Otto SP, Lenormand T. Resolving the paradox of sex and recombination. Nat Rev Genet. 2002;3:252–261. doi: 10.1038/nrg761. [DOI] [PubMed] [Google Scholar]
- Pawlowska TE. Genetic processes in arbuscular mycorrhizal fungi. FEMS Microbiol Lett. 2005;251:185–192. doi: 10.1016/j.femsle.2005.08.007. [DOI] [PubMed] [Google Scholar]
- Pawlowska TE, Taylor JW. Organization of genetic variation in individuals of arbuscular mycorrhizal fungi. Nature. 2004;427:733–737. doi: 10.1038/nature02290. [DOI] [PubMed] [Google Scholar]
- Poggeler S. Genomic evidence for mating abilities in the asexual pathogen Aspergillus fumigatus. Curr Genet. 2002;42:153–160. doi: 10.1007/s00294-002-0338-3. [DOI] [PubMed] [Google Scholar]
- Ramesh MA, Malik SB, Logsdon JM., Jr A phylogenomic inventory of meiotic genes: evidence for sex in Giardia and an early eukaryotic origin of meiosis. Curr Biol. 2005;15:185–191. doi: 10.1016/j.cub.2005.01.003. [DOI] [PubMed] [Google Scholar]
- Ronquist F, Huelsenbeck JP. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics. 2003;19:1572–1574. doi: 10.1093/bioinformatics/btg180. [DOI] [PubMed] [Google Scholar]
- San-Segundo PA, Roeder GS. Pch2 links chromatin silencing to meiotic checkpoint control. Cell. 1999;97:313–324. doi: 10.1016/s0092-8674(00)80741-2. [DOI] [PubMed] [Google Scholar]
- Schurko AM, Logsdon JM., Jr. Using a meiosis detection toolkit to investigate ancient asexual “scandals” and the evolution of sex. Bioessays. 2008;30:579–589. doi: 10.1002/bies.20764. [DOI] [PubMed] [Google Scholar]
- Schurko AM, Neiman M, Logsdon JM., Jr Signs of sex: what we know and how we know it. Trends Ecol Evol. 2009;24:208–217. doi: 10.1016/j.tree.2008.11.010. [DOI] [PubMed] [Google Scholar]
- Stamatakis A. RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics. 2006;22:2688–2690. doi: 10.1093/bioinformatics/btl446. [DOI] [PubMed] [Google Scholar]
- Stukenbrock EH, Rosendahl S. Clonal diversity and population genetic structure of arbuscular mycorrhizal fungi (Glomus spp.) studied by multilocus genotyping of single spores. Mol Ecol. 2005;14:743–752. doi: 10.1111/j.1365-294X.2005.02453.x. [DOI] [PubMed] [Google Scholar]
- Vandenkoornhuyse P, Leyval C, Bonnin I. High genetic diversity in arbuscular mycorrhizal fungi: evidence for recombination events. Heredity. 2001;87:243–253. doi: 10.1046/j.1365-2540.2001.00941.x. [DOI] [PubMed] [Google Scholar]
- Villeneuve AM, Hillers KJ. Whence meiosis? Cell. 2001;106:647–650. doi: 10.1016/s0092-8674(01)00500-1. [DOI] [PubMed] [Google Scholar]
- Whelan S, Goldman N. A general empirical model of protein evolution derived from multiple protein families using a maximum-likelihood approach. Mol Biol Evol. 2001;18:691–699. doi: 10.1093/oxfordjournals.molbev.a003851. [DOI] [PubMed] [Google Scholar]
- Wong S, Fares MA, Zimmermann W, Butler G, Wolfe KH. Evidence from comparative genomics for a complete sexual cycle in the “asexual” pathogenic yeast Candida glabrata. Genome Biol. 2003;4:R10. doi: 10.1186/gb-2003-4-2-r10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright S, Finnegan D. Genome evolution: sex and the transposable element. Curr Biol. 2001;11:R296–R299. doi: 10.1016/s0960-9822(01)00168-3. [DOI] [PubMed] [Google Scholar]