Abstract
We have developed a novel multimodal microscopy system that incorporates confocal Raman, confocal reflectance, and quantitative phase microscopy (QPM) into a single imaging entity. Confocal Raman microscopy provides detailed chemical information from the sample, while confocal reflectance and quantitative phase microscopy show detailed morphology. Combining these intrinsic contrast imaging modalities makes it possible to obtain quantitative morphological and chemical information without exogenous staining. For validation and characterization, we have used this multi-modal system to investigate healthy and diseased blood samples. We first show that the thickness of a healthy red blood cell (RBC) shows good correlation with its hemoglobin distribution. Further, in malaria infected RBCs, we successfully image the distribution of hemozoin (malaria pigment) inside the cell. Our observations lead us to propose morphological screening by QPM and subsequent chemical imaging by Raman for investigating blood disorders. This new approach allows monitoring cell development and cell-drug interactions with minimal perturbation of the biological system of interest.
OCIS codes: (180.3170) Interference Microscopy, (180.5655) Raman microscopy, (170.1530) Cell analysis
1. Introduction
Imaging a live cell without staining is a major challenge. The most widely used non-staining technique for visualizing transparent living cells is the phase contrast method. Sub-cellular components induce changes in optical path length, and the resulting phase variation can be transformed into intensity variation. Our laboratory has developed several forms of quantitative phase microscopy (QPM) based on interferometry techniques. QPM has provided the quantitative morphological structure of a living cell in 2D [1,2]. An imaging detector, such as a CCD, is used to capture distorted interferograms from samples, and optical phase delays are calculated. Phase delays can be used to directly compute important physical cellular properties. For samples that are fairly homogeneous, it is reasonably inferred that phase delays provide direct information about cell shape. A healthy red blood cell (RBC), which can be considered to have internally uniform hemoglobin distributions, is an important example of this.
However, most biological samples have a complex intra-cellular structure. Even the relatively simple, homogeneous healthy RBC becomes quite heterogeneous when diseased (e.g. upon malaria infection). Two-dimensional projections of phase delays contain information about cell shape and about sub-cellular structure. It is important to decouple these two features when studying either different eukaryotic cells or the same cell when pathology simultaneously alters both cell shape and sub-cellular structure. One way to achieve this separation is to measure phase delays from different angles (tomographic phase microscopy) [3]. From such measurements, variations in refractive index (which is indicative of sub-cellular structure) can be determined. Another approach is to use high-resolution confocal reflectance microscopy to independently measure cell shape [4]. Although this approach does not provide as high quality morphological information as does tomographic measurement [3], it offers reasonably good decoupling in a much simpler and faster manner.
Other investigators have also developed methods based on digital holography, itself an extensive form of interferometric measurement. The QPM techniques from our laboratory can be considered as off-axis digital holography at the focal plane. In digital holography, object information can be retrieved by numerical reconstruction from the digitally captured hologram. In particular, digital holography [5] has been used for quantitative microscopy. In-line digital holography is implemented with phase-shifting methods [6] and applied to microscopic imaging [7]. High axial resolution [8] is achieved with multiple phase-shifted holograms. For faster imaging, researchers have also developed off-axis holography in which phase information is calculated from a single hologram [9]. Off-axis digital holographic microscopy has been successfully applied in cell studies [10,11].
Overall, QPM benefits from its inherent fast and full-field characteristics. It provides quantitative morphology from untreated cells at faster than video rate. However, its usefulness for addressing realistic biological situations has been limited due to its inherent lack of chemical specificity. A refractive index variation inside of a cell is caused by the heterogeneous distribution of chemicals such as nucleic acids, proteins, and lipids. Researchers have long-desired to look at chemical information to monitor cell development or cell-drug interaction in the cell’s natural state. Since phase measurements are indirect, phase contrast images come with a fundamental (and often confounding) question: “What causes the variations in the cell’s refractive index?”
Since its first observation in 1928 [12], Raman scattering has been widely used as an analytical tool in many fields. Small amounts of inelastically scattered light from a sample carry “fingerprint” vibrational information that can be used for both qualitative and quantitative analysis. Incorporating confocal microscopy [13], which is extensively used to obtain three-dimensional information, into conventional Raman spectroscopy provides an exciting opportunity for obtaining accurate chemical and morphological information with high spatial resolution [14]. Raman spectroscopy can distinguish macromolecules as well as large structural features such as organelles by their characteristic Raman spectra [15–17]. Several types of bacteria can be easily identified with Raman spectroscopy [18]. Despite its great promise, confocal Raman microscopy, in contrast to fluorescence imaging, has not been widely used for cellular imaging because the signals of Raman scattering are intrinsically weak. Recently, Xie and associates used coherence-based enhancement techniques to produce Raman signals strong enough for high-speed imaging [19–21]. These techniques have gained widespread acceptance and have been successfully applied in biological investigations. Furthermore, despite the weak signal of spontaneous Raman spectroscopy, it has been successfully used in clinical applications of a point measurement device. The success of this method has been due to its relatively simple instrumentation and ability to track multiple components. Also, even with a weak Raman signal, the spectroscopic information from a laboratory-based Raman microscope can be used to characterize tissues in disease states such as breast cancer [22] and arteriosclerosis [23].
The relatively simple structures of RBCs make them an almost ideal cell type for both QPM and Raman microscopy. Since their internal structure is relatively homogeneous, the shape of an RBC can be directly inferred from phase delay information. Perturbations to this homogenous internal structure can be measured and interpreted as changes in cell morphology [24–26]. For example, RBCs infected with the malaria parasite Plasmodium falciparum undergo significant changes in internal structure as the parasite develops. In previous studies, the various stages of parasite development were distinguished by the observed changes in the infected RBC’s shape and membrane fluctuations [27]. Sickle cell anemia has also been studied [28]. Fundamental RBC biology has been explored using QPM [29,30]. However, the lack of molecular specificity inherent in QPM prevents unambiguous and detailed quantitative analyses when substantial changes in chemical composition and morphology occur simultaneously.
It is precisely the case that both kinds of change occur as P. falciparum develops within the RBC. Large quantities of initially uniformly distributed hemoglobin are taken up by the parasite and digested. The released heme is then polymerized to hemozoin, which is concentrated within a sub-cellular parasite compartment. The chemical environment of the heme present in hemoglobin is different from that in hemozoin, and the two environments can be detected, distinguished, and quantitatively characterized by Raman spectroscopy [31,32,35]. Raman microscopy has been previously used for such purposes in studies of parasite-infected RBCs. However, wide field imaging (imaging multiple RBCs) is still challenging due to the intrinsically weak Raman signal. Multimodal approaches to address this problem have been attempted, but quantitative morphological data could not be obtained [33]. The lack of such data limits the development of automated screening to study the time-resolved impact of different anti-malarial drugs on hemozoin formation in developing parasites.
In the work reported in this paper, we combined confocal Raman and quantitative phase microscopy. Both imaging modalities use intrinsic contrast mechanisms. The combined system provides a powerful tool for obtaining both morphological and chemical information from unlabeled samples. Several interesting biological studies are in progress using this unique capability of the system. For example, we are using this system to investigate different blood disorders. In such a study, QPM is first used as a screening tool to select abnormal RBCs from a wide field of view. Next, we use Raman to map the selected cells and gather detailed chemical information. In the present work, we obtained multimodal images from healthy blood smears in which the thickness of each RBC shows good correlation with the measured hemoglobin distribution. Furthermore, the hemoglobin and hemozoin distributions are clearly distinguished in the parasite-infected RBCs.
2. Methods
2.1. Instrumentation
The setup (Fig. 1 ) is based on a home-built inverted microscope. Variable intensity-controlled white LEDs (not shown) are used as an illumination source for bright field imaging. The CMOS camera (BCN-B050-U, MightTex) is used to capture the image. Because the cells are transparent, oblique illumination (diffracted-light contrast enhancement, DLC) is used to enhance the cellular features [34]. A frequency-doubled Nd:YAG laser (Millennia 5sJ, Spectra-Physics) at 532 nm is the pump source for a CW tunable Ti:sapphire Laser (3900S, Spectra-Physics). The 785 nm output is used as the excitation source for both the confocal reflectance and confocal Raman systems. For high resolution imaging, a water immersion objective lens (Olympus UPLSAPO60XWIR 60X/1.20) is used. A flip mirror, placed after the tube lens, allows the image focused at the sample plane from the incoherent transmission source to be observed at the video camera with ~67 × magnification. The collimated beam is band pass filtered (BPF, LL01-785-12.5, Semrock) and redirected to dual-axis galvanometer mirrors (CT-6210, Cambridge Technology) by a dichroic mirror (DM1, LPD01-785RU-25, Semrock). After the galvanometer mirrors, the beam size is adjusted by a telescope and focused at the sample plane by an objective lens. XY positioning is achieved by a micrometer-controlled stage (406, New Focus). Coarse and fine adjustment of sample focus is done using a piezo actuator combined with a differential micrometer (DRV517, Thorlabs).
Fig. 1.
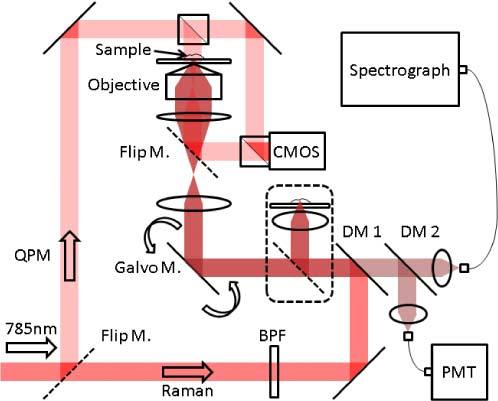
Home-built multimodal microscopy system. The system incorporates confocal Raman, quantitative phase contrast, confocal reflectance and bright field microscopes. By combining four intrinsic imaging modalities, quantitative morphological and chemical information can be acquired from unstained live cells. The high-throughput stage (in the dashed line) is for characterizing liquid or powder samples that do not require spatial resolution.
At the sample plane, refractive index gradients in the sample give rise to the confocal reflectance signal, and the molecular bonds in the sample give rise to the inelastically scattered spontaneous Raman signal. Both signals retrace the excitation beam path until the first dichroic mirror (DM1). Raman scattered light passes through the first dichroic mirror while the major portion of the elastically scattered Rayleigh beam is rejected. The remaining Rayleigh light after the first dichroic mirror is filtered again by the second dichroic mirror (DM2, LPD01-785RU-25, Semrock), and is subsequently measured as the confocal reflectance signal. Both confocal reflectance and confocal Raman signals are collected by standard multimode fibers which are replaceable depending on the resolution and signal strength from the sample. It is worth mentioning that these two imaging modalities provide sample information from exactly co-registered sample points, which cannot be achieved from bright field imaging. In order to analyze liquid or powder samples that do not require spatially-resolved imaging, an additional high-throughput arm is included with minimal optical elements for single point measurements.
The confocal Raman signal is delivered to an imaging spectrograph (HoloSpec f/1.8i, Kaiser Optical Systems) and spectra are captured by a liquid nitrogen cooled CCD (LN/CCD-1340/400-EB, Roper Scientific). The confocal reflectance signal is delivered to a photomultiplier tube (PMT, H9656-20, Hamamatsu) and amplified by a PMT controller (C7169, Hamamatsu). Labview 8.2 software (National Instruments) and a data acquisition board (PCI-6251, National Instruments) are used to control the devices.
As a simple form of QPM instrumentation, Hilbert phase microscopy [1] is included in the setup. A Mach-Zehnder interferometer is integrated into the scanning microscope. The laser beam is divided into object and reference beams and superimposed in front of the camera, which is also used for bright field imaging. Pixel to pixel distance of the camera is 2.2 µm.
2.2. Analysis
The off-axis interferogram from the Mach-Zehnder interferometer is processed using the standard Hilbert transform [1]. Briefly, the interferogram is Fourier transformed and the first order signal is selected in Fourier space. The phase from the inverse transformed field gives the optical phase delays in the sample plane. To extract phase delay for the sample only, we separately measure the reference phase information without a sample in the field of view and subtract it from the phase delays for the sample. The phase resolution measured from the empty area without sample was 0.067 radians (~λ/100). The RBC has a relatively uniform sub-cellular structure. Assuming that the RBC is predominantly composed of hemoglobin, optical phase delays can be directly converted into RBC thickness.
Dominant spectra are calculated by principal component analysis and Raman images are reconstructed from their spatial distribution. For healthy RBCs, the hemoglobin Raman signal mainly dominates the spectra. Unlike healthy RBCs, malaria-infected RBCs contain additional major components such as malaria pigment (the hemozoin crystal), which can be used to indicate the progression of the disease. Composed of polymerized heme molecules, hemozoin has distinct iron-carboxylate bonds yielding a Raman signature different from that of hemoglobin. From the Raman images, hemoglobin and hemozoin regions can be clearly distinguished.
2.3. Sample preparation
A drop of blood was collected from a healthy volunteer and used for the blood smear. After being dried for 30 minutes in the air, the sample was installed on the microscope sample stage.
P. falciparum strain 3D7 was cultured in leukocyte-free human RBCs (Research Blood Components, Brighton, MA) under an atmosphere of 5% O2, 5% CO2 and 90% N2, at 2% hematocrit in RPMI culture medium 1640 (Gibco Life Technologies) supplemented with 25 mM HEPES (Sigma), 200 mM hypoxanthine (Sigma), 0.20% NaHCO3 (Sigma) and 0.25% Albumax II (Gibco Life Technologies). Parasites were synchronized by treatment with 5% sorbitol at least 12 hours before sample collection. For microscopy, RBCs infected with late-stage parasites were pelleted and resuspended at ~2% hematocrit in Alsever's acid citrate-dextrose buffer (114 mM glucose, 27 mM sodium citrate, 72 mM NaCl, pH adjusted to 6.1 with citric acid).
3. Results and Discussions
We have used a system combining QPM and confocal Raman microscopy to investigate both healthy and P. falciparum-infected RBCs. Bright field images (Fig. 2(a) ) were used to find a proper sample position. For QPM measurement, an off-axis interferogram is captured from the same field of view. The Hilbert transformation was used to calculate phase delays caused by the sample. Assuming hemoglobin is uniformly distributed inside of RBC, this phase image can be converted into the thickness of the RBC (Fig. 2(c)). The confocal reflectance, which is reconstructed from rejected Rayleigh light, has exactly the same field of view as the confocal Raman image. Since Raman mapping is a time-consuming process, confocal reflectance (Fig. 2(b)) provides the guidance for confocal Raman image. The sample is illuminated with 3.5mW of laser power and the Raman signal is integrated for 3 seconds from each pixel. It took about 45 minutes to acquire the 30 × 30-pixel Raman images in Fig. 2(d).
Fig. 2.
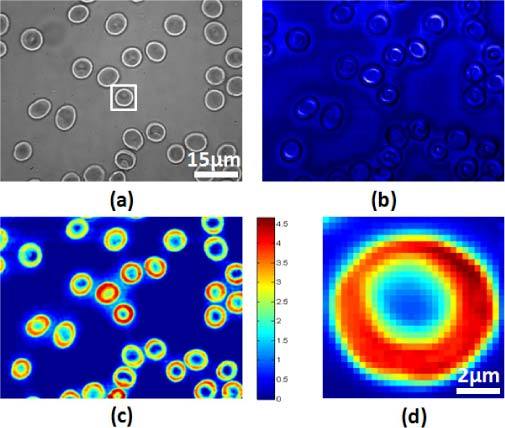
(a) Bright field, (b) confocal reflectance, and (c) quantitative phase contrast images of a healthy blood smear. The color bar indicates phase variation in radians. (d) Hemoglobin distribution from Raman mapping for the boxed RBC in (a).
Previously [27], we have shown that QPM can distinguish healthy RBCs from the abnormally-shaped P. falciparum-infected RBCs. Although it does not have the desirable molecular specificity, QPM has features that make it an attractive screening tool. First, three-dimensional cellular structure in a full field of view can be acquired very rapidly. Depending on the instrument’s configuration (e.g. objective lens magnification and imaging device acquisition speeds), more than a million cells can be screened per second. For comparison, the most commonly used current diagnosis technique is manual inspection of a Giemsa-stained blood smear by light microscopy, and this requires a trained technician who can only score fewer than 100 slides per hour. As the fraction of P. falciparum-infected RBCs may be quite low (≤ few percent), rapid screening to identify infected cells for detailed chemical characterization is critical for maintaining overall high efficiency of the imaging process. We anticipate that with quantitative morphological information, the screening process can be automated in the future.
Abnormal RBC morphology is associated with, but not specific to, P. falciparum infection. Acanthocytes, which are abnormally shaped RBCs, can also be observed in other diseases. For example, liver disease and uremia generate spiky RBCs which can be mis-classified as P. falciparum-infected RBCs. QPM in screening mode identifies suspicious RBCs for Raman mapping, which provides the detailed chemical information needed to distinguish different RBC pathologies. Figure 2(d) shows the distribution of hemoglobin in the selected RBC in Fig. 2(a). From the principal component analysis, the dominant spectrum corresponds to hemoglobin and Fig. 2(d) shows its distribution.
To illustrate that our system is capable of providing quantitative data on P. falciparum-infected RBCs, trophozoite-stage RBCs were analyzed. At this developmental stage, RBC hemoglobin concentrations have declined and parasite hemozoin content has increased. By measuring the hemozoin accumulation, therefore, parasite development can be directly monitored. Direct monitoring is especially relevant because some potent anti-malarial drugs are known to bind heme and disrupt hemozoin formation in vitro. This disruption is proposed to increase the labile heme concentrations to levels that are toxic to the parasite [36]. Being able to directly image changes in both hemoglobin and hemozoin content at the same time provides an important opportunity for connecting the in vitro and in vivo biochemistry of hemozoin formation, particularly during anti-malarial drug perturbation.
With the wide field of view, we located suspicious RBCs and QPM was used to confirm the abnormal shape and refractive index from the infected cells. Figure 3(a) shows the suspect RBC in bright field image. QPM image in Fig. 3(b) shows morphological feature on the RBC surface which is a potential indicator for malaria infection. Raman mapping was performed on a selected RBC. It took about 30 minutes to acquire the 25 × 25-pixel Raman images in Fig. 3. As for the case of healthy RBCs in Fig. 2, we applied principal component analysis. Figure 3(c) shows the distribution of the first principal component with the corresponding spectrum shown in Fig. 3(d). The spectrum in Fig. 3(d) closely resembles the hemoglobin spectrum. Figure 3(e) shows the distribution of the second principal component with the corresponding spectrum shown in Fig. 3(f). The characteristic hemozoin peak at 1374cm−1 (ν4 band of porphyrin enhanced by hemozoin formation) is clearly shown and confirms that hemozoin is localized in the region indicated by the arrow. Furthermore, hemozoin localization has good correlation with the small morphological feature in the QPM image in Fig. 3(b). Compared to the healthy RBC (Fig. 2), which has only one dominating chemical component (hemoglobin), the malaria-infected RBC has two major components (hemoglobin and hemozoin).
Fig. 3.
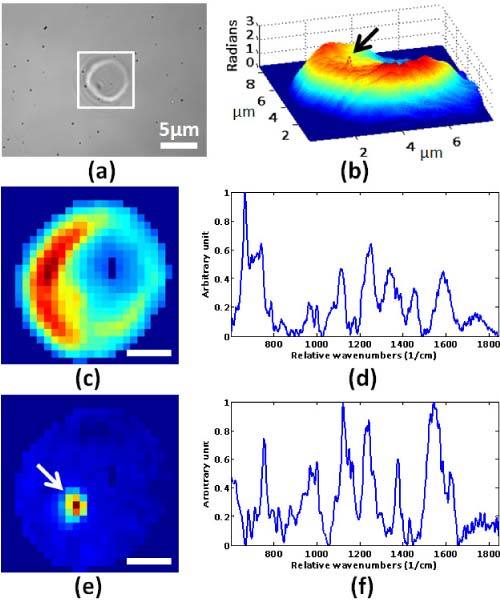
(a) Bright field image for blood smear. (b) QPM image for the boxed RBC in (a). (c) Raman image reconstructed from the first principal component. Scale bar represents 2µm. (d) Spectrum of the first principal component. (e) Raman image reconstructed from the second principal component. Scale bar represents 2µm. (f) Spectrum of the second principal component. The characteristic hemozoin peak at 1374 cm−1 is clearly shown.
4. Conclusions
We have developed a novel multimodal microscopy system. Combining quantitative phase microscopy (QPM) with confocal Raman microscopy provides label-free, quantitative morphological and chemical information from unfixed cells. The combination offers an excellent system for monitoring cell development and cell-drug interactions.
To validate this new system, we analyzed both normal and P. falciparum-infected RBCs. The thickness of an RBC determined by QPM correlates well with its hemoglobin distribution obtained by Raman imaging. Furthermore, we observed that the distribution of hemozoin distribution in a malaria-infected RBC in the trophozoite stage is localized inside the host RBC. In the near future we will use this system to investigate P. falciparum development in RBCs to better understand how different anti-malarial drugs perturb this process and impact the process of the degradation of hemoglobin and consequent accumulation of hemozoin.
Although we used RBCs as samples for system validation, the usefulness of the new multimodal system is not limited to the study of blood disorders. It can be used for a large variety of cellular studies, and many interesting projects are currently under way with this new system.
Acknowledgments
This work was performed at the MIT Laser Biomedical Research Center and was supported by the NIH National Center for Research Resources, grant P41-RR02594. The authors would like to thank Prof. Charles H. Holbrow for proofreading the manuscript. I. B. acknowledges the support of the Lester Wolfe Fellowship from the Laser Biomedical Research Center. S. J. G. is supported by the NIEHS Predoctoral Training Grant [grant 5-T32-ES007020].
References and links
- 1.Ikeda T., Popescu G., Dasari R. R., Feld M. S., “Hilbert phase microscopy for investigating fast dynamics in transparent systems,” Opt. Lett. 30(10), 1165–1167 (2005). 10.1364/OL.30.001165 [DOI] [PubMed] [Google Scholar]
- 2.Popescu G., Ikeda T., Dasari R. R., Feld M. S., “Diffraction phase microscopy for quantifying cell structure and dynamics,” Opt. Lett. 31(6), 775–777 (2006). 10.1364/OL.31.000775 [DOI] [PubMed] [Google Scholar]
- 3.Choi W., Fang-Yen C., Badizadegan K., Oh S., Lue N., Dasari R. R., Feld M. S., “Tomographic phase microscopy,” Nat. Methods 4(9), 717–719 (2007). 10.1038/nmeth1078 [DOI] [PubMed] [Google Scholar]
- 4.Lue N., Choi W., Popescu G., Yaqoob Z., Badizadegan K., Dasari R. R., Feld M. S., “Live cell refractometry using Hilbert phase microscopy and confocal reflectance microscopy,” J. Phys. Chem. A 113(47), 13327–13330 (2009). 10.1021/jp904746r [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Schnars U., Jüptner W., “Direct recording of holograms by a CCD target and numerical reconstruction,” Appl. Opt. 33(2), 179–181 (1994). 10.1364/AO.33.000179 [DOI] [PubMed] [Google Scholar]
- 6.Yamaguchi I., Zhang T., “Phase-shifting digital holography,” Opt. Lett. 22(16), 1268–1270 (1997). 10.1364/OL.22.001268 [DOI] [PubMed] [Google Scholar]
- 7.Zhang T., Yamaguchi I., “Three-dimensional microscopy with phase-shifting digital holography,” Opt. Lett. 23(15), 1221–1223 (1998). 10.1364/OL.23.001221 [DOI] [PubMed] [Google Scholar]
- 8.Kang J. W., Hong C. K., “Phase-contrast microscopy by in-line phase-shifting digital holography: shape measurement of a titanium pattern with nanometer axial resolution,” Opt. Eng. 46(4), 040506–040508 (2007). 10.1117/1.2731317 [DOI] [Google Scholar]
- 9.Cuche E., Bevilacqua F., Depeursinge C., “Digital holography for quantitative phase-contrast imaging,” Opt. Lett. 24(5), 291–293 (1999). 10.1364/OL.24.000291 [DOI] [PubMed] [Google Scholar]
- 10.Marquet P., Rappaz B., Magistretti P. J., Cuche E., Emery Y., Colomb T., Depeursinge C., “Digital holographic microscopy: a noninvasive contrast imaging technique allowing quantitative visualization of living cells with subwavelength axial accuracy,” Opt. Lett. 30(5), 468–470 (2005). 10.1364/OL.30.000468 [DOI] [PubMed] [Google Scholar]
- 11.Mann C., Yu L., Lo C. M., Kim M., “High-resolution quantitative phase-contrast microscopy by digital holography,” Opt. Express 13(22), 8693–8698 (2005). 10.1364/OPEX.13.008693 [DOI] [PubMed] [Google Scholar]
- 12.Raman C. V., Krishnan K. S., “A new type of secondary radiation,” Nature 121(3048), 501–502 (1928). 10.1038/121501c0 [DOI] [Google Scholar]
- 13.T. Wilson, Confocal Microscopy (Academic, London; 1990). [Google Scholar]
- 14.Puppels G. J., de Mul F. F. M., Otto C., Greve J., Robert-Nicoud M., Arndt-Jovin D. J., Jovin T. M., “Studying single living cells and chromosomes by confocal Raman microspectroscopy,” Nature 347(6290), 301–303 (1990). 10.1038/347301a0 [DOI] [PubMed] [Google Scholar]
- 15.Uzunbajakava N., Lenferink A., Kraan Y., Volokhina E., Vrensen G., Greve J., Otto C., “Nonresonant confocal Raman imaging of DNA and protein distribution in apoptotic cells,” Biophys. J. 84(6), 3968–3981 (2003). 10.1016/S0006-3495(03)75124-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Krafft C., Knetschke T., Siegner A., Funk R. H. W., Salzer R., “Mapping of single cells by near infrared Raman microspectroscopy,” Vib. Spectrosc. 32(1), 75–83 (2003). 10.1016/S0924-2031(03)00049-3 [DOI] [Google Scholar]
- 17.Matthäus C., Chernenko T., Newmark J. A., Warner C. M., Diem M., “Label-free detection of mitochondrial distribution in cells by nonresonant Raman microspectroscopy,” Biophys. J. 93(2), 668–673 (2007). 10.1529/biophysj.106.102061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rösch P., Harz M., Schmitt M., Peschke K. D., Ronneberger O., Burkhardt H., Motzkus H. W., Lankers M., Hofer S., Thiele H., Popp J., “Chemotaxonomic identification of single bacteria by micro-Raman spectroscopy: application to clean-room-relevant biological contaminations,” Appl. Environ. Microbiol. 71(3), 1626–1637 (2005). 10.1128/AEM.71.3.1626-1637.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zumbusch A., Holtom G. R., Xie X. S., “Three-dimensional vibrational imaging by coherent anti-Stokes Raman scattering,” Phys. Rev. Lett. 82(20), 4142–4145 (1999). 10.1103/PhysRevLett.82.4142 [DOI] [Google Scholar]
- 20.Cheng J. X., Jia Y. K., Zheng G., Xie X. S., “Laser-scanning coherent anti-Stokes Raman scattering microscopy and applications to cell biology,” Biophys. J. 83(1), 502–509 (2002). 10.1016/S0006-3495(02)75186-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Freudiger C. W., Min W., Saar B. G., Lu S., Holtom G. R., He C., Tsai J. C., Kang J. X., Xie X. S., “Label-free biomedical imaging with high sensitivity by stimulated Raman scattering microscopy,” Science 322(5909), 1857–1861 (2008). 10.1126/science.1165758 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Haka A. S., Shafer-Peltier K. E., Fitzmaurice M., Crowe J., Dasari R. R., Feld M. S., “Diagnosing breast cancer by using Raman spectroscopy,” Proc. Natl. Acad. Sci. U.S.A. 102(35), 12371–12376 (2005). 10.1073/pnas.0501390102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Motz J. T., Fitzmaurice M., Miller A., Gandhi S. J., Haka A. S., Galindo L. H., Dasari R. R., Kramer J. R., Feld M. S., “In vivo Raman spectral pathology of human atherosclerosis and vulnerable plaque,” J. Biomed. Opt. 11(2), 021003 (2006). 10.1117/1.2190967 [DOI] [PubMed] [Google Scholar]
- 24.Rappaz B., Barbul A., Emery Y., Korenstein R., Depeursinge C., Magistretti P. J., Marquet P., “Comparative study of human erythrocytes by digital holographic microscopy, confocal microscopy, and impedance volume analyzer,” Cytometry A 73A(10), 895–903 (2008). 10.1002/cyto.a.20605 [DOI] [PubMed] [Google Scholar]
- 25.Brazhe A. R., Brazhe N. A., Maksimov G. V., Ignatyev P. S., Rubin A. B., Mosekilde E., Sosnovtseva O. V., “Phase-modulation laser interference microscopy: an advance in cell imaging and dynamics study,” J. Biomed. Opt. 13(3), 034004 (2008). 10.1117/1.2937213 [DOI] [PubMed] [Google Scholar]
- 26.Yusipovich A. I., Parshina E. Y., Brysgalova N. Y., Brazhe A. R., Brazhe N. A., Lomakin A. G., Levin G. G., Maksimov G. V., “Laser interference microscopy in erythrocyte study,” J. Appl. Phys. 105(10), 102037 (2009). 10.1063/1.3116609 [DOI] [Google Scholar]
- 27.Park Y. K., Diez-Silva M., Popescu G., Lykotrafitis G., Choi W., Feld M. S., Suresh S., “Refractive index maps and membrane dynamics of human red blood cells parasitized by Plasmodium falciparum,” Proc. Natl. Acad. Sci. U.S.A. 105(37), 13730–13735 (2008). 10.1073/pnas.0806100105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shaked N. T., Satterwhite L. L., Telen M. J., Truskey G. A., Wax A., “Quantitative microscopy and nanoscopy of sickle red blood cells performed by wide field digital interferometry,” J. Biomed. Opt. 16(3), 030506 (2011). 10.1117/1.3556717 [DOI] [PubMed] [Google Scholar]
- 29.Park Y. K., Best C. A., Badizadegan K., Dasari R. R., Feld M. S., Kuriabova T., Henle M. L., Levine A. J., Popescu G., “Measurement of red blood cell mechanics during morphological changes,” Proc. Natl. Acad. Sci. U.S.A. 107(15), 6731–6736 (2010). 10.1073/pnas.0909533107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Park Y. K., Best C. A., Auth T., Gov N. S., Safran S. A., Popescu G., Suresh S., Feld M. S., “Metabolic remodeling of the human red blood cell membrane,” Proc. Natl. Acad. Sci. U.S.A. 107(4), 1289–1294 (2010). 10.1073/pnas.0910785107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wood B. R., Langford S. J., Cooke B. M., Glenister F. K., Lim J., McNaughton D., “Raman imaging of hemozoin within the food vacuole of Plasmodium falciparum trophozoites,” FEBS Lett. 554(3), 247–252 (2003). 10.1016/S0014-5793(03)00975-X [DOI] [PubMed] [Google Scholar]
- 32.Frosch T., Koncarevic S., Zedler L., Schmitt M., Schenzel K., Becker K., Popp J., “In situ localization and structural analysis of the malaria pigment hemozoin,” J. Phys. Chem. B 111(37), 11047–11056 (2007). 10.1021/jp071788b [DOI] [PubMed] [Google Scholar]
- 33.Wood B. R., Hermelink A., Lasch P., Bambery K. R., Webster G. T., Khiavi M. A., Cooke B. M., Deed S., Naumann D., McNaughton D., “Resonance Raman microscopy in combination with partial dark-field microscopy lights up a new path in malaria diagnostics,” Analyst (Lond.) 134(6), 1119–1125 (2009). 10.1039/b822603b [DOI] [PubMed] [Google Scholar]
- 34.Piekos W. B., “Diffracted-light contrast enhancement: a re-examination of oblique illumination,” Microsc. Res. Tech. 46(4-5), 334–337 (1999). [DOI] [PubMed] [Google Scholar]
- 35.Pagola S., Stephens P. W., Bohle D. S., Kosar A. D., Madsen S. K., “The structure of malaria pigment β-haematin,” Nature 404(6775), 307–310 (2000). 10.1038/35005132 [DOI] [PubMed] [Google Scholar]
- 36.Slater A. F. G., Cerami A., “Inhibition by chloroquine of a novel haem polymerase enzyme activity in malaria trophozoites,” Nature 355(6356), 167–169 (1992). 10.1038/355167a0 [DOI] [PubMed] [Google Scholar]


