Abstract
Optical coherence tomography (OCT) allows imaging dynamic structures and fluid flow within scattering tissue, such as the beating heart and blood flow in murine embryos. For any given system, the frame rate, spatial resolution, field-of-view (FOV), and signal-to-noise ratio (SNR) are interconnected: favoring one aspect limits at least one of the others due to optical, instrumentation, and software constraints. Here we describe a spatio-temporal mosaicing technique to reconstruct high-speed, high spatial-resolution, and large-field-of-view OCT sequences. The technique is applicable to imaging any cyclically moving structure and operates on multiple, spatially overlapping tiled image sequences (each sequence acquired sequentially at a given spatial location) and effectively decouples the (rigid) spatial alignment and (non-rigid) temporal registration problems. Using this approach we reconstructed full-frame OCT sequences of the beating embryonic rat heart (11.5 days post coitus) and compared it to direct imaging on the same system, demonstrating a six-fold improvement of the frame rate without compromising spatial resolution, FOV, or SNR.
OCIS codes: (110.4500) Optical coherence tomography, (180.1655) Coherence tomography, (110.4155) Multiframe image processing, (170.4500) Optical coherence tomography, (100.0100) Image processing
1. Introduction
Optical coherence tomography (OCT) is increasingly being adopted as an accurate imaging method to study embryonic cardiovascular anatomy and dynamics. It allows for non-destructive imaging through several millimeters of biological tissue with single cell resolution. OCT has been used for a wide array of biomedical applications, including structural imaging of whole mammalian and avian embryos [1,2], flow measurements [3,4], and 4D imaging of mammalian and avian embryonic hearts [5–7]. High frame rate, signal-to-noise ratio (SNR), and spatial resolution as well as the ability to image over a large field of view are all equally important factors for accurate visualization and analysis.
Improvements in light sources, scanner efficiency, data transmission protocols, and storage capacity have collectively produced improvements in the acquisition frame rate for OCT [8, 9]. The time over which the signal is gathered to produce one image limits the spatial resolution when imaging dynamic samples since sample motion introduces blurring if the image capture time is too long [10]. Increasing the frame rate can be achieved through faster scanning but decreasing the dwell time also reduces the SNR, as fewer photons contribute to the signal. For cardiovascular systems, which exhibit velocities of the order of millimeters per second, framerates of 100 frames per second or more are required to achieve a spatial resolution of 10 μm and below. Thus, there is a need for advanced methods that improve the frame rate while preserving the spatial resolution.
For optical and electron microscopy of static or nearly static samples, tiled imaging, followed by mosaic reconstruction, has become a central tool to extend the limited field of view of high-numerical aperture microscope objectives. Several methods have been proposed to automatically or semi-automatically spatially register (align) individual microscopy tiles to form an image mosaic [11–14] extending methods initially developed for panoramic photography [15, 16]. These technique, however, produce static images and are therefore not directly applicable in our case.
Recently, we demonstrated that dynamic 3D images of the embryonic zebrafish heart can be reconstructed by post-acquisition synchronization of nongated slice sequences acquired on a fast confocal microscope [17, 18]. Building upon our ability to accurately register image sequences in time, we here present a combined (rigid) spatial and (non-rigid) temporal registration method that allows us to extend the scope of static mosaicing methods to improve the field of view and resolution of dynamic 2D OCT by combining tiled image sequences of cyclically moving structures in the mammalian embryonic heart.
2. Method
Our method operates as follows. We assume the object to be imaged has a local scattering intensity that is periodic, that is
| (1) |
where T is the period of the heart beat (assuming heart imaging). We acquire N image sequences (see Fig. 1(a) for an example with N = 2) that are spatially tiled such that
| (2) |
for 0 ≤ x < Lx, 0 ≤ y < Ly, and 0 ≤ t < Lt, where Lx and Ly are the width and height of the tiles, respectively, Lt is the duration of each sequence and where xn is the spatial offset in the x-direction of the nth tile, and tn is the time at which recording of the nth sequence started. The duration of the sequence, Lt, must be at least two periods long, so that one full heart beat period, starting at any arbitrary time in the cardiac cycle, is included in the sequence.
Fig. 1.
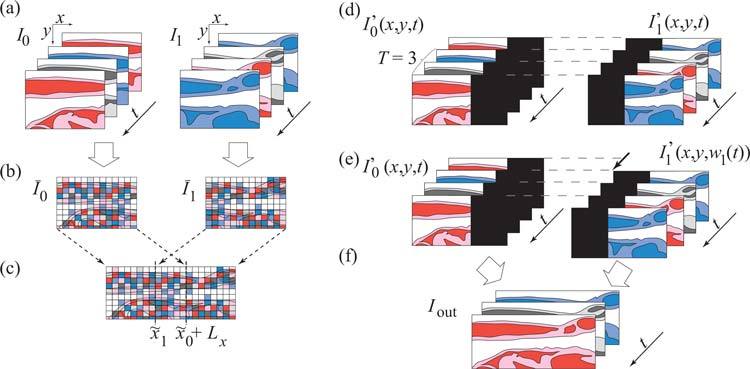
Tiled acquisition and automatic mosaicing procedure for dynamic images. (a) Overlapping cardiac image series, are acquired sequentially. (b) Each sequence is collapsed to a static image (temporal maximum). (c) Static images are spatially aligned. (d) Image sequences are spatially transformed and padded with zeros. (e) Spatially aligned sequences are temporally synchronized. (f) Synchronized image sequences are blended. (Color online).
We assume that the spatial and temporal offsets, xn and tn, respectively, are unknown. Our task is to estimate them, which is an image registration (and synchronization) problem (we assume, here, that tiles are aligned in the y direction, but this is not a fundamental requirement of our approach, which could be generalized to include registration in both the x and y directions). The periodicity assumption allows us to decouple the spatial and temporal registration problems as follows. We first collapse each sequence along the temporal direction (Fig. 1(b)) to obtain a spatial signature of the tile that is independent of time
| (3) |
We next pair-wise align the N static tiles Īn, that is, we estimate the tiling-offsets xn as follows
| (4) |
with x0 = 0 (Fig. 1(c)). In words, we find the horizontal position for which the mean-squared difference between the previous and current tiles’ grayscale values (calculated over the region of overlap of the two tiles) is minimal. In practice, we use a rigid-body, pyramid-based, registration implementation proposed by Thévenaz et al. [19], which we constrain to register tiles along the horizontal direction.
We next define zero-padded image sequences
| (5) |
in which the original sequences have been placed at their estimated spatial offset (see Fig. 1(d)). These sequences are spatially, but not temporally aligned. Proceeding recursively for n = 1,...,N – 1, and choosing the sequence I′0 as a reference, we align pairs of sequences I′n and I′n–1 via a non-uniform time-warping procedure in which we minimize the following cost functions (1 ≤ n < N):
| (6) |
where wn−1 (with w0(t) = t, the identity function) is the warping function applied to the sequence I′n−1, and w is a candidate cost function to warp In so that it matches one period of the warped version of I′n−1. The first integral compares the two neighboring, temporally warped sequences (sequences whose time-axis has been deformed), while the second integral keeps the extent of this deformation in check by penalizing candidate warping functions w that stretch or compress the time axis. These two contributions to the cost function are balanced by the parameter 0 < λ ≤ 1 to favor either good matching of the warped and reference sequences (λ = 1) or the temporal integrity of the warped sequence (λ → 0). We constrain the search for warping functions wn to the set ℳ = {w ∈ C1([0, T))|0 ≤ w(t) < Lt and w(t1) < w(t2), t1 < t2} of continuous, non-negative, and strictly increasing functions bounded by Lt and defined over the interval [0, T). To determine the warping function wn that minimizes the cost function, we use a previously described dynamic programming algorithm [20]. The optimal warping function wn produced by this algorithm is such that wn ∈ {w̄n ∈ ℳ |Qn{w̄n} = minw∈ℳ Qn{w}}. Applying this warping function to the sequence I′n(x, y,t) results in sequence I′n(x, y, wn(t)), which is synchronized to all previously warped tiles, I′i(x, y, wi(t)), i = 0,..., n – 1 (see Fig. 1(e)).
Once all warping functions are determined, we blend the time-warped sequences according to
| (7) |
where, Ba,b(x) is a function defined for a < b such that
| (8) |
and
| (9) |
The final output of our algorithm (see Fig. 1(f)) is a blended, synchronized image series that covers one full period
| (10) |
3. Experiments and Results
To verify our method in practice, we acquired image sequences of a live rat embryo (11.5 days post coitus) using a swept-source OCT (SS-OCT) system as described previously in [3]. Figure 2 shows a representative frame of an SS-OCT image series of the embryonic heart and yolk sac acquired at different frame rates and spatial resolutions. Increasing the frame rate limits the spatial resolution over the region of interest (Fig. 2(a); 512 x 64 pixels, FOV=2.8 mm × 1 mm, 150 frames per second, SNR=15.76dB ). Improving the spatial resolution decreases the frame rate over the same region (Fig. 2(b); 512 × 500 pixels, FOV=2.8 mm × 0.97 mm, 25 frames per second, SNR=15.93dB ). Figure 2(c) shows the series of consecutive image sequences with smaller FOV (each 512 × 64 pixels, FOV=2.8 mm × 0.125 mm, 150 frames per second, 1000 frames (6.7 s, about 13 periods), SNR=15.76dB ) stitched together. The overlaps of the series are marked at the bottom. Figure 2(d) and Media 1 (5.7MB, MOV) show the result of the synchronization of the same series of sequences. Importantly, the latter reconstructed sequence has the same spatial resolution as the series acquired at low frame rate, Fig. 2(b) (512 × 500 pixels, FOV=2.8 mm × 0.97 mm), but with the same temporal resolution as the low spatial resolution sequence in Fig. 2(a) (150 frames per second) and a better signal to noise ratio (SNR=16.30dB ), thereby demonstrating that we can jointly achieve high spatial and temporal resolutions while preserving the field of view. The increase in SNR is due to the fact that in the areas of overlap the resulting signal is a weighted average of two series.
Fig. 2.
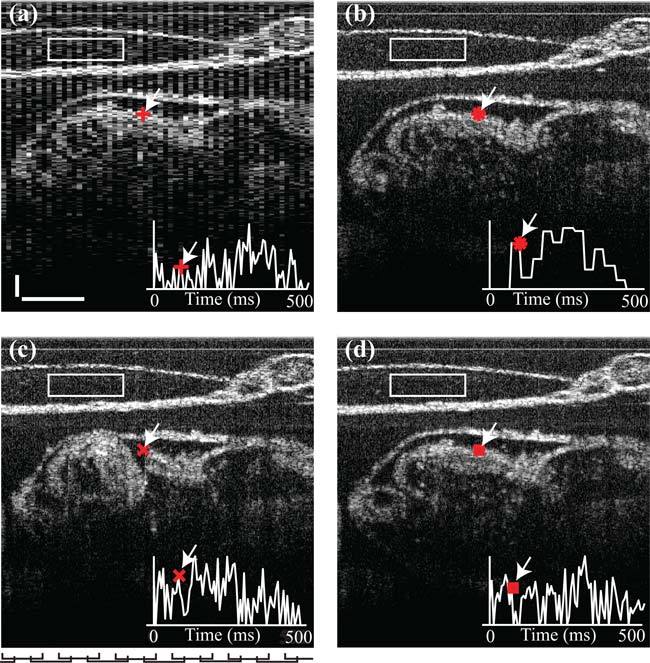
Dynamic mosaicing overcomes frame rate, resolution, and FOV interdependence for fast SS-OCT cardiac imaging. (a) High frame rate, low spatial resolution, large FOV: 150 fps, 512×64 pixels (b) Low frame rate, high spatial resolution, large FOV, 25fps, 512×500 pixels. (c) High frame rate, high spatial resolution, small FOV (12 tiles, before dynamic mosaicing, each 150 fps, 512×64 pixels). Marks at bottom of panel indicate lateral position of individual tiles. (d) High frame rate, high spatial resolution, high FOV (after dynamic mosaicing), 150 fps, 512×500 pixels. Inserts represent time-course (500 ms total duration) of pixel intensity (arbitrary units) at location marked by arrows. SNRs were computed over rectangular boxes. Scale bars are 0.2 mm. ( Media 1 (5.7MB, MOV) ).
To evaluate the accuracy of our method, we generated test datasets by extracting subsequences from each of N = 11 measured tile image sequence (512 × 64 pixels, FOV=2.8 mm × 0.125 mm, 150 frames per second, 1000 frames; see above and Fig. 2(c)). Specifically, we first split each measured sequence into two overlapping, left and right tile sequences, both of size 512 × 42 pixels and same duration as the original sequence (Figs. 3(b) and 3(c)). We further extracted frames covering two nonoverlapping time intervals (one interval at the beginning and one toward the end of the sequence, both spanning at least two heartbeat periods) from both the original sequence and the left and right sequences. Each measured image sequence thereby resulted in six subsequences labeled A through F (Figs. 3(a)–3(c)).
Fig. 3.

Accuracy evaluation of tiled acquisition and automatic mosaicing procedure. (a) Original sequence (one of the tiles in Fig. 2(c)) is split into (b) a left tile sequence and (c) a right tile sequence. (d)–(g) Sequence subsets are registered in space and/or time. (d) Temporal alignment of first period in subsequence A to subsequence B. (e) Spatio-temporal alignment of first period in subsequence C to subsequence F. (f) Spatio-temporal alignmnent of first period in subsequence E to subsequence D. (g) Spatio-temporal alignment of first period in subsequence C to subsequence E.
Given the six subsequences generated from any one of the 11 measured sequences, we registered the first cardiac cycle in subsequence A to subsequence B (an operation we denote by A→B, see also Fig. 3(d)), and similarly, the first cardiac cycle in subsequences C, E, and C to subsequences F, D, and E, respectively (i.e. C→F, E→D, and C→E, see Figs. 3(e), 3(f), and 3(g), respectively).
We first verified that the registration algorithm produced similar reconstructions when merging left or right tiles from an early cardiac cycle (subsequence C or F, respectively) to right or left tiles, respectively, extracted from later cardiac cycles (subsequence F or D, respectively) or when synchronizing non-tiled sequences directly (subsequences A and B). The temporal registration results for C→F and E→D were therefore compared against each other and against those obtained when directly synchronizing non-tiled frames of the same early cardiac cycle (subsequence A) to the same later cardiac cycles (subsequence B), i.e. A→B.
Assuming the notation W, X, Y, or Z for arbitrary subsequence labels A through F, we compared temporal warping function , obtained when synchronizing subsequences W and X, with , obtained when synchronizing subsequences Y and Z (with all four subsequences, W, X, Y, and Z, extracted from the same tile n), by computing a discrete equivalent of
| (11) |
which captures the average temporal discrepancy between the two synchronization operations.
Also, we verified that the spatial match of left and right tiles was found correctly (assuming that the sample’s spatial motion was identical from one heartbeat to the next). We calculated the spatial registration error incurred when aligning subsequence W to subsequence X (both extracted from tile n), given the overlap xn (known by construction) and the estimated offset , as
| (12) |
The average spatial error and temporal discrepancies were computed in each case for N = 11 tiles, i.e.,
| (13) |
and
| (14) |
Numerical results are reported in the first three columns of Table 1 showing discrepancies of less than a frame in time and spatial registration accuracy of the order of one pixel.
Table 1.
Registration Evaluation*
| Registration Experiment | ||||
| (W→X) | C→F | E→D | C→F | C→E |
| Spatial Registration Evaluation | ||||
| Average spatial discrepancy | ||||
| Δx̃W→X (pixels) | 0.4 ± 0.1 | 1.7 ± 0.5 | 0.4 ± 0.1 | 0.0 ± 0.00 |
| Temporal Registration Evaluation | ||||
| Reference transform (Y→Z) | A→B | A→B | E→D | Identity |
| Average temporal discrepancy | ||||
| ΔtW→X,Y→Z or ΔtC→E (frames) | 0.2 ± 0.1 | 0.6 ± 0.4 | 0.6 ± 0.4 | 0.0 ± 0.00 |
Each experiment was carried out on N = 11 tiles and the average result is provided. The sequence labels A–F correspond to those in Fig. 3. The notation W→X, stands for “First cardiac cycle of subsequence W is registered to subsequence X”, where symbols W, X, Y, and Z stand for labels in the range A–F.
Finally, as a control to ensure that the spatial registration procedure was accurate and did not affect the accuracy of the temporal registration procedure, we also merged tile-pairs generated from the same cardiac period. To this end, we registered subsequence C to subsequence E (within the same tile, see Fig. 3(g)). The spatial error was obtained as above, while the temporal error was obtained as
| (15) |
Both spatial and temporal registration errors, reported in the last column of Table 1, show that registration was perfect in this case for all 11 tiles tested.
4. Conclusion
In summary, we have demonstrated the applicability of an image acquisition protocol and reconstruction algorithm for OCT images of cardio-vascular structures whose motion is cyclical, as dictated by the cyclic contraction of the heart. Dynamic mosaicing overcomes the limitations that arise from the frame rate, resolution, and FOV being interdependent for fast OCT cardiac imaging. Although we believe our space-time registration method is well adapted to the task at hand as it provides a simple solution to separately address the spatial and temporal registration problems (which considerably reduces computational complexity) other methods for space-time registration developed in the context of human cardiac imaging in adults using magnetic resonance imaging, single-photon emission computed tomography, or X-ray computer tomography could be used for similar purposes. In particular, use of registration techniques that are not only nonrigid in time but also in space [21, 22] could likely improve our mosaic reconstructions in cases where the spatial motion of the embryonic heart is not conserved from one heartbeat to the next.
Future work will include investigating possible improvements that could result from using such methods as well as combining our procedure with previously developed noise removal strategies [23] or techniques to reconstruct high frame rate 3D+time volumes from 2D+time series [17].
Acknowledgments
This research was supported by DOD ONR YIP (K.V.L.), NIH HL077187, HL095586, and T32HL007676, grants from UCSB/Santa Barbara Cottage Hospital (M.L), AHA 10SDG3830006 (I. V. L.), and a Hellman Family Faculty fellowship (M.L.). We thank Sandeep Bhat for help in preparing Fig. 2 and Movie 1 (5.7MB, MOV) .
References and links
- 1.Yelbuz T., Choma M., Thrane L., Kirby M., Izatt J., “A new high-resolution imaging technology to study cardiac development in chick embryos,” Circulation 106, 2771–2774 (2002). 10.1161/01.CIR.0000042672.51054.7B [DOI] [PubMed] [Google Scholar]
- 2.Larina I. V., Carbajal E. F., Tuchin V. V., Dickinson M. E., Larin K. V., “Enhanced OCT imaging of embryonic tissue with optical clearing,” Laser Phys. Lett. 5, 476–479 (2008). 10.1002/lapl.200810019 [DOI] [Google Scholar]
- 3.Larina I., Sudheendran N., Ghosn M., Jiang J., Cable A., Larin K., Dickinson M., “Live imaging of blood flow in mammalian embryos using Doppler swept-source optical coherence tomography,” J. Biomed. Opt. 13, 060506 (2008). 10.1117/1.3046716 [DOI] [PubMed] [Google Scholar]
- 4.Larina I., Ivers S., Syed S., Dickinson M., Larin K., “Hemodynamic measurements from individual blood cells in early mammalian embryos with Doppler swept source OCT,” Opt. Lett. 34, 986–988 (2009). 10.1364/OL.34.000986 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Larin K. V., Larina I. V., Liebling M., Dickinson M. E., “Live imaging of early developmental processes in mammalian embryos with optical coherence tomography,” J. Innov. Opt. Health Sci. 2, 253–259 (2009). 10.1142/S1793545809000619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gargesha M., Jenkins M. W., Wilson D. L., Rollins A. M., “High temporal resolution OCT using image-based retrospective gating,” Opt. Express 17, 10786–10799 (2009). 10.1364/OE.17.010786 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Liu A., Wang R., Thornburg K., Rugonyi S., “Efficient postacquisition synchronization of 4-D nongated cardiac images obtained from optical coherence tomography: application to 4-D reconstruction of the chick embryonic heart,” J. Biomed. Opt. 14, 044020 (2009). 10.1117/1.3184462 [DOI] [PubMed] [Google Scholar]
- 8.Schmoll T., Kolbitsch C., Leitgeb R. A., “Ultra-high-speed volumetric tomography of human retinal blood flow,” Opt. Express 17, 4166–4176 (2009). 10.1364/OE.17.004166 [DOI] [PubMed] [Google Scholar]
- 9.Wieser W., Biedermann B. R., Klein T., Eigenwillig C. M., Huber R., “Multi-megahertz OCT: High quality 3D imaging at 20 million A-scans and 4.5 GVoxels per second,” Opt. Express 18, 14685–14704 (2010). 10.1364/OE.18.014685 [DOI] [PubMed] [Google Scholar]
- 10.Vermot J., Fraser S. E., Liebling M., “Fast fluorescence microscopy for imaging the dynamics of embryonic development,” HFSP J. 2, 143–155 (2008). 10.2976/1.2907579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Can A., Stewart C., Roysam B., Tanenbaum H., “A feature-based technique for joint, linear estimation of high-order image-to-mosaic transformations: Mosaicing the curved human retina,” IEEE Trans. Pattern Anal. 24, 412–419 (2002). 10.1109/34.990145 [DOI] [Google Scholar]
- 12.Thévenaz P., Unser M., “User-friendly semiautomated assembly of accurate image mosaics in microscopy,” Microsc. Res. Tech. 70, 135–146 (2007). 10.1002/jemt.20393 [DOI] [PubMed] [Google Scholar]
- 13.Emmenlauer M., Ronneberger O., Ponti A., Schwarb P., Griffa A., Filippi A., Nitschke R., Driever W., Burkhardt H., “XuvTools: free, fast and reliable stitching of large 3D datasets,” J. Microsc. 233, 42–60 (2009). 10.1111/j.1365-2818.2008.03094.x [DOI] [PubMed] [Google Scholar]
- 14.Zupanc J., Dobnikar A., Drobne D., Valant J., Erdogmus D., Bas E., “Biological reactivity of nanoparticles: mosaics from optical microscopy videos of giant lipid vesicles,” J. Biomed. Opt. 16, 026003 (2011). 10.1117/1.3533319 [DOI] [PubMed] [Google Scholar]
- 15.Sawhney H., Kumar R., “True multi-image alignment and its application to mosaicing and lens distortion correction,” IEEE Trans. Pattern Anal. 21, 235–243 (1999). 10.1109/34.754589 [DOI] [Google Scholar]
- 16.Shum H., Szeliski R., “Systems and experiment paper: Construction of panoramic image mosaics with global and local alignment,” Int. J. Comput. Vis. 36, 101–130 (2000). 10.1023/A:1008195814169 [DOI] [Google Scholar]
- 17.Liebling M., Forouhar A. S., Gharib M., Fraser S. E., Dickinson M. E., “Four-dimensional cardiac imaging in living embryos via postacquisition synchronization of nongated slice sequences,” J. Biomed. Opt. 10, 054001 (2005). 10.1117/1.2061567 [DOI] [PubMed] [Google Scholar]
- 18.Liebling M., Forouhar A. S., Wolleschensky R., Zimmerman B., Ankerhold R., Fraser S. E., Gharib M., Dickinson M. E., “Rapid three-dimensional imaging and analysis of the beating embryonic heart reveals functional changes during development,” Dev. Dynam. 235, 2940–2948 (2006). 10.1002/dvdy.20926 [DOI] [PubMed] [Google Scholar]
- 19.Thévenaz P., Ruttimann U. E., Unser M., “A pyramid approach to subpixel registration based on intensity,” IEEE Trans. Image Process. 7, 27–41 (1998). 10.1109/83.650848 [DOI] [PubMed] [Google Scholar]
- 20.Liebling M., Vermot J., Forouhar A., Gharib M., Dickinson M., Fraser S., “Nonuniform temporal alignment of slice sequences for four-dimensional imaging of cyclically deforming embryonic structures,” in “Proc. ISBI 2006,”(2006), pp. 1156–1159
- 21.Frangi A., Rueckert D., Schnabel J., Niessen W., “Automatic construction of multiple-object three-dimensional statistical shape models: Application to cardiac modeling,” IEEE Trans. Med. Imaging 21, 1151–1166 (2002). 10.1109/TMI.2002.804426 [DOI] [PubMed] [Google Scholar]
- 22.Perperidis D., Mohiaddin R. H., Rueckert D., “Spatio-temporal free-form registration of cardiac MR image sequences,” Med. Image Anal. 9, 441–456 (2005). 10.1016/j.media.2005.05.004 [DOI] [PubMed] [Google Scholar]
- 23.Bhat S., Larina I. V., Larin K. V., Dickinson M. E., Liebling M., “Multiple-cardiac-cycle noise reduction in dynamic optical coherence tomography of the embryonic heart and vasculature,” Opt. Lett. 34, 3704–3706 (2009). 10.1364/OL.34.003704 [DOI] [PMC free article] [PubMed] [Google Scholar]


