Abstract
Background
Skin lesion color is an important feature for diagnosing malignant melanoma. In previous research, skin lesion color was investigated for discriminating malignant melanoma lesions from benign lesions in clinical images. Colors characteristics of melanoma were determined using color histogram analysis over a training set of images. Percent melanoma color and color clustering ratio features were used to quantify the presence of melanoma-colored pixels within skin lesions for skin lesion discrimination.
Methods
In this research, the relative color histogram analysis technique is used to evaluate skin lesion discrimination based on color feature calculations in different regions of the skin lesion in dermoscopy images. The histogram analysis technique is examined for varying training set sizes from the set of 113 malignant melanomas and 113 benign dysplastic nevi images.
Results
Experimental results show improved discrimination capability for feature calculations focused in the interior lesion region. Recognition rates for malignant melanoma and dysplastic nevi as high as 87.7% and 74.9%, respectively, are observed for the color clustering ratio computed using the outer 75% uniformly distributed area with a 10% offset within the boundary.
Conclusions
Experimental results appear to indicate that the melanoma color feature information is located in the interior of the lesion, excluding the 10% central-most region. The techniques presented here including the use of relative color and the determination of benign and malignant regions of the relative color histogram may be applicable to any set of images of benign and malignant lesions.
Keywords: image processing, malignant melanoma, color analysis, skin lesion, dysplastic nevus, color histogram
Early detection of malignant melanoma can be lifesaving (1–3) and may be aided by digital image analysis (4–10). Certain colors are associated with melanoma in dermoscopy images, including blue and gray in veil structures, dark gray, blue and black in peppering associated with regression, white in scar-like regression, erythema associated with inflammation, brown in dots, globules and blotches as well as the pigment network, and black in dots (11, 12). Automatic detection of colors in clinical images (13–18) and dermoscopy images (5, 6, 19) using a number of techniques has been presented, but delineation of the range of benign vs. malignant coloring in digital dermoscopy images has not previously been attempted. In previous research, a data-driven relative color histogram analysis technique was examined for determining colors characteristic of melanomas in clinical images (16, 17). The use of relative color (13), in which the background color is subtracted from the image color, has been proposed as a technique to avoid color distortion in the imaging process as well as a method of accounting for variations in normal skin color.
This research presents the relative color histogram technique that is applied to discrimination of benign and malignant colors in dermoscopy images. An overview of the color feature analysis algorithm used in the present study is presented in Fig. 1. Experiments presented here compare detection of clustered colors, as seen in the dermoscopy image lesion structures listed above, with detection of colors without regard to structure. Additional experiments are performed to evaluate the histogram analysis technique based on diagnostic accuracy achieved with: (1) different numbers of lesion images used as training data to identify melanoma colors and (2) different areas of the lesion selected for color analysis.
Fig. 1.

Overview of algorithm for color feature calculations and lesion discrimination.
The remaining sections of this paper include: (1) the techniques used to calculate the percent melanoma color and color clustering ratio features, (2) experiments performed for melanoma and benign lesion discrimination, (3) experimental results, (4) discussion, and (5) conclusions from the study.
Methods
The general method of dermoscopy skin lesion color image analysis for melanoma discrimination is presented in this section, including steps to: (1) construct the relative color histogram over a training set of images, (2) compute color features over the training set of lesion images and determine feature thresholds for lesion discrimination, (3) compute features and determine diagnostic accuracy over a test set of images, and (4) determine regions of the lesion for which step (3) is performed. The following sections present the steps given above in greater detail.
Procedure to construct relative color histogram
Dermoscopy image data and procedure to determine skin lesion boundaries
This research uses dermoscopy images consisting of digitized red, green, blue (RGB) color skin lesion images, with a typical size of 1024 × 768 pixels. The data set includes 113 malignant melanoma images, confirmed by histopathology and 113 dysplastic nevus (benign) dermoscopy images selected at random from 2000 lesions in the EDRA interactive Atlas of Dermoscopy (4). The melanomas are all invasive malignant melanomas, with no melanomas in situ.
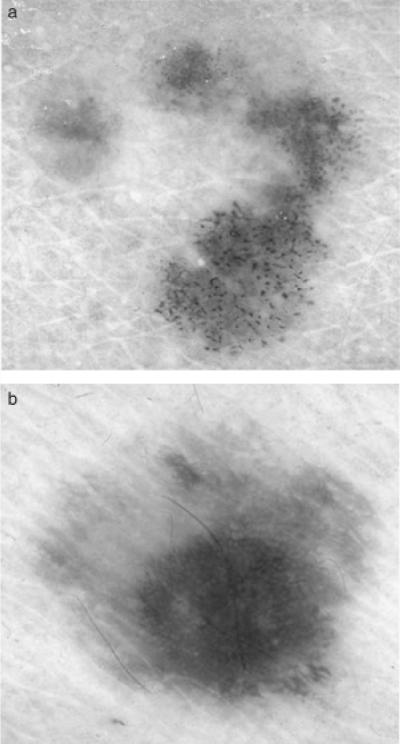
Skin lesion borders were found using manually chosen points along the border that were joined with a least mean squares distance second-order spline curve. A dermatologist working with a group of students found all borders, attempting to find as accurate borders as possible without a known gold standard for border accuracy. Manual borders obtained using this approach have been applied in several skin lesion feature analysis studies (13, 16–18, 20, 21). Figure 2 shows image examples from the image data set of a malignant melanoma (a) and a benign (dysplastic nevus) lesion (b).
Fig. 2.
Dermoscopy image examples of melanoma and benign lesions: (a) melanoma image, (b) benign image (dysplastic nevus).
Relative color and surrounding skin color
A full-color image of a lesion was represented as an array of pixels, with each pixel having three integer RGB color values, each ranging from 0 to 255, with a subset of the array, generally contiguous, comprising the lesion. The image was normalized to the average background skin color by subtracting the background skin color, determined uniquely for that image, from the color value for each pixel of the image array within the lesion boundary, generating a new relative color image (13, 16, 17).
The background skin color for each image was determined by averaging all non-lesion pixels that were within the skin-colored region of RGB space. In the absence of significant color distortion, normal living skin falls within the quadrant of RGB space that satisfies R>B and R>G (22). In the absence of shadow, R>90. Examples of dermoscopy regions that do not satisfy these requirements include those that are in shadow, such as areas not in direct contact with the dermatoscope, black and white ruler markings, and reflections from bubbles.
A distance transform-based approach (23) was used to find the region outside of the lesion for computing the surrounding skin color. Using Euclidean distance, the rounded distance of each pixel outside of the lesion to the lesion boundary is determined. Let U denote the total number of pixels in the non-lesion region with skin colors that satisfy the RGB constraints presented above for a distance less than or equal to D from the lesion boundary. Starting with D = 1 (pixels adjacent to the lesion), D is incremented by unit steps until U ≥ 4AL, where AL is the area of the lesion. This approach was used for small lesions that are less than approximately 5 mm in diameter. However, larger lesions in dermoscopy images extended over 1/4 of the total image area and calculations for the surrounding skin were computed over the entire dermoscopy image except for the lesion.
Relative color histogram quantization
For an Y row by X column RGB image, I, the actual value of each pixel is given by I(x, y) = (r(x,y), g(x,y), b(x,y)), where 1 ≤ x ≤ X and 1 ≤ y ≤ Y. Let O denote the skin lesion region within the color image, defined as O = {(x,y)|(x,y)∈I and is inside the closed skin lesion boundary}. Then, the relative color for all skin lesion pixels is given as Orel(x,y) = (rrel(x,y), grel(x,y), brel(x,y)) = (r(x,y) − rskin, g(x,y) − gskin, b(x,y) − bskin), where − 255 ≤ rrel(x,y), grel(x,y), brel(x,y) ≤ 255. rskin, gskin, and bskin are the average RGB values computed from the surrounding skin. Thus, the relative color space is of size 511 × 511 × 511.
In order to reduce memory requirements and processing time and to facilitate grouping of similar colors, the relative color histogram may be quantized into cubic bins, each bin measuring approximately 2c on each axis and containing approximately (2c)3 different relative colors. Let H represent the relative color histogram and H(i,j,k) denote a relative color histogram bin, where i = trunc(255 + r(x,y) − rskin/2c), j = trunc(255 + g(x,y) − gskin/2c) and k = trunc(255 + b(x,y) − bskin/2c). For c = 0, the quantized histogram is the original histogram, where each bin contains one relative color. For c = 9, the relative color histogram contains only one bin and cannot discriminate between benign and malignant lesions. Previous research showed that color features are preserved with c = 2 (16). For c = 2, all bins (except for a single smaller bin H(− 63, − 63, − 63)) represent color pixels that have one of (2c)3 = 64 different relative colors for each bin H(i,j,k), − 63 ≤ i, j, k ≤ 64. Partitioning the relative color histogram such that the bin H(− 63, − 63, − 63) contains a smaller number (27) of different relative colors causes no problem because those relative colors are infrequently observed in skin lesions.
Melanoma color mapping and relative color histogram analysis
A training set with MTotal melanoma and BTotal benign images is selected for determining relative colors, as quantized in the relative color histogram, associated with melanoma and benign lesions. For each training image, the pixels inside of the lesion are mapped to the relative color histogram bins. Let N(i,j,k) represent the number of pixels within the training image lesion that map to bin H(i,j,k). H(i,j,k) is considered populated by the training image if N(i,j,k) ≥ K, where the threshold K=0.00125AL was selected based on clinical image analysis (16). K was chosen to reduce noise without interfering with the mapping. See Omission of threshold for 100% boundary area percentge for experiments performed using K=0.
After bin population for each melanoma and benign training image is completed, the relative color histogram bins are initially labeled. Let M̂(i,j,k) and B̂(i,j,k) represent the number of melanoma and benign images, respectively, that populated bin H(i,j,k). Then, H(i,j,k) is labeled as a melanoma bin if M̂i,j,k/MTotal > B̂i,j,k/BTotal a benign bin if B̂i,j,k/BTotal > M̂i,j,k/MTotal, an uncertain bin if M̂i,j,k/MTotal = B̂i,j,k/BTotal > 0, and an unpopulated bin if M̂i,j,k/MTotal = B̂i,j,k/BTotal = 0. The mapping of images to histogram bins was performed rather than mapping pixels from the images to the histogram bins so that large lesions or images at higher resolutions would not be over represented for bin labeling.
Relative color histogram bin labeling extrapolation
A large number of bins H(i,j,k) remain unpopulated or uncertain, even after training runs with large image sets have populated H. Many of the unlabeled bins lie within uniformly labeled regions, either benign or malignant, and it is reasonable to expect that these regions are likely to contain more pixels from the same type of lesion. Thus, extrapolation is reasonably expected to increase diagnostic accuracy. In order to assess colors from test images that may map into unpopulated or uncertain bins, extrapolation was performed to label those bins that are close to melanoma or benign bins the same as the nearby known bins.
The following iterative extrapolation algorithm was used. For each uncertain or unpopulated bin H(i,j,k), the total number of benign and melanoma bins are found in the adjacent neighboring bins, denoted as CB and CM, respectively. Note that neighbors are considered in three dimensions. For bins on the interior of H, there are 26 adjacent neighbors. There are 17 neighbors for H(i,j,k) on a face of H, 11 neighbors H(i,j,k) on an edge of H, and seven neighbors for H(i,j,k) on a corner of H. Let δM represent the count of melanoma neighbors and δB refer to the count of benign neighbors. An uncertain or unpopulated bin H(i,j,k) is changed to a melanoma bin if δM ≥ 1 and δB=0 or δM–δB ≥ 3. An uncertain or unpopulated bin is relabeled as a benign bin if δB ≥ 1 and δM=0 or δB–δM ≥ 5. There is a stricter criterion for re-labeling uncertain or unpopulated bins as benign rather than as melanoma because of the need to avoid false negative lesion classifications. The iterative process is stopped after additional conversions are negligible, which occurs after extending extrapolation 10 bins from the mapped bins. An example of the quantized histogram H showing benign and malignant bins after this process is completed is shown in Fig. 3.
Fig. 3.
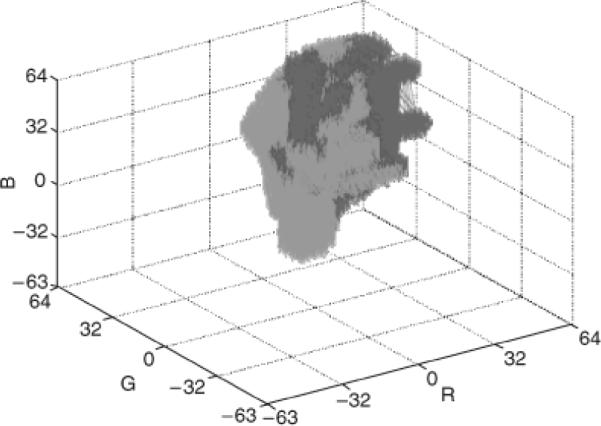
Example of three-dimensional relative color histogram bin labeling for a training set of images. The melanoma-labeled bins are red regions. The benign-labeled bins are green regions.
Color feature calculations and lesion discrimination
Two color features were computed using the labeled relative color histogram bins from the training set of images: the percentage of melanoma color and color clustering ratio. This section presents the two features and the approaches used for skin lesion discrimination based on the individual features.
Percentage of melanoma color
The percentage of melanoma color feature P is the percent of the lesion with pixels with relative colors that map into melanoma bins. Formally, the percentage of melanoma color within a lesion is given as P = 100R/AL, where R is the total number of pixels within the lesion with relative colors that map into melanoma bins. For lesion evaluation, P is computed over all training images. A lesion is scored as malignant if P ≥ α. Otherwise, the lesion is benign. α is a threshold determined from the percentage of melanoma color features found over the training set of images. The threshold α is selected such that tpα ≥ tnα is minimized for α ε [0,100], where α is evaluated in increments of 1, and tpα (true positive rate) and tnα (true negative rate) are the percentage of correctly classified melanoma and benign lesions at threshold α, respectively.
Color clustering ratio feature
The color clustering ratio feature (17, 18) provides a quantitative measure of the grouping of melanoma pixels within the lesion. Let V denote the set of relative color values that map into melanoma bins from the training set of images. Define a lesion region of interest as a contiguous subset of the lesion or the lesion itself. Let L denote the set of pixel locations within the skin lesion region of interest with relative color O that map into melanoma bins, formally L={(x, y)|Orel(x, y)εV}. For every pixel (x,y)εL, let W(x,y) denote the number of eight-connected neighbors and WM(x,y) denote the number of eight-connected neighbors with relative colors that map into melanoma bins. The eight-connected neighbors for (x,y)εL that lie outside of the lesion region of interest are excluded from calculating W(x,y) and WM(x,y). Then, S = Σ(x;y)εLWM(x;y) represents the total number of neighbors with relative colors that map into melanoma bins for all pixels within the lesion region of interest with relative colors that map into melanoma bins. The cumulative total number of eight-connected neighbors for all (x,y)εL is denoted as T = Σ(x;y)εLW(x;y). T includes all neighbors of melanoma color pixels within the skin lesion regardless of whether the neighbor is mapped to a melanoma color. The color clustering ratio for a lesion is given as C=S/T. If C ≥ β, the lesion is scored as a melanoma. Otherwise, the lesion is called benign. β is a threshold determined from the color clustering ratio features found over the training set of images. The threshold β is selected such that tpβ ≥ tnβ is minimized for βε[0,1], where β is evaluated in increments of 0.001, and tpβ and tnβ are the percentage of correctly classified melanoma and benign lesions at threshold β, respectively.
Lesion region analysis
Two types of lesion interior regions are examined for color analysis, including: (1) the boundary area percentage and (2) the offset boundary area percentage (17). The boundary area percentage is defined as the skin lesion region with uniform distance from the boundary to the interior that contains a specified percentage of the lesion area. The boundary area region is found by iterative inward region growing from the lesion boundary toward the lesion centroid. Starting with the lesion boundary, all eight-connected neighbors of each boundary pixel located in the lesion interior are identified. A cumulative count of all uniquely identified interior pixels is maintained. The resulting cumulative count is compared with the desired area percentage. If the area percentage is met or exceeded, region growing is stopped. Otherwise, the uniquely identified interior pixels become the new inner lesion border. From the new inner border, the unique eight-connected interior border pixels are found and are added to the cumulative count of boundary area pixels. The process continues until the area percentage criterion has been satisfied. Figure 4 shows an example with the boundary area percentage (white) of 25% of the lesion area (lesion from Fig. 2(a)). This area is used for feature calculations, and the remaining 75% of the lesion interior is excluded from feature calculations.
Fig. 4.

Boundary area percentage example using 25% of the lesion area for analysis (white region). (This is the same lesion as in Fig. 2(a).)
The offset boundary area percentage is defined as the outermost skin lesion region within a uniform distance of the boundary containing the specified percentage lesion area. The boundary area percentage determination then starts from the inner boundary of the specified offset boundary area region. When the boundary is offset, the boundary area percentages are applied as percentages of the original lesion area, computed using the inner border of the offset boundary area as the starting boundary. Figure 5 presents an example of a 10% offset boundary area percentage (gray area) and 75% boundary area percentage (white area) (lesion from Fig. 2(a)). Note that the features are calculated in the boundary area (white area in Fig. 5) only.
Fig. 5.

Offset boundary area example using 10% of the lesion area as offset (gray region) and 75% of the lesion area for analysis (white region). (This is the same lesion as in Fig. 1(a).)
Percent melanoma color feature and color clustering ratio feature for specified regions
The percent of melanoma color feature P and color clustering ratio feature C are computed based on the specified boundary area percentage and offset area percentage regions. For lesion discrimination, the threshold calculations for the percentage of melanoma color and color clustering ratio features from the training set of images are made using the features determined over the same specified lesion region. Note that eight-connected neighbors examined for the color clustering ratio are only included if they are within the specified region of the lesion.
Experiments Performed
For the experiments performed, 80% of the image data was used in the training set (BTotal = 90 benign lesions and MTotal = 90 melanomas), with the remaining 20% of the images comprising the test set (23 benign lesions and 23 melanomas). Lesion discrimination experiments were performed using 18 randomly chosen training and test sets. For each training set of images, the entire skin lesion was used to generate the relative color histogram and the labeling of melanoma and benign bins. The relative color histogram bin labels obtained for that training set were used to compute the percent melanoma color and color clustering ratio features for different lesion regions. In order to determine the effectiveness of the percent melanoma color and color clustering ratio features, the following boundary area percentage and offset boundary area percentage experiments were performed. Note that the same 18 randomly chosen training/test sets were used for all experiments to enable direct comparison of the results obtained.
Boundary area percentage experiments
The percentage of melanoma color and color clustering features were computed for the boundary area percentages, including 100%, 90%, 75%, 50%, 25%, and 10% of the lesion interior. Starting at the lesion boundary, the interior of the lesion is traversed uniformly to contain the area percentage specified.
Offset boundary area percentage experiments
The percentage of melanoma color and color clustering ratio features were calculated for the offset boundary area percentage case with offset percentage of 10% for lesion interior area percentages of 10%, 25%, 50%, 75%, and 90%. The offset percentage of 10% was chosen based on high correct melanoma discrimination results in clinical images (17).
Omission of threshold for 100% boundary area percentage
The percentage of melanoma color feature for the 100% boundary area case was computed based on constructing the relative color histogram for each training image using a bin population threshold of K=0. This experiment was performed in order to compare lesion discrimination results using no noise removal (K=0) for histogram bin population in individual lesion training images with the noise removal threshold (K=0.00125AL) determined in previous research (16) and applied to the experiments performed in parts `Boundary area percentage experiments, Offset boundary area percentage experiments, and Incomplete training set results for 100% boundary area percentage.'
Incomplete training set results for 100% boundary area percentage
This set of experiments examined the impact on melanoma discrimination using different numbers of images in the training set. Using the same 18 randomly generated training/test sets from the experiments in `Boundary area percentage experiments and Offset boundary area percentage experiments,' relative color histogram (H) bin labeling was performed using 25%, 50%, 75%, and 100% of the total training images. For the 50% case, there were 45 melanoma and 45 benign images, respectively, in the training set of images that were randomly selected from the 90 melanoma and 90 benign images, respectively, in the original training set. Note that the test sets were unchanged. The percent melanoma color and color clustering ratio features were computed over the training and test sets of images for the 100% boundary area percentage case (entire lesion).
Experimental Results
Relative color histogram labeling example
Figure 3 shows a three-dimensional representation of the relative color bin melanoma and benign labels using the technique presented in `Methods' for one of the 18 randomly generated training sets of images. The red boxes correspond to the melanoma-labeled bins, and the green boxes represent the benign-labeled bins. The melanoma- and benign-labeled bins have large non-overlapping regions in the relative color histogram when viewing the three-dimensional plot. There are also overlapping regions in the relative color histogram, where it is difficult to clearly distinguish melanoma colors from benign colors. Note that the indices for each relative color are from −63 to 64. For the training set of images used to generate the histogram bin mapping shown in Fig. 3, the actual number of bins labeled as melanoma is larger than the number labeled as benign, 54,148 and 48,527, respectively.
Boundary area percentage experimental results
For the boundary area percentage experiments performed, lesion discrimination was examined for percent melanoma color and color clustering ratio features. Table 1 presents the average and standard deviation (SD) results over 18 training/test sets for the percent melanoma color feature for the 75% boundary area percentage case. Table 2 gives the average and SD results over 18 training/test sets for the color clustering ratio feature for the 75% boundary area percentage case.
TABLE 1.
Average and standard deviation results over 18 training/test sets for the 75% boundary area case for the percent melanoma color feature
| Training results (% correct) |
Test results (% correct) |
||||
|---|---|---|---|---|---|
| Iteration | Threshold α | Melanoma | Benign | Melanoma | Benign |
| 1 | 20.3 | 85.6 | 85.6 | 78.3 | 78.3 |
| 2 | 20.9 | 87.8 | 87.8 | 91.3 | 91.3 |
| 3 | 28.4 | 84.4 | 84.4 | 82.6 | 82.6 |
| 4 | 22.1 | 85.6 | 85.6 | 87.0 | 87.0 |
| 5 | 21.9 | 86.7 | 86.7 | 91.3 | 78.3 |
| 6 | 27.9 | 82.2 | 82.2 | 82.6 | 78.3 |
| 7 | 24.2 | 83.3 | 83.3 | 60.9 | 87.0 |
| 8 | 21.1 | 86.7 | 85.6 | 82.6 | 82.6 |
| 9 | 23.0 | 84.4 | 84.4 | 87.0 | 82.6 |
| 10 | 24.5 | 86.7 | 86.7 | 87.0 | 69.6 |
| 11 | 20.5 | 86.7 | 85.6 | 78.3 | 78.3 |
| 12 | 26.8 | 85.6 | 85.6 | 65.2 | 82.6 |
| 13 | 18.7 | 87.8 | 86.7 | 87.0 | 65.2 |
| 14 | 24.1 | 88.9 | 88.9 | 82.6 | 78.3 |
| 15 | 23.8 | 86.7 | 86.7 | 69.6 | 73.9 |
| 16 | 20.4 | 87.8 | 86.7 | 82.6 | 73.9 |
| 17 | 20.2 | 85.6 | 85.6 | 82.6 | 73.9 |
| 18 | 22.3 | 86.7 | 86.7 | 78.3 | 82.6 |
| Average | 86.0 | 85.8 | 80.9 | 79.2 | |
| Standard deviation | 1.7 | 1.6 | 8.3 | 6.4 | |
The threshold α is shown for each training/test set.
TABLE 2.
Average and standard deviation results over 18 training/test sets for the 75% boundary area case for the color clustering ratio feature
| Training results (% correct) |
Test results (% correct) |
||||
|---|---|---|---|---|---|
| Iteration | Threshold β | Melanoma | Benign | Melanoma | Benign |
| 1 | 66.7 | 86.7 | 86.7 | 95.7 | 78.3 |
| 2 | 67.1 | 90.0 | 88.9 | 82.6 | 82.6 |
| 3 | 69.1 | 87.8 | 86.7 | 95.7 | 82.6 |
| 4 | 66.9 | 86.7 | 85.6 | 87.0 | 78.3 |
| 5 | 69.2 | 85.6 | 84.4 | 95.7 | 73.9 |
| 6 | 72.2 | 85.6 | 85.6 | 87.0 | 73.9 |
| 7 | 69.7 | 86.7 | 86.7 | 78.3 | 82.6 |
| 8 | 66.1 | 86.7 | 86.7 | 87.0 | 82.6 |
| 9 | 66.2 | 86.7 | 86.7 | 87.0 | 82.6 |
| 10 | 69.7 | 85.6 | 85.6 | 91.3 | 60.9 |
| 11 | 66.5 | 86.7 | 86.7 | 87.0 | 73.9 |
| 12 | 70.4 | 88.9 | 87.8 | 73.9 | 82.6 |
| 13 | 68.0 | 87.8 | 87.8 | 87.0 | 73.9 |
| 14 | 67.5 | 85.6 | 85.6 | 95.7 | 78.3 |
| 15 | 70.2 | 83.3 | 82.2 | 82.6 | 69.6 |
| 16 | 66.9 | 84.4 | 83.3 | 82.6 | 73.9 |
| 17 | 69.1 | 88.9 | 87.8 | 87.0 | 87.0 |
| 18 | 69.2 | 88.9 | 88.9 | 82.6 | 73.9 |
| Average | 86.8 | 86.3 | 87.0 | 77.3 | |
| Standard deviation | 1.7 | 1.7 | 6.1 | 6.2 | |
The threshold β is shown for each training/test set.
Figure 6 presents the average percent melanoma color and color clustering ratio correct melanoma and benign lesion test recognition rates over 18 random test sets for the boundary area percentage cases of 10%, 25%, 50%, 75%, 90%, and 100%. The horizontal axis shows the boundary area percentage, labeled as the percentage of the skin lesion starting from the lesion boundary used for feature calculations. The vertical axis gives the percentage correct discrimination. The results for the percent melanoma color and color clustering ratio methods are shown for each boundary area percentage with plus and minus one SD shown for the average correct percentage discrimination. The key for interpreting the recognition results presented for each boundary area percentage case is: PMC - melanoma refers to the recognition rates of the percent melanoma color method applied to the melanoma test sets, CCR - melanoma denotes the classification rates of the color clustering ratio method applied to the melanoma test sets, PMC - benign refers to the recognition rates of the percent melanoma color method applied to the benign lesion test sets and CCR - benign denotes the classification rates of the color clustering ratio methods applied to the benign lesion test sets. Note that the average and SD training rates are not shown because the thresholds a and b are chosen based on the approximate equality between the true positive and true negative rates.
Fig. 6.
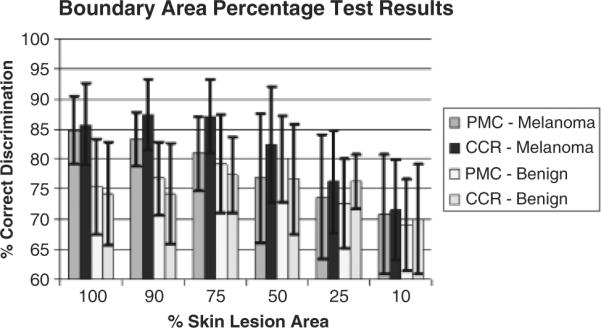
Average and standard deviation melanoma test results over 18 test sets for the boundary area percentage cases of 100%, 90%, 75%, 50%, 25%, and 10%. PMC and CCR refer to the percent melanoma color and color clustering ratio features, respectively.
In Fig. 6, it can be seen that the color clustering ratio feature achieves higher true positive test rates than the corresponding rates for the percentage melanoma color feature for all boundary area percentage cases tested, with no significant differences in SD results, over the 18 training/test sets. The 90% boundary area percentage case for the color clustering ratio feature produces the highest true positive rate (87.4%) of the cases examined. Inspecting Fig. 6 shows that the true positive rate increases for both features as the central portion of the skin lesion used for color feature analysis increases, except for the 100% (entire lesion) boundary area percentage case for the color clustering ratio.
Offset boundary area percentage experimental results
In the second set of experiments, 10% of the lesion boundary region was excluded for feature calculations, referenced as the offset boundary area percentage. Figure 7 presents the average and SD test results for the percent melanoma color and color clustering ratio features for the different interior lesion regions using 10% offset from the boundary. The notations for the percent melanoma color and color clustering ratio melanoma and benign average and SD results are the same as for Fig. 6.
Fig. 7.
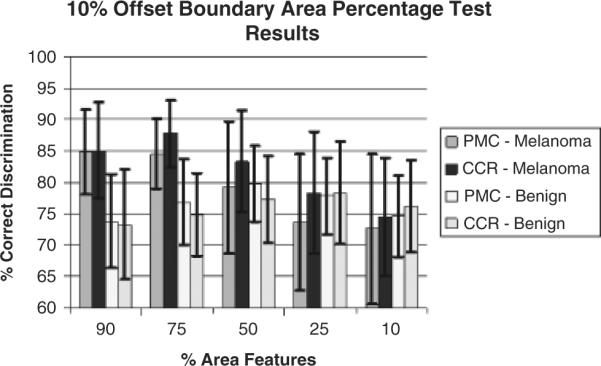
Ten percent offset boundary area percentage average and standard deviation test results for 90%, 75%, 50%, 25%, and 10% lesion area cases starting from the inner boundary of the offset region over 18 test sets. The horizontal axis shows the percentage of the lesion area used for feature calculations (% area features).
Based on experimental results obtained for the boundary area percentage cases presented in Fig. 7, the 10% and 25% area boundaries yield the worst melanoma discrimination capability for the percent melanoma color and color clustering ratio features. Furthermore, the melanoma discrimination capability improves with increased boundary area percentage, with the best melanoma discrimination results for the percent melanoma color and color clustering ratio features obtained at 90% and 75% boundary area percentages, respectively.
Omission of threshold for 100% boundary area percentage
In the third set of experiments, the percentage of a training lesion's pixels with relative colors mapped to a histogram bin for that bin to be considered populated was set to zero (K=0). This affects the labeling of melanoma and benign bins over the training set of images for color feature calculations. In previous research with clinical images, requiring a small percentage of the lesion to be mapped to a histogram bin improved lesion discrimination by focusing melanoma and benign color labeling on color ranges with a moderate frequency of occurrence in the training set of images (16–18). The no-threshold requirement (K=0) and the small percentage of the lesion requirement (K=0.000125AL) methods of individual training image histogram bin population were evaluated for the 100% boundary area percentage case for the percentage of melanoma color feature. Figure 8 shows the average and SD test results over the 18 training/test sets. From Fig. 8, there is 17.1% decline in correct melanoma recognition for K=0. The SDs of the melanoma and benign classification results are substantially higher for K=0, providing for less consistent results.
Fig. 8.
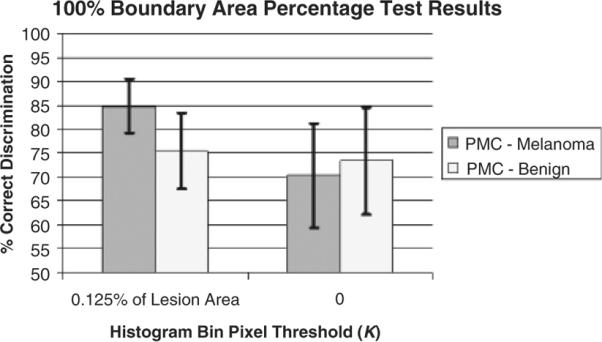
Hundred percent boundary area percentage case average and standard deviation test results over 18 test sets for percentage of melanoma color feature using histogram bin thresholds K=0.00125AL and K=0.
100 boundary area percentage results for different numbers of training images
The final set of experiments examined the impact on melanoma discrimination using different numbers of images in the training set. For these experiments, relative color histogram bin labeling was performed using 25%, 50%, 75%, and 100% of the training images utilized for generating the experimental results presented in `Relative color histogram labeling example, Boundary area percentage experimental results, and Offset boundary area percentage experimental results.' The percent melanoma color and color clustering ratio features were computed over the 100% boundary area percentage case (entire lesion). Figure 9 shows the average and SD results over the 18 training/test sets.
Fig. 9.
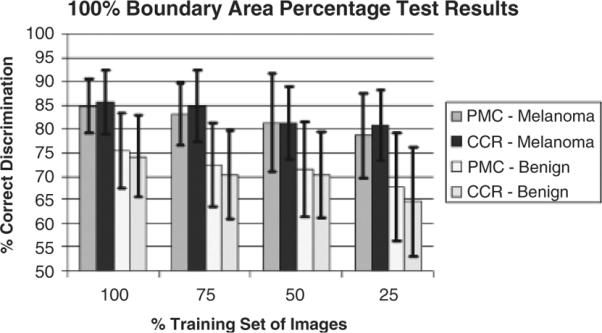
Average and standard deviation melanoma test results over 18 test sets for the 100% boundary area percentage case using 25%, 50%, 75%, and 100% of the training images.
From Fig. 9, the melanoma and benign lesion classification rates decrease as the number of training images decreases, as expected. However, the melanoma correct classification rates for the percentage of melanoma color and color clustering ratio features are not significantly reduced as the number of training images decreases, falling from 84.8% to 78.5% and 85.7% to and 80.8%, respectively, as the training set is reduced to only 25% of its original size. The corresponding benign lesion correct classification rates are somewhat more dependent on training set size, with the percentage of melanoma color and color clustering ratio classification rates falling from 75.4% to 67.6% and from 74.2% to 64.7%, respectively.
Using fewer images in the training set leads to higher SDs over the 18 training/test sets for each feature, meaning the results are less consistent than for using greater numbers of images in the training set. The experimental results show that the region growing algorithm performs reasonably well in histogram bin assignment, even with relatively few images used to generate the relative color histogram.
Discussion
Relative color vs. absolute color
Relative color (13) has been proposed as a method to avoid color distortion in imaging processes, because of varying film types and lighting, as well as a method of accounting for variations in normal skin color. The relative color technique equalizes color changes because of widely varying skin types. No amount of calibration can over-come this physiological variation. In one study of clinical images, relative color indices had higher Pearson's correlation with the diagnosis of melanoma than absolute color indices (24).
Analysis of boundary area percentage and offset boundary area percentage results
There were two reasons for comparing results with and without the offset boundary. The first was the lack of a gold standard for determining skin lesion borders, creating concern that some surrounding normal skin might be included in the feature calculations. The second was to compare results with and without the peripheral-most areas, which are important in clinical lesion determination. In comparing Figs 6 and 7, no loss was observed excluding these areas for feature calculations in the 10% offset case. The best melanoma discrimination results were obtained for the percent melanoma color and color clustering ratio features obtained at 100% (entire lesion) and 90% boundary area percentages, respectively. This contrasts with previous research with clinical images, where the outer boundary areas (10% and 25% boundary area percentage cases) yielded the best melanoma discrimination capability (17). These results suggest that the color discrimination information for both color features lies in the interior of dermoscopy images, rather than in the periphery, in contrast to clinical lesion images. This is in agreement with the relative difficulty in discerning image colors in the center of lesions when viewing clinical images, as well as the presence of a significant fraction of critical features of melanoma close to the center of the lesion in dermoscopy images.
Comparison of features: percentage of melanoma color vs. color clustering ratio
Note that in all experiments comparing the percentage of melanoma color and color clustering ratio features (Figs 6 and 7), the color clustering ratio provides higher discrimination results than the percentage of melanoma color for melanoma. This is not true for benign lesions. This result may indicate the relatively greater presence of structured color features in melanoma. The presence of a greater number of known colored structures such as branch streaks, brown globules, and black dots in melanoma is consistent with the greater discrimination power of the color clustering ratio feature for melanoma as compared with benign lesions.
Relative color histogram construction process and sources of error
There are two reasons that the entire lesion was used in the training images for relative color histogram construction and bin labeling. First, using the entire lesion allows a direct comparison of the feature calculations performed using different portions of the lesion in the test set. Second, better melanoma discrimination was obtained in the test sets when training was performed over the entire lesion area for the dermoscopy image experiments performed in this research and in clinical image analysis (16, 17).
Sources of error in the methods investigated have been considered. For varying lesion size, there would be a bias toward representation of larger lesions if pixels from benign and malignant lesions were simply counted as they mapped to bins. Similarly, for varying resolution, the contribution of numbers of pixels would vary, for a Y × X image, the larger Y and X would contribute more pixels to the histogram and over-represent a given lesion. A method was used where the pixels of a given color from one image yield one point in a given color bin to avoid these sources of over- and under-representation. Another source of error could be a non-optimal bin size. Bins containing 64 relative colors in the quantized histogram H were satisfactory for lesion discrimination in clinical images (16–18) and were not altered for this research. A third source of error may be a non-optimal training lesion area threshold for histogram bin population (K). Experimental results presented in `Omission of threshold for 100% boundary area percentage' that requiring a minimal percentage of the training lesion area (0.125%) within a histogram bin for bin population contributed to improved lesion discrimination over having no lesion area requirement for histogram bin population. The value of K used in this research was based on previous clinical image color feature analysis (16).
Conclusion
In this research, unstructured color information via percentage of melanoma color and structured color information via the color clustering ratio were compared for discriminating melanoma and benign skin lesions in dermoscopy images. For the experiments performed, the color clustering ratio overall produced slightly better melanoma discrimination results than the percent melanoma color, suggesting the importance of structured color information. Additionally, different regions were compared with respect to discriminating the benign and malignant classes. These results appear to indicate that the melanoma color feature information is located in the interior of the lesion, excluding the 10% central-most region. Techniques presented here including the use of relative color and the determination of benign and malignant regions of the relative color histogram may be applicable to any set of images of benign and malignant lesions.
Acknowledgement
This research was supported by NIH-SBIR grants CA 60294-03 and CA101639-01.
References
- 1.Friedman RJ, Rigel DS, Kopf AW. Early detection of malignant melanoma: the role of physician examination and self-examination of the skin. Ca-A Cancer J Clin. 1985;35:130–151. doi: 10.3322/canjclin.35.3.130. [DOI] [PubMed] [Google Scholar]
- 2.Rigel DS. Malignant melanoma: incidence issues and their effect on diagnosis and treatment in the 1990s. Mayo Clin Proc. 1997;72:367–371. doi: 10.4065/72.4.367. [DOI] [PubMed] [Google Scholar]
- 3.Jemal A, Murray T, Samuels A, Ghafoor A, Ward E, Thun MJ. Cancer statistics 2003. Ca-A Cancer Clin. 2003;53:5–26. doi: 10.3322/canjclin.53.1.5. [DOI] [PubMed] [Google Scholar]
- 4.Argenziano G, Soyer HP, De Giorgi V, Piccolo D. Dermoscopy, an interactive atlas. EDRA Medical Publishing; Milan, Italy: 2000. [Google Scholar]
- 5.Elbaum M, Kopf AW, Rabinovitz HS, Langley RGB, Kamino H, Mihm MC, Sober AJ, Peck GL, Bogdan A. Automatic differentiation of melanoma from melanocytic nevi with multispectral digital dermoscopy: a feasibility study. J Am Acad Dermatol. 2001;44:207–218. doi: 10.1067/mjd.2001.110395. [DOI] [PubMed] [Google Scholar]
- 6.Moncrieff M, Cotton S, Claridge E, Hall P. Spectrophotometric intracutaneous analysis: a new technique for imaging pigmented lesions. Br J Dermatol. 2002;146:448–457. doi: 10.1046/j.1365-2133.2002.04569.x. [DOI] [PubMed] [Google Scholar]
- 7.Jamora MJ, Wainwright BD, Meehan SA, Bystryn J-C. Improved identification of potentially dangerous pigmented skin lesions by computerized image analysis. Arch Dermatol. 2003;139:195–198. doi: 10.1001/archderm.139.2.195. [DOI] [PubMed] [Google Scholar]
- 8.Rosado B, Menzies S, Harbauer A, Pehamberger H, Wolff K, Binder M, Kittler H. Accuracy of computer diagnosis of melanoma: a quantitative meta-analysis. Arch Dermatol. 2003;139:361–367. doi: 10.1001/archderm.139.3.361. [DOI] [PubMed] [Google Scholar]
- 9.Fleming MG, Steger C, Zhang J, Gao J, Cognetta AB, Pollak I, Dyer CR. Techniques for a structural analysis of dermatoscopic imagery. Comput Med Imaging Graph. 1998;22:375–389. doi: 10.1016/s0895-6111(98)00048-2. [DOI] [PubMed] [Google Scholar]
- 10.Ganster H, Pinz A, Rohrer R, Wilding E, Binder M, Kittler H. Automated melanoma recognition. IEEE Trans Med Imag. 2001;20:233–238. doi: 10.1109/42.918473. [DOI] [PubMed] [Google Scholar]
- 11.Argenziano G, Fabbrocini G, Carli P, De Giorgi V, Delfino M. Epiluminescence microscopy for the diagnosis of doubtful melanocytic skin lesions. Arch Dermatol. 1998;134:1563–1570. doi: 10.1001/archderm.134.12.1563. [DOI] [PubMed] [Google Scholar]
- 12.Stolz W, Braun-Falco O, Bilek P, Landthaler M, Burgdorf WHC, Cognetta AB. Color atlas of dermatoscopy. Blackwell Science Ltd.; Cambridge, MA: 1988. [Google Scholar]
- 13.Umbaugh SE, Moss RH, Stoecker WV. Automatic color segmentation of images with application to detection of variegated coloring in skin tumors. IEEE Eng Med Biol. 1989;8:43–52. doi: 10.1109/51.45955. [DOI] [PubMed] [Google Scholar]
- 14.Green A, Martin N, Pfitzner P, O'Rourke M, Knight N. Computer image analysis in the diagnosis of melanoma. J Am Acad Dermatol. 1994;31:958–964. doi: 10.1016/s0190-9622(94)70264-0. [DOI] [PubMed] [Google Scholar]
- 15.Aitken JF, Pfitzner J, Battistutta D, O'Rourke PK, Green AC, Martin NG. Reliability of computer image analysis of pigmented skin lesions of Australian adolescents. Cancer. 1996;78:252–257. doi: 10.1002/(SICI)1097-0142(19960715)78:2<252::AID-CNCR10>3.0.CO;2-V. [DOI] [PubMed] [Google Scholar]
- 16.Faziloglu Y, Stanley RJ, Moss RH, Stoecker WV, Mclean RP. Color histogram analysis for melanoma discrimination in clinical images. Skin Res Technol. 2003;9:147–155. doi: 10.1034/j.1600-0846.2003.00030.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chen J, Stanley R, Moss RH, Stoecker WV. Color analysis of skin lesion regions for melanoma discrimination in clinical images. Skin Res Technol. 2003;9:94–104. doi: 10.1034/j.1600-0846.2003.00024.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Stanley RJ, Moss RH, Stoecker WV, Aggarwal C. A fuzzy-based histogram analysis technique for skin lesion discrimination in dermatology clinical images. Comput Med Imag Graph. 2003;27:387–396. doi: 10.1016/s0895-6111(03)00030-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rubegni P, Cevenini G, Burroni M, Dell'Eva G, Sbano P, Cuccia A, Andreassi L. Digital dermoscopy analysis of atypical pigmented skin lesions: a stepwise logistic discriminant analysis approach. Skin Res Technol. 2002;8:276–281. doi: 10.1034/j.1600-0846.2001.00350.x. [DOI] [PubMed] [Google Scholar]
- 20.Stoecker WV, Li WW, Moss RH. Automatic detection of asymmetry in skin tumors. Comput Med Imag Graph. 1992;16:191–197. doi: 10.1016/0895-6111(92)90073-i. [DOI] [PubMed] [Google Scholar]
- 21.Ercal F, Chawla A, Stoecker WV, Lee HC, Moss RH. Neural network diagnosis of malignant melanoma from color images. IEEE Trans Biomed Eng. 1994;41:837–845. doi: 10.1109/10.312091. [DOI] [PubMed] [Google Scholar]
- 22.Moss RH, Stoecker WV, Lin S-J. Skin cancer recognition by computer vision. Comput Med Imag Graph. 1989;13:31–36. doi: 10.1016/0895-6111(89)90076-1. [DOI] [PubMed] [Google Scholar]
- 23.Haralick RM, Shapiro LG. Computer and robot vision. vol. 1. Addison-Wesley; New York: 1992. [Google Scholar]
- 24.Stoecker WV, Moss RH, Madsen RW, Lee HC, Ercal F, Umbaugh SE. Which computer-detected features of pigmented lesions best predict a diagnosis of malignant melanoma?. Proceedings of Melonoma Research, 4th World Conference on Melanoma; Sidney, Australia. 1997. pp. S13–S14. [Google Scholar]