Abstract
Background:
miR-15a and miR-16-1 have been identified as tumor suppressor genes in prostate cancer, but their safe and effective delivery to target cells is key to the successful use of this therapeutic strategy. RNA aptamer A10 has been used as a ligand, targeting prostate cancer cells that express prostate-specific membrane antigen (PSMA). Compared with A10, the binding of the second-generation RNA aptamer, A10-3.2, to PSMA is more efficient.
Methods:
A10-3.2 was investigated as a PSMA-targeting ligand in the design of a polyamidoamine (PAMAM)-based microRNA (miR-15a and miR-16-1) vector to prostate cancer cells. Using polyethyleneglycol (PEG) as a spacer, PAMAM was conjugated to aptamer (PAMAM-PEG-APT) and used as a vehicle for miRNA target delivery.
Results:
Luciferase assays of pGL-3 expression against PC3 (PSMA−) and LNCaP (PSMA+) cells demonstrated that the transfection efficiency of the synthesized DNA/PAMAM-PEG-APT complex was higher than that of the DNA/PAMAM-PEG complex. In addition, cell viability assays of LNCaP (PSMA+) cells showed that, with a N/P ratio of 15:1, the IC50 value of miRNA/PAMAM-PEG-APT was approximately 4.7-fold lower than that of miRNA/PAMAM-PEG.
Conclusion:
This PSMA-targeted system may prove useful in widening the therapeutic window and allow for selective killing of prostate cancer cells.
Keywords: miRNA, aptamer, polyamidoamine, prostate-specific membrane antigen, targeted delivery, prostate cancer
Introduction
Aptamers, ie, RNA or DNA oligonucleotides,1,2 are ligands that can bind target antigens with high affinity and specificity3 by folding into unique three-dimensional conformations.4,5 Aptamer A106 is a RNA aptamer that can bind to the extracellular domain of the prostate-specific membrane antigen (PSMA) commonly found on the cell surface of prostate cancer cells. Its capability for active binding and uptake by targeted cancer cells in vitro has been demonstrated.7–10 However, aptamer A10 has some shortcomings, such as difficult chemical synthesis, low stability, and low activity in vivo, which limit its efficacy for tumor-specific treatment. The second-generation aptamer, A10-3.2, has been synthesized recently to optimize aptamer A10.11 Aptamer A10-3.2 is obtained by truncating aptamer A10 from 71 to 39 nucleotides. Compared with aptamer A10, aptamer A10-3.2 is easily synthesized chemically, has enhanced silencing activity and specificity, and its in vivo kinetics can be optimized by modification.
A10-polyethylene glycol (PEG)-polyethyleneimine (PEI) has been synthesized recently and used as a target-specific intracellular delivery carrier for small interfering RNA (siRNA) in the treatment of prostate cancer.12 Although PEG-modified PEI has lower toxicity, its biological toxicity in vivo remains a difficult problem to tackle.13–16 For this reason, we chose a new material polyamidoamine (PAMAM) to replace PEI. PAMAM is a novel class of nanoscopic and spherical polymer that has been proven to be an efficient gene carrier.17 Positively charged PAMAM with primary amino surface groups has the inherent ability to associate with and condense RNA, and has been used for biocompatible and highly effective siRNA delivery.18,19 PAMAM encapsulated gold nanorods can enhance charge transfer and biocompatibility.20–22 Therefore, we used aptamer A10-3.2 as the PSMA-targeting ligand in our study to construct an aptamer-conjugated PAMAM in an attempt to achieve an efficiently targeted gene delivery to prostate cancer cells.
siRNA has been widely used for the treatment of prostate cancer. However, given that prostate cancer is the combined effect of disordered expression of multiple genes, single use of siRNA targeting specific genes is often ineffective.23–25 There are obvious difficulties using the RNA interference technique, but microRNA can solve these problems effectively. Recent reports26–29 show that miR-15a and miR-16-1 act as tumor suppressor genes in prostate cancer, and associate with the expression of BCL2, CCND1, and WNT3 A, thus promoting survival, proliferation, and invasion in prostate cancer cells. Therefore, the delivery of miR-15a and miR-16-1 could be useful for the treatment of prostate cancer.
In our study, aptamer A10-3.2 was used as a new ligand conjugated with PAMAM via bifunctional PEG to carry miR-15a and miR-16-1 and form an efficient PSMA-targeting system (miRNA/PAMAM-PEG-APT). A PSMA-positive prostate cancer cell line, LNCaP (PSMA+), was used as the model cell line, and a PSMA-negative prostate cancer cell line, PC3 (PSMA−), was used as a control to determine the specificity of the delivery system. The characteristics and anticancer effect of miRNA/PAMAM-PEG-APT were investigated in vitro.
Materials and methods
Materials
Materials used in this study were: PAMAM dendrimer (generation 5, 5% w/w solution in methyl alcohol, containing 128 surface primary amino groups [molecular weight 28,826]); dithiothreitol (DTT, Sigma-Aldrich, St Louis, MO); α-malemidyl-ω-N-hydroxysuccinimidyl polyethyleneglycol (NHS-PEG-MAL, molecular weight 3400, Nektar Therapeutics, Huntsville, AL); DyLight-633 NHS ester and micro-bicinchoninic acid protein assay kit (Pierce, Rockford, IL); plasmid pEGFP-N2 (Clontech, Palo Alto, CA) and a pGL3-control vector (Promega, Madison, WI) purified from bacteria using a Qiagen plasmid mega kit (Qiagen GmbH, Hilden, Germany); system and reporter lysis buffer (Promega); prostate carcinoma PC3 and LNCaP cell lines (American Tissue Type Culture); anti-PSMA aptamer (sequence 5′-GGGA GGACGAUGCGGAUCAGCCAUGUUUACGUCACUCC U-(CH2)6-S-S-(CH2)6-OH-3′ with 2′-fluoro pyrimidines) and nuclease-free water (RiboBio, Guangzhou, China); miRNA-15a (sequence 5′-UAGCAGCACAUAAUGGUUUGUG-3′ with 2′-fluoro pyrimidines); miRNA-16-1 (sequence 5′-UAGCAGCACGUAAAUAUUGGCG-3′ with 2′-fluoro pyrimidines), and negative control-miRNA (sequence 5′-UUCUCCGAACGUGUCACGUUTT-3′ with 2′-fluoropyrimidines, GenePharma, Shanghai, China). All the other materials were reagent grade and obtained from commercial sources.
Synthesis of PAMAM-PEG and PAMAM-PEG-APT
A 1:2 conjugate of PAMAM and NHS-PEG3400-MAL was prepared by a specific reaction between the primary amino groups on the surface of PAMAM and the NHS groups of the bifunctional PEG derivative (nPAMAM:nNHS-PEG3400-MAL = 1:2). The reaction was then performed in phosphate-buffered solution (pH 8.0) for 15 minutes at room temperature. The resulting conjugate, PAMAM-PEG, was purified by ultrafiltration using a 5 kDa molecular weight cutoff membrane to remove unreacted PEG, and the buffer was exchanged into phosphate-buffered solution (pH 7.0). The purified PAMAM-PEG obtained was lyophilized and stored at 4°C for use.
To prepare 3′-SH-modified aptamer (SH-APT), 0.1 μmol OH-(CH2)6-S-S-(CH2)6-APT was dissolved in 2.5 mL 100 mM DTT (pH 8.0), and incubated at room temperature for 30 minutes. The entire sample was loaded onto a Glen Gel-Pak™ 2.5 (Glen Research, Sterling, VA) desalting column, equilibrated with 25 mL 50 mM sodium phosphate (pH 6.0), allowed to drip through, and eluted with 2.5 mL sodium phosphate (pH 6.0). Finally, the purified conjugation of SH-APT was collected.
To conjugate the anti-PSMA aptamer with PAMAM-PEG, 0.2 μmol MAL-activated PAMAM-PEG was dissolved in 3 mL nuclease-free phosphate-buffered solution (pH 7.0), reacted with 0.1 μmol SH-APT for 12 hours at 4°C, and the resulting product, PAMAM-PEG-APT, was purified by ultrafiltration using a 20 kDa molecular weight cutoff membrane to remove unreacted aptamer and suspended in nuclease-free water. The anti-PSMA aptamer-conjugated PAMAM-PEG (PAMAM-PEG-APT) was stored at 4°C. For the synthesis of DyLight-633-labeled conjugate, PAMAM was first reacted with DyLight-633 in 100 mM NaHCO3 for 12 hours at 4°C, purified and identified as described previously.26 The corresponding conjugate, DyLight-633-labeled PAMAM-PEG-APT, was synthesized as described previously.
Characterization of PAMAM derivatives
The characteristics of PAMAM derivatives were determined by nuclear magnetic resonance (NMR) spectroscopy. PAMAM, PAMAM-PEG, and PAMAM-PEG-APT were solubilized in D2O and determined in a 300 mHz spectrometer (Varian, Palo Alto, CA).
Cellular uptake of DyLight-633-labeled dendrimers in PC3 and LNCaP
PC3 and LNCaP were seeded at a density of 8 × 104 cells/well in six-well plates (Corning-Coaster, Tokyo, Japan), cultured for 72 hours, and checked under the microscope for confluency and morphology. Wells with a similar cell density were chosen for the experiments. PC3 and LNCaP were then incubated with DyLight-633-labeled PAMAM-PEG-APT in a PAMAM concentration range of 0.04–1.20 μM for 60 minutes. Cells were washed three times with phosphate-buffered solution (pH 7.4) and visualized under a DMIL fluorescent microscope (Leica Camera AG, Solms, Germany). For quantitative analysis, PC3 and LNCaP treated as described above was trypsinized and centrifuged at 1600 rpm for 8 minutes to obtain a cell pellet, which was subsequently resuspended in phosphate-buffered solution (pH 7.4) and analyzed using a flow cytometer (FACSCalibur, San Jose, CA) equipped with an argon ion laser (638 nm) as the excitation source. The fluorescence intensity of DyLight-633 was collected at 658 nm. For each sample, 10,000 events were collected and data were analyzed by CellQuest software. Untreated PC3 and LNCaP served as the control. Living cells were defined by gating the major population of cells, and only cells within this gate were analyzed. The mean fluorescence intensity of the cells was calculated using a histogram plot.
Preparation of DNA/PAMAM-PEG-APT complexes
DNA/PAMAM-PEG-APT complexes were prepared by gently mixing (6 μg/pEGFP/500 μL DI water) with various amounts of polymer stock solution, corresponding to N/P ratios ranging from 1 to 25, into a final volume of 1000 μL. The complexes were incubated for 30 minutes at room temperature prior to analysis. The sizes and zeta potentials of DNA/PAMAM-PEG-APT complexes formed in phosphate-buffered saline (10 mM, pH 7.4) at various N/P ratios (1–25) were measured in triplicate using a Zetasizer Nano-ZS90 (Malvern Instruments Ltd, Worcestershire, UK).
Efficiency of gene expression in PC3 and LNCaP
PC3 and LNCaP cell lines were cultured in RPMI 1640 medium (Gibco, Invitrogen, Carlsbad, CA) supplemented with 10% fetal bovine serum and 1% antibiotics at 37°C in a humidified atmosphere with 5% CO2. PC3 and LNCaP cells were seeded into six-well plates (Corning-Coaster, Tokyo, Japan) at a density of 3 × 105 cells per well and incubated at 37°C for 24 hours (to reach 70% confluence) prior to transfection. Complexes containing 3 μg DNA, with PAMAM to DNA at various N/P ratios (1–25), were added to the cells in fetal bovine serum-free medium, and the mixture was incubated at 37°C for 1 hour. After 48 hours, fluorescence images of the gene expression products, ie, green fluorescent protein, were acquired and photographed under a fluorescence microscope. The efficiency of gene transfer was quantitatively determined by luciferase activity using the pGL3-control vector. After 48 hours of transfection, activity of the gene expression product, luciferase, was quantified using a luciferase assay system (Promega) following the manufacturer’s instructions. Briefly, cells were firstly lysed in 150 μL 1 × cell culture lysis reagent for 30 minutes at room temperature, the luciferase activity was measured by a luminometer (Promega E6080), and finally the protein content was measured using a micro-bicinchoninic acid protein assay kit.
Western blot analysis
To assess the downregulation of Bcl-2, Cyclin D1, and Wnt3a gene in LNCaP cells, cells were transfected with miRNA(miRNA-15a:miRNA-16-1 = 1:1)/PAMAM-PEG-APT complexes and negative control-miRNA (NC-miRNA)/PAMAM-PEG-APT complexes. Cells were harvested at 24, 48, and 72 hours post-transfection and lysed in cold RIPA buffer (50 mM Tris, pH 8.0, 150 mM NaCl, 1 mM EGTA, and 0.25% sodium deoxycholate), and then the lysate was incubated for 15 minutes at 4°C and removed by centrifugation at 12,000 rpm for 15 minutes. The supernatant was analyzed for protein concentrations using the Bradford assay (Bio-Rad, Hercules, CA). An equal amount (10 mg) of protein was subjected to electrophoresis on sodium dodecyl sulfate-polyacrylamide gels and then transferred to polyvinylidene fluoride membrane (Millipore, Billerica, MA). The blotted membrane was immunostained with antibodies specific to anti-Bcl-2 (BioWorld, Visalia, CA), anti-Cyclin D1 (BioWorld) and anti-Wnt3a antibodies (R&D Systems Inc, Minneapolis, MN). Blots were developed by the enhanced chemiluminescence reagents (Amersham Pharmacia, Buckinghamshire, UK) and visualized by the Gene-Genius Imaging System (Syngene, Frederick, MD).
In vitro anticancer effect
The antiproliferation effect of miRNA/PAMAM-PEG-APT on LNCaP and PC3 cells was evaluated by a cell counting kit-8 (CCK-8, Dojindo, Japan). LNCaP cells and PC3 cells were seeded in 96-microwell plates at a density of 1 × 104 cells per well and incubated at 37°C for 24 hours. Cells were treated with various concentrations (0.02–50 μM) of complexes: miRNA(miRNA-15-a:miRNA-16-1 = 1:1)/PAMAM-PEG-APT, NC-miRNA/PAMAM-PEG-APT and miRNA (miRNA-15-a:miRNA-16-1 = 1:1)/PAMAM-PEG for 72 hours. After that, 10 μL of CCK-8 was added to each well and incubated for another 2 hours. The absorbance of each well was determined with a microplate reader (Thermo Scientific, Rockford, IL) and a standard curve was drawn according to the absorbance and cell number, and each sample was tested in triplicate. The antiproliferation effect was expressed as the IC50 value as the concentration that inhibits cell growth by 50% as compared with nontreated controls. IC50 value was calculated according to the equation: y = bottom × (top-bottom)/[1 + 10(logIC50−x) × Hill slope], where bottom and top are the minimum and maximum y-axis values, respectively, of a plateau in the curve, and Hill slope is the steepness of the curve using GraphPad 5.0 PRISM® (GraphPad Software Inc, San Diego, CA).
Statistical analysis
The data were presented as means ± standard deviation. Statistically significant differences were determined using two-sample t-tests and analysis of variance, with P < 0.05 as the significance level.
Results and discussion
Characterization of PAMAM derivatives
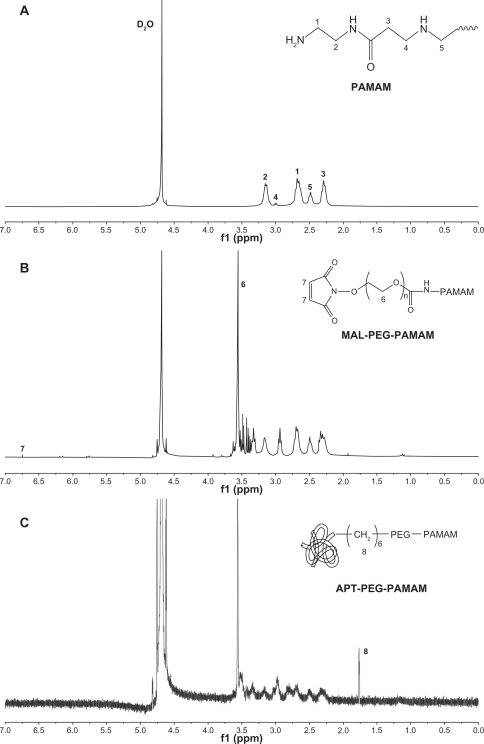
The solvent peak of D2O was found at 4.65 ppm (Figure 1). The methylene protons of branching units of PAMAM had multiple peaks between 2.2 and 3.4 ppm (Figure 1A), which is consistent with the NMR spectrum of PAMAM (generation = 5) reported previously.30 The NMR spectrum of PAMAM-PEG had multiple peaks of the repeat units in PEG at 3.5 ppm and a characteristic peak of the MAL group in PEG at 6.7 ppm (Figure 1B), confirming conjugation of PEG to PAMAM. However, the MAL peak disappeared in the NMR spectrum of PAMAM-PEG-APT, even when the repeat units of PEG still presented a sharp peak at 3.5 ppm (Figure 1C), indicating that the MAL group had reacted with the thiol group of SH-APT. The NMR spectra demonstrated successful synthesis of PAMAM-PEG-APT. In addition, the integrated areas of NMR peaks were used to quantify the number of PEG chains per PAMAM, with the assumption of 280 methylene protons per PEG and 2032 per PAMAM. As shown in Figure 1B, PAMAM-PEG had a PAMAM/PEG proton ratio of 0.74, implying an average of 2.1 PEG chains per PAMAM. In our study, we used the small hairpin method to modify aptamers so that aptamers could connect to PAMAM under mild conditions, thus maintaining optimal biological activity of the aptamers and avoiding loss of the target. The synthesis was made simple by using PEG as a linker. In addition, a long circulation was ensured, because parts of the PEG were not connected to the aptamers.
Figure 1.
NMR spectra of (A) PAMAM, (B) PAMAM-PEG, and (C) PAMAM-PEG-APT in D2O at 300 mHz.
Abbreviations: PAMAM, polyamidoamine; PEG, polyethylene glycol; APT, aptamer.
Cellular uptake of DyLight-633-labeled dendrimers in PC3 and LNCaP
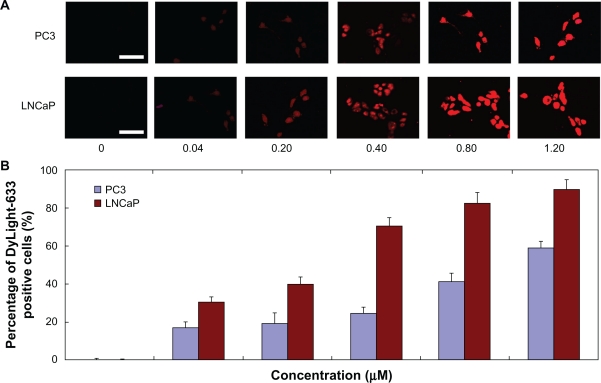
The results of cellular uptake for DyLight-633-labeled dendrimers are shown using fluorescent images and quantified with a percentage of DyLight-633 positive cells. The fluorescence intensity corresponded to that of the DyLight-633-labeled PAMAM-PEG-APT concentrations (Figure 2). After exposure of PC3 and LNCaP to different concentrations of DyLight-633-labeled PAMAM-PEG-APT (0.04–1.20 μM) for 60 minutes, the percentage of DyLight-633-positive PC3 and LNCaP cells increased from 16.97% to 59.07% and from 30.61% to 89.91%, respectively (Figure 2B). The percentage of DyLight-633-positive LNCaP cells of PAMAM-PEG-APT was much higher than that of PC3 cells at each concentration (Figure 2B), suggesting that conjugation of aptamer facilitated uptake of the vector by LNCaP more efficiently. Because the surfaces of the PC3 cells had low expression of PSMA, aptamers could not be mediated by PAMAM-PEG-APT into the cells, resulting in lower efficiency than for LNCaP.
Figure 2.
(A) Fluorescence microscopy images (scale bar 50 μm) and (B) fluorescent-activated cell sorting analysis (n = 3, error bars represent a standard deviation) after a 60-minute incubation of DyLight-633-labeled PAMAM-PEG-APT as a function of concentration range against PC3 and LNCaP cells, respectively.
Abbreviations: PAMAM, polyamidoamine; PEG, polyethylene glycol; APT, aptamer.
Characterization of DNA/PAMAM-PEG-APT complexes
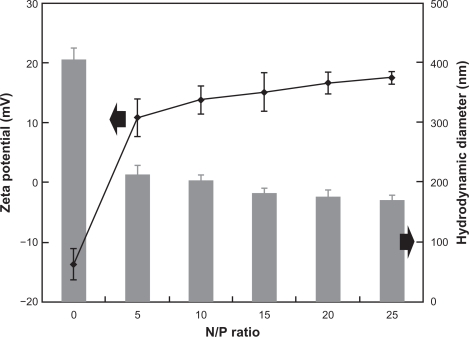
In our study, we used PEG as a spacer between PAMAM and aptamer to disperse dendrimer molecules by increasing hydrophilicity and reducing nonspecific interactions with the cellular membrane.31 Aptamer was coupled to the remote end of the PEG chain, triggering the receptor-mediated mechanism to increase the accumulation of DNA/PAMAM-PEG-APT in prostate cancer. The sizes and zeta potentials of the pEGFP/PAMAM-PEG-APT complexes were analyzed using the Zetasizer Nano-ZS90 over a range of N/P ratios. As shown in Figure 3, the average size of the pEGFP/PAMAM-PEG-APT complexes decreased and the zeta potential increased with increasing N/P ratios. At higher N/P ratios, the smaller size resulted from formation of more compact structures, owing to the higher ionic interactions and the net electrostatic repulsive forces between the complexes.
Figure 3.
Sizes and zeta potentials of pEGFP/PAMAM-PEG-APT complexes at various N/P ratios.
Abbreviations: PAMAM, polyamidoamine; PEG, polyethylene glycol; APT, aptamer.
Efficiency of gene expression in PC3 (PSMA−) and LNCaP (PSMA+)
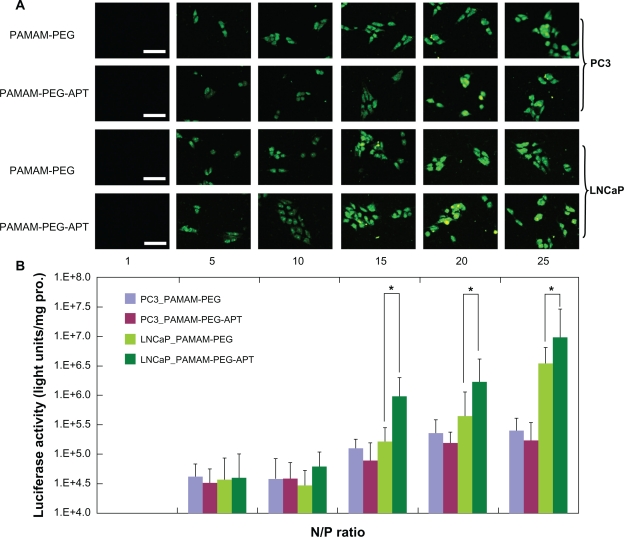
To observe the targeted delivery and transfection efficiency of PAMAM-PEG-APT in PSMA-overexpressing cells, LNCaP cells and PC3 cells were treated with DNA/PAMAM-PEG and DNA/PAMAM-PEG-APT complexes in series of N/P ratios using 3 μg pEGFP (Figure 4A) or pGL3-Control Vector (Figure 4B). The result of luciferase activity assay indicated that DNA/PAMAM-PEG-APT complexes resulted in higher gene expression than the DNA/PAMAM-PEG complexes in LNCaP cells, whereas the gene expression was rarely observed in PC3 cells transfected by DNA/PAMAM-PEG-APT complexes at various N/P ratios. As shown in Figure 4B, the luciferase activity of PAMAM-PEG-APT (1.195906 × 106 units/mg protein) was six-fold higher than that of PAMAM-PEG (1.82086 × 105 units/mg protein) in LNCaP cells at a N/P ratio of 15, but no difference was observed in PC3 cells. This might attribute to the targeting head group aptamer, which could enhance the transfection efficiency by selectively binding to PSMA-overexpressing cells through the aptamer-mediated approach.32 The strong positive charges of the complexes provided higher transfection efficiency but also could induce cytotoxicity by disrupting and solubilizing cell membranes.33 To balance the high gene expression with cytotoxicity, we set the optimal N/P ratio at 15 for the synthesis of DNA/PAMAM-PEG-APT complexes.
Figure 4.
(A) Fluorescence microscopy images (scale bar 50 μm) and (B) luciferase activity analysis (n = 4, error bars represent the standard deviation) after transfection of pGL-3/PAMAM-PEG or pGL-3/PAMAM-PEG-APT as a function of N/P ratios against PC3 and LNCaP cells, respectively. *P < 0.05.
Abbreviations: PAMAM, polyamidoamine; PEG, polyethylene glycol; APT, aptamer.
Western blot analysis
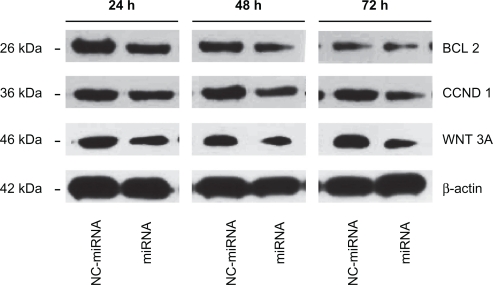
Figure 5 shows the results of Western blot of Bcl-2, Cyclin D1, and Wnt3a, the direct target of miRNA-15a and miRNA-16-1,26 which were determined at 24, 48, and 72 hours following transfection. Their expression began decreasing after 48 hours of miRNA transfection, and decreased significantly after 72 hours, especially for Wnt3a, whereas this phenomenon was rarely observed in NC-miRNA transfection. Thus, downregulation of Bcl-2, Cyclin D1, and Wnt3a by the delivery of miRNA, whose functions are directly associated with cancer progression and invasion,26 is expected to increase cell apoptosis. It has been demonstrated that PAMAM-PEG-APT can carry miRNA effectively into cells to have an inhibitory effect, so use of miR-15a and miR-16-1 in the treatment of prostate cancer may become a reality.
Figure 5.
Bcl-2, Cyclin D1, and Wnt3a knock down following miRNA/PAMAM-PEG-APT complexes delivery to LNCaP cells. The cells were treated with miRNA/PAMAM-PEG-APT (miRNA) and NC-miRNA/PAMAM-PEG-APT (NC-miRNA) complexes and processed for immunoblotting with anti-Bcl-2, anti-Cyclin D1, and anti-Wnt3a antibodies at 24, 48, and 72 hours post-transfection. β-actin was a loading control.
Abbreviations: PAMAM, polyamidoamine; PEG, polyethylene glycol; APT, aptamer.
In vitro anticancer effect
To evaluate the anticancer potential of miRNA/PAMAM-PEG-APT, we performed a series of in vitro cytotoxicity assays using the CCK8 assay to determine the IC50 value. LNCaP cells were treated with miRNA/PAMAM-PEG-APT, NC-miRNA/PAMAM-PEG-APT, and miRNA/PAMAM-PEG complexes (0.02–50 μM). The results show that cell growth was inhibited in a dose-dependent manner (Figure 6). The calculated IC50 value was 0.0144 μM, 0.788 μM, and 556.47 μM, respectively. Therefore, miRNA/PAMAM-PEG-APT had an approximately 5.47-fold lower IC50 value than miRNA/PAMAM-PEG. It could be concluded from the experimental results of cellular uptake of DyLight-633-labeled dendrimers and efficiency of gene expression in LNCaP as described earlier that PAMAM-PEG-APT could raise the gene transfection rate in LNCaP, so we may infer that PAMAM-PEG-APT could carry more miRNA into LNCaP. In addition, the results of Western blot analysis showed that the more miRNA was carried into LNCaP, the further Bcl-2, Cyclin D1, and Wnt3a were downregulated, which explains why miRNA/PAMAM-PEG-APT had a lower IC50 value. Thus, there is reason to believe that miRNA/PAMAM-PEG-APT could become a novel nanotargeted drug delivery system for the treatment of prostate cancer.
Figure 6.
Cell viabilities of LNCaP cells treated with miRNA/PAMAM-PEG-APT, NC-miRNA/PAMAM-PEG-APT, miRNA/PAMAM-PEG complexes, respectively, (n = 3, error bars represent the standard deviation).
Abbreviations: PAMAM, polyamidoamine; PEG, polyethylene glycol; APT, aptamer.
Conclusion
In this study, we successfully constructed an efficient target gene delivery system mediated by the second-generation aptamer (PAMAM-PEG-APT), which could synergistically induce selective cell death of prostate cancer cells by loading miR-15a and miR-16-1. PAMAM-PEG-APT could effectively deliver miRNA to prostate cancer cells overexpressing PSMA, resulting in tumoricidal efficacy as verified by cell viability assays. It should be emphasized that because PAMAM-PEG-APT uses the second-generation APT A10-3.2, synthesis of the system becomes much easier and biological targeting becomes even stronger. In addition, use of PAMAM decreases toxicity of the system and endows it with better biocompatibility. Replacement of siRNA with miRNA provides multiple-target inhibition of prostate cancer and enhances the anticancer effect of the system. Therefore, this research may prove to be a significant contribution to the use of nanocarrier-based targeted delivery of miRNAs for the treatment of cancer.
Acknowledgments
We acknowledge the financial support from the National Natural Science Foundation of China (Project No. 30973459 and 81072100).
Footnotes
Disclosure
The authors report no conflicts of interest in this work.
References
- 1.Ellington AD, Szostak JW. In vitro selection of RNA molecules that bind specific ligands. Nature. 1990;346(6287):818–822. doi: 10.1038/346818a0. [DOI] [PubMed] [Google Scholar]
- 2.Tuerk C, Gold L. Systematic evolution of ligands by exponential enrichment: RNA ligands to bacteriophage T4 DNA polymerase. Science. 1990;249(4968):505–510. doi: 10.1126/science.2200121. [DOI] [PubMed] [Google Scholar]
- 3.Wu F, Wuensch SA, Azadniv M, Ebrahimkhani MR, Crispe IN. Galactosylated LDL nanoparticles: A novel targeting delivery system to deliver antigen to macrophages and enhance antigen specific T cell responses. Mol Pharm. 2009;6(5):1506–1517. doi: 10.1021/mp900081y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ishida S, Usui T, Yamashiro K, et al. VEGF164-mediated inflammation is required for pathological, but not physiological, ischemia-induced retinal neovascularization. J Exp Med. 2003;198(3):483–489. doi: 10.1084/jem.20022027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brody EN, Gold L. Aptamers as therapeutic and diagnostic agents. J Biotechnol. 2000;74(1):5–13. doi: 10.1016/s1389-0352(99)00004-5. [DOI] [PubMed] [Google Scholar]
- 6.Lupold SE, Hicke BJ, Lin Y, Coffey DS. Identification and characterization of nuclease-stabilized RNA molecules that bind human prostate cancer cells via the prostate-specific membrane antigen. Cancer Res. 2002;62(14):4029–4033. [PubMed] [Google Scholar]
- 7.Cheng J, Teply BA, Sherifi I, et al. Formulation of functionalized PLGA-PEG nanoparticles for in vivo targeted drug delivery. Biomaterials. 2007;28(5):869–876. doi: 10.1016/j.biomaterials.2006.09.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Farokhzad OC, Jon S, Khademhosseini A, Tran TN, Lavan DA, Langer R. Nanoparticle-aptamer bioconjugates: A new approach for targeting prostate cancer cells. Cancer Res. 2004;64(21):7668–7672. doi: 10.1158/0008-5472.CAN-04-2550. [DOI] [PubMed] [Google Scholar]
- 9.Dhar S, Gu FX, Langer R, Farokhzad OC, Lippard SJ. Targeted delivery of cisplatin to prostate cancer cells by aptamer functionalized Pt(IV) prodrug-PLGA-PEG nanoparticles. Proc Natl Acad Sci USA. 2008;105(45):17356–17361. doi: 10.1073/pnas.0809154105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tong R, Yala L, Fan TM, Cheng J. The formulation of aptamer-coated paclitaxel-polylactide nanoconjugates and their targeting to cancer cells. Biomaterials. 2010;31(11):3043–3053. doi: 10.1016/j.biomaterials.2010.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dassie JP, Liu XY, Thomas GS, et al. Systemic administration of optimized aptamer-siRNA chimeras promotes regression of PSMA-expressing tumors. Nat Biotechnol. 2009;27(9):839–849. doi: 10.1038/nbt.1560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kim E, Jung Y, Choi H, et al. Prostate cancer cell death produced by the co-delivery of Bcl-xL shRNA and doxorubicin using an aptamer-conjugated polyplex. Biomaterials. 2010;31(16):4592–4599. doi: 10.1016/j.biomaterials.2010.02.030. [DOI] [PubMed] [Google Scholar]
- 13.Choi YJ, Kang SJ, Kim YJ, Lim YB, Chung HW. Comparative studies on the genotoxicity and cytotoxicity of polymeric gene carriers polyethylenimine (PEI) and polyamidoamine (PAMAM) dendrimer in Jurkat T-cells. Drug Chem Toxicol. 2010;33(4):357–366. doi: 10.3109/01480540903493507. [DOI] [PubMed] [Google Scholar]
- 14.Mortimer M, Kasemets K, Heinlaan M, Kurvet I, Kahru A. High throughput kinetic Vibrio fischeri bioluminescence inhibition assay for study of toxic effects of nanoparticles. Toxicol In Vitro. 2008;22(5):1412–1417. doi: 10.1016/j.tiv.2008.02.011. [DOI] [PubMed] [Google Scholar]
- 15.Jang SH, Choi SJ, Oh JH, et al. Nonviral gene delivery to human ovarian cancer cells using arginine-grafted PAMAM dendrimer. Drug Dev Ind Pharm. 2011;37(1):41–46. doi: 10.3109/03639045.2010.489563. [DOI] [PubMed] [Google Scholar]
- 16.Beyerle A, Irmler M, Beckers J, Kissel T, Stoeger T. Toxicity pathway focused gene expression profiling of PEI-based polymers for pulmonary applications. Mol Pharm. 2010;7(3):727–737. doi: 10.1021/mp900278x. [DOI] [PubMed] [Google Scholar]
- 17.Zhang XQ, Wang XL, Huang SW, et al. In vitro gene delivery using polyamidoamine dendrimers with a trimesyl core. Biomacromolecules. 2005;6(1):341–350. doi: 10.1021/bm040060n. [DOI] [PubMed] [Google Scholar]
- 18.Luo D, Haverstick K, Belcheva N, Han E, Saltzman WM. Poly(ethylene glycol)-conjugated PAMAM dendrimer for biocompatible, high-efficiency DNA delivery. Macromolecules. 2002;35(9):3456–3462. [Google Scholar]
- 19.Braun CS, Vetro JA, Tomalia DA, Koe GS, Koe JG, Middaugh CR. Structure/function relationships of polyamidoamine/DNA dendrimers as gene delivery vehicles. J Pharm Sci. 2005;94(2):423–436. doi: 10.1002/jps.20251. [DOI] [PubMed] [Google Scholar]
- 20.Ding H, Yong KT, Roy I, et al. Gold nanorods coated with multilayer polyelectrolyte as contrast agents for multimodal imaging. J Phys Chem C. 2007;111(34):12552–12557. [Google Scholar]
- 21.Bonoiu AC, Mahajan SD, Ding H, et al. Nanotechnology approach for drug addiction therapy: Gene silencing using delivery of gold nanorod-siRNA nanoplex in dopaminergic neurons. Proc Natl Acad Sci U S A. 2009;106(14):5546–5550. doi: 10.1073/pnas.0901715106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhang ZQ, Pan BF, Wang K, et al. Electrochemical property and cell toxicity of gold electrode modified by monolayer PAMAM encapsulated gold nanorods. Nano Biomed Eng. 2010;2(3):182–188. [Google Scholar]
- 23.Kawasaki H, Wadhwa R, Taira K. World of small RNAs: From ribozymes to siRNA and miRNA. Differentiation. 2004;72(2–3):58–64. doi: 10.1111/j.1432-0436.2004.07202006.x. [DOI] [PubMed] [Google Scholar]
- 24.Pontes O, Pikaard CS. siRNA and miRNA processing: New functions for Cajal bodies. Curr Opin Genet Dev. 2008;18(2):197–203. doi: 10.1016/j.gde.2008.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lin SL, Chang D, Ying SY. Asymmetry of intronic pre-miRNA structures in functional RISC assembly. Gene. 2005;356:32–38. doi: 10.1016/j.gene.2005.04.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bonci D, Coppola V, Musumeci M, et al. The miR-15a-miR-16-11 cluster controls prostate cancer by targeting multiple oncogenic activities. Nat Med. 2008;14(11):1271–1277. doi: 10.1038/nm.1880. [DOI] [PubMed] [Google Scholar]
- 27.Clevers H. Wnt/beta-catenin signaling in development and disease. Cell. 2006;127(3):469–480. doi: 10.1016/j.cell.2006.10.018. [DOI] [PubMed] [Google Scholar]
- 28.Dhanasekaran SM, Barrette TR, Ghosh D, et al. Delineation of prognostic biomarkers in prostate cancer. Nature. 2001;412(6849):822–826. doi: 10.1038/35090585. [DOI] [PubMed] [Google Scholar]
- 29.Sherr CJ. Cancer cell cycles. Science. 1996;274(5293):1672–1677. doi: 10.1126/science.274.5293.1672. [DOI] [PubMed] [Google Scholar]
- 30.Huang R, Ke W, Liu Y, Jiang C, Pei Y. The use of lactoferrin as a ligand for targeting the polyamidoamine-based gene delivery system to the brain. Biomaterials. 2008;29(2):238–246. doi: 10.1016/j.biomaterials.2007.09.024. [DOI] [PubMed] [Google Scholar]
- 31.Nama HY, Nama K, Hahn HJ, et al. Biodegradable PAMAM ester for enhanced transfection efficiency with low cytotoxicity. Biomaterials. 2009;30(4):665–673. doi: 10.1016/j.biomaterials.2008.10.013. [DOI] [PubMed] [Google Scholar]
- 32.Bagalkot V, Farokhzad OC, Langer R, Jon S. An aptamer-doxorubicin physical conjugate as a novel targeted drug-delivery platform. Angew Chem Int Ed Engl. 2006;45(48):8149–8152. doi: 10.1002/anie.200602251. [DOI] [PubMed] [Google Scholar]
- 33.Fischer D, Li Y, Ahlemeyer B, Krieglstein J, Kissel T. In vitro cytotoxicity testing of polycations: Influence of polymer structure on cell viability and hemolysis. Biomaterials. 2003;24(7):1121–1131. doi: 10.1016/s0142-9612(02)00445-3. [DOI] [PubMed] [Google Scholar]