Abstract
Background
Asthma is a common disease of children with a complex genetic origin. Understanding the genetic basis of asthma susceptibility will allow disease prediction and risk stratification.
Objective
We sought to identify asthma susceptibility genes in children.
Methods
A nested case-control genetic association study of children of Caucasian European ancestry from a birth cohort was conducted. Single nucleotide polymorphisms (SNPs, n=116,024) were genotyped in pools of DNA samples from cohort children with physician-diagnosed asthma (n=112) and normal controls (n=165). A genomic region containing the ATPAF1 gene was significantly associated with asthma. Additional SNPs within this region were genotyped in individual samples from the same children and in eight independent study populations consisting of Caucasian, African American, Hispanic, or other ancestries. SNPs were also genotyped or imputed in two consortia control populations. ATPAF1 expression was measured in bronchial biopsies from asthmatics and controls.
Results
Asthma was associated with a cluster of SNPs and SNP haplotypes containing the ATPAF1 gene with two SNPs achieving significance at a genome-wide level (p=2.26×10−5 to 2.2×10−8). Asthma severity was also associated with SNPs and haplotypes in the primary population. SNP and/or gene-level associations were confirmed in the four non-Hispanic populations. Haplotype associations were confirmed in the non-Hispanic populations (p=0.045 to 0.0009). ATPAF1 total RNA expression was significantly (p<0.01) higher in bronchial biopsies from asthmatics than controls.
Conclusion
Genetic variation in the ATPAF1 gene predisposes children of different ancestry to asthma.
Keywords: asthma, ATPAF1, children, gene, genetic, genome-wide association, purinergic, respiratory, single nucleotide polymorphism, SNP
INTRODUCTION
Asthma is a debilitating chronic inflammatory disease of the conducting airways whose symptoms often manifest early in childhood. Many affected children struggle with this disease throughout their lives. Asthma is remarkably common, with the prevalence in children exceeding 30% in some parts of the world.1, 2 Indeed, it is a disease of concern world-wide with particularly high incidence in Northwestern Europe, the USA, and in some populations of Hispanic ancestry.2–4 Gene variations, in tandem with environmental factors, are believed to be the primary drivers behind asthma development and symptom exacerbations. Therefore, we undertook a study to determine susceptibility genes for asthma in a birth cohort of children from the UK with the rationale that understanding genetic factors will allow us to predict disease risk. Despite numerous studies, few genes have emerged as underlying asthma in the majority of populations examined5–7 thus, the hunt for asthma susceptibility genes continues.
There are a growing number of populations thoroughly characterized for asthma and related phenotypes and several of these have been extensively genotyped. Consequently, these populations provide a powerful means for assessing the genetic contributions to asthma. The Isle of Wight birth cohort was established over 20 years ago for the purpose of investigating asthma during childhood8 and is the primary population in this study. The replication populations that were examined for this report are similarly well-established, providing access to children with asthma and their families. The more recent genetic data generated in these populations can provide a new dimension to our understanding of asthma in children.
Here we report a multi-stage genetic association study for childhood asthma in the Isle of Wight birth cohort as the primary population with replication studies conducted in eight additional groups. Support for an associated gene was pursued by comparing expression levels between asthmatics and controls.
MATERIAL AND METHODS
Study design
The objective of our study was to identify asthma susceptibility genes in the Isle of Wight birth cohort of children.8 We used an efficient and sequential strategy to optimize the search for asthma susceptibility genes. We first examined approximately 100K SNPs across the genome using pooled DNA samples from a case-control subset of the cohort. A linkage disequilibrium (LD) block on chromosome 1p33-p32.31 was significantly associated with asthma. Additional SNPs within this region were then genotyped in individual samples from the same children. The region of interest was next examined in additional populations that had been characterized for asthma and genotyped genome-wide or were able to specifically genotype selected SNPs for replication purposes. These SNPs were also genotyped or imputed in two consortia control populations. Finally, functional relevance of the associated gene, ATPAF1 (ATP synthase mitochondrial F1 complex assembly factor 1), was assessed by gene expression studies.
Subjects
The primary study population was a case-control subset of children from the Isle of Wight 1989–1990 birth cohort study. Asthma diagnosis was assessed at age 10 years and controls randomly selected from among cohort children who had never wheezed nor been given a diagnosis of asthma in their life. In addition, asthma severity in the Isle of Wight subjects was categorized based on the Global Initiative for Asthma (Update 2007: GINA Report) classification scheme. The additional populations of asthmatic children that were examined for replication purposes have been described elsewhere9–14 and their key characteristics are summarized in Table I. Briefly, the Wessex population consisted of families with at least two biologic siblings with a current physician’s diagnosis of asthma.14 The publically available SHARP data used were the trios from the Childhood Asthma Research and Education (CARE) network15, 16 and the Childhood Asthma Management Program (CAMP) project.9, 13, 16 The two populations from the Genetics of Asthma in Latino Americans (GALA) study10 consisted of asthma cases and both parents. The Mexico Childhood Asthma Study11, 17 was also a case-parent trio design. To enhance the power of our study additional consortia controls were acquired from the Wellcome Trust Case Control Consortium (WTCCC)18 and the Genetics of Systemic Lupus Erythematosus (SLEGEN) study.19 The asthma status of the consortia controls was unknown, therefore, their sample sizes were adjusted to account for potential misclassification based on a predicted asthma prevalence rate of 15%.
Table I.
Characteristics of the Isle of Wight primary population, replication populations, and consortia controls
| Isle of Wight 1989–1990 Birth Cohort (IOW) | Wessex family study | Childhood Asthma Management Program (CAMP) | Childhood Asthma Research and Education (CARE) | Genetics of Asthma in Latino Americans (GALA) | Mexico Childhood Asthma Study | International Consortium for SLE Genetics (SLEGEN) | Wellcome Trust Case Control Consortium (WTCCC) | ||
|---|---|---|---|---|---|---|---|---|---|
| Location | UK | UK | US | US | PR & US | MX & US | MX | UK & US | UK |
| Ancestry | Caucasian | Caucasian | Caucasian, African American, Hispanic, other | Caucasian, African American, Hispanic, other | Puerto Rican | Mexican | Mexican | Caucasian | Caucasian |
| Population structure | Nested case-control | Family-based | Case-parent trios | Case-parent trios | Case-parent trios | Case-parent trios | Case-parent trios | Controls | Controls |
| Numbers | 112 cases; 165 controls | 341 families; 1,481 individuals | 442 triosa | 131 triosb | 395 trios | 298 trios | 492 trios | 3,471 controls | 3,004 controls |
| Age of cases | 10 yr | 5–21 yr | 5–12 yr | 6–17 yr | 8–40 yrc | 8–40 yrc | 5–17 yr | N/A | N/A |
| Asthma criteria | PD asthma + symptoms | PD asthma + medications | asthma symptoms or medications | asthma symptoms | PD asthma + symptoms | PD asthma + symptoms | PD asthma | N/A | N/A |
Number of CAMP affected offspring trios per ancestral group: Caucasian n=334, African American n=42, Hispanic n=30, other n=36.
Number of CARE affected offspring trios per ancestral group: Caucasian n= 95, African American n=10, Hispanic n=16, other n=10.
Asthma onset during childhood
PD=Physician diagnosed
Gene expression studies were conducted on bronchial biopsy samples collected from asthmatic and control individuals from the Montreal area of Quebec, Canada.20, 21
Ethics approval was granted by local research ethics committees for the Isle of Wight population and written (parental) consent was obtained. Ethics approval for each of the other populations has been reported previously9–14 and permission for access to the consortia and SHARP data was obtained (study accession: phs000166.v2.p1).22
Statistical analyses
Hybridization intensity comparisons of the case and control pools were used to identify significant allele frequency differences for each SNP.23 Z2 p-values were calculated and then ordered as a means for ranking all SNPs. A 40 kb sliding window identified clusters of significant SNPs. An allelic model was used in the analysis of pooled SNP data as individual genotypes were not available.
Asthma associations with individual polymorphisms in Isle of Wight subjects were determined by chi-square tests of an allelic model implemented with PLINK software (v.1.0).24 Odds ratios for SNPs were calculated via SNP.assoc (v.1.4)25 implemented in R (v.2.5.1).26 Chi-square p-values determined with PLINK software were also utilized to detect asthma associations with haplotypes in Isle of Wight and Wessex subjects. Regression models were used to compute odds ratios for the haplotypes for the case-control data using SNPassoc (v.1.4) 25 and haplo.stats (v.1.3.8)27 programs implemented in R (v.2.5.1), 26 incorporating haplotype ambiguity.
SNPs in the region of interest were examined for associations with asthma in the replication populations (methods details in Table II and online repository). Genotype data from prior genome wide association studies (GWAS) for families in the CAMP9, CARE15, GALA10, and Mexico Childhood Asthma studies28 were analyzed across the region of interest in or near the ATPAF1-C1ORF223-KIAA0494 LD block. GWAS data were not available for the Wessex study, thus, specifically genotyped in the Wessex subjects were the tag SNPs examined in the Isle of Wight individuals.
Table II.
Genetic associations for asthma in the Isle of Wight and replication populations
| Primary population | Replication populations | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Population | IOW pooled samples |
IOW individual samples |
Wessex | CAMP + CARE | GALA | Mexico Childhood Asthma Study |
|||||
| Genotyping platform | Affymetrix 100K GeneChip array |
Custom Illumina Goldengate |
Kbiosciences Kaspar Competitive PCR |
Affymetrix 6.0 | ABI Taqman and Affymetrix 6.0† |
Illumina 550 |
|||||
| Model | Allelic | Allelic | Allelic | Allelic | Allelic | Allelic | |||||
| Software | LatteThunder | PLINK | PLINK | PDTphase | PBAT | PLINK | |||||
| Statistical test | Z2 p-valueb | χ2 p-value | TDT p-value | PDT p-value | FBAT p-valuec | TDT p- value |
|||||
| SNP | Chr 1 positionsa |
Caucasian European |
Caucasian European |
Caucasian European |
Caucasian American |
African American |
Hispanic | Other ethnicity |
Puerto Rican |
Mexican | Mexican |
| rs1258000 | 46866854 | - | 0.0362 | 0.1113 | - | - | - | - | - | - | 0.4503 |
| rs2289447 | 46890755 | 2.20×10−8 | 0.0180 | 0.1216 | - | - | - | - | (0.5789) | (0.6307) | - |
| rs620431 | 46890776 | - | 0.0063 | 0.2476 | - | - | - | - | - | - | - |
| rs1150068 | 46891505 | 0.0034 | 0.0099 | 0.2286 | - | - | - | - | (0.6289) | (0.7501) | - |
| rs629412 | 46893260 | - | - | - | 0.03389 | 0.2568 | 0.1573 | 0.8084 | - | - | - |
| rs654509 | 46899758 | - | - | - | 0.01141 | 1 | 0.3173 | 1 | - | - | - |
| rs601060 | 46912167 | - | - | - | 0.1246 | 0.3532 | 0.5485 | 0.6831 | 0.4244 | 0.8297 | - |
| rs1048380 | 46915125 | 0.0006 | 0.0084 | 0.0926 | - | - | - | - | (0.6126) | (0.9178) | - |
| rs12048954 | 46918995 | - | - | - | 0.0859 | 0.0495 | 0.2858 | 0.6015 | 0.2119 | 0.5893 | - |
| rs2275380 | 46920315 | 0.0124 | 0.0437 | 0.0312 | 0.1225 | 0.09558 | 0.2382 | 0.7518 | 0.1802 | 0.6596 | 0.5525 |
| rs1150064 | 46920631 | 0.0053 | 0.0099 | 0.1576 | - | - | - | - | (0.4746) | (0.6892) | - |
| rs6665021 | 46925107 | - | - | - | 1 | 1 | 1 | 1 | (0.8522) | (0.1577) | - |
| rs4660956 | 46931073 | - | - | - | 0.1741 | 0.3458 | 0.5637 | 0.5316 | 0.62877 | 0.64696 | - |
| rs6671124 | 46937193 | - | - | - | 0.5050 | 0.0065 | 0.5002 | 0.6911 | 0.3718 | 0.8732 | - |
| rs1440487 | 46939662 | 0.4309 | 0.7883 | 0.2399 | 0.1593 | 0.0896 | 0.6831 | 0.6831 | 0.5611 | 0.7763 | 0.3173 |
| rs1440486 | 46939825 | 2.26×10−5 | 0.0124 | 0.0926 | - | - | - | - | (0.5806) | (0.9011) | - |
| rs10749863 | 46946915 | - | - | - | 0.1122 | 0.2513 | 0.5485 | 0.6831 | 0.5680 | 0.7829 | - |
| rs2218189 | 46949405 | - | 0.0116 | 0.1576 | - | - | - | - | (0.5127) | (0.5738) | 0.6152 |
| rs6670495 | 46960495 | - | 0.0078 | 0.2222 | - | - | - | - | (0.4414) | (0.6293) | - |
| rs11582403 | 46986540 | - | - | - | 0.9287 | 0.2568 | 0.3173 | 0.03251 | 0.13033 | 0.34826 | - |
| rs6662321 | 46992646 | - | - | - | 0.4817 | 0.4111 | 0.5271 | 0.0009 | (0.0841) | (0.6181) | - |
Positions from HapMap Data Rel 28 PhaseII+III, August 10, on NCBI B36 assembly, dbSNP b126 CEU data
False discovery rate cut-off for α=0.05 is Z2 p-value=2.27×10−5
Data in parentheses were imputed
We further explored associations with GINA classification of asthma severity in Isle of Wight subjects using PLINK to examine allelic tests for asthma severity quantitative scores. Examined were individual SNPs, the 11-SNP haplotype, and 3-SNP sliding window haplotypes with significance indicated by Wald test asymptotic p-values.
The initial association analysis done in the Isle of Wight population was performed on a genome-wide level and correction for multiple testing was performed. The genome-wide false discovery rate cut off of alpha=0.05 was Z2 p-value of 2.27×10−5. The follow-up studies in the Isle of Wight and replication populations were performed on a limited number of SNPs that were located almost exclusively within a single LD block, thus, multiple testing corrections for these studies were not applied.
Gene expression
Expression of the asthma-associated gene, ATPAF1, along with S9 ribosomal protein gene were measured by the StepOnePlus PCR system (Applied Biosystems Inc, Foster City, CA, USA) in 17 bronchial biopsy samples from normal, mild asthmatic, and severe asthmatic subjects (see online repository for more methods details). In brief, endoscopic bronchial biopsies were obtained, tissue RNA was extracted, and total RNA was reverse transcribed. ATPAF1 levels were normalized using ribosomal protein S9 gene expression. Primers spanned at least one intron. Expression data were analyzed with Kruskal-Wallis followed by Dunn’s multiple comparison tests.
RESULTS
SNP associations
The gene discovery study of the 100K arrays yielded 98,921 SNPs of sufficient quality for analysis. The sliding window analysis yielded 60 clusters of SNPs with Z2 p-value<0.005 throughout the genome. SNP rs2289447 (Z2 p-value=2.2×10−8, Table II) ranked 6th among all SNPs on the microarray and was located in a SNP cluster on chromosome 1p33-p32.31 containing the ATPAF1 and neighbouring C1ORF223 and KIAA0494 genes (Fig 1, Table E1). This region had sustained significance (Z2 p-value range=0.0124 to 2.2×10−8, Table II) across a cluster of 6 SNPs (rs2289447, rs1150068, rs1048380, rs2275380, rs1150064, and rs1440486) and was therefore selected for further study.
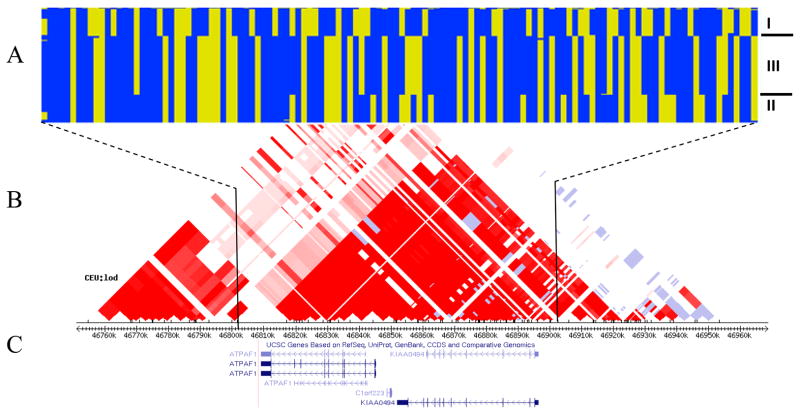
Figure 1.
ATPAF1, C1ORF223, KIAA0494 genes HapMap phase data illustrating the three main haplotypes (a), the CEU LOD linkage disequilibrium plot with block pattern based on confidence intervals (Gabriel et al)53 (b), and the UCSC gene track38, 48 (c).
This region was next targeted for focused genotyping in individual samples; the same subjects (n=227) were used as in the pools. Nine of eleven informative SNPs in the LD block containing ATPAF1, C1ORF223, and KIAA0494 were significantly associated with asthma in the Isle of Wight population (Table II) and the minor alleles were found to be protective (OR=0.44, CI 0.27–0.73 to OR=0.52, CI 0.33–0.82; Table E2). The use of consortia controls in our association analysis, assuming a 15% misclassification rate, increased power at least 20% and supported the original associations in the Isle of Wight cohort (nine SNPs) and added support for an additional SNP, rs2275380 (Table E3).
Transmission disequilibrium tests (TDT) in the Wessex replication population confirmed the association of rs2275380 with asthma diagnosis (Table II). Due to genotyping platform differences, identical SNPs were not examined in the other replication populations. However, gene-level replication was found in the Caucasian American, African American, and other ethnicity populations within the CAMP and CARE studies (Table II). No SNPs reached significance in any of the populations of Hispanic descent (CAMP and CARE Hispanic, GALA Mexican or Puerto Rican, and Mexico Childhood Asthma Study families) (Table II).
Haplotype associations
Common haplotypes were found in the Isle of Wight and Wessex populations (Fig 1, Table III). In the Isle of Wight cohort haplotypes I and III were found to confer asthma risk (Chi-square p=0.035) and protection (Chi-square p=0.0048), respectively (Table III). Odds ratios for the haplotypes indicated more than two-fold decreased risk of asthma associated with haplotype III (OR 0.45, 95% CI 0.26–0.78, p=0.0042) as compared to haplotype I as a reference. Transmission disequilibrium tests in the Wessex population confirmed the association of haplotype I with increased risk of asthma diagnosis (Chi-square p=0.0156). Several two- and three-marker haplotype associations with asthma were seen in the CAMP and CARE Caucasian American, African American, and other ethnicity families (p=0.0008 to 0.045, Table E4).
Table III.
Haplotype associations for asthma in European children
| Haplotype | rs1258000 | rs2289447 | rs620431 | rs1150068 | rs1048380 | rs2275380 | rs1150064 | rs1440487 | rs1440486 | rs2218189 | rs6670495 | Isle of Wight Case-Control Studya | Wessex Family-Based Study | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Haplotype Frequency Controls |
Haplotype Frequency Cases |
χ2 | χ2 p- value |
Haplotype Frequency Parents |
Transmitted Haplotypes |
Untransmitted Haplotypes |
χ2 | χ2 p- value |
||||||||||||
| I | A | C | C | T | C | A | A | C | G | A | T | 0.4796 | 0.5736 | 4.4450 | 0.0350 | 0.4807 | 225.9 | 204 | 5.8520 | 0.0156 |
| II | A | C | C | T | C | G | A | T | G | A | T | 0.2243 | 0.2158 | 0.0524 | 0.8190 | 0.2210 | 154.1 | 185 | 2.8100 | 0.0937 |
| III | G | T | A | C | T | G | T | C | A | G | A | 0.2452 | 0.1436 | 7.9600 | 0.0048 | 0.2140 | 147 | 168 | 1.4000 | 0.2367 |
Isle of Wight data based on individual genotypes
Asthma severity associations
Asthma severity scores of 0 to 4 were represented in the Isle of Wight population (Table E5). Nine SNPs were significantly associated with asthma severity (p=0.02 to 0.005, Table IV), the same SNPs associated with asthma. In addition, haplotype III, which is the 11-SNP haplotype of 34124342131 (A=1, C=2, G=3, T=4) for SNPs rs1258000| rs2289447| rs620431| rs1150068| rs1048380| rs2275380| rs1150064| rs1440487| rs1440486| rs2218189| rs6670495 (as shown in Fig 1 and Table III) was significantly associated with asthma severity (p=0.00763). Finally, multiple 3-SNP sliding window haplotypes were significant for asthma severity in the Isle of Wight with Wald asymptotic p-values of 0.02 to 0.007 (data not shown).
Table IV.
Quantitative allelic association of asthma severity scores in the Isle of Wight population
| SNP | Regression coefficient (beta) | Standard error | Regression r-squared | Wald test (based on t-distribution) | Wald test asymptotic p-value |
|---|---|---|---|---|---|
| rs4518838 | −0.1044 | 0.1694 | 0.001407 | −0.6167 | 0.538 |
| rs1258000 | −0.4084 | 0.1648 | 0.02233 | −2.479 | 0.01381 |
| rs1273237 | −1.203 | 1.6290 | 0.002016 | −0.7385 | 0.4609 |
| rs631840 | −1.203 | 1.6290 | 0.002016 | −0.7385 | 0.4609 |
| rs2289447 | −0.4007 | 0.1735 | 0.01951 | −2.309 | 0.0217 |
| rs620431 | −0.3949 | 0.1611 | 0.02201 | −2.451 | 0.01488 |
| rs1150068 | −0.4452 | 0.1717 | 0.02428 | −2.592 | 0.01005 |
| rs1048380 | −0.4824 | 0.1721 | 0.02839 | −2.804 | 0.005421 |
| rs2275380 | −0.06084 | 0.1383 | 0.000716 | −0.4399 | 0.6604 |
| rs1150064 | −0.4319 | 0.1719 | 0.02283 | −2.512 | 0.01259 |
| rs1440487 | 0.2744 | 0.1588 | 0.01102 | 1.728 | 0.08516 |
| rs1440486 | −0.4259 | 0.1724 | 0.02218 | −2.47 | 0.01414 |
| rs2218189 | −0.4431 | 0.1724 | 0.02405 | −2.57 | 0.01071 |
| rs6670495 | −0.4598 | 0.1792 | 0.0238 | −2.566 | 0.01083 |
n=269–272 genotypes per SNP
ATPAF1 relevance to asthma
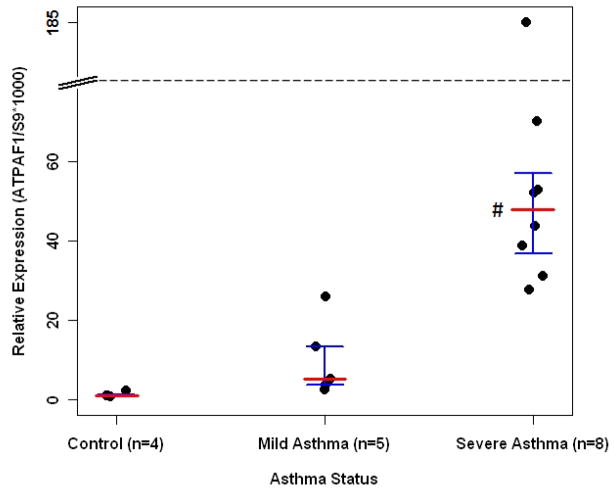
The expression of ATPAF1 in bronchial biopsy samples obtained from subjects with severe asthma was markedly (50-fold) elevated as compared to controls (p<0.01, Fig 2).
Figure 2.
Gene expression study of ATPAF1 from biopsied bronchial tissue.20, 21 # indicates statistical significance for severe asthma versus no asthma (p< 0.01). Median expression value indicated by red horizontal line; and first and third quantiles indicated by brackets.
DISCUSSION
A genomic region on chromosome 1q33-q32.31 met genome-wide significance for asthma in the Isle of Wight cohort. Subsequent detailed examination using a combination of targeted genotyping and haplotype analysis in the primary population, along with genotyping plus imputation in consortia controls, confirmed the association with an LD block containing ATPAF1, C1ORF223, and KIAA0494 genes. Furthermore, asthma severity was associated with SNPs and haplotypes in this LD block. Replication studies pursued in eight independent populations revealed instances of strict replication with specific SNPs and haplotypes in a Caucasian European population. Further gene-level association was found with additional SNPs and haplotypes (genotyped using other platforms) in the other three non-Hispanic replication populations. Thus, while not all significant findings in the primary population were replicated, major trends in association were identified across the LD block in all but the populations of Hispanic descent. Finally, data demonstrating differential upregulation of ATPAF1 expression in asthmatics as compared to control subjects lend support for a role for this gene, which is novel to asthma.
While no previous asthma linkage or association has been as finely mapped on 1p3 as the current study, our work supports previous studies that implicated the 1p31 to 1p36 region.29–38 Indeed, the chromosome region 1p has been consistently identified in genome screens for asthma and related phenotypes in populations of different races and ethnicities.29–31, 33 Thus, our finding of association of asthma with a 115 kb LD block at 1p33-p32.31 both support and extend these earlier studies.
ATPAF1, C1ORF223, and KIAA0494 are adjacent genes on chromosome 1p33-p32.31 and occupy much of a 115 kb LD block (Fig 1).38 Little is known about C1ORF223 (open reading frame for protein LOC37497, which is expressed predominately in testes)39 and KIAA0494 (widely expressed inferred calcium binding ion protein)39 genes and due to the overlap of coding and regulatory sequence among the three genes, additional studies on each gene are warranted. While we cannot exclude C1ORF223 or KIAA0494 from having a role in asthma, we chose to prioritize further study of ATPAF1 because 1) there is a well-established relationship between puringeric (ATP and adenosine) signalling and bronchoconstriction,40, 41 and 2) epithelial cell expression of the Th2 promoting cytokine IL-33 is regulated by purinergic signalling.42
ATPAF1 is a nuclear gene encoding ATPAF1 (ATP11), which is a soluble mitochondria protein that binds to unassembled β subunits of the F1-ATP synthase43 and prevents the F1 alpha and beta subunits from aggregating in the matrix.44 The mechanism of correct assembly of the ATP synthase F1 complex requires ATPAF145 and is preserved in all eukaryotic lineages capable of ATP synthesis via oxidative phosphorylation.46 ATPAF1 is widely expressed, including in whole lung tissue.47
Functional significance is predicted for several of the SNPs associated with asthma in this study.38, 48 Specifically, sequence encompassing rs1258000 is typical of regulatory elements.38, 49 Similarly, rs620431 has high regulatory potential and also lies 60 bp downstream of the exon 6/intron 6 boundary making it a potential splicing modulating element for the alternatively spliced exon 7.38
The most direct evidence of functional relevance of the ATPAF1 gene in asthma comes from its differential expression in bronchial tissue between asthmatics and controls (Fig 2). ATPAF1 was highly expressed in bronchial biopsies from those with severe asthma. Not only does this suggest a mechanism by which the gene may modify asthma risk, but it is also consistent with the findings of Chen et al50 in which they report that genes that are differentially expressed have a greater likelihood of containing variants that cause disease. Furthermore, the elevated ATPAF1 expression in bronchial tissue from severe asthmatics is consistent with and builds upon our findings of SNP and haplotype associations with asthma severity among the Isle of Wight children. Indeed, the importance of ATP-signalling in bronchoconstriction makes the link that we have identified between asthma severity and ATPAF1 expression and genetic variants all the more compelling.
The risk of reporting statistical significance merely by chance is a major concern in association studies in which a high number of tests are conducted. However, this is unlikely in the present study as several SNPs retained significance at the genome-wide level after correction for multiple testing, the outcomes were consistent across individual SNP and haplotype analyses, and SNPs and haplotypes showed association with asthma in the replication populations of the same race. In addition, data showing functional relevance of ATPAF1 further reinforce the validity of our findings.
For replication, we chose populations that had previously been studied for asthma genetics. We did not limit our selection to populations of the same ancestry as the primary cohort because inclusion of diverse populations broadens the relevance of the information generated. Indeed, variability in replication between cohorts has been a feature of studies of asthma genetics.7, 51, 52 We found significant associations in all the Caucasian populations, along with the one African American population, and one population of other ethnicity (not Caucasian, African American, or Hispanic). Interestingly, none of the populations of Hispanic descent (Hispanic, Puerto Rican and Mexican) showed association, indicating a race-specific trend. As expected, there were allele frequency differences between the populations of different races (Table E6). The allele frequencies across the four populations of Hispanic origin were fairly consistent with one another, with their minor allele frequencies in most cases ≥10% higher than in the three Caucasian populations. However, the minor allele frequency differences tended to be greatest between the Caucasian and African populations, which would not explain the observed trend. Other possible explanations for lack of replication in the Hispanic ancestry populations include type II error due to lack of power and differences in environmental exposures between cohorts.
In conclusion, our sequential strategy, as well as the use of well-phenotyped populations, led to identification of an association between ATPAF1 region variants and asthma. Studies to further understand the mechanistic role of this gene in asthma are being pursued.
Supplementary Material
KEY MESSAGES.
Variants in and around the ATPAF1 gene modify the risk of asthma in children.
ATPAF1 –related genetic susceptibility to asthma occurred in different ancestral groups of children, but not those of Hispanic descent.
Asthma severity is associated with variants in and around ATPAF1 and ATPAF1 expression is upregulated in severe asthmatics as compared to controls.
Acknowledgments
Funding support: This study was funded by the National Institutes of Health, grants R01 AI061471, R01 HL67736, P01 HL076383, T32 GM063483, Asthma UK (364) and the Asthma, Allergy and Inflammation Research Charity. The Wessex Family Cohort was originally recruited in collaboration with Genome Therapeutics Corporation and Schering-Plough. Richard and Edith Strauss Foundation of Canada and Dr. Ron Olivenstein supported the severe asthma program and collection of bronchial biopsies. The GALA studies were supported by HL078885, HL088133, AI077439, ES015794, Robert Wood Johnson Foundation Amos Medical Faculty Development Program, Flight Attendant Medical Research Institute (FAMRI). The Mexico Childhood Asthma Study was supported by the Intramural Research Program of the National Institutes of Health, National Institute of Environmental Health Sciences (Z01 ES49019). Subject enrollment was also supported in part by the National Council of Science and Technology (grant 26206-M), Mexico. Dr. Romieu was supported in part by the National Center for Environmental Health at the Centers for Disease Control. The CAMP study was supported by contracts with the National Heart, Lung, and Blood Institute (NO1-HR-16044, NO1-HR-16045, NO1-HR-16046, NO1-HR-16047, NO1-HR-16048, NO1-HR-16049, NO1-HR-16050, NO1-HR-16051, and NO1-HR-16052) and by General Clinical Research Center grants from the National Center for Research Resources (M01RR00051, M01RR0099718-24, M01RR02719-14, and RR00036). The CARE study was supported by grants (HL071742-01, HL004519-04, 5U10HL064287, 5U10HL064288, 5U10HL064295, 5U10HL064307, 5U10HL064305, and 5U10HL064313) from the National Heart, Lung, and Blood Institute. This study was carried out in part in the General Clinical Research Centers at Washington University School of Medicine (M01 RR00036) sponsored by the National Institutes of Health and the National Jewish Medical and Research Center (M01 RR00051). The data analyses for the CAMP, CARE, Wellcome Trust Case Control Consortium, and the Consortium for Systemic Lupus Erythematosus Genetics populations were supported by the Wake Forest School of Medicine Center for Public Health Genomics.
The authors thank Dennis Shubitowski and David Hutchings for technical assistance and Beth Cobb for administrative support. Mrs. Sharon Matthews helped with Isle of Wight phenotype data collection at age 10 years. Andrea Mogas, Susan Foley, and Alejandro Vazquez extracted RNA and made cDNA from the biopsy tissues. The authors acknowledge the NIH GWAS Data Repository (dbGAP) and the investigators involved in the National Heart, Lung, and Blood Institute SNP Health Association Resource (SHARe) Asthma Resource Project (SHARP) for use of their genotype and phenotype data. The authors thank Hao Wu and Grace Chiu for the analysis of the Mexico City Childhood Asthma Study data.
ABBREVIATIONS
- CAMP
Childhood Asthma Management Program
- CARE
Childhood Asthma Research and Education network
- CI
Confidence interval
- GALA
Genetics of Asthma in Latino Americans
- GWAS
Genome wide association study
- IOW
Isle of Wight
- LD
Linkage disequilibrium
- MAF
Minor allele frequency
- OR
Odds ratio
- RNA
Ribonucleic acid
- SLEGEN
International Consortium for Systemic Lupus Erythematosus Genetics
- SNP
Single nucleotide polymorphism
- WTCCC
Wellcome Trust Case Control Consortium
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Eder W, Ege MJ, von Mutius E. The asthma epidemic. N Engl J Med. 2006;355:2226–35. doi: 10.1056/NEJMra054308. [DOI] [PubMed] [Google Scholar]
- 2.Pearce N, Ait-Khaled N, Beasley R, Mallol J, Keil U, Mitchell E, et al. Worldwide trends in the prevalence of asthma symptoms: phase III of the International Study of Asthma and Allergies in Childhood (ISAAC) Thorax. 2007;62:757–65. doi: 10.1136/thx.2006.070169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Carter-Pokras OD, Gergen PJ. Reported asthma among Puerto Rican, Mexican-American, and Cuban children, 1982 through 1984. Am J Public Health. 1993;83:580–2. doi: 10.2105/ajph.83.4.580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Homa DM, Mannino DM, Lara M. Asthma Mortality in U.S. Hispanics of Mexican, Puerto Rican, and Cuban Heritage, 1990–1995. Am J Respir Crit Care Med. 2000;161:504–9. doi: 10.1164/ajrccm.161.2.9906025. [DOI] [PubMed] [Google Scholar]
- 5.Moffatt MF, Kabesch M, Liang LM, Dixon AL, Strachan D, Heath S, et al. Genetic variants regulating ORMDL3 expression contribute to the risk of childhood asthma. Nature. 2007;448:470–U5. doi: 10.1038/nature06014. [DOI] [PubMed] [Google Scholar]
- 6.Vercelli D. Discovering susceptibility genes for asthma and allergy. Nature Reviews Immunology. 2008;8:169–82. doi: 10.1038/nri2257. [DOI] [PubMed] [Google Scholar]
- 7.Holloway JW, Koppelman GH. Identifying novel genes contributing to asthma pathogenesis. Current Opinion in Allergy and Clinical Immunology. 2007;7:69–74. doi: 10.1097/ACI.0b013e328013d51b. [DOI] [PubMed] [Google Scholar]
- 8.Kurukulaaratchy RJ, Fenn MH, Waterhouse LM, Matthews SM, Holgate ST, Arshad SH. Characterization of wheezing phenotypes in the first 10 years of life. Clinical and Experimental Allergy. 2003;33:573–8. doi: 10.1046/j.1365-2222.2003.01657.x. [DOI] [PubMed] [Google Scholar]
- 9.Long-term effects of budesonide or nedocromil in children with asthma. The Childhood Asthma Management Program Research Group. N Engl J Med. 2000;343:1054–63. doi: 10.1056/NEJM200010123431501. [DOI] [PubMed] [Google Scholar]
- 10.Burchard EG, Avila PC, Nazario S, Casal J, Torres A, Rodriguez-Santana JR, et al. Lower bronchodilator responsiveness in Puerto Rican than in Mexican subjects with asthma. Am J Respir Crit Care Med. 2004;169:386–92. doi: 10.1164/rccm.200309-1293OC. [DOI] [PubMed] [Google Scholar]
- 11.David GL, Romieu I, Sienra-Monge JJ, Collins WJ, Ramirez-Aguilar M, del Rio-Navarro BE, et al. Nicotinamide adenine dinucleotide (phosphate) reduced:quinone oxidoreductase and glutathione S-transferase M1 polymorphisms and childhood asthma. Am J Respir Crit Care Med. 2003;168:1199–204. doi: 10.1164/rccm.200305-684OC. [DOI] [PubMed] [Google Scholar]
- 12.Hancock DB, Romieu I, Shi M, Sienra-Monge JJ, Wu H, Chiu GY, et al. Genome-wide association study implicates chromosome 9q21. 31 as a susceptibility locus for asthma in mexican children. PLoS Genet. 2009;5:e1000623. doi: 10.1371/journal.pgen.1000623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Himes BE, Hunninghake GM, Baurley JW, Rafaels NM, Sleiman P, Strachan DP, et al. Genome-wide association analysis identifies PDE4D as an asthma-susceptibility gene. Am J Hum Genet. 2009;84:581–93. doi: 10.1016/j.ajhg.2009.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Van Eerdewegh P, Little RD, Dupuis J, Del Mastro RG, Falls K, Simon J, et al. Association of the ADAM33 gene with asthma and bronchial hyperresponsiveness. Nature. 2002;418:426–30. doi: 10.1038/nature00878. [DOI] [PubMed] [Google Scholar]
- 15.Guilbert TW, Morgan WJ, Krawiec M, Lemanske RF, Jr, Sorkness C, Szefler SJ, et al. The Prevention of Early Asthma in Kids study: design, rationale and methods for the Childhood Asthma Research and Education network. Control Clin Trials. 2004;25:286–310. doi: 10.1016/j.cct.2004.03.002. [DOI] [PubMed] [Google Scholar]
- 16.Mailman MD, Feolo M, Jin Y, Kimura M, Tryka K, Bagoutdinov R, et al. The NCBI dbGaP database of genotypes and phenotypes. Nat Genet. 2007;39:1181–6. doi: 10.1038/ng1007-1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hancock DB, Romieu I, Shi M, Sienra-Monge J-J, Wu H, Chiu GY, et al. Genome-Wide Association Study Implicates Chromosome 9q21. 31 as a Susceptibility Locus for Asthma in Mexican Children. PLoS Genet. 2009;5:e1000623. doi: 10.1371/journal.pgen.1000623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Genome-wide association study of 14,000 cases of seven common diseases and 3, 000 shared controls. Nature. 2007;447:661–78. doi: 10.1038/nature05911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Harley JB, Alarcon-Riquelme ME, Criswell LA, Jacob CO, Kimberly RP, Moser KL, et al. Genome-wide association scan in women with systemic lupus erythematosus identifies susceptibility variants in ITGAM, PXK, KIAA1542 and other loci. Nat Genet. 2008;40:204–10. doi: 10.1038/ng.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Foley SC, Mogas AK, Olivenstein R, Fiset PO, Chakir J, Bourbeau J, et al. Increased expression of ADAM33 and ADAM8 with disease progression in asthma. Journal of Allergy and Clinical Immunology. 2007;119:863–71. doi: 10.1016/j.jaci.2006.12.665. [DOI] [PubMed] [Google Scholar]
- 21.Shannon J, Ernst P, Yamauchi Y, Olivenstein R, Lemiere C, Foley S, et al. Differences in airway cytokine profile in severe asthma compared to moderate asthma. Chest. 2007:chest.07–1881. doi: 10.1378/chest.07-1881. [DOI] [PubMed] [Google Scholar]
- 22.The database of Genotypes and Phenotypes (dbGaP). [Cited 2010.] Available from http://www.ncbi.nlm.nih.gov/gap
- 23.Yang HC, Pan CC, Lu RCY, Fann CSJ. New adjustment factors and sample size calculation in a DNA-pooling experiment with preferential amplification. Genetics. 2005;169:399–410. doi: 10.1534/genetics.104.032052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MAR, Bender D, et al. PLINK: A tool set for whole-genome association and population-based linkage analyses. American Journal of Human Genetics. 2007;81:559–75. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gonzalez JR, Armengol L, Sole X, Guino E, Mercader JM, Estivill X, et al. SNPassoc: an R package to perform whole genome association studies. Bioinformatics. 2007;23:644–5. doi: 10.1093/bioinformatics/btm025. [DOI] [PubMed] [Google Scholar]
- 26.R Development Core Team. R: A Language and Environment for Statistical Computing. Vienna, Austria: 2007. [Google Scholar]
- 27.Lake SL, Lyon H, Tantisira K, Silverman EK, Weiss ST, Laird NM, et al. Estimation and tests of haplotype-environment interaction when linkage phase is ambiguous. Human Heredity. 2003;55:56–65. doi: 10.1159/000071811. [DOI] [PubMed] [Google Scholar]
- 28.Wu H, Romieu I, Shi M, Hancock DB, Li HL, Sienra-Monge JJ, et al. Evaluation of candidate genes in a genome-wide association study of childhood asthma in Mexicans. Journal of Allergy and Clinical Immunology. 2010;125:321–7. doi: 10.1016/j.jaci.2009.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Blumenthal MN, Langefeld CD, Beaty TH, Bleecker ER, Ober C, Lester L, et al. A genome-wide search for allergic response (atopy) genes in three ethnic groups: Collaborative Study on the Genetics of Asthma. Human Genetics. 2004;114:157–64. doi: 10.1007/s00439-003-1030-5. [DOI] [PubMed] [Google Scholar]
- 30.Bouzigon E, Dizier MH, Krahenbuhl C, Lemainque A, Annesi-Maesano I, Betard C, et al. Clustering patterns of LOD scores for asthma-related phenotypes revealed by a genome-wide screen in 295 French EGEA families. Human Molecular Genetics. 2004;13:3103–13. doi: 10.1093/hmg/ddh340. [DOI] [PubMed] [Google Scholar]
- 31.Bouzigon E, Siroux V, Dizier MH, Lemainque A, Pison C, Lathrop M, et al. Scores of asthma and asthma severity reveal new regions of linkage in EGEA study families. European Respiratory Journal. 2007;30:253–9. doi: 10.1183/09031936.00162206. [DOI] [PubMed] [Google Scholar]
- 32.Colilla S, Nicolae D, Pluzhnikov A, Blumenthal MN, Beaty TH, Bleecker ER, et al. Evidence for gene-environment interactions in a linkage study of asthma and smoking exposure. Journal of Allergy and Clinical Immunology. 2003;111:840–6. doi: 10.1067/mai.2003.170. [DOI] [PubMed] [Google Scholar]
- 33.Dizier MH, Besse-Schmittler C, Guilloud-Bataille M, Annesi-Maesano I, Boussaha M, Bousquet J, et al. Genome screen for asthma and related phenotypes in the French EGEA study. American Journal of Respiratory and Critical Care Medicine. 2000;162:1812–8. doi: 10.1164/ajrccm.162.5.2002113. [DOI] [PubMed] [Google Scholar]
- 34.Haagerup A, Bjerke T, Schiotz PO, Binderup HG, Dahl R, Kruse TA. Asthma and atopy - a total genome scan for susceptibility genes. Allergy. 2002;57:680–6. doi: 10.1034/j.1398-9995.2002.23523.x. [DOI] [PubMed] [Google Scholar]
- 35.Mathias RA, Freidhoff LR, Blumenthal MN, Meyers DA, Lester L, King R, et al. Genome-wide linkage analyses of total serum IgE using variance components analysis in asthmatic families. Genetic Epidemiology. 2001;20:340–55. doi: 10.1002/gepi.5. [DOI] [PubMed] [Google Scholar]
- 36.Xu JF, Meyers DA, Ober C, Blumenthal MN, Mellen B, Barnes KC, et al. Genomewide screen and identification of gene-gene interactions for asthma-susceptibility loci in three US populations: Collaborative study on the genetics of asthma. American Journal of Human Genetics. 2001;68:1437–46. doi: 10.1086/320589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.A genome-wide search for asthma susceptibility loci in ethnically diverse populations. Nat Genet. 1997;15:389–92. doi: 10.1038/ng0497-389. [DOI] [PubMed] [Google Scholar]
- 38.Kent WJ, Sugnet CW, Furey TS, Roskin KM, Pringle TH, Zahler AM, et al. The human genome browser at UCSC. Genome Research. 2002;12:996–1006. doi: 10.1101/gr.229102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rebhan M, Chalifa-Caspi V, Prilusky J, Lancet D. GeneCards: integrating information about genes, proteins and diseases. Trends Genet. 1997;13:163. doi: 10.1016/s0168-9525(97)01103-7. [DOI] [PubMed] [Google Scholar]
- 40.Idzko M, Hammad H, van Nimwegen M, Kool M, Willart MAM, Muskens F, et al. Extracellular ATP triggers and maintains asthmatic airway inflammation by activating dendritic cells. Nature Medicine. 2007;13:913–9. doi: 10.1038/nm1617. [DOI] [PubMed] [Google Scholar]
- 41.Schnurr M, Then F, Galambos P, Scholz C, Siegmund B, Endres S, et al. Extracellular ATP and TNF-alpha synergize in the activation and maturation of human dendritic cells. Journal of Immunology. 2000;165:4704–9. doi: 10.4049/jimmunol.165.8.4704. [DOI] [PubMed] [Google Scholar]
- 42.Hudson CA, Christophi GP, Gruber RC, Wilmore JR, Lawrence DA, Massa PT. Induction of IL-33 expression and activity in central nervous system glia. J Leukoc Biol. 2008;84:631–43. doi: 10.1189/jlb.1207830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wang ZG, Ackerman SH. The assembly factor Atp11p binds to the beta-subunit of the mitochondrial F-1-ATPase. Journal of Biological Chemistry. 2000;275:5767–72. doi: 10.1074/jbc.275.8.5767. [DOI] [PubMed] [Google Scholar]
- 44.Sheluho D, Ackerman SH. An accessible hydrophobic surface is a key element of the molecular chaperone action of Atp11p. Journal of Biological Chemistry. 2001;276:39945–9. doi: 10.1074/jbc.M107252200. [DOI] [PubMed] [Google Scholar]
- 45.Wang ZG, White PS, Ackerman SH. Atp11p and Atp12p are assembly factors for the F-1-ATPase in human mitochondria. Journal of Biological Chemistry. 2001;276:30773–8. doi: 10.1074/jbc.M104133200. [DOI] [PubMed] [Google Scholar]
- 46.Pickova A, Potocky M, Houstek J. Assembly factors of F1F0-ATP synthase across genomes. Proteins-Structure Function and Bioinformatics. 2005;59:393–402. doi: 10.1002/prot.20452. [DOI] [PubMed] [Google Scholar]
- 47.Su AI, Wiltshire T, Batalov S, Lapp H, Ching KA, Block D, et al. A gene atlas of the mouse and human protein-encoding transcriptomes. Proceedings of the National Academy of Sciences of the United States of America. 2004;101:6062–7. doi: 10.1073/pnas.0400782101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Gibbs RA, Belmont JW, Hardenbol P, Willis TD, Yu FL, Yang HM, et al. The International HapMap Project. Nature. 2003;426:789–96. doi: 10.1038/nature02168. [DOI] [PubMed] [Google Scholar]
- 49.King DC, Taylor J, Elnitski L, Chiaromonte F, Miller W, Hardison RC. Evaluation of regulatory potential and conservation scores for detecting cis-regulatory modules in aligned mammalian genome sequences. Genome Res. 2005;15:1051–60. doi: 10.1101/gr.3642605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Chen R, Morgan AA, Dudley J, Deshpande T, Li L, Kodama K, et al. FitSNPs: highly differentially expressed genes are more likely to have variants associated with disease. Genome Biol. 2008;9:R170. doi: 10.1186/gb-2008-9-12-r170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Holloway JW, Yang IA, Holgate ST. Genetics of allergic disease. Journal of Allergy and Clinical Immunology. 2010;125:S81–S94. doi: 10.1016/j.jaci.2009.10.071. [DOI] [PubMed] [Google Scholar]
- 52.Rogers AJ, Raby BA, Lasky-Su JA, Murphy A, Lazarus R, Klanderman BJ, et al. Assessing the reproducibility of asthma candidate gene associations, using genome-wide data. Am J Respir Crit Care Med. 2009;179:1084–90. doi: 10.1164/rccm.200812-1860OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Barrett JC, Fry B, Maller J, Daly MJ. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics. 2005;21:263–5. doi: 10.1093/bioinformatics/bth457. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.