Abstract
The systemic pharmacokinetics and pharmacodynamics of small molecules are determined by subcellular transport phenomena. Although approaches used to study the subcellular distribution of small molecules have gradually evolved over the past several decades, experimental analysis and prediction of cellular pharmacokinetics remains a challenge. In this article, we surveyed the progress of subcellular distribution research since the 1960s, with a focus on the advantages, disadvantages and limitations of the various experimental techniques. Critical review of the existing body of knowledge pointed to many opportunities to advance the rational design of organelle-targeted chemical agents. These opportunities include: 1) development of quantitative, nonfluorescence-based, whole cell methods and techniques to measure the subcellular distribution of chemical agents in multiple compartments; 2) exploratory experimentation with nonspecific transport probes that have not been enriched with putative, organelle-targeting features; 3) elaboration of hypothesis-driven, mechanistic and modeling-based approaches to guide experiments aimed at elucidating subcellular distribution and transport; and 4) introduction of revolutionary conceptual approaches borrowed from the field of synthetic biology combined with cutting edge experimental strategies. In our laboratory, state-of-the-art subcellular transport studies are now being aimed at understanding the formation of new intracellular membrane structures in response to drug therapy, exploring the function of drug-membrane complexes as intracellular drug depots, and synthesizing new organelles with extraordinary physical and chemical properties.
Keywords: drug transport, pharmacokinetics, biodistribution, drug targeting, databases, mathematical modeling, drug delivery, drug-membrane aggregates, unnatural organelles, synthetic organelles
Introduction
Many drugs require entrance into specific subcellular organelles to reach their targets, or they have side effects associated with unwanted accumulation in non-target sites within cells. Therefore, novel drug targeting strategies to improve compound efficiency in reaching specific organelles have been sought to increase a molecule’s potency and decrease undesired side effects. For example, small molecules are being targeted to mitochondria by conjugating these molecules to cell-penetrating, lipophilic peptides, oligoguanidinium, or triphenylphosphonium moieties 1–6. To fulfill this organelle targeting drug design strategy, there have been many efforts aimed at characterizing the physiological properties of the most important intracellular organelles and identifying key physicochemical features that determine the accumulation of exogenous chemical agents inside these organelles 7–10. More importantly, the mechanisms driving the distribution kinetic of chemical agents within the cell and the dynamic cellular response to these chemical agents are being revealed with the aid of new experimental strategies and conceptual approaches.
To put the state-of-the-art of subcellular transport research in perspective, information about the subcellular distribution of 967 unique compounds was compiled from a total of 448 scientific publications dating back to the 1960s. Current knowledge of the physiological properties and general principles of targeted delivery to the major intracellular organelles were summarized (Table 1). The criteria of article collection and the qualitative description of subcellular localization patterns were described in a separate paper (11, accepted in Molecular Pharmaceutics). From the outset, it is noteworthy that most compounds were reported to localize within a single organelle, with few reports of compounds localized to multiple sites (Table 2). Based on these references, we review the historical progress of subcellular biodistribution research, focusing on the evolution of experimental, theoretical and conceptual approaches used to analyze the organelle-targeting features of small molecule chemical agents.
Table 1.
Features of major subcellular compartments that affect the intracellular distribution pattern of small chemicals.
| Major subcellular compartments |
Major biological function |
Morphological features |
Physiological features | General mechanisms of accumulation |
|---|---|---|---|---|
| Lysosomes & endosomes |
Degradation of excessive cellular proteins, lipids and organelles |
Membrane-bound, vesicles |
Acidic luminal environment (pH<6) Contain unique, lysosomal protease and hydrolyses. |
Active transport; ion trapping; interaction with organelle resident proteins; receptor endocytosis; fluid phase pinocytosis; intracellular membrane trafficking. |
| Mitochondria | Energy conversion and storage of calcium ions |
Membrane-bound, with internal membrane structure |
Negative membrane potential. Weakly basic luminal pH (~8). Two membranes. Contain DNA. |
Active transport; trapping by chemoelectrical potential; interaction with mtDNA andorganelle resident proteins |
| Nucleus | Storage of genetic materials |
Membrane-bound with large protein pores |
Composed of phosphate-rich DNA, RNA, and a large variety of proteins |
Active transport; partitioning to nuclear envelope; interaction with DNA, RNA (nucleolar) and organelle resident proteins |
| Plasma membrane |
Separation of the cytosol form the outside environment |
Membrane protein-rich phospholipid bilayer membrane |
Fluid mosaic, lipid bilayer with selective permeability. |
Lipophilic partitioning; absorption to phospholipids; interaction with membrane proteins |
| Endoplasm reticulum |
Facilitation of protein folding and transport of synthesized proteins |
Interconnected network of membrane tubules. |
Intracellular membrane trafficking; interaction with phospholipids or organelle resident proteins |
|
| Golgi apparatus |
Process and package of macromolecules |
Stacks of semicircular or planar membrane-bound compartments |
Intracellular membrane trafficking; interaction with phospholipids or organelle resident proteins |
|
| Cytosol | Intracellular solution-like matrix |
Interaction with cytosolic components such as the cytoskeleton |
Table 2. Number and chemical diversity of small drug-like molecules with reported subcellular localization(s).
Chemical structures and reference articles can be found in the supporting information.
| Localization | Number of compounds |
% | Number of references |
|---|---|---|---|
| Total | 967 | 100 | 448 |
| Endo-lysosomes | 226 | 23 | 96 |
| Mitochondria | 259 | 27 | 136 |
| Nucleus | 123 | 13 | 67 |
| Plasma membrane | 162 | 17 | 75 |
| Endoplasmic reticulum & Golgi apparatus |
37 | 4 | 26 |
| Cytosol | 59 | 6 | 36 |
| Multiple sites | 101 | 10 | 71 |
Pharmacological effects as evidence for specific organelle accumulation
Eukaryotic cells have highly organized subcellular compartments with distinct structural and functional features. Pharmacological effects, i.e. changes in these features, especially changes in organelle morphology (swelling, rupture, shrinkage, etc.) upon drug treatment have been used in a large number of studies as evidence for localization in specific organelles. Surveying the literature, a large number of subcellular localization reports were based on evidence that exogenous chemicals induced changes in the structure or function of specific organelles (Table 3).
Table 3. Methods used for subcellular localization studies.
Chemical structures and reference articles can be found in the supporting information.
| Evidence | Count | % Population |
|---|---|---|
| Pharmacological effects | 171 | 18 |
| Chemical analysis | ||
| Uptake/binding studies | 67 | 7 |
| Cell fractionation | 54 | 6 |
| Microscopic imaging studies | ||
| Fluorescence microscopy | 633 | 65 |
| Histochemistry methods | 9 | < 1 |
| Others | 6 | < 1 |
| Not mentioned | 25 | < 3 |
Prior to the widespread adoption of cell-based uptake and transport assays in small molecule drug development, morphological changes were commonly used as evidence for organelle accumulation. From the 1960s to the 1980s, similar numbers of studies were based on observations with light microscopy, fluorescence microscopy and transmission electron microscopy 12–14. Light microscopy was the preferred tool in detecting expansion in the endolysosomal compartment, visible as a massive, cytoplasmic vacuolation phenomenon. With transmission electron microscopy, morphological changes of the major organelles could be observed directly, with or without the aid of specific organelle tracers. In the early 1980s, fluorescence microscopy became increasingly applied to the detection of organelle swelling and shrinkage using fluorescent probes. In the endolysosomal compartment, the observed morphological changes have been shown to reflect the accumulation of weakly basic compounds inside these organelles, or the inhibitory effects of cations on the activity of lysosomal proteins 15–17.
Morphological changes in lysosomes and mitochondria often coincided with changes in membrane potential, pH gradients or membrane permeability 18–22. Under some circumstances such changes resulted in the release of a resident, organelle-specific enzyme into the cytosol or into the extracellular compartment. Thus the detection of fluctuation in voltage or pH gradients, or the detection of released organelle components, was used as evidence for accumulation of exogenous small molecules in specific organelles, from the 1970s 23–26 and continuing to this day27–30.
Also since the 1970s, analytical measurements using thin layer chromatography and HPLC to detect alterations in organelle composition, including changes in lipid content, protein concentrations and metabolic changes, have been used as evidence to infer accumulation of small molecules in specific organelles 31–33. For example, significant increases of phospholipids in the renal cortex of gentamicin- or netilmicin-treated rats 34 were ascribed to impaired lysosomal degradation of phospholipids due to inhibition of lysosomal phospholipase C by accumulation of said molecules in lysosomes. Ammonia, amiodarone and some other compounds that interfere with degradation of proteins or phospholipids in lysosomes 35–37 were also associated with inhibition of lysosomal proteases and phospholipases due to the accumulation of these weakly basic compounds in the lysosomes, resulting in intralysosomal pH changes with consequent effects on lysosomal enzyme activities 38–42.
Nevertheless, claims that a molecule “accumulates in” an organelle based on a change in organelle structure (or function) are circumstantial and prone to misinterpretation and experimental artifacts. For example, in the case of toxic compounds, inhibition of organelle function may not require direct interaction, or accumulation within a specific organelle. For instance, apoptosis signal transduction pathways lead to mitochondrial membrane permeabilization, loss of mitochondrial membrane potential and the release of cytochorome c from the mitochondria, as well as nonspecific effects on other organelles. Therefore, induced changes in mitochondrial volume, membrane potential, or permeability do not necessarily reflect a direct interaction with mitochondria. The same is true for other organelles 43.
Chemical analysis as evidence for specific organelle accumulation
Pharmacokinetics gradually became part of drug development from the 1960s through the 1980s. However, only since the 1990s, there has been an increasing recognition of the importance of cellular pharmacokinetics as a determinant of systemic pharmacokinetics. In the process, quantitative measurement of chemical uptake in vivo or in vitro became increasingly important as direct evidence supporting the actual localization of a molecule in a specific subcellular compartment. Irrespective of the experimental strategy, analytical measurements were increasingly applied in cellular uptake or distribution studies, providing direct evidence for accumulation in specific organelles. However, only a relative small fraction of the molecules whose intracellular localization has been reported in the scientific literature is supported by such evidence (Table 3).
In uptake experiments, researchers measure the mass of exogenous chemical agents accumulating in intact cells or in isolated organelles after in vitro or in vivo administration of the compound. In some cases, a known, organelle-targeting compound was used to compete for the interaction or otherwise inhibit the organelle-specific accumulation mechanism. For instance, in a report of the subcellular localization sites of weakly basic molecules, the reduced cellular uptake after the disruption of trans-membrane pH gradients was used as evidence for endolysosomal accumulation 44, 45. Less commonly, ion-selective electrodes have been used to study the uptake of positively charged, lipophilic compounds in isolated mitochondria 25, 46–48. Binding to resident organelle-specific components including protein, lipids or nucleic acids has also been measured as direct evidence to demonstrate organelle or cytosolic accumulation 49–52.
Starting in the 1990s, there was an increase in the number of investigations looking at the qualitative or semi-qualitative (relative) distribution of a compound in all subcellular compartments, simultaneously, featuring analytical measurements following cell fractionation (Table 3). In cell fractionation studies, the basic experimental strategy has been to isolate the various organelles by differential centrifugation 53, followed by measurements of the absolute amount of a compound in each organelle fraction 54–56 and/or comparing that amount relative to the total accumulation of the compound in the cell 57–59. Reliable separation of distinct subcellular organelles is critical to the evaluation of subcellular distribution profile. While the presence or activity of organelle-specific marker proteins in each fraction can be readily determined, effective separation of subcellular compartments requires little overlap in protein markers between the fractions. Then the fractions can be subjected to chemical analysis of organelle associated compound accumulation by means of spectrophotometry 60, HPLC 61–63, LC/MS 64, and most commonly, by scintillation counting of radiolabeled compounds 65–68.
Many significant advances in distribution studies were achieved through the development of cell fractionation techniques. Organelle separation and analysis techniques such as immunoisolation, fluorescence activated sorting and electromigration analysis were developed. However their use in subcellular distribution studies remains infrequent, possibly because they are technically demanding. For organelle immunoisolation, cell homogenates were exposed to organelle-specific antibodies attached to solid supports and the cell fractions of interest were concentrated by binding to antibody 69–71. Fluorescent activated cell sorting was applied to separate multiple intracellular organelles stained with membrane dyes or labeled with fluorescent antibodies that bind to organelle membrane proteins 72–75. Since around the early 2000s’ electrophoresis has been used to separate different subcellular organelle fractions from cell homogenates 76–84. Most recently, magnetic chromatography methods were developed to isolate and enrich lysosomes from cells that internalize iron-containing particles 85–87.
As a caveat, organelle isolation procedures are not necessarily free from experimental artifacts: organelle isolation procedures are inherently disruptive 48. During the multi-step procedures that are necessary to attain organelle purity for further analysis, organelle damage and compound leakage from one or more subcellular organelles are inevitable and difficult to control 53.
Whole cell based microscopic imaging studies as evidence for intracellular localization
Whole cell based microscopic imaging studies using intrinsically fluorescent or fluorescently - tagged molecules accounted for over half of all articles reporting a molecule’s subcelllar localization (Table 3) and have been most common over the past decade. Less commonly, electron microscopy combined with immunocytochemical methods was used to obtain high resolution information of intracellular distribution of small molecular weight compounds that precipitated out at their sites of accumulation 88, 89 or that were tagged with a specific epitope for antibody recognition 90, 91 (Table 3).
Compared to pharmacological and chemical analyses which are tedious and prone to artifacts, microscopic imaging has generally been preferred as an efficient and reliable method to obtain real-time intracellular distribution data. Microscopic visualization of intrinsically fluorescent or fluorescently-tagged molecules provided the evidence for establishing subcellular localization in the majority of published, subcellular localization studies (Table 3). While the intracellular accumulation sites of fluorescent compounds can be determined directly based on the characteristic morphology of stained compartments 92–98, the use of resident, reference fluorescent markers 99–103 has enabled determination of subcellular distribution by analysis of colocalization patterns. Following advances in location proteomics 104–108, machine vision-based quantitative image analysis has been used to establish the degree of co-localization between compounds of interest with an organelle-specific reference marker 109–111. Furthermore, large combinatorial libraries of fluorescent probes and automated high content screening instruments have facilitated analysis of chemical motifs associated with specific intracellular distribution patterns 110–112. More recently, fluorescence resonance energy transfer has been used to study the trafficking and distribution of xenobiotics 113.
Although fluorescence-based imaging techniques offer many advantages over other detection methods, evidence for subcellular localization based on a molecule’s fluorescence is generally criticized due to well-known artifacts. For example, environmental factors such as binding status 114, ionic strength 115, solvent polarity 116–118, pH 119–121 and temperature 122–124 can affect a molecule’s fluorescence spectrum or quantum yield. If fluorescence is dependent on environmental factors, interpretation of subcellular distribution patterns might not be entirely accurate or complete as molecules may not be fluorescent in every compartment they localize to125. For non-fluorescent molecules to be detectable using fluorescence microscopy, a fluorescent tag needs to be conjugated to the compound. This tag can alter the distribution of the original compound. Thus, claims about the subcellular localization of a tagged compound are only valid in the context of the entire small molecule-fluorescent probe conjugate.
Over the past decade, more sensitive and general imaging methods, such as the confocal Raman microscopy and secondary ion mass spectroscopy, have been used to monitor the distribution of non-fluorescent compounds in cells 126. To date, a few pioneering studies based on these techniques have been reported 127, 128 (Table 3). Nevertheless, significant breakthroughs have been achieved. For example, the major challenge in conventional Raman imaging is how to amplify and quantify the weak resonance signal in live cell environment. The application of coherent anti-stokes Raman scattering has led to improvement in signal detection and has allowed tracking the intracellular distribution of endogenous lipids, virus RNA and organelles129–131.
Yet another recent advance was the application of secondary ion mass spectrometry (SIMS) for analyzing subcellular localization sites of chemical agents. SIMS is a sensitive technique traditionally used in material sciences to analyze the elemental, isotopic or molecular composition of thin films 132–134. Beginning in the late 1990s’ SIMS has been used to monitor the phospholipid composition of biological membranes 135 and map the distribution of isotope labeled chemical agents after in vitro or in vivo dosing 136–139. Though still in its infancy, SIMS is garnering attention in subcellular distribution studies because of its high sensitivity and outstanding resolution.
Computational models to frame quantitative hypotheses and predict subcellular distribution patterns
Since the 2000s,cheminformatics and computational modeling-based approaches have become essential to drug discovery and development. In parallel, cheminformatics and computational approaches are increasingly being used to generate and test hypothesis about intracellular distribution patterns and chemical-organelle interactions 7–9, 110, 140–142. However, evaluation of computational models is inherently dependent on the quantity and quality of subcellular localization measurements.
Computational models for predicting biodistribution can be classified into two categories: statistically-based regression models and mechanism-based physiological models. Typical statistical models use experimental observations of small molecule subcellular localization as a training dataset. With calculated physicochemical properties as input parameters, regression or multivariate statistical analysis methods can be applied to make qualitative (yes/no) or quantitative (how much) descriptions of the compound distribution pattern in the training set. If the fit between the model and the training set is acceptable, the model can be applied to a different test set of molecules with overlapping physicochemical properties, to make predictions about the molecules’ localization. In cheminformatics research 143, this is referred to as a quantitative structure-activity relationship (QSAR) study.
Several published articles have analyzed compound subcellular accumulation sites using decision trees and other QSAR tools 7–9, 144, 145. However, the success of QSAR-based models depends largely on the accurate calculation of molecular properties and quality of input data. Ideally the observations used for predictive QSAR models should be derived from the same experiments, based on the same mechanism of study, assessed with the same criteria, and performed with similar methods so as to avoid intra-laboratory variations in the manner the measurements are made and defined. QSAR models also benefit from large data sets of compounds. Therefore, QSAR models based on scant published data obtained with different methods and experimental approaches are more descriptive than predictive.
Physiologically-based predictive subcellular localization models have been derived from theoretical computation of mass transfer between different organelles according to Fick’s Law of diffusion and Nernst-Plank equations. Physiologically-based models have been developed to calculate the intracellular accumulation and organelle distribution of molecules in cells suspended in homogeneous extracellular concentration of a chemical agent, or in cells exposed to a transcellular concentration gradient 146, 147. Using combinations of input values, simulations can be performed to mimic the steady state distribution of small molecules in lysosomes, mitochondria and cytosol of millions of virtual molecules differing in molecular properties10. To demonstrate the potential of this approach, a predictive, multi-scale, cell-based model was constructed to simulate the distribution properties of pulmonary drugs in different cell types and anatomical regions of the lung 148. Translated to the in vivo realm, validation of whole organ models will require detailed experimental measurements and kinetic analysis of small molecule distribution at multiple scales, in a manner that exceeds the capabilities of state of the art experimental approaches 128 by many orders of magnitude.
From Pharmacokinetics to Synthetic Biology: Conclusion and Future Outlook
Thus far, we have presented a comprehensive survey of the past and present state of the art of subcellular transport knowledge, focused on the evolution of experimental approaches and methods. Our understanding of small molecule distribution inside cells has been shaped (and is being reshaped) by the application and limitations associated of each one of these approaches and methods, and the invention of new ones (Table 4). Analytical measurements following precise cell fractionation can be considered the most quantitative and convincing evidence for the preferential accumulation of chemical agents in specific subcellular compartments. However, fractionation studies are low throughput and labor intensitive. Accordingly, live cell-based imaging with fluorescence microscopes has become the most common method for documenting the subcellular distribution of small molecule chemical agents.
Table 4.
A comparison of experimental methodologies.
| Methods | Experimental systems |
Instruments | Pros. & Cons. | |
|---|---|---|---|---|
| Pharmacological effect | Dead or living cells |
LM; FM; TEM; analytical instruments such as HPLC, LCMS, and GE |
Does not provide sufficient evidence to ascertain localization Only provides indirect evidence for effect on a specific organelle. |
|
| Analytical measurements | ||||
| Uptake/binding experiments |
Isolated organelles or cell culture |
FM; FS; radiometer, or analytical instruments |
Provide adequate evidence for localization to a specific organelle. Cannot assess the accumulation in all subcellular organelles at one time |
|
| Distribution studies | Dead cells | Centrifuge, FACS, CC, CE, and analytical instruments |
Depict the relative distribution pattern in all cellular organelles |
Separation of cellular organelles is difficult Not suitable if compound undergoes rapid efflux |
| Whole cell based imaging studies | ||||
| Immune-/ Histochemistry |
Dead cells | TEM |
Sample processing steps may cause redistribution Detection of small amounts is challenging. |
|
| Fluorescence microscopy |
Live cells | FM |
May miss localization information if fluorescence intensity changes with environmental factors |
|
| Raman imaging | Live cells | Raman microscopy | Signals are weak and require amplification | |
| Secondary ion mass spectrometry |
Dead cells | SIMS device | ||
| Computational predictions |
in silico | Computers | ||
Abreactions: LM - light microscopy; FM - fluorescence microscopy; FS - fluorescence spectrometer; FACS - Fluorescence-activated cell sorting; CC – column chromtography; CE - capillary electrophoresis TEM - transmission electron microscopy; HPLC - high performance liquid chromatography; LC/MS - Liquid chromatography-mass spectrometry; GE - gel electrophoresis.
The application of fluorescence-based techniques in subcellular distribution studies has had two major consequences on the current state of knowledge in this field: 1) much of what is presently known about the subcellular localization properties of small molecules is biased towards fluorescent compounds, with either intrinsic fluorescence or fluorescent molecular tags; and 2) the majority of compounds with reported subcellular localizations are either highly specific, organelle-targeting transport probes or their subcellular distribution has been analyzed only in the context of a specific organelle (Table 2). Thus, the development of methods to quantify the distribution pattern of non-fluorescent, non-targeted molecules at the whole cell level will be necessary to expand our current understanding of the subcellular distribution properties of small molecules. For this reason, in addition to whole cell experimental analysis methods, physiologically-based modeling efforts aimed at predicting cellular pharmacokinetics are contributing positively towards formulating a hypothesis-driven framework for experimental, quantitative analysis of cellular biodistribution phenomena. Although still at its inception phase, whole-cell, mechanism-based computational modeling is a promising tool in terms of providing quantitative hypotheses for guiding the design of experiments aimed at furthering understanding of subcellular distribution and transport phenomena, without focusing on a particular location.
In the future, combinations of experimental methods will be used to study cells loaded with concentrated solutions of small molecules, which should facilitate analyses and provide new insights into the interaction of cells with exogenous chemical agents. For example, by combining computational modeling, Raman confocal microscopy, fluorescence microscopy, electron microscopy and chemical analysis 128, we found that incubating cells with concentrated chloroquine solutions (such as those found in the urine of patients undergoing chloroquine therapy) drives the formation of intralysosomal drug-membrane complexes that bind to other weakly basic molecules (Figure 1) 128. With clofazimine, combining biochemical, microscopy and molecular imaging techniques, revealed that continuous exposure of cells to supersaturated drug solutions resulted in the synthesis of intracellular, autophagosome-like drug inclusions, new organelle-like cytoplasmic structures formed by condensed drug-membrane aggregates derived from mitochondria and possibly other organelles 149. While such drug-membrane complexes may form and accumulate inside cells, such complexes may also form at the plasma membrane and can be shed by cells into the extracellular medium150.
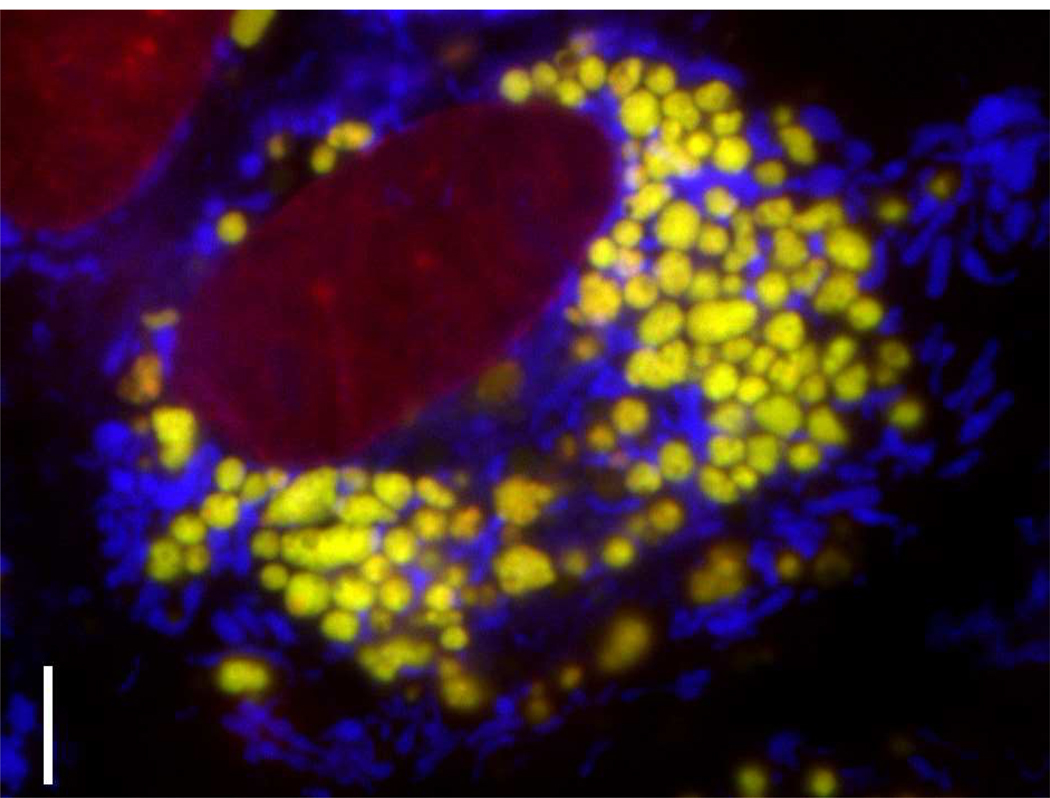
Figure 1.
MDCK cells treated with 50 µM chloroquine concentration for 4 hours prior to staining with Lysotracker Green (yellow, lysosomes), Mitotracker Red (blue, mitochondria) and Hoechst (red, nuclei). Cells exposed to high concentrations of chloroquine undergo profound changes in endolysosomal membrane organization. Continuous accumulation of chloroquine leads to the formation of multilamellar drug-membrane complexes that visibly bind to Lysotracker Green within the lumen of the expanded lysosomes 128. Scale bar: 5 µm.
It is important to realize that cells tend to deplete exogenous chemical agents from the extracellular medium. By adding supersaturating solutions of chemical agents to cells and replenishing the chemical agents as they are depleted from the medium, it is possible to drive the formation of drug-membrane aggregates, which in turn could be used to drive the formation of unnatural, semi-synthetic organelles 110, 128, 149, 151–153. While harnessing the natural subcellular transport pathways, supersaturated solutions of chemical agents (which can be prepared from insoluble molecules pre-solubilized in concentrated DMSO stock solutions before diluting in aqueous media) can drive the formation of atypical supramolecular architectures formed by complexation of chemical agents with cellular membranes. In turn, these supramolecular architectures can aggregate and grow by accretion to form microscopic cytoplasmic inclusions endowed with new, interesting chemical, physical and biological features. The formation of these “unnatural organelles” parallels an emerging interest in the test-tube synthesis of artificial organelles 154–156.
To conclude, traditional approaches aimed at studying the subcellular disposition of small molecules can now be applied towards altering the internal membrane organization of the cell. In our laboratory, this has motivated us to study the synthesis of “unnatural” supramolecular architectures inside cells, derived from complexes of chemical agents with cellular membranes. Continued investigation of subcellular transport phenomena will lead to fundamental insights into the chemistry of life. The ability to predict and optimize the subcellular transport and biodistribution properties of small molecules at the cellular level is now appreciated as a stepping stone towards predicting and optimizing systemic pharmacokinetics and pharmacodynamics. In the future, we envision that today’s research may culminate in the development of new drug delivery strategies and therapeutic modalities. While accurate, quantitative assessment of the microscopic distribution of small molecules inside cells remains a challenge, subcellular transport research is being re-energized by revolutionary concepts derived from systems and synthetic biology. Progress in this field will continue to accelerate with the development of an increasingly sophisticated combination of methods and analytical techniques.
Acknowledgement
This work was funded by NIH grant 1RO1GM078200-01 to G. Rosania. H.N. Tsai was supported with funds from the Center for Molecular Drug Targeting at the University of Michigan, College of Pharmacy.
Footnotes
Supporting information
An excel database file with all the literature references and information of compounds with reported subcellular localizations or compounds from PubChem and DrugBank databases was deposited at the University of Michigan’s Deep Blue data repository and can be accessed through the following URL: http://hdl.handle.net/2027.42/84659
This data has been converted to tabular form and is also provided in the accompanying article 11, under review in Molecular Pharmaceutics.
References
- 1.Zhao K, Zhao GM, Wu D, Soong Y, Birk AV, Schiller PW, Szeto HH. Cell-permeable peptide antioxidants targeted to inner mitochondrial membrane inhibit mitochondrial swelling, oxidative cell death, and reperfusion injury. J Biol Chem. 2004;279:34682–34690. doi: 10.1074/jbc.M402999200. [DOI] [PubMed] [Google Scholar]
- 2.Fernandez-Carneado J, Van Gool M, Martos V, Castel S, Prados P, de Mendoza J, Giralt E. Highly efficient, nonpeptidic oligoguanidinium vectors that selectively internalize into mitochondria. J Am Chem Soc. 2005;127:869–874. doi: 10.1021/ja044006q. [DOI] [PubMed] [Google Scholar]
- 3.Horton KL, Stewart KM, Fonseca SB, Guo Q, Kelley SO. Mitochondria-penetrating peptides. Chem Biol. 2008;15:375–382. doi: 10.1016/j.chembiol.2008.03.015. [DOI] [PubMed] [Google Scholar]
- 4.Yousif LF, Stewart KM, Kelley SO. Targeting mitochondria with organelle-specific compounds: strategies and applications. Chembiochem. 2009;10:1939–1950. doi: 10.1002/cbic.200900185. [DOI] [PubMed] [Google Scholar]
- 5.Rajendran L, Knolker HJ, Simons K. Subcellular targeting strategies for drug design and delivery. Nat Rev Drug Discov. 2010;9:29–42. doi: 10.1038/nrd2897. [DOI] [PubMed] [Google Scholar]
- 6.Fonseca SB, Pereira MP, Mourtada R, Gronda M, Horton KL, Hurren R, Minden MD, Schimmer AD, Kelley SO. Rerouting chlorambucil to mitochondria combats drug deactivation and resistance in cancer cells. Chem Biol. 2011;18:445–453. doi: 10.1016/j.chembiol.2011.02.010. [DOI] [PubMed] [Google Scholar]
- 7.Colston J, Horobin RW, Rashid-Doubell F, Pediani J, Johal KK. Why fluorescent probes for endoplasmic reticulum are selective: an experimental and QSAR-modelling study. Biotech Histochem. 2003;78:323–332. doi: 10.1080/10520290310001646659. [DOI] [PubMed] [Google Scholar]
- 8.Horobin RW, Stockert JC, Rashid-Doubell F. Fluorescent cationic probes for nuclei of living cells: why are they selective? A quantitative structure-activity relations analysis. Histochem Cell Biol. 2006;126:165–175. doi: 10.1007/s00418-006-0156-7. [DOI] [PubMed] [Google Scholar]
- 9.Horobin RW, Trapp S, Weissig V. Mitochondriotropics: a review of their mode of action, and their applications for drug and DNA delivery to mammalian mitochondria. J Control Release. 2007;121:125–136. doi: 10.1016/j.jconrel.2007.05.040. [DOI] [PubMed] [Google Scholar]
- 10.Zhang X, Zheng N, Rosania GR. Simulation-based cheminformatic analysis of organelle-targeted molecules: lysosomotropic monobasic amines. J Comput Aided Mol Des. 2008;22:629–645. doi: 10.1007/s10822-008-9194-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zheng N, Tsai HN, Zhang X, Shedden K, Rosania GR. The Subcellular Distribution of Small Molecules: A Meta-Analysis. Mol Pharmaceutics. doi: 10.1021/mp200093z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lippincott-Schwartz J, Yuan LC, Bonifacino JS, Klausner RD. Rapid redistribution of Golgi proteins into the ER in cells treated with brefeldin A: evidence for membrane cycling from Golgi to ER. Cell. 1989;56:801–813. doi: 10.1016/0092-8674(89)90685-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Journey LJ, Goldstein MN. The effect of terramycin on the fine structure of HeLa cell mitochondria. Cancer Res. 1963;23:551–554. [PubMed] [Google Scholar]
- 14.Lucocq J, Warren G, Pryde J. Okadaic acid induces Golgi apparatus fragmentation and arrest of intracellular transport. J Cell Sci. 1991;100(Pt 4):753–759. doi: 10.1242/jcs.100.4.753. [DOI] [PubMed] [Google Scholar]
- 15.Falgueyret JP, Desmarais S, Oballa R, Black WC, Cromlish W, Khougaz K, Lamontagne S, Masse F, Riendeau D, Toulmond S, Percival MD. Lysosomotropism of basic cathepsin K inhibitors contributes to increased cellular potencies against off-target cathepsins and reduced functional selectivity. J Med Chem. 2005;48:7535–7543. doi: 10.1021/jm0504961. [DOI] [PubMed] [Google Scholar]
- 16.Wibo M, Poole B. Protein degradation in cultured cells. II. The uptake of chloroquine by rat fibroblasts and the inhibition of cellular protein degradation and cathepsin B1. J Cell Biol. 1974;63:430–440. doi: 10.1083/jcb.63.2.430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Homewood CA, Warhurst DC, Peters W, Baggaley VC. Lysosomes, pH and the anti-malarial action of chloroquine. Nature. 1972;235:50–52. doi: 10.1038/235050a0. [DOI] [PubMed] [Google Scholar]
- 18.Mitchell P, Moyle J. Estimation of membrane potential and pH difference across the cristae membrane of rat liver mitochondria. Eur J Biochem. 1969;7:471–484. doi: 10.1111/j.1432-1033.1969.tb19633.x. [DOI] [PubMed] [Google Scholar]
- 19.Verity MA, Brown WJ. Membrane permeability of hepatic mitochondria and lysosomes studied by structure-linked enzyme changes. Exp Mol Pathol. 1973;19:1–14. doi: 10.1016/0014-4800(73)90035-x. [DOI] [PubMed] [Google Scholar]
- 20.Beatrice MC, Palmer JW, Pfeiffer DR. The relationship between mitochondrial membrane permeability, membrane potential, and the retention of Ca2+ by mitochondria. J Biol Chem. 1980;255:8663–8671. [PubMed] [Google Scholar]
- 21.Scarlett JL, Sheard PW, Hughes G, Ledgerwood EC, Ku HH, Murphy MP. Changes in mitochondrial membrane potential during staurosporine-induced apoptosis in Jurkat cells. FEBS Lett. 2000;475:267–272. doi: 10.1016/s0014-5793(00)01681-1. [DOI] [PubMed] [Google Scholar]
- 22.Kornhuber J, Henkel AW, Groemer TW, Stadtler S, Welzel O, Tripal P, Rotter A, Bleich S, Trapp S. Lipophilic cationic drugs increase the permeability of lysosomal membranes in a cell culture system. J Cell Physiol. 2010;224:152–164. doi: 10.1002/jcp.22112. [DOI] [PubMed] [Google Scholar]
- 23.Takayama S, Ojima Y. Photosensitizing Activity of Carcinogenic and Noncarciogenic Polycyclic Hydrocarbons on Cultured Cells. Japan. J. Genetics. 1969;44:231–240. [Google Scholar]
- 24.Kowaltowski AJ, Turin J, Indig GL, Vercesi AE. Mitochondrial effects of triarylmethane dyes. J Bioenerg Biomembr. 1999;31:581–590. doi: 10.1023/a:1005421112345. [DOI] [PubMed] [Google Scholar]
- 25.Filipovska A, Kelso GF, Brown SE, Beer SM, Smith RA, Murphy MP. Synthesis and characterization of a triphenylphosphonium-conjugated peroxidase mimetic. Insights into the interaction of ebselen with mitochondria. J Biol Chem. 2005;280:24113–24126. doi: 10.1074/jbc.M501148200. [DOI] [PubMed] [Google Scholar]
- 26.Tian E, Landowski TH, Stephens OW, Yaccoby S, Barlogie B, Shaughnessy JD., Jr Ellipticine derivative NSC 338258 represents a potential new antineoplastic agent for the treatment of multiple myeloma. Mol Cancer Ther. 2008;7:500–509. doi: 10.1158/1535-7163.MCT-07-0524. [DOI] [PubMed] [Google Scholar]
- 27.Szulc ZM, Bielawski J, Gracz H, Gustilo M, Mayroo N, Hannun YA, Obeid LM, Bielawska A. Tailoring structure-function and targeting properties of ceramides by site-specific cationization. Bioorg Med Chem. 2006;14:7083–7104. doi: 10.1016/j.bmc.2006.07.016. [DOI] [PubMed] [Google Scholar]
- 28.Dindo D, Dahm F, Szulc Z, Bielawska A, Obeid LM, Hannun YA, Graf R, Clavien PA. Cationic long-chain ceramide LCL-30 induces cell death by mitochondrial targeting in SW403 cells. Mol Cancer Ther. 2006;5:1520–1529. doi: 10.1158/1535-7163.MCT-05-0513. [DOI] [PubMed] [Google Scholar]
- 29.Novgorodov SA, Szulc ZM, Luberto C, Jones JA, Bielawski J, Bielawska A, Hannun YA, Obeid LM. Positively charged ceramide is a potent inducer of mitochondrial permeabilization. J Biol Chem. 2005;280:16096–16105. doi: 10.1074/jbc.M411707200. [DOI] [PubMed] [Google Scholar]
- 30.Bielawska A, Bielawski J, Szulc ZM, Mayroo N, Liu X, Bai A, Elojeimy S, Rembiesa B, Pierce J, Norris JS, Hannun YA. Novel analogs of D-e-MAPP and B13. Part 2: signature effects on bioactive sphingolipids. Bioorg Med Chem. 2008;16:1032–1045. doi: 10.1016/j.bmc.2007.08.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Doherty WP, Campbell TC. Aflatoxin inhibition of rat liver mitochondria. Chem Biol Interact. 1973;7:63–77. doi: 10.1016/0009-2797(73)90016-1. [DOI] [PubMed] [Google Scholar]
- 32.Kalina M, Bubis JJ. Myeloid bodies formation in triparanol treated cultured cells. Virchows Arch B Cell Pathol. 1975;19:349–357. doi: 10.1007/BF02889378. [DOI] [PubMed] [Google Scholar]
- 33.Byczkowski JZ. The mode of action of p,p'=DDT on mammalian mitochondria. Toxicology. 1976;6:309–314. doi: 10.1016/0300-483x(76)90034-2. [DOI] [PubMed] [Google Scholar]
- 34.Feldman S, Wang MY, Kaloyanides GJ. Aminoglycosides induce a phospholipidosis in the renal cortex of the rat: an early manifestation of nephrotoxicity. J Pharmacol Exp Ther. 1982;220:514–520. [PubMed] [Google Scholar]
- 35.Seglen PO. Protein degradation in isolated rat hepatocytes is inhibited by ammonia. Biochemical and Biophysical Research Communications. 1975;6:42–52. doi: 10.1016/s0006-291x(75)80292-0. [DOI] [PubMed] [Google Scholar]
- 36.Seglen PO, Gordon PB. Effects of lysosomotropic monoamines, diamines, amino alcohols, and other amino compounds on protein degradation and protein synthesis in isolated rat hepatocytes. Mol Pharm. 1980;18:468–475. [PubMed] [Google Scholar]
- 37.Martin WJ, 2nd; Kachel DL, Vilen T, Natarajan V. Mechanism of phospholipidosis in amiodarone pulmonary toxicity. J Pharmacol Exp Ther. 1989;251:272–278. [PubMed] [Google Scholar]
- 38.de Duve C, de Barsy T, Poole B, Trouet A, Tulkens P, Van Hoof F. Commentary. Lysosomotropic agents. Biochem Pharmacol. 1974;23:2495–2531. doi: 10.1016/0006-2952(74)90174-9. [DOI] [PubMed] [Google Scholar]
- 39.Luiken JJ, Aerts JM, Meijer AJ. The role of the intralysosomal pH in the control of autophagic proteolytic flux in rat hepatocytes. Eur J Biochem. 1996;235:564–573. doi: 10.1111/j.1432-1033.1996.00564.x-i2. [DOI] [PubMed] [Google Scholar]
- 40.Huang SS, Koh HA, Huang JS. Suramin enters and accumulates in low pH intracellular compartments of v-sis-transformed NIH 3T3 cells. FEBS Lett. 1997;416:297–301. doi: 10.1016/s0014-5793(97)01213-1. [DOI] [PubMed] [Google Scholar]
- 41.Holz FG, Schutt F, Kopitz J, Eldred GE, Kruse FE, Volcker HE, Cantz M. Inhibition of lysosomal degradative functions in RPE cells by a retinoid component of lipofuscin. Invest Ophthalmol Vis Sci. 1999;40:737–743. [PubMed] [Google Scholar]
- 42.Schutt F, Bergmann M, Kopitz J, Holz FG. [Mechanism of the inhibition of lysosomal functions in the retinal pigment epithelium by lipofuscin retinoid component A2-E] Ophthalmologe. 2001;98:721–724. doi: 10.1007/s003470170078. [DOI] [PubMed] [Google Scholar]
- 43.Boya P, Gonzalez-Polo RA, Poncet D, Andreau K, Vieira HL, Roumier T, Perfettini JL, Kroemer G. Mitochondrial membrane permeabilization is a critical step of lysosome-initiated apoptosis induced by hydroxychloroquine. Oncogene. 2003;22:3927–3936. doi: 10.1038/sj.onc.1206622. [DOI] [PubMed] [Google Scholar]
- 44.Ishizaki J, Yokogawa K, Ichimura F, Ohkuma S. Uptake of imipramine in rat liver lysosomes in vitro and its inhibition by basic drugs. J Pharmacol Exp Ther. 2000;294:1088–1098. [PubMed] [Google Scholar]
- 45.Daniel WA, Wojcikowski J, Palucha A. Intracellular distribution of psychotropic drugs in the grey and white matter of the brain: the role of lysosomal trapping. Br J Pharmacol. 2001;134:807–814. doi: 10.1038/sj.bjp.0704319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kamo N, Muratsugu M, Hongoh R, Kobatake Y. Membrane potential of mitochondria measured with an electrode sensitive to tetraphenyl phosphonium and relationship between proton electrochemical potential and phosphorylation potential in steady state. J Membr Biol. 1979;49:105–121. doi: 10.1007/BF01868720. [DOI] [PubMed] [Google Scholar]
- 47.Davey GP, Tipton KF, Murphy MP. Uptake and accumulation of 1-methyl-4-phenylpyridinium by rat liver mitochondria measured using an ion-selective electrode. Biochem J. 1992;288(Pt 2):439–443. doi: 10.1042/bj2880439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.James AM, Blaikie FH, Smith RA, Lightowlers RN, Smith PM, Murphy MP. Specific targeting of a DNA-alkylating reagent to mitochondria. Synthesis and characterization of [4-((11aS)-7-methoxy-1,2,3,11a-tetrahydro-5H-pyrrolo[2,1-c][1,4]benzodiaze pin-5-on-8-oxy)butyl]-triphenylphosphonium iodide. Eur J Biochem. 2003;270:2827–2836. doi: 10.1046/j.1432-1033.2003.03660.x. [DOI] [PubMed] [Google Scholar]
- 49.Prasad AR, Luduena RF, Horowitz PM. Detection of energy transfer between tryptophan residues in the tubulin molecule and bound bis(8-anilinonaphthalene-1-sulfonate), an inhibitor of microtubule assembly, that binds to a flexible region on tubulin. Biochemistry. 1986;25:3536–3540. doi: 10.1021/bi00360a010. [DOI] [PubMed] [Google Scholar]
- 50.Wagner E, Cotten M, Mechtler K, Kirlappos H, Birnstiel ML. DNA-binding transferrin conjugates as functional gene-delivery agents: synthesis by linkage of polylysine or ethidium homodimer to the transferrin carbohydrate moiety. Bioconjug Chem. 1991;2:226–231. doi: 10.1021/bc00010a006. [DOI] [PubMed] [Google Scholar]
- 51.Schneider K, Naujok A, Zimmermann HW. Influence of trans-membrane potential and of hydrophobic interactions on dye accumulation in mitochondria of living cells. Photoaffinity labelling of mitochondrial proteins, action of potential dissipating drugs, and competitive staining. Histochemistry. 1994;101:455–461. doi: 10.1007/BF00269496. [DOI] [PubMed] [Google Scholar]
- 52.Ward LD, Timasheff SN. Cooperative multiple binding of bisANS and daunomycin to tubulin. Biochemistry. 1994;33:11891–11899. doi: 10.1021/bi00205a027. [DOI] [PubMed] [Google Scholar]
- 53.Michelsen U, von Hagen J. Isolation of subcellular organelles and structures. Methods Enzymol. 2009;463:305–328. doi: 10.1016/S0076-6879(09)63019-6. [DOI] [PubMed] [Google Scholar]
- 54.Mellett LB, Woods LA. The intra cellular distribution of N-C14-methyl levorphanol in brain, liver and kidney tissue of the rat. J Pharmacol Exp Ther. 1959;125:97–104. [PubMed] [Google Scholar]
- 55.Nair PP, Bucana C. Intracellular distribution of vitamin D in rat liver. Biochim Biophys Acta. 1966;124:254–259. doi: 10.1016/0304-4165(66)90187-5. [DOI] [PubMed] [Google Scholar]
- 56.Albert AE, Warwick GP. The subcellular distribution of tritiated 4-dimethylaminoazobenzene and 2-methyl-4-dimethylaminoazobenzene in rat liver and spleen following a single oral administration. Chem Biol Interact. 1972;5:65–68. doi: 10.1016/0009-2797(72)90048-8. [DOI] [PubMed] [Google Scholar]
- 57.Choie DD, del Campo AA, Guarino AM. Subcellular localization of cis-dichlorodiammineplatinum(II) in rat kidney and liver. Toxicol Appl Pharmacol. 1980;55:245–252. doi: 10.1016/0041-008x(80)90086-1. [DOI] [PubMed] [Google Scholar]
- 58.Yoshida H, Okumura K, Hori R. Subcellular distribution of basic drugs accumulated in the isolated perfused lung. Pharm Res. 1987;4:50–53. doi: 10.1023/a:1016481911538. [DOI] [PubMed] [Google Scholar]
- 59.Miniati M, Paci A, Cocci F, Ciarimboli G, Monti S, Pistolesi M. Mitochondria act as a reservoir for the basic amine HIPDM in the lung. Eur Respir J. 1996;9:2306–2312. doi: 10.1183/09031936.96.09112306. [DOI] [PubMed] [Google Scholar]
- 60.Vickers AE, Sipes IG, Brendel K. Metabolism-related spectral characterization and subcellular distribution of polychlorinated biphenyl congeners in isolated rat hepatocytes. Biochem Pharmacol. 1986;35:297–306. doi: 10.1016/0006-2952(86)90529-0. [DOI] [PubMed] [Google Scholar]
- 61.Martin WJ, 2nd; Standing JE. Amiodarone pulmonary toxicity: biochemical evidence for a cellular phospholipidosis in the bronchoalveolar lavage of human subjects. J Pharmacol Exp Ther. 1988;244:774–779. [PubMed] [Google Scholar]
- 62.Duvvuri M, Feng W, Mathis A, Krise JP. A cell fractionation approach for the quantitative analysis of subcellular drug disposition. Pharm Res. 2004;21:26–32. doi: 10.1023/b:pham.0000012148.12516.3f. [DOI] [PubMed] [Google Scholar]
- 63.Saito Y, Fukuhara A, Nishio K, Hayakawa M, Ogawa Y, Sakamoto H, Fujii K, Yoshida Y, Niki E. Characterization of cellular uptake and distribution of coenzyme Q10 and vitamin E in PC12 cells. J Nutr Biochem. 2009;20:350–357. doi: 10.1016/j.jnutbio.2008.04.005. [DOI] [PubMed] [Google Scholar]
- 64.Liu A, Pajkovic N, Pang Y, Zhu D, Calamini B, Mesecar AL, van Breemen RB. Absorption and subcellular localization of lycopene in human prostate cancer cells. Mol Cancer Ther. 2006;5:2879–2885. doi: 10.1158/1535-7163.MCT-06-0373. [DOI] [PubMed] [Google Scholar]
- 65.Bell RG, Matschiner JT. Intracellular distribution of vitamin K in the rat. Biochim Biophys Acta. 1969;184:597–603. doi: 10.1016/0304-4165(69)90274-8. [DOI] [PubMed] [Google Scholar]
- 66.Woo Y, Argus MF, Arcos JC. Tissue and subcellular distribution of 3H-dioxane in the rat and apparent lack of microsome-catalyzed covalent binding in the target tissue. Life Sci. 1977;21:1447–1456. doi: 10.1016/0024-3205(77)90199-0. [DOI] [PubMed] [Google Scholar]
- 67.Dial LD, Anestis DK, Kennedy SR, Rankin GO. Tissue distribution, subcellular localization and covalent binding of 2-chloroaniline and 4-chloroaniline in Fischer 344 rats. Toxicology. 1998;131:109–119. doi: 10.1016/s0300-483x(98)00122-x. [DOI] [PubMed] [Google Scholar]
- 68.Ross MF, Prime TA, Abakumova I, James AM, Porteous CM, Smith RA, Murphy MP. Rapid and extensive uptake and activation of hydrophobic triphenylphosphonium cations within cells. Biochem J. 2008;411:633–645. doi: 10.1042/BJ20080063. [DOI] [PubMed] [Google Scholar]
- 69.Gruenberg JE, Howell KE. Reconstitution of vesicle fusions occurring in endocytosis with a cell-free system. EMBO J. 1986;5:3091–3101. doi: 10.1002/j.1460-2075.1986.tb04615.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Howell KE, Gruenberg J, Ito A, Palade GE. Immuno-isolation of subcellular components. Prog Clin Biol Res. 1988;270:77–90. [PubMed] [Google Scholar]
- 71.Pasquali C, Fialka I, Huber LA. Subcellular fractionation, electromigration analysis and mapping of organelles. J Chromatogr B Biomed Sci Appl. 1999;722:89–102. doi: 10.1016/s0378-4347(98)00314-4. [DOI] [PubMed] [Google Scholar]
- 72.Murphy RF. Processing of Endocytosed Material. Advances in Molecular and Cell Biology. 1988;2:159–180. [Google Scholar]
- 73.Bock G, Steinlein P, Huber LA. Cell biologists sort things out: Analysis and purification of intracellular organelles by flow cytometry. Trends Cell Biol. 1997;7:499–503. doi: 10.1016/S0962-8924(97)01160-4. [DOI] [PubMed] [Google Scholar]
- 74.Rajotte D, Stearns CD, Kabcenell AK. Isolation of mast cell secretory lysosomes using flow cytometry. Cytometry A. 2003;55:94–101. doi: 10.1002/cyto.a.10065. [DOI] [PubMed] [Google Scholar]
- 75.Nasirudeen AM, Tan KS. Isolation and characterization of the mitochondrion-like organelle from Blastocystis hominis. J Microbiol Methods. 2004;58:101–109. doi: 10.1016/j.mimet.2004.03.008. [DOI] [PubMed] [Google Scholar]
- 76.Stefaner I, Klapper H, Sztul E, Fuchs R. Free-flow electrophoretic analysis of endosome subpopulations of rat hepatocytes. Electrophoresis. 1997;18:2516–2522. doi: 10.1002/elps.1150181405. [DOI] [PubMed] [Google Scholar]
- 77.Tulp A, Verwoerd D, Benham A, Neefjes J. High-resolution density gradient electrophoresis of proteins and subcellular organelles. Electrophoresis. 1997;18:2509–2515. doi: 10.1002/elps.1150181404. [DOI] [PubMed] [Google Scholar]
- 78.Tulp A, Fernandez-Borja M, Verwoerd D, Neefjes J. High-resolution density gradient electrophoresis of subcellular organelles and proteins under nondenaturing conditions. Electrophoresis. 1998;19:1288–1293. doi: 10.1002/elps.1150190812. [DOI] [PubMed] [Google Scholar]
- 79.Weber PJ, Weber G, Eckerskorn C. Isolation of organelles and prefractionation of protein extracts using free-flow electrophoresis. Curr Protoc Protein Sci. 2004 doi: 10.1002/0471140864.ps2205s32. Chapter 22, Unit 22 5. [DOI] [PubMed] [Google Scholar]
- 80.Xiong G, Chen Y, Arriaga EA. Measuring the doxorubicin content of single nuclei by micellar electrokinetic capillary chromatography with laser-induced fluorescence detection. Anal Chem. 2005;77:3488–3493. doi: 10.1021/ac0500378. [DOI] [PubMed] [Google Scholar]
- 81.Chen Y, Arriaga EA. Individual acidic organelle pH measurements by capillary electrophoresis. Anal Chem. 2006;78:820–826. doi: 10.1021/ac051513x. [DOI] [PubMed] [Google Scholar]
- 82.Johnson RD, Navratil M, Poe BG, Xiong G, Olson KJ, Ahmadzadeh H, Andreyev D, Duffy CF, Arriaga EA. Analysis of mitochondria isolated from single cells. Anal Bioanal Chem. 2007;387:107–118. doi: 10.1007/s00216-006-0689-6. [DOI] [PubMed] [Google Scholar]
- 83.Kostal V, Arriaga EA. Recent advances in the analysis of biological particles by capillary electrophoresis. Electrophoresis. 2008;29:2578–2586. doi: 10.1002/elps.200700917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Wang Y, Arriaga EA. Monitoring incorporation, transformation and subcellular distribution of N-l-leucyl-doxorubicin in uterine sarcoma cells using capillary electrophoretic techniques. Cancer Lett. 2008;262:123–132. doi: 10.1016/j.canlet.2007.11.034. [DOI] [PubMed] [Google Scholar]
- 85.Diettrich O, Mills K, Johnson AW, Hasilik A, Winchester BG. Application of magnetic chromatography to the isolation of lysosomes from fibroblasts of patients with lysosomal storage disorders. FEBS Lett. 1998;441:369–372. doi: 10.1016/s0014-5793(98)01578-6. [DOI] [PubMed] [Google Scholar]
- 86.Duvvuri M, Krise JP. A novel assay reveals that weakly basic model compounds concentrate in lysosomes to an extent greater than pH-partitioning theory would predict. Mol Pharmaceutics. 2005;2:440–448. doi: 10.1021/mp050043s. [DOI] [PubMed] [Google Scholar]
- 87.Kaufmann AM, Goldman SD, Krise JP. A fluorescence resonance energy transfer-based approach for investigating late endosome-lysosome retrograde fusion events. Anal Biochem. 2009;386:91–97. doi: 10.1016/j.ab.2008.11.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Hoff SF, MacInnis AJ. Ultrastructural localization of phenothiazines and tetracycline: a new histochemical approach. J Histochem Cytochem. 1983;31:613–625. doi: 10.1177/31.5.6841967. [DOI] [PubMed] [Google Scholar]
- 89.Muller T. Electron microscopic demonstration of intracelluar promethazine accumulation sites by a precipitation technique: application to the cerebellar cortex of the mouse. J Histochem Cytochem. 1996;44:531–535. doi: 10.1177/44.5.8627010. [DOI] [PubMed] [Google Scholar]
- 90.Ivanova S, Batliwalla F, Mocco J, Kiss S, Huang J, Mack W, Coon A, Eaton JW, Al-Abed Y, Gregersen PK, Shohami E, Connolly ES, Jr, Tracey KJ. Neuroprotection in cerebral ischemia by neutralization of 3-aminopropanal. Proc Natl Acad Sci U S A. 2002;99:5579–5584. doi: 10.1073/pnas.082609299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Li W, Yuan XM, Ivanova S, Tracey KJ, Eaton JW, Brunk UT. 3-Aminopropanal, formed during cerebral ischaemia, is a potent lysosomotropic neurotoxin. Biochem J. 2003;371:429–436. doi: 10.1042/BJ20021520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Bain JA, Mayer SE. The intracellular localization of fluorescent convulsants. J Pharmacol Exp Ther. 1956;118:1–16. [PubMed] [Google Scholar]
- 93.Egorin MJ, Clawson RE, Ross LA, Schlossberger NM, Bachur NR. Cellular accumulation and disposition of aclacinomycin A. Cancer Res. 1979;39:4396–4400. [PubMed] [Google Scholar]
- 94.Terasaki M, Song J, Wong JR, Weiss MJ, Chen LB. Localization of endoplasmic reticulum in living and glutaraldehyde-fixed cells with fluorescent dyes. Cell. 1984;38:101–108. doi: 10.1016/0092-8674(84)90530-0. [DOI] [PubMed] [Google Scholar]
- 95.Bucana C, Saiki I, Nayar R. Uptake and accumulation of the vital dye hydroethidine in neoplastic cells. J Histochem Cytochem. 1986;34:1109–1115. doi: 10.1177/34.9.2426339. [DOI] [PubMed] [Google Scholar]
- 96.Steinberg SF, Bilezikian JP, Al-Awqati Q. Fura-2 fluorescence is localized to mitochondria in endothelial cells. Am J Physiol. 1987;253:C744–C747. doi: 10.1152/ajpcell.1987.253.5.C744. [DOI] [PubMed] [Google Scholar]
- 97.Jeon CJ, Masland RH. Selective accumulation of diamidino yellow and chromomycin A3 by retinal glial cells. J Histochem Cytochem. 1993;41:1651–1658. doi: 10.1177/41.11.8409373. [DOI] [PubMed] [Google Scholar]
- 98.Deng Y, Bennink JR, Kang HC, Haugland RP, Yewdell JW. Fluorescent conjugates of brefeldin A selectively stain the endoplasmic reticulum and Golgi complex of living cells. J Histochem Cytochem. 1995;43:907–915. doi: 10.1177/43.9.7543914. [DOI] [PubMed] [Google Scholar]
- 99.Chalfie M, Tu Y, Euskirchen G, Ward WW, Prasher DC. Green fluorescent protein as a marker for gene expression. Science. 1994;263:802–805. doi: 10.1126/science.8303295. [DOI] [PubMed] [Google Scholar]
- 100.Heim R, Prasher DC, Tsien RY. Wavelength mutations and posttranslational autoxidation of green fluorescent protein. Proc Natl Acad Sci U S A. 1994;91:12501–12504. doi: 10.1073/pnas.91.26.12501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Huang M, Chalfie M. Gene interactions affecting mechanosensory transduction in Caenorhabditis elegans. Nature. 1994;367:467–470. doi: 10.1038/367467a0. [DOI] [PubMed] [Google Scholar]
- 102.Rizzuto R, Brini M, Pizzo P, Murgia M, Pozzan T. Chimeric green fluorescent protein as a tool for visualizing subcellular organelles in living cells. Curr Biol. 1995;5:635–642. doi: 10.1016/s0960-9822(95)00128-x. [DOI] [PubMed] [Google Scholar]
- 103.Heim R, Tsien RY. Engineering green fluorescent protein for improved brightness, longer wavelengths and fluorescence resonance energy transfer. Curr Biol. 1996;6:178–182. doi: 10.1016/s0960-9822(02)00450-5. [DOI] [PubMed] [Google Scholar]
- 104.Chen X, Murphy RF. Objective clustering of proteins based on subcellular location patterns. J Biomed Biotechnol. 2005;2005:87–95. doi: 10.1155/JBB.2005.87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Chen X, Velliste M, Murphy RF. Automated interpretation of subcellular patterns in fluorescence microscope images for location proteomics. Cytometry A. 2006;69:631–640. doi: 10.1002/cyto.a.20280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Chen SC, Zhao T, Gordon GJ, Murphy RF. Automated image analysis of protein localization in budding yeast. Bioinformatics. 2007;23:66–71. doi: 10.1093/bioinformatics/btm206. [DOI] [PubMed] [Google Scholar]
- 107.Garcia Osuna E, Murphy RF. Automated, systematic determination of protein subcellular location using fluorescence microscopy. Subcell Biochem. 2007;43:263–276. doi: 10.1007/978-1-4020-5943-8_12. [DOI] [PubMed] [Google Scholar]
- 108.Newberg J, Hua J, Murphy RF. Location proteomics: systematic determination of protein subcellular location. Methods Mol Biol. 2009;500:313–332. doi: 10.1007/978-1-59745-525-1_11. [DOI] [PubMed] [Google Scholar]
- 109.Chen VY, Khersonsky SM, Shedden K, Chang YT, Rosania GR. System dynamics of subcellular transport. Mol Pharmaceutics. 2004;1:414–425. doi: 10.1021/mp049916t. [DOI] [PubMed] [Google Scholar]
- 110.Shedden K, Li Q, Liu F, Chang YT, Rosania GR. Machine vision-assisted analysis of structure-localization relationships in a combinatorial library of prospective bioimaging probes. Cytometry A. 2009;75:482–493. doi: 10.1002/cyto.a.20713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Shedden K, Rosania GR. Chemical address tags of fluorescent bioimaging probes. Cytometry A. 2010;77A:429–438. doi: 10.1002/cyto.a.20847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Rosania GR, Crippen G, Woolf P, States D, Shedden K. A cheminformatic toolkit for mining biomedical knowledge. Pharm Res. 2007;24:1791–1802. doi: 10.1007/s11095-007-9285-5. [DOI] [PubMed] [Google Scholar]
- 113.Phelps MA, Foraker AB, Gao W, Dalton JT, Swaan PW. A novel rhodamine-riboflavin conjugate probe exhibits distinct fluorescence resonance energy transfer that enables riboflavin trafficking and subcellular localization studies. Mol Pharm. 2004;1:257–266. doi: 10.1021/mp0499510. [DOI] [PubMed] [Google Scholar]
- 114.Pagano RE, Martin OC, Kang HC, Haugland RP. A novel fluorescent ceramide analogue for studying membrane traffic in animal cells: accumulation at the Golgi apparatus results in altered spectral properties of the sphingolipid precursor. J Cell Biol. 1991;113:1267–1279. doi: 10.1083/jcb.113.6.1267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Baxter DF, Kirk M, Garcia AF, Raimondi A, Holmqvist MH, Flint KK, Bojanic D, Distefano PS, Curtis R, Xie Y. A novel membrane potential-sensitive fluorescent dye improves cell-based assays for ion channels. J Biomol Screen. 2002;7:79–85. doi: 10.1177/108705710200700110. [DOI] [PubMed] [Google Scholar]
- 116.Kotaki A, Yagi K. Fluorescence properties of flavins in various solvents. J Biochem. 1970;68:509–516. doi: 10.1093/oxfordjournals.jbchem.a129381. [DOI] [PubMed] [Google Scholar]
- 117.Haidekker MA, Brady TP, Lichlyter D, Theodorakis EA. Effects of solvent polarity and solvent viscosity on the fluorescent properties of molecular rotors and related probes. Bioorg Chem. 2005;33:415–425. doi: 10.1016/j.bioorg.2005.07.005. [DOI] [PubMed] [Google Scholar]
- 118.Gustavsson T, Sarkar N, Banyasz A, Markovitsi D, Improta R. Solvent effects on the steady-state absorption and fluorescence spectra of uracil, thymine and 5-fluorouracil. Photochem Photobiol. 2007;83:595–599. doi: 10.1111/j.1751-1097.2007.00052.x. [DOI] [PubMed] [Google Scholar]
- 119.White A. Effect of pH on fluorescence of tryosine, tryptophan and related compounds. Biochem J. 1959;71:217–220. doi: 10.1042/bj0710217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Weiner ID, Hamm LL. Use of fluorescent dye BCECF to measure intracellular pH in cortical collecting tubule. Am J Physiol. 1989;256:F957–F964. doi: 10.1152/ajprenal.1989.256.5.F957. [DOI] [PubMed] [Google Scholar]
- 121.Jiang XJ, Lo PC, Yeung SL, Fong WP, Ng DK. A pH-responsive fluorescence probe and photosensitiser based on a tetraamino silicon(IV) phthalocyanine. Chem Commun (Camb) 2010;46:3188–3190. doi: 10.1039/c000605j. [DOI] [PubMed] [Google Scholar]
- 122.Sundbom E, Strand M, Hallgren JE. Temperature-induced fluorescence changes: a screening method for frost tolerance of potato (solanum sp.) Plant Physiol. 1982;70:1299–1302. doi: 10.1104/pp.70.5.1299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Ogawa H, Inouye S, Tsuji FI, Yasuda K, Umesono K. Localization, trafficking, and temperature-dependence of the Aequorea green fluorescent protein in cultured vertebrate cells. Proc Natl Acad Sci U S A. 1995;92:11899–11903. doi: 10.1073/pnas.92.25.11899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Gota C, Okabe K, Funatsu T, Harada Y, Uchiyama S. Hydrophilic fluorescent nanogel thermometer for intracellular thermometry. J Am Chem Soc. 2009;131:2766–2767. doi: 10.1021/ja807714j. [DOI] [PubMed] [Google Scholar]
- 125.Ohulchanskyy TY, Pudavar HE, Yarmoluk SM, Yashchuk VM, Bergey EJ, Prasad PN. A monomethine cyanine dye Cyan 40 for two-photon-excited fluorescence detection of nucleic acids and their visualization in live cells. Photochem Photobiol. 2003;77:138–145. doi: 10.1562/0031-8655(2003)077<0138:amcdcf>2.0.co;2. [DOI] [PubMed] [Google Scholar]
- 126.Olson KJ, Ahmadzadeh H, Arriaga EA. Within the cell: analytical techniques for subcellular analysis. Anal Bioanal Chem. 2005;382:906–917. doi: 10.1007/s00216-005-3135-2. [DOI] [PubMed] [Google Scholar]
- 127.Ling J, Weitman SD, Miller MA, Moore RV, Bovik AC. Direct Raman imaging techniques for study of the subcellular distribution of a drug. Appl Opt. 2002;41:6006–6017. doi: 10.1364/ao.41.006006. [DOI] [PubMed] [Google Scholar]
- 128.Zheng N, Zhang X, Rosania GR. Effect of phospholipidosis on the cellular pharmacokinetics of chloroquine. J Pharmacol Exp Ther. 2011;336:661–671. doi: 10.1124/jpet.110.175679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Nan X, Potma EO, Xie XS. Nonperturbative chemical imaging of organelle transport in living cells with coherent anti-stokes Raman scattering microscopy. Biophys J. 2006;91:728–735. doi: 10.1529/biophysj.105.074534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Nan X, Tonary AM, Stolow A, Xie XS, Pezacki JP. Intracellular imaging of HCV RNA and cellular lipids by using simultaneous two-photon fluorescence and coherent anti-Stokes Raman scattering microscopies. Chembiochem. 2006;7:1895–1897. doi: 10.1002/cbic.200600330. [DOI] [PubMed] [Google Scholar]
- 131.Lyn RK, Kennedy DC, Sagan SM, Blais DR, Rouleau Y, Pegoraro AF, Xie XS, Stolow A, Pezacki JP. Direct imaging of the disruption of hepatitis C virus replication complexes by inhibitors of lipid metabolism. Virology. 2009;394:130–142. doi: 10.1016/j.virol.2009.08.022. [DOI] [PubMed] [Google Scholar]
- 132.Galle P. Tissue localization of stable and radioactive nuclides by secondary-ion microscopy. J Nucl Med. 1982;23:52–57. [PubMed] [Google Scholar]
- 133.Boxer SG, Kraft ML, Weber PK. Advances in imaging secondary ion mass spectrometry for biological samples. Annu Rev Biophys. 2009;38:53–74. doi: 10.1146/annurev.biophys.050708.133634. [DOI] [PubMed] [Google Scholar]
- 134.Kilburn MR, Jones DL, Clode PL, Cliff JB, Stockdale EA, Herrmann AM, Murphy DV. Application of nanoscale secondary ion mass spectrometry to plant cell research. Plant Signal Behav. 2010;5:760–762. doi: 10.4161/psb.5.6.11775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Pacholski ML, Cannon DM, Jr, Ewing AG, Winograd N. Static time-of-flight secondary ion mass spectrometry imaging of freeze-fractured, frozen-hydrated biological membranes. Rapid Commun Mass Spectrom. 1998;12:1232–1235. doi: 10.1002/(SICI)1097-0231(19980930)12:18<1232::AID-RCM319>3.0.CO;2-G. [DOI] [PubMed] [Google Scholar]
- 136.Oba K, Gong H, Amemiya T, Baba K, Takaya K. Applying secondary ion mass spectrometry to the analysis of elements in goblet cells of conjunctiva. J Electron Microsc (Tokyo) 2001;50:325–332. doi: 10.1093/jmicro/50.4.325. [DOI] [PubMed] [Google Scholar]
- 137.Chandra S, Lorey ID, Smith DR. Quantitative subcellular secondary ion mass spectrometry (SIMS) imaging of boron-10 and boron-11 isotopes in the same cell delivered by two combined BNCT drugs: in vitro studies on human glioblastoma T98G cells. Radiat Res. 2002;157:700–710. doi: 10.1667/0033-7587(2002)157[0700:qssims]2.0.co;2. [DOI] [PubMed] [Google Scholar]
- 138.Chehade F, de Labriolle-Vaylet C, Moins N, Moreau MF, Papon J, Labarre P, Galle P, Veyre A, Hindie E. Secondary ion mass spectrometry as a tool for investigating radiopharmaceutical distribution at the cellular level: the example of I-BZA and (14)C-I-BZA. J Nucl Med. 2005;46:1701–1706. [PubMed] [Google Scholar]
- 139.Clode PL, Stern RA, Marshall AT. Subcellular imaging of isotopically labeled carbon compounds in a biological sample by ion microprobe (NanoSIMS) Microsc Res Tech. 2007;70:220–229. doi: 10.1002/jemt.20409. [DOI] [PubMed] [Google Scholar]
- 140.Li Q, Kim Y, Namm J, Kulkarni A, Rosania GR, Ahn YH, Chang YT. RNA-selective, live cell imaging probes for studying nuclear structure and function. Chem Biol. 2006;13:615–623. doi: 10.1016/j.chembiol.2006.04.007. [DOI] [PubMed] [Google Scholar]
- 141.Trapp S, Rosania GR, Horobin RW, Kornhuber J. Quantitative modeling of selective lysosomal targeting for drug design. Eur Biophys J. 2008;37:1317–1328. doi: 10.1007/s00249-008-0338-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Balaz S. Modeling kinetics of subcellular disposition of chemicals. Chem Rev. 2009;109:1793–1899. doi: 10.1021/cr030440j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Selassie CD. History of Quantitative Structure-Activity Relationships. In: Abraham DJ, editor. Burger’s Medicinal Chemistry and Drug Discovery. 6th ed. New York: John Wiley & Sons, Inc; 2003. pp. 1–48. [Google Scholar]
- 144.Balaz S, Sturdik E, Augustin J. Subcellular distribution of compounds in biosystems. Bull Math Biol. 1988;50:367–378. doi: 10.1007/BF02459706. [DOI] [PubMed] [Google Scholar]
- 145.Dvorsky R, Balaz S, Sawchuk RJ. Kinetics of subcellular distribution of compounds in simple biosystems and its use in QSAR. J Theor Biol. 1997;185:213–222. doi: 10.1006/jtbi.1996.0308. [DOI] [PubMed] [Google Scholar]
- 146.Trapp S, Horobin RW. A predictive model for the selective accumulation of chemicals in tumor cells. Eur Biophys J. 2005;34:959–966. doi: 10.1007/s00249-005-0472-1. [DOI] [PubMed] [Google Scholar]
- 147.Zhang X, Shedden K, Rosania GR. A cell-based molecular transport simulator for pharmacokinetic prediction and cheminformatic exploration. Mol Pharmaceutics. 2006;3:704–716. doi: 10.1021/mp060046k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Yu JY, Rosania GR. Cell-based multiscale computational modeling of small molecule absorption and retention in the lungs. Pharm Res. 2010;27:457–467. doi: 10.1007/s11095-009-0034-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Baik J, Rosania GR. Capsular Xenobiotic Inclusions within Autophagosome-like Bodies; American Association of Pharmaceutical Scientists Annual Meeting; New Orleans, LA, USA: 2010. [Google Scholar]
- 150.Shedden K, Xie XT, Chandaroy P, Chang YT, Rosania GR. Expulsion of small molecules in vesicles shed by cancer cells: association with gene expression and chemosensitivity profiles. Cancer Res. 2003;63:4331–4337. [PubMed] [Google Scholar]
- 151.Pais AV, Pereira S, Garg I, Stephen J, Antony M, Inchara YK. Intra-abdominal, crystal-storing histiocytosis due to clofazimine in a patient with lepromatous leprosy and concurrent carcinoma of the colon. Lepr Rev. 2004;75:171–176. [PubMed] [Google Scholar]
- 152.Chatman LA, Morton D, Johnson TO, Anway SD. A strategy for risk management of drug-induced phospholipidosis. Toxicol Pathol. 2009;37:997–1005. doi: 10.1177/0192623309352496. [DOI] [PubMed] [Google Scholar]
- 153.de Souza W, Rodrigues JC. Sterol Biosynthesis Pathway as Target for Anti-trypanosomatid Drugs. Interdiscip Perspect Infect Dis. 2009;2009:642502. doi: 10.1155/2009/642502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Tanner P, Baumann P, Enea R, Onaca O, Palivan C, Meier W. Polymeric Vesicles: From Drug Carriers to Nanoreactors and Artificial Organelles. Acc Chem Res. 2011 doi: 10.1021/ar200036k. [DOI] [PubMed] [Google Scholar]
- 155.Ben-Haim N, Broz P, Marsch S, Meier W, Hunziker P. Cell-specific integration of artificial organelles based on functionalized polymer vesicles. Nano Lett. 2008;8:1368–1373. doi: 10.1021/nl080105g. [DOI] [PubMed] [Google Scholar]
- 156.Tanner P, Egli S, Balasubramanian V, Onaca O, Palivan CG, Meier W. Can polymeric vesicles that confine enzymatic reactions act as simplified organelles? FEBS Lett. 2011;585:1699–1706. doi: 10.1016/j.febslet.2011.05.003. [DOI] [PubMed] [Google Scholar]