Abstract
Poly(ADP)ribose polymerase (PARP) inhibitors modify the enzymatic activity of PARP1/2. When certain PARP inhibitors are used either alone or in combination with DNA damage agents they may cause a G2/M mitotic arrest and/or apoptosis in a susceptible genetic context. PARP1 interacts with the cell cycle checkpoint proteins Ataxia Telangectasia Mutated (ATM) and ATM and Rad3-related (ATR) and therefore may influence growth arrest cascades. The PARP inhibitor PJ34 causes a mitotic arrest by an unknown mechanism in certain cell lines, therefore we asked whether PJ34 conditionally activated the checkpoint pathways and which downstream targets were necessary for mitotic arrest. We found that PJ34 produced a concentration dependent G2/M mitotic arrest and differentially affected cell survival in cells with diverse genetic backgrounds. p53 was activated and phosphorylated at Serine15 followed by p21 gene activation through both p53-dependent and -independent pathways. The mitotic arrest was caffeine sensitive and UCN01 insensitive and did not absolutely require p53, ATM or Chk1, while p21 was necessary for maintaining the growth arrest. Significantly, by using stable knockdown cell lines, we found that neither PARP1 nor PARP2 were required for any of these effects produced by PJ34. These results raise questions and cautions for evaluating PARP inhibitor effectiveness, suggesting that not only should effects on PARP’s diverse ADP-ribosylation independent protein interactions be considered, but also effects on homologous proteins that may be producing either overlapping or distinct effects.
Keywords: poly(ADP)-ribose polymerase, PARP inhibitors, p21, breast cancer, cell cycle checkpoint
1. Introduction
Poly(ADP)-ribose polymerase1 (PARP1) inhibitors are touted as a breakthrough for cancer treatment in solid tumors such as triple-negative breast cancer and ovarian cancer through their effects on PARP1’s enzymatic ADP ribosylation function; however, there are less well-characterized effects on other PARP1 interactions and reported functions that may also be critical for successful PARP inhibitor therapy.
PARP1 is the most abundant of 18 predicted mammalian PARPs, has some overlapping activity with PARP2 [1] and interacts with DNA and other proteins to affect replication, DNA repair or recombination and gene transcription by both its enzymatic activity and protein-protein/DNA interactions [2, 3]. Cancer sensitivity to PARP inhibitor monotherapy likely relies on a permissive cellular genetic context or lesion such as BRCA1/2 mutations in breast cancer cells [4, 5], or by causing sensitization to alkylating agents and ionizing radiation for additive lethality [6]. When used alone, the PARP inhibitor PJ34 caused cell cycle arrest in breast cancer (MCF7) [7], leukemia [8] and melanoma cell lines (M14) [9], an effect shared by only a few PARP inhibitors suggesting that specific effects on PARP1 and subsequently the checkpoint pathways are responsible. The structural heterogeneity of PARP inhibitors suggests a high probability for pleiotropic secondary effects on PARP1, other PARP family members or NAD+ pocket containing proteins and ADP-ribosyltransferases; therefore, a critical question for this field is what PARP specific non-enzymatic and PARP non-specific effects are caused by PARP inhibitors, and how do they affect both normal and cancerous cells?
Cell cycle checkpoint activation and growth arrest in response to external and internal DNA damage relies on the ATM and ATR kinases and their downstream targets, Chk1, Chk2 and p53 [10, 11]. ATM and ATR activation results in Chk2(Thr68) [12], Chk1(Ser317, 345) [13] and p53(Ser15) [14] phosphorylation, inactivation of cdc25c and subsequently CyclinB/Cdk1 [15]. In general, ATM-Chk2 regulates the G1/S checkpoint (sometimes through p53) [16] or the G2/M checkpoint [17], and ATR-Chk1 regulates the S and G2/M checkpoints [18], although cross talk is known [19]. PARP1 with both ATR [20] and ATM [21] and interestingly ATM/PARP1 double mutant mice are embryonic lethal [22], suggesting another susceptible pathway for PARP inhibitor induced apoptosis. Certain PARP inhibitors including PJ34 may induce growth arrest when used in conjunction with irradiation [23] and methylating agents [24], or cause a G2/M arrest by themselves [4, 7], highlighting potentially different outcomes for the inhibition of activated PARP versus the effects from inhibitor occupied un-activated PARP. The complex functional and physical relationship between PARP1, DNA repair and ATM/ATR suggest that PARP inhibitors could contextually affect the checkpoint kinase cascade, however, the up-and downstream mechanisms are poorly understood.
Following checkpoint activation, one target for both p53-dependent G1-arrest [25] and p53-independent G2 arrest [26] is p21waf1/cip1, whose expression is regulated by diverse molecules and regulatory complexes [27]. P21 directly inhibits the CDK1/2 (cdc2) kinases [28], participates in p53-dependent transcriptional repression of cdc25c, cdc2, cyclin B and Chk1 [29] and binds PARP1 [30]. PARP inhibitors can inhibit p53 activation, delay the phosphorylation of p53 and γ-H2A.X [21] and p53-dependent and -independent p21 expression following a DNA damage stimulus [31, 32], possibly through repressive effects on p21 transcription [33]. Regulating p21 expression is one model by which PARP inhibitors may cause mitotic arrest following activation of different checkpoint pathways.
In the current study we demonstrate that the PARP inhibitor PJ34 produced a concentration dependent, G2/M growth mitotic arrest in cells with different genetic backgrounds. PJ34 activated the ATM/ATR checkpoint pathways producing a mitotic arrest that was attenuated by caffeine, but not UCN01. ATM and Chk1 were each not required for PJ34 mitotic arrest, but the absence of both ATM/ATR attenuated the effect. Following PJ34 treatment, rapid p21 gene expression occurred by both p53-dependent and independent mechanisms and although p53 was activated and phosphorylated, it was not absolutely required for mitotic arrest whereas p21 was necessary, at least in part, for full growth arrest. Time and dose limited PJ34 exposure resulted in survivability differences, within and between different cell lines. Most importantly, we show by stable cell line PARP knockdown that these effects do not require PARP1, raising questions and cautions to improve our understanding for both the non-enzymatic, PARP specific and off-target effects of PARP inhibitors.
2. Materials and Methods
2.1 Cell lines
All cell lines were maintained as sub-confluent monolayers and freshly passaged for each experiment. MCF-7, U2OS, TAT3, H1299, HeLa cell lines were grown in Dulbecco’s Minimal Essential Medium (DMEM) supplemented with 10% fetal calf serum (FCS) and antibiotics (100 U/ml penicillin and 100 µg/ml streptomycin) while primary human skin fibroblasts (HSF) were supplemented with 15% FCS. PARP knockdown (KD) cell lines (MCF7, HeLa, H1299) were maintained in 10% FCS/DMEM plus appropriate selection antibiotics. Construction of the HeLa:PARPKD lines was previously described [33]; the H1299 and MCF7 PARPKD cell lines were created in the same manner. In brief, stable shRNA KD cell lines were obtained by selecting resistant cell pools transfected with lentiviruses containing shRNAs for p53 [34], Chk1 (5’-AAGCGTGCCGTAGACTGTCCA), PARP1, PARP2, co-transfected PARP1+2 or a non-specific control sequence (AAACTACCGTTGTTATAGGTG) utilizing the human H1 promoter, T6 terminator and selection markers. Multiple lines were screened and single cell lines were established and used for these experiments.
2.2. Antibodies
Primary antibodies used: PARP1, phospho(Ser15)-p53, Caspase-3 (3G2), p21 (DCS60), phospho(Serine345)-Chk1 (133D3), phospho(Thr68)-Chk2 (C13C1), Akt, cyclin D1 (DSC6), cyclin D3 (DSC22), p15INK4B, and p27Kip1 were from Cell Signaling; PARP1 (C2-10; BD Pharmigen); phospho(Ser10)-HistoneH3, phospho(Ser139)-γH2A.X were from Millipore; tubulin (b-5-1-2; Sigma); p53 (Bp53-12); Chk1 (G4); cdc25c (C20), cdc2 (F4); cyclin B (H20) were from Santa Cruz. Secondary antibodies were conjugated to either fluorophores (Invitrogen) or horseradish peroxidase (Bio-Rad).
2.3 Drug Treatments
All compounds were dissolved and stored per the manufacturers recommendations. PJ34 ([N-(6-oxo-5,6-dihydrophenanthridin-2-yl)-N, N-dimethylacetamide. HCl]), EB47, 3-aminobenzamide (3AB) (EMD Biosciences), TIQ-A (Thieno[2,3-c]isoquinolin-5-one) and 4-AN (4-amino-1,8-napthalimide) (Sigma) were diluted into cell culture media at the indicated concentrations. For withdrawal experiments, cells were treated for the indicated time and then either left in the same media (no wash) or washed 3 times (wash) and maintained in fresh growth media without drug for 24 or 48 hours. Hydroxyurea (10 mM; Sigma), caffeine (5 mM, Sigma), UCN01 (7-hydroxystaurosporine, 300 nM; Alexis) or Galardin (GM6001; (R)-N4-Hydroxy-N1-[(S)-2-(1H-indol-3-yl)-1-methylcarbamoyl-ethyl]-2-isobutyl-succinamide; Sigma) were either co-applied with PJ34 or diluted into media alone.
2.4 Mitotic Assay
Cells were plated in 12 well dishes and treated while in growth phase for either 6 or 24 hours as indicated. Tubulin and phospho(Ser10)HistoneH3 immunofluorescence were performed by fixation in 4% paraformaldehyde, then the cells were washed, blocked and incubated with anti-phospho(Ser10)-Histone H3 (pH3; 1:400) and anti-tubulin (1:5000) in 3% Normal Goat Serum (NGS)/0.1%TX100/PBS, washed and incubated with secondary fluorophore antibodies. Staining in each well was quantified on an Odyssey Imager (Licor). Data were collected by acquiring an image within each well boarder and processed by creating a binary grayscale image using NIH-ImageJ software, background subtracted, calculating pH3/tubulin signal ratio for each triplicate condition and a normalized mitotic index was calculated relative to the control condition. Statistical analysis (SEM or t-tests, 95% confidence interval [CI]) and graphing was performed with Prism software.
2.5 Western blotting
Whole cell extracts were prepared in cold lysis buffer (50 mM HEPES pH7.4, 250 mM NaCl, 10 mM NaF, 0.2 mM EDTA, 0.5% NP-40 plus protease inhibitors (Sigma)), cellular debris pelleted and the supernatant quantified and used for western blots. For some experiments, antibodies were incubated in a 1:1 dilution of Aqua Block (East Coast Bio) and 0.5% Tween-20/TBS and detection by fluorophore secondary antibody using an Odyssey Imager. Phospho-specific antibodies were incubated in 5%BSA/0.5% Tween-20/TBS and HRP conjugated secondary antibodies were used.
2.6 Semi-quantitative and real-time PCR
RNA isolation, sq-PCR and rtPCR for p21 and β-actin were performed as previously described [33]. Real-time PCR results were analyzed by calculating the relative, normalized target gene expression in the low exponential range [35] using separately treated triplicate samples, graphed relative to control samples. Statistical analyses utilized two-tailed t-test, 95% CI.
2.7 Clonegenic Assays
Clonegenic survival experiments or mitotic index analysis following PJ34 treatment was done as follows. Survival assays used cells treated in growth phase as indicated for each experiment. For recovery experiments, the cells were treated and then either left in the same media (no wash) or washed with growth media three times, the growth media replaced and cells incubated for the time indicated prior to analysis or re-plating. Treated cells were trypsinized and plated in triplicate at optimized densities for each cell line. Colonies were stained with 0.5%(w/v) crystal violet in 6%(v/v) gluteraldehyde, excluding colonies of less than 50 cells from the analysis. The survival fraction was calculated as a mean of the ratio of surviving colonies per plated cells multiplied by the plating efficiency of the control cells [36].
2.8 Immunocytochemistry
Cells were fixed in 4% paraformaldehyde, blocked in 3%NGS/PBS/0.1%Triton X-100 and incubated with primary antibodies for 16hrs at 4°C, washed and incubated with appropriate secondary fluorescent antibodies for 2 hrs, counter-stained with DAPI (12.5 µg/ml) and viewed by UV fluorescence on a Zeiss Axiophot microscope.
2.9 Flow Cytometry
Cells were harvested after the treatment and times indicated by trypsinizing and washing in 0.1%BSA/PBS, fixed in cold 4°C ethanol, washed in PBS and stained with propidium iodide (75 µg/ml propidium iodide; 0.5 µg/ml RNAseA, 3.8 mM Sodium Citrate in PBS) overnight. Flow cytometry was performed on a FACSCalibur (Becton-Dickinson) instrument using a minimum of 104 events and analyzed with FCS Express and MODFitLT software.
2.10 siRNA treatment for p21 silencing
Cells were plated in growth phase in 12 well plates. First, cells were transfected with either a non-specific control or two different p21 siRNAs (100 nM; Cell Signaling) using Lipofectamine 2000 (Invitrogen) and incubated for 16 hours. The wells were washed and growth media replaced without or with 50 µM PJ34 for experiments.
3. Results
3.1 PJ34 Induces a Dose Dependent Mitotic Arrest
To examine the growth arrest produced by PJ34 we established a quantitative imager based mitotic assay, similar to other non-counting phospho-H3 (pH3) immunostaining methods [37] (Fig. S1). A concentration dependent PJ34 inhibition of pH3 immunostaining representing a mitotic arrest was observed in U2OS cells (Fig. 1(A), graph), primary human skin fibroblasts (HSF, Fig. S2) as well as LnCAP, MCF7, H1299 and HeLa cell lines (Figs 2, 3 and data not shown), similar to other cell lines [7, 9].
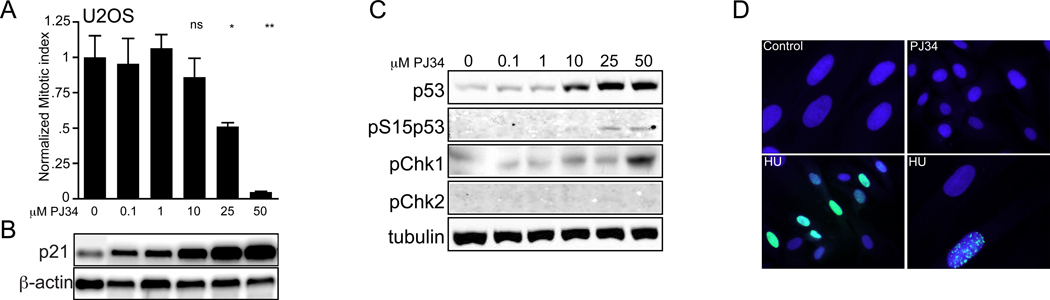
Figure 1.
PJ34 induces a dose dependent mitotic arrest, activating both p53 and p21. (A) A normalized mitotic index plotted as a ratio of (% phospho-H3 signal / tubulin), graphed relative to untreated, growth phase controls in U2OS cells treated with increasing PJ34 concentrations for 24 hours. Graphed is a single, representative experiment with triplicate samples showing a significant reduction in pH3 positive mitotic nuclei with 25 (*p=0.04) and 50 µM PJ34 (**p=0.003) compared to untreated cells (Cont) (error bars, SEM; ns = not significant).
(B) Semi-quantitative PCR and ethidium gel electrophoresis for p21 and β-actin control on parallel treated U2OS cells.
(C) Western blots on PJ34 treated U2OS cells for the indicated proteins. pChk1 = phospho(Serine345)-Chk1, pChk2 = phospho(Threonine68)-Chk2, pS15p53 = phospho(serine15)-p53. Tubulin is shown as a loading control.
(D) Indirect immunofluorescence for γ-H2A.X with DAPI nuclear co-staining in HSF cells treated with 50 µM PJ34 (24 hours) or 10 mM HU (18 hours). The images are merged; anti-γ-H2A.X staining (green) is seen in most HU treated nuclei (blue), but not in PJ34 treated cells. Higher power magnification shows the γ-H2A.X positive foci (lower right panel).
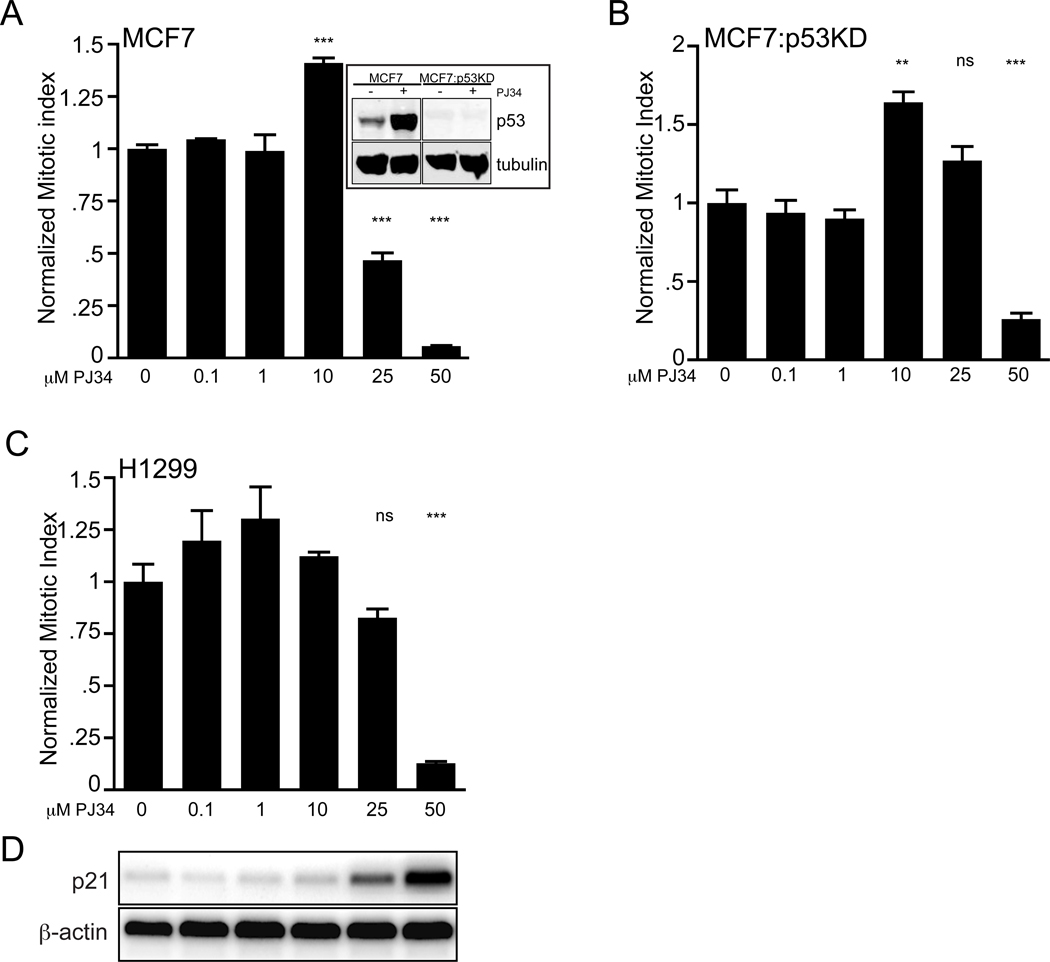
Figure 2.
PJ34 cell cycle arrest does not require p53. (A) Inset, western blot for p53 showing both the parent MCF7 and the lentiviral shRNA MCF7 p53-knockdown (MCF7:p53KD) cell lines with p53 activation by 50 µM PJ34 in MCF7 cells alone. Tubulin is shown as a loading control.
(A, B, C) Cell cycle arrest in MCF7, MCF7:p53KD and H1299 cell lines treated with PJ34 (24 hrs) and the data plotted as a normalized mitotic index as in (Fig. 1(A)). Representative triplicate experiments for (A) MCF7 (relative to untreated cells, ***p<0.002), (B) MCF7:p53KD (**p=0.004; *** p =0.001) or (C) H1299 cells (***p<0.001) are shown; error bars, SEM. (D) Semi-quantitative PCR for p21 and β-actin control on parallel PJ34 treated H1299 cultures.
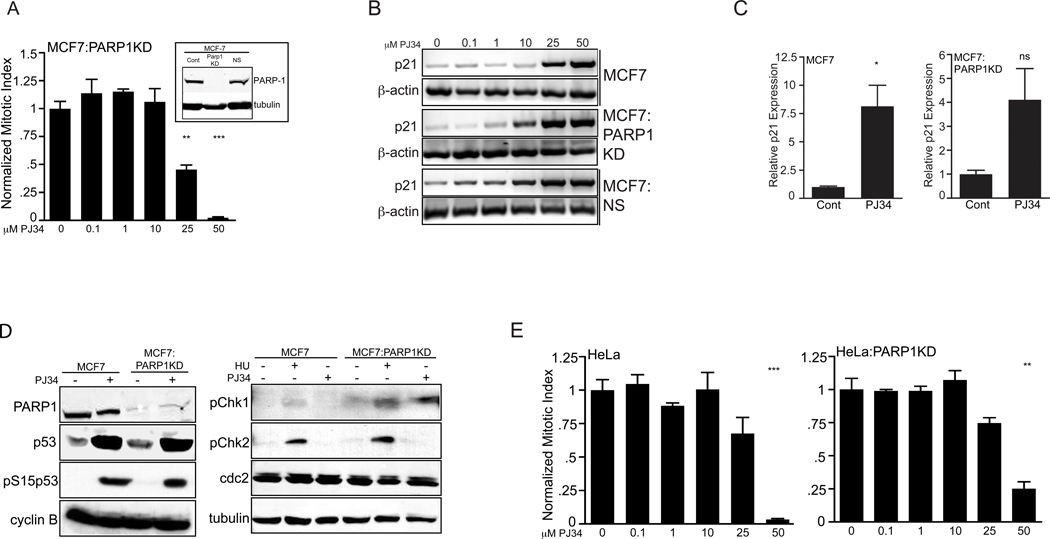
Figure 3.
PARP1 is not required for PJ34 induced mitotic arrest. (A) Growth arrest from PJ34 treated (24 hrs) MCF7:PARP1KD cells plotted as a normalized mitotic index from a single representative triplicate experiment; error bars, SEM. Inset, western blot for PARP1 and tubulin loading control for MCF7 (Cont), MCF7:PARP1KD, and MCF7:NS (non-specific shRNA lentivirus) cell lines. Relative to untreated cells, **p=0.002, ***p<0.001.
(B) Semi-quantitative PCR of p21 and β-actin control on parallel PJ34 treated (24 hrs) samples in MCF7, MCF7:PARP1KD and MCF7:NS cell lines.
(C) Real-time PCR in MCF7 and MCF7:PARP1KD cells for p21 and β–actin in control (untreated, growth phase) and 50 µM PJ34 treated cells (6 hrs; MCF7 (*p=0.01), MCF7:PARP1KD (p=0.07)). The quantitative data are graphed as the relative, normalized target gene expression in the low exponential range. A single representative experiment on triplicate, parallel treated samples is shown. Error bars, SEM.
(D) Western blots for the proteins indicated at left treated with either 50 µM PJ34 for 24 hrs or 10 mM HU for 4 hrs. Abbreviations are as in Fig. 1(C). Tubulin is shown as a loading control.
(E) Mitotic arrest in PJ34 treated HeLa and HeLa:PARP1KD cells plotted as a normalized mitotic index from a single representative triplicate experiment. HeLa 25 µM p=0.08, 50 µM ***p<0.001; HeLaPARP1KD 25 µM p=0.06, 50 µM **p=0.002 ; error bars, SEM.
p21 is necessary for both G2- and G1-phase arrest [30]; we therefore asked if PJ34 activated p21 expression. A concentration dependent increase in p21 gene expression was seen (Fig. 1(B)), mirroring the effective range for mitotic arrest at 50 µM PJ34 (***p=0.003). Furthermore, PJ34 resulted in activated and phosphorylated p53(Ser15) (Fig. 1(C)), phospho-Chk1(Ser345) and minimal phospho-Chk2(Thr68). Next, we asked if the accumulation of γ-H2A.X foci indicating stalled replication was the stimulus for checkpoint activation. Hydroxyurea treated HSF cells produced the expected nuclear γ-H2A.X immunostaining, whereas 50 µM PJ34 did not (Fig. 1(D)). These results show that the PARP inhibitor PJ34 activates checkpoint pathways and p21 expression in both cancer cell lines and primary fibroblasts, but the mechanism is likely not exclusively derived from double strand breaks or stalled replication forks.
3.2 Neither PARP nor p53 is absolutely required for PJ34 cell cycle arrest
Many carcinomas acquire functional p53 mutations [38] and p21 functions either as a survival or senescence factor [39] in metastatic breast cancer [40]; therefore, we asked if PJ34 caused growth arrest and p21 activation in a p53 null background. PJ34 activated p53 (Fig. 2(A), inset) in MCF7 cells and produced a mitotic arrest. Knockdown of p53 in the same background (MCF7:p53KD; Lentiviral Knockdown) or the p53-null H1299 cell line failed to abrogate the mitotic arrest (Figs. 2(A), (B), (C)). An increase in the percent mitotics is seen between 1 and 10 µM PJ34, suggesting a possible transient block followed by a synchronous escape. Interestingly, p53-independent p21 activation was also induced by PJ34, corresponding to the onset of mitotic arrest (Fig. 2(D)). These results demonstrate that p53 is not absolutely required for PJ34 mitotic arrest and that PJ34 can activate p53-independent p21 gene expression.
PARP inhibitors target PARPs enzymatic function but may modify non-(ADP)ribosylation functions and/or have PARP independent effects, although specific data are lacking [3]; therefore, we asked whether PARP1 or PARP2 were required for PJ34 growth arrest. Using the MCF7:PARP1KD stable cell line (Fig. 3A, inset) no difference in mitotic arrest was observed (Fig. 3(A)) compared to MCF7 cells (Fig. 2(A)). Similar results were seen in MCF7:NS (a non-specific control Lentiviral siRNA cell line), MCF7:PARP2 or MCF7:PARP1+2KD cell lines (data not shown). The expression of p21 by 24 hours was increased slightly in the MCF7:PARP1KD cells compared to the MCF7 and MCF7:NS cell lines (Figs. 3(B), 5(B)), while the overall onset of gene expression was rapid (6 hrs; Fig. 3(C)), suggesting direct transcriptional activation. Both MCF7 and MCF7:PARP1KD cells had similar p53 activation and Serine15 phosphorylation (Fig. 3(D), left panel). MCF7:PARP1KD cells phosphorylated Chk1 to a greater degree in response to a hydroxyurea (HU) control and PJ34 without significant Chk2 phosphorylation (Fig. 3(D), right). No change in cyclin B or cdc2 was seen. To confirm that PARP1 was dispensable we examined a second and more rapidly dividing cell line. Wild type HeLa, HeLa:PARP1KD (Fig. 3(E)) and HeLa:PARP1+2KD (data not shown) cells exhibited a similar PJ34 induced mitotic arrest to each other and to the other cell lines examined (Figs. 1, 2, 3(A), S2). These data demonstrate that PJ34 activation of the checkpoint cascade, p21 expression and mitotic arrest are not exclusively dependent upon PARP1 or PARP2.
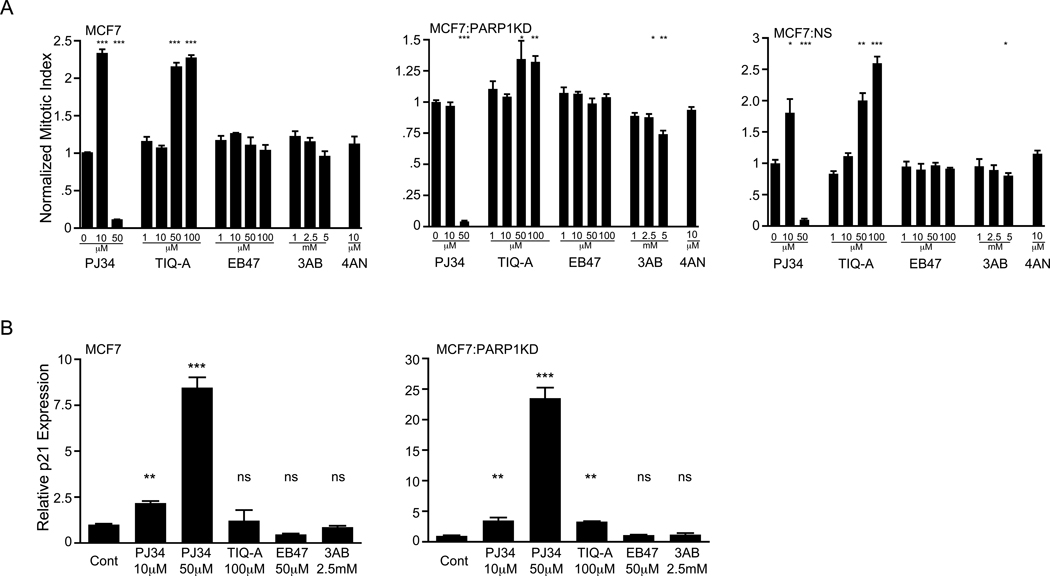
Figure 5.
PJ34 caused the most significant mitotic arrest amongst 5 tested PARP inhibitors. (A) Normalized mitotic index in PARP inhibitor treated MCF7, MCF7:PARP1KD and MCF7:NS cells (24 hours) from a single representative, triplicate experiment in parallel treated cultures; error bars, SEM. MCF7 (***p<0.001). MCF7:PARP1KD PJ34 50 µM (***p<0.001), TIQ 50 µM (*p=0.05) and 100 µM (**p=0.003), 3AB 2.5mM (*p=0.02) and 5 mM (**p=0.001). MCF7:NS PJ34 10 µM (*p<0.02) and 50 µM (***p<0.001), TIQ 50 µM (**p<0.002) and 100 µM (***p<0.001), 3AB 5 mM (*p=0.05).
(B) Real-time PCR in triplicate parallel cultured MCF7 and MCF7:PARP1KD cells for p21 and β–actin in control (untreated, growth phase) and PARP inhibitor treated cells at the indicated concentrations (note the scale differences). The data were acquired and plotted as in Fig. 3(C) compared relative to the controls (ns = not significant, ** p<0.01, *** p<0.001). Error bars, SEM.
3.3 PARP knockdown does not affect cell cycle distribution in response to PJ34
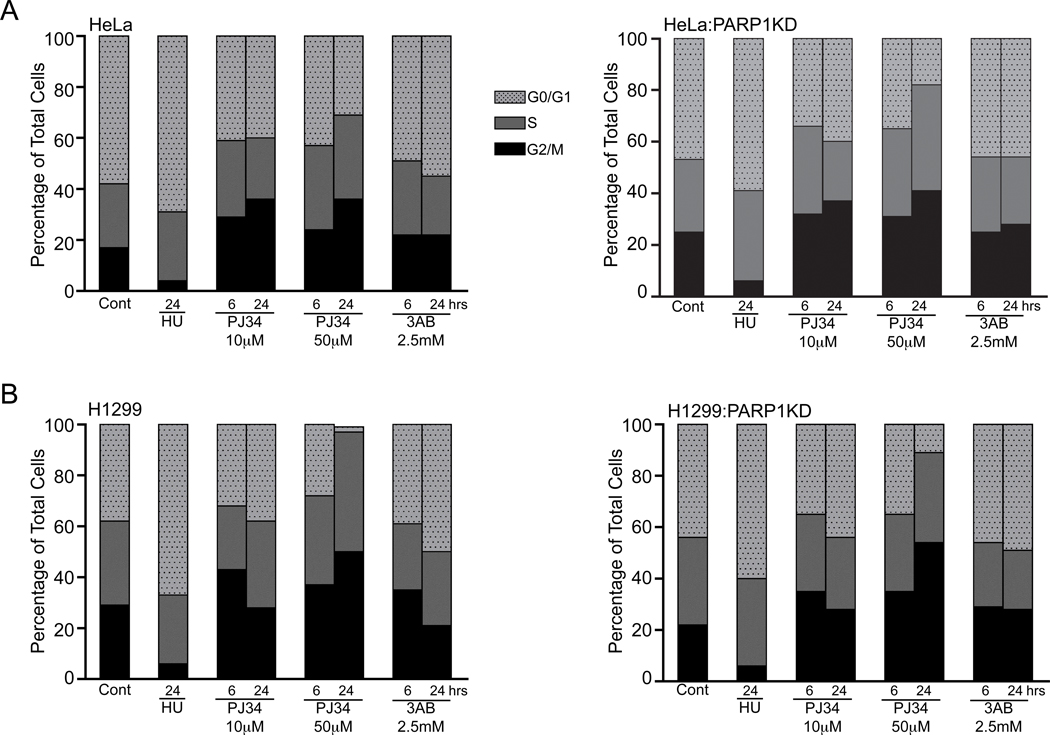
We examined the effect of PJ34 and PARP knockdown on the cell cycle distribution using 10 and 50 µM PJ34 treatments at 6 and 24 hours with HU as a G1/S arrest control. PARP1 knockdown did not significantly change the cell cycle distributions, similar to previous work in MCF7 cells (Figs. S3 and S4) [41]. Since HeLa cell divide more rapidly making cell cycle re-distribution more apparent we also examined those lines demonstrating G2/M accumulation with either dose of PJ34 evident by 6 hours (Figs. 4(A)). The G2/M fractional changes in control or treated cells were not significantly altered by PARP1 knockdown. By 24 hours, PJ34 (50 µM) increased the G2/M fraction in HeLa cells 12% (31% total cells), in HeLa:PARP1KD 26% (41% total cells), as compared with MCF7 14% (28% total cells) and MCF7:PARP1KD cells 5% (22% total cells) (Figs. 4(A), S3, S4). Treatment with 10 µM PJ34 also caused an increase in the G2/M populations after 6 and 24 hours and altered the G0/G1 populations. At known ADP-ribosylation inhibitory concentrations (2.5 mM) the PARP inhibitor 3AB did not significantly change the G2/M fraction. The dual absence of p53 and PARP1 also did not abrogate the mitotic arrest (Figs. 4(B), S5, S6) when the H1299 and H1299:PARP1KD cell lines were compared. 50 µM PJ34 steadily increased the G2/M fraction (H1299 0 hrs 29%, 6 hrs 37%, 24 hrs 50%) with a substantial change in the inter-S phase population. Interestingly, 10 µM PJ34 appeared to cause an increase in the G2/M population at 6 hrs from which the cells escaped by 24 hours, a contrast to the results seen in HeLa or MCF7 cells. Collectively, these data demonstrate that PJ34 causes a G2/M cell cycle arrest that is independent from PARP1 or 2. Additionally, these results show that p53 can participate in the mitotic arrest and its absence changes the cell cycle distributions in response to PJ34, but does not attenuate the G2/M mitotic arrest.
Figure 4.
Cell cycle distributions determined by FACS analysis in response to 10mM hydroxyurea (HU), PJ34 and 3-aminobenzamide (3AB) in (A) HeLa and HeLa:PARP1KD and (B) H1299 and H1299:PARP1KD cell lines treated for the time and at the concentrations indicated and graphed as a percentage of total cells for each phase.
3.4 Cell cycle arrest is not a generalized effect of some experimental PARP inhibitors
We asked if other commonly used PARP inhibitors also caused mitotic arrest and p21 activation. Four PARP inhibitors of varying EC50 and aqueous solubility were tested in MCF7, MCF7:PARP1KD and MCF7:NS cells. PJ34 (50 µM) produced a significant mitotic arrest and higher concentrations of 3AB (5 mM) showed a slight trend, while 10 µM PJ34 and TIQ-A caused a relative increase in the mitotic index in MCF7 parent and derived cell lines (Fig. 5(A)). PJ34 (50 µM) activated p21 expression more significantly in MCF7:PARP1KD cells compared to MCF7 cells (Fig. 5(B)). Both PJ34 (10 µM; both cell lines) and TIQ-A (MCF7:PARP1KD cells) caused small increases in p21 expression. We conclude that among these compounds, PJ34 causes the most significant cell cycle arrest and p21 activation.
3.5 Checkpoint pathway protein analysis
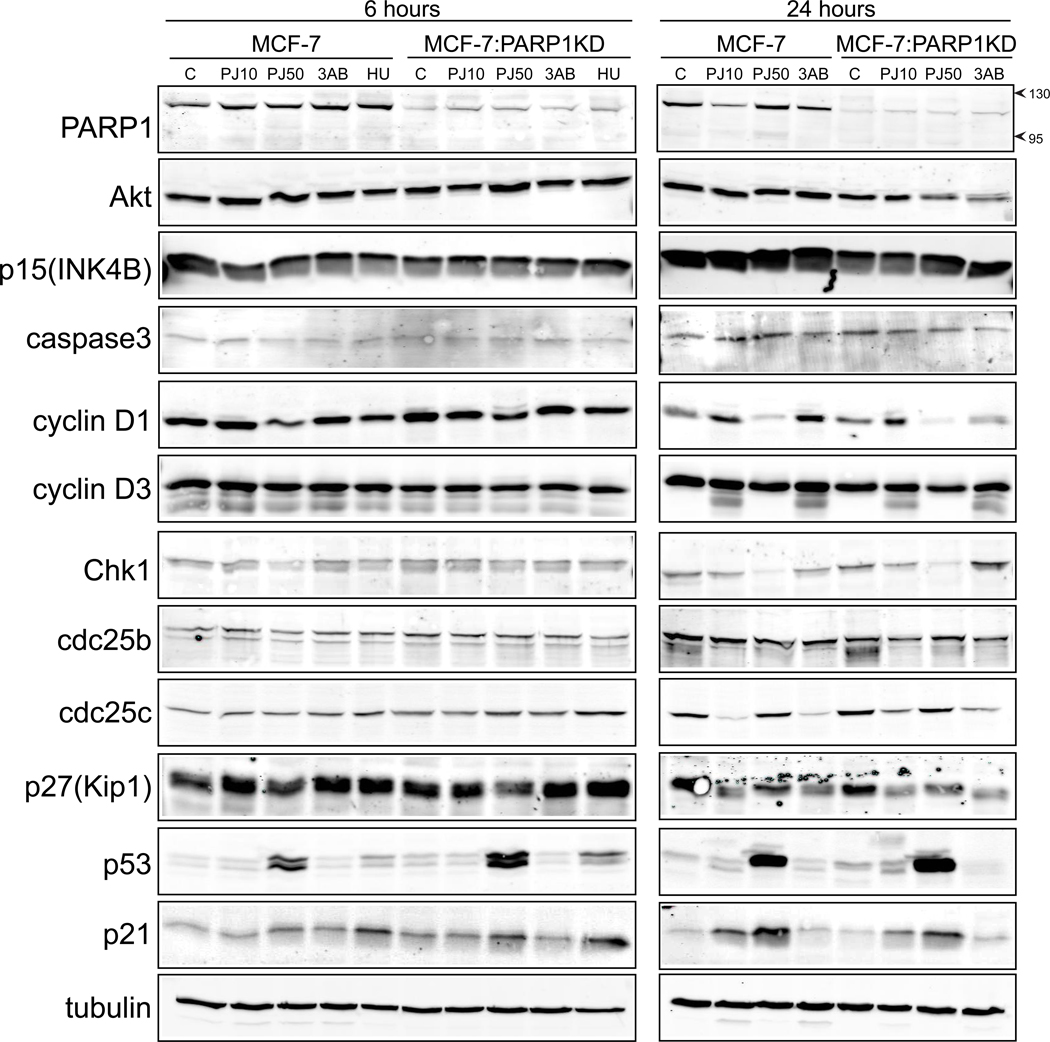
We examined other checkpoint pathway proteins to determine if there were any significant differences in the absence of PARP1 or following PJ34 treatment. Western blots were performed on MCF7 and MCF7:PARP1KD cells following either 6 or 24 hours treatment with 10 or 50 µM PJ34, 3AB (2.5 mM) or HU (10 mM) (Fig. 6). The absence of PARP1 did not significantly alter any of the proteins analyzed. At 6 hours, both 50 µM PJ34 and HU increased p53 and p21 in both cell types. By 24 hours both 10 and 50 µM PJ34 p21 accumulated further with decreases in cyclin D1, p27Kip1 and Chk1. There was no increased cleavage of Caspase3. Interestingly, 10 µM but not 50 µM PJ34 reduced cdc25c at 24 hours. We conclude that PJ34 activated p21 is likely one mechanism required for the G2/M mitotic arrest and that changes in cyclin D1 and p27Kip1 may have related roles.
Figure 6.
Western blots in MCF7 and MCF7:PARP1KD cells for checkpoint pathway proteins. The cells were treated with 10 mM hydroxyurea (HU), 10 µM PJ34 (PJ10), 50 µM PJ34 (PJ50) or 2.5 mM 3-aminobenzamide (3AB) for either 6 or 24 hours and analyzed for the proteins at the left. Arrows on the PARP1 blot indicate Mr to show no significant appearance of the 85kDa PARP cleavage product.
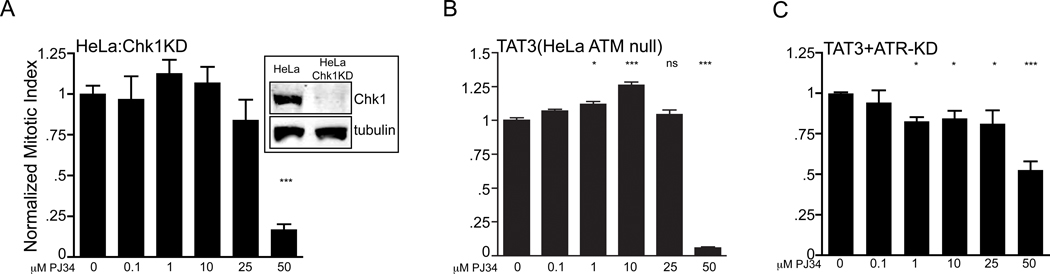
3.6 PJ34 mitotic arrest requires ATR but does not depend upon Chk1 or ATM
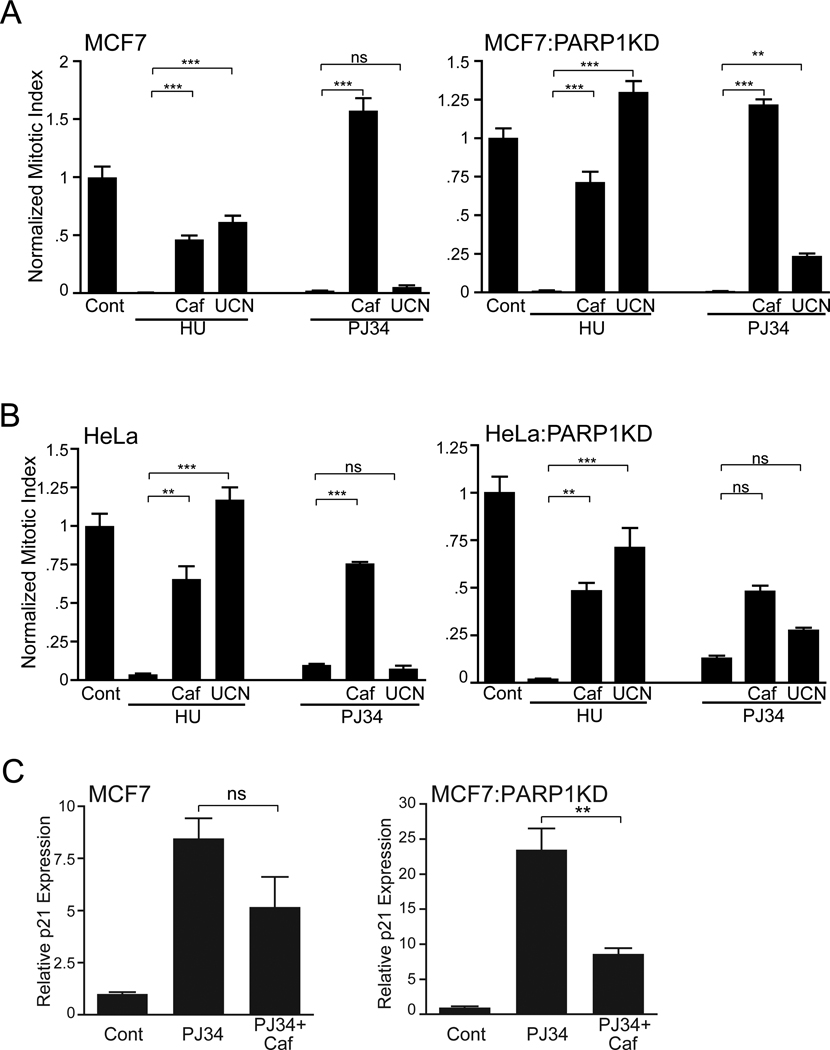
Since PARP1 is known to interact with both ATM and ATR and PJ34 induced phosphorylation of Chk1 (Fig. 1(B)) and activation of a downstream target in p21, we asked if PJ34 mitotic arrest required ATM/ATR and Chk1? Since an ATM knockdown cell line is available in a HeLa background (TAT3) we used both TAT3:ATRKD (clone 19) [34] and HeLa:Chk1KD cell lines. The absence of either Chk1 (Fig. 7(A)) or ATM (Fig. 7(B)) did not affect the mitotic arrest while the dual absence of ATR/ATM caused a ~50% attenuation of 50 µM PJ34 (Fig. 7(C)). Next we asked if either the ATM/ATR inhibitor caffeine [42] or the Chk1 inhibitor UCN01 attenuated PJ34 mitotic arrest. Both 10 mM caffeine and 300 nM UCN01 attenuated a G1/S HU arrest in PARP1 containing or knockdown MCF7 or HeLa cell lines. Caffeine also attenuated or nearly attenuated the PJ34 mitotic arrest in either MCF7 or HeLa wild type and their respective PARP1KDs (Figs. 8A, B; in the HeLa:PARP1KD PJ34+caffeine p=0.06). UCN01 was ineffective in wild type cells and exhibited partial attenuation (~25%) in the PARP1KD cells. Similar results were obtained in the absence of p53 in either H1299 or MCF7:p53KD cells (Fig. S7). Caffeine also partially attenuated PJ34 stimulated p21 expression in MCF7 and MCF7:PARP1KD cells (Fig. 8C). We conclude that PJ34 mitotic arrest and, in part, p21 activation requires checkpoint activation through ATR, while Chk1 and ATM are not absolutely required, suggesting either potential cross talk or redundancy between ATM/ATR and Chk1/2 or an additional participating caffeine sensitive ATM/ATR pathway separate from Chk1/2 and p53 that is responsive to PJ34 resulting in downstream p21 gene activation.
Figure 7.
ATR is necessary but ATM and Chk1 are not required for PJ34 induced mitotic arrest. A normalized mitotic index plotted from a single representative triplicate experiment with PJ34 treated (A) HeLa:Chk1KD, (B) TAT3 (HeLa:ATM null) or (C) TAT3:ATR-KD cells (ATMnull+ATRsiRNA, clone 19 [34]); error bars, SEM. Inset, western blot for Chk1 and tubulin showing the Chk1 knockdown. HeLa:Chk1KD PJ34 50 µM (***p<0.001); TAT3 PJ34 1 µM (*p=0.01), PJ34 10 µM (***p<0.001), PJ34 50 µM (***p<0.001); TAT3:ATR-KD (*p<0.03; ***p<0.001).
Figure 8.
Caffeine but not UCN01 abrogated PJ34 growth arrest and was not dependent upon PARP1 while a HU growth arrest was abrogated by both caffeine and UCN01 without dependence upon PARP1. A normalized mitotic index from a single representative triplicate experiment is shown. (A) MCF7 and MCF7:PARP1KD cells were analyzed in growth phase (Cont) or treated with HU (10 mM) or 50 µM PJ34 alone or with the addition of either caffeine or UCN01 for 24 hrs. (***p<0.0003; **p<0.001) (B) HeLa and HeLa:PARP1KD cells analyzed as in (A) (***p<0.0004; **p<0.002; ns – not significant); error bars, SEM.
(C) MCF7 and MCF7:PARP1KD cells were treated as in (8 (A)) and analyzed by real-time PCR for p21 and β–actin in control, PJ34 or PJ34+caffeine treated cells. The data are plotted and analyzed as in Fig 3(C). PJ34+caf compared to PJ34 alone (MCF7, p=0.13; MCF7:PARP1KD, p= 0.008).
3.7 p21 and PARP1 are required for G2/M mitotic arrest following PJ34
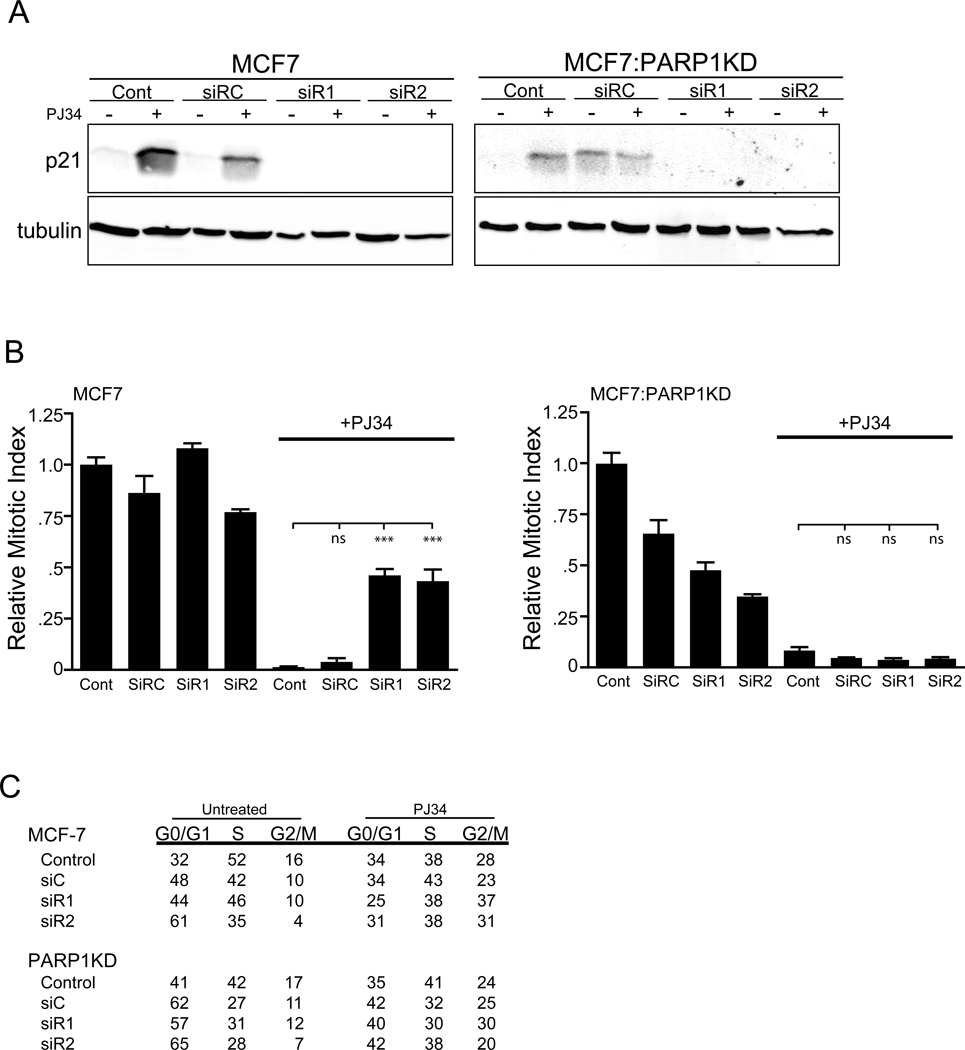
To determine if p21 activation is necessary for PJ34 mitotic arrest we used 2 different siRNAs to abrogate PJ34 activated p21 gene expression. Transfection of siRNAs against p21 but not a non-specific control abrogated p21 expression in response to PJ34 (Fig. 9(A)) in both MCF7 and MCF7:PARP1KD cells. Depletion of p21 attenuated the growth arrest ~50% in MCF7 cells but not in MCF7:PARP1KD cells (Fig. 9(B)). Both the transfection and the p21 siRNA alone caused a reduction in the percent pH3 positive cells in the MCF7:PARP1KD cell line, possibly through increased synchrony by eliminating p21 mediated growth maintenance and a failure of PARP mediated DNA repair. FACS analysis on these treated cells did not reveal a fractional G2/M difference in the untreated cells while the siRNAs increased the G0/G1 fractions in each line. Following PJ34 there was an 11% increase in the G2/M fraction in the p21 siRNA plus PJ34 treated MCF7 cells compared to 6% for the MCF7:PARP1KD cells (Fig. 9(C)). We conclude that in the presence of PARP1, p21 is a required component for the growth arrest induced by PJ34.
Figure 9.
Elimination of PJ34 activated p21 expression attenuates growth arrest. (A) Western blot for p21 and tubulin loading control in MCF7 and MCF7:PARP1KD cells untreated (Cont), transfected with either a non-specific siRNA (siRC), or siRNAs against p21 (siR1, siR2) for 18 hours followed by treatment with (+) or without (−) 50 µM PJ34 for 24 hours.
(B) Normalized mitotic index plotted from parallel cultures treated as in (9 (A)). A single representative triplicate experiment is shown for Control or siRNA transfected cultures with (+) or without (−) 50 µM PJ34. Error bars, SEM (ns = not significant, ***p<0.001).
(C) FACS analysis on parallel treated cultures from (9 (A)). The table shows the percentage of total cells represented in G0/G1, S or G2/M in either MCF7 or MCF7:PARP1KD cells.
3.8. PJ34 mitotic arrest is conditionally survivable
Prior studies showed that PJ34 caused both time and concentration dependent significant cell death in MCF7 cells [7], although shorter exposures were not significantly lethal in other cells [9]. The activation of p21 suggests that under conditional exposure, cells may either choose to repair and re-enter the cell cycle or undergo apoptosis; therefore, we asked if the mitotic arrest was reversible and survivable.
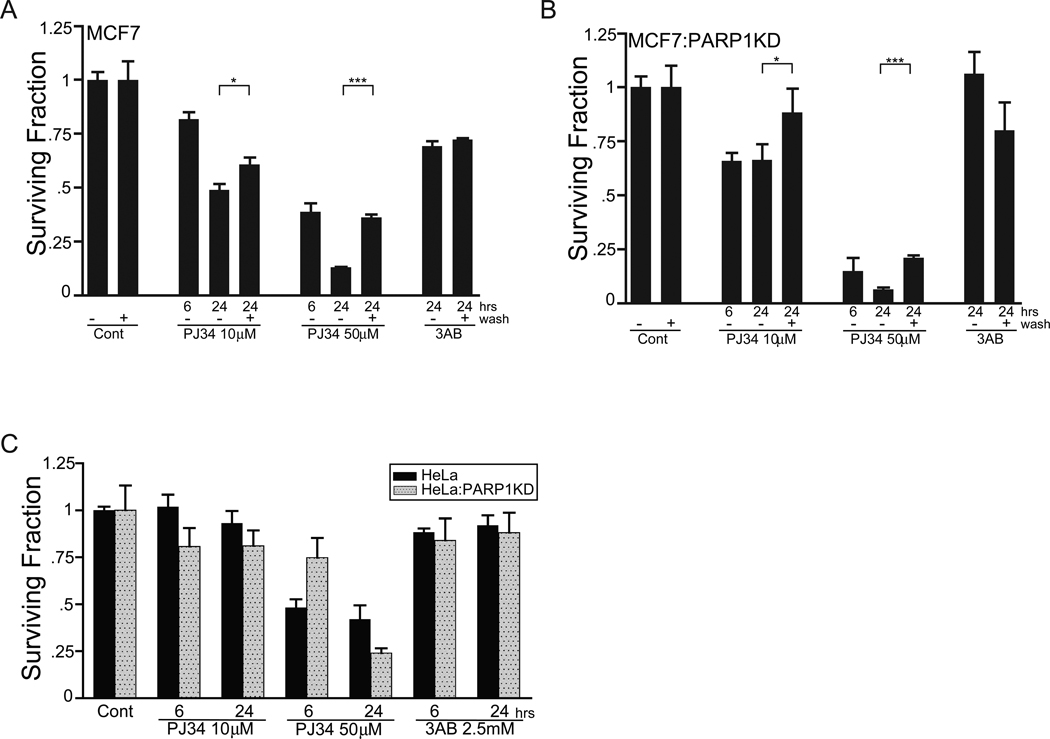
We determined if PJ34 concentration and exposure time differentially affected survival using a clonegenic assay. In MCF7 cells, treatment with 50 µM PJ34 for 6 (39% and 15% survival, MCF7 and MCF7:PARP1KD cells, respectively) and 24 hours (13% and 6%) produced greater lethality than treatment with 10 µM PJ34 for 6 (82% and 66%) and 24 hours (49% and 66%; Figs. 10(A), (B)). Recovery in fresh media prior to re-plating (+ wash) significantly increased survival in both MCF7 (36%; p<0.05) and MCF7:PARP1KD (21%, p<0.001) cells following 24 hour treatment with 50 µM PJ34. There were no significant differences in survival following treatment with 2.5 mM 3-AB. HeLa and HeLa:PARP1KD cells demonstrated greater survival (50 µM PJ34 at 24 hours; 42% and 24%, respectively) than MCF7 cell lines for both PJ34 concentrations and exposure times (Fig. 10(C)). Collectively, these experiments suggest that PJ34 induced growth arrest and cellular lethality will be influenced by cellular genetic background, dose and exposure time.
Figure 10.
PJ34 exposure is conditionally lethal. (A) MCF7 or (B) MCF7:PARP1KD cells were treated for 6 or 24 hours with either 10 or 50 µM PJ34 or 2.5 mM 3AB as a negative control and a clonegenic survival assay performed. One set of samples received a media wash and 24-hour recovery period (+ wash) following treatment and prior to harvesting and re-plating. Data are plotted as a ratio of colonies/plated cells compared to the plating efficiency for the control condition and are representative triplicate, parallel treated samples from a single experiment performed at least twice. Error bars, SEM, (* = p<0.05; *** p<0.001).
(C) HeLa (solid bars) and HeLa:PARP1KD (stippled bars) cells were treated for 6 or 24 hours with PJ34 or 3AB as shown and a clonegenic survival assay performed as in 10(A, B).
4. Discussion
In this study we demonstrate that the PARP inhibitor PJ34 causes a concentration dependent, caffeine sensitive and UCN01 insensitive cell cycle checkpoint pathway activation followed by a p21 dependent G2/M arrest. Significantly, neither the mitotic arrest nor p21 activation is exclusively PARP1 or -2 dependent. p53 was activated and both p53(Ser15) and Chk1(Ser345) were phosphorylated; however, p53, ATM and Chk1 were not required for mitotic arrest. Our data suggest a hypothesis of ATM/ATR cross talk with direct ATR-Chk1 or ATM-Chk2 activation to initiate and maintain the arrest. Alternatively, either an ATM/ATR dependent, Chk1/2 and p53 independent target may be responsible or more likely there are separable, PARP dependent and independent effects on the checkpoint cascade and p21 activation. The reproducible growth arrest in cells from different genetic backgrounds highlights the potential utility of PARP inhibitor monotherapy; however, these data also suggest significant caution when examining PARP inhibitor effects since both ADP-ribosylation inhibition independent and PARP independent effects occur. Our results highlight the point made by some investigators for PARP inhibitor results to be confirmed by genetic or small molecule comparisons [3].
Why do some PARP inhibitors cause growth and mitotic arrest? Only certain PARP inhibitors affect cell cycle progression including PJ34 [7, 9], KU0058948 [8], KU00586864 [4] and the breast cancer drugs Olaparib [43] and Iniparib [44]. Heterogeneous responses to these compounds are predictable due to structural variability potentially directing differential effects on both ADP-ribosylation and non-ADP-ribosylation functions, including protein-protein interactions and transcription [2] and non-PARP targets sharing NAD+-binding site homology. PARP inhibitors primarily synergize with DNA damage agents [3, 29] or ionizing radiation [23, 45] for synthetic lethality, a premise driving their clinical use. Genetic aberrations in carcinomas such as BRCA1/2 mutations [5], ATM mutations [46], or deficiencies in homologous recombination [47] sensitize cells to PARP inhibitors. Promoter response experiments comparing PARP1 knockdown versus PARP inhibition underscore PARP1 functions potentially apart from activated PARP1 and ADP-ribosylation, yet affected by PARP inhibitors [48, 49]. By being caffeine sensitive, PJ34 growth arrest likely works through ATM/ATR, but UCN01 insensitivity and resistance to Chk1 knockdown (Figs. 7, 8) suggest that either Chk2 or a different target is required, although the downstream targets leading to p21 activation are not clear. Mechanisms of PARP inhibitor growth arrest are likely influenced by cellular genetics and due to multiple, simultaneous effects on different pathways dependent upon inhibitor specific effects on PARP1 interactions with ATM/ATR, replication and gene expression.
Little is known about off-target effects of PARP inhibitors. In our experiments, PARP1 knockdown in multiple cell lines derived from different tissues and primary human fibroblast cells failed to abrogate any of the PJ34 specific effects. PJ34 binds zinc activated enzyme matrix-metalloproteinase2 (MMP2) and PARP1 and MMP2 may functionally interact [50, 51]. MMP2 inhibition by Galardin did not abrogate PJ34 mitotic arrest (Fig. S8) making it unlikely that binding to MMP2 is responsible for its growth effects. PJ34 activated p53-independent p21 expression (Fig. 2), suggesting that this mechanism may be useful for PARP inhibitor therapy in metastatic transformed carcinomas lacking functional p53 and subsequently, p21 growth arrest. Distinct differences were seen in lethality (Fig. 10) using a single 24hr exposure with 10 or 50 µM PJ34 and although both concentrations caused redistribution within the cell cycle (Figs. 4, S3), only the 50 µM concentration produced an extended mitotic arrest. This may represent the inflection point between PARP specific and PARP independent effects by PJ34 leading to mitotic arrest. Whether other PARP inhibitors that produce mitotic arrest also possess these PARP independent effects is not known, but should be studied to elucidate the mechanism behind both the PARP-independent and PARP dependent components of the growth arrest. Potentially, molecules such as PJ34 that have PARP ADP-ribosylation inhibiting activity may also possess these other anti-neoplastic functions, either as separate or complimentary mechanisms.
Previous work described PJ34 as being lethal following 10 µM exposure for 48–72 hours in MCF7, but not MCF10A cells [7]. In contrast, we found a shorter 24-hour exposure produced a G2/M mitotic arrest only with 50 µM PJ34. Cell cycle distribution was similar with both concentrations (Fig. 4) but interestingly 10 µM PJ34 and the similar, phenanthridine derived TIQ-A caused an increase in phospho-H3 positive cells (Figs. 5(A), 11(A)) suggesting an initial block, synchronizing and escape effect since 10 µM PJ34 treated MCF7 and HeLa cells had significantly greater survival than 50 µM PJ34 treated cells (Fig. 10). We did not see an increase in γ-H2A.X following PJ34 treatment (Fig. 1(D)) as has been seen with other PARP inhibitors [47] potentially due to shorter exposure time. Survival potential is critical in understanding both the dose toxicity and the PARP specific and non-specific effects following a single PARP inhibitor pulse since a washout experiment showed increased survival (Fig. 10). Multiple effects on distinct PARP pathways or targets and off-target proteins from different doses form a complex mechanism suggesting the potential for pharmacologically separable targeting of these effects.
How is p21 involved in PJ34 activated mitotic arrest? The p21 promoter is a therapeutic target in many cancers [53]. P21 is functionally linked to PARP1 [30] and p21 activation by ionizing radiation or other DNA damage signals is repressed following pre-treatment with PARP inhibitors [31, 33] suggesting PARP1 participates in p21 transcription, similar to direct transcription effects at other promoters [49]. How the two mechanisms of p21 expression response to PJ34 after DNA damage compared to PJ34 alone differ is not clear, highlighting the differences between PARP inhibitor effects following DNA damage and PARP activation versus PARP inhibitors alone. P21 may influence the G1 and G2 checkpoints through cdk1/cdk2 [54, 55] and by repressing transcription of Cdc25c, Chk1 and cyclin B1 [56]. 10 µM PJ34 activates p21 (Figs. 5, 6) and caused a reduction in cdc25c (Fig. 6), possibly signifying a transition from arrest where activated Chk1/2 have caused increased degradation of cdc25c with inactivation of cyclinB/cdk1 preceding the resumption of M phase. Only 50 µM PJ34 significantly activated p53-independent p21 expression, indicating de-repression effects at the p21 promoter and also caused a reduction in absolute Chk1 levels (Fig. 6). PARP1 dependent attenuation of mitotic arrest by p21 siRNA (Fig. 8) suggested that at least in part, p21 is required for the G2/M growth arrest and that there is a functional role for PARP1 in growth arrest maintenance. How p21 affects the mitotic arrest and potentially transitions the cell into repair and mitotic re-entry or apoptosis in response to different concentrations of PJ34 may be directly influenced by its PARP dependent and PARP-independent activities.
Collectively, this study highlights both the utility and perils of PARP inhibitors, showing differential activation of checkpoint pathways, p21 expression and cell survival that is dependent upon dose, exposure time and cellular genetic background. The PARP independent effects may be super-imposed over the PARP dependent effects at certain concentrations. Our data suggest a hypothesis that although PJ34 may have PARP specific effects relevant to mitotic arrest that there are clearly other targets for this compound that are also involved in the checkpoint cascade and p21 activation suggesting that this molecule and possibly others in its PARP inhibitor subclass, possess other anti-neoplastic activity apart from PARP inhibition. Further investigation into the mitotic arrest mechanisms activated by PARP inhibitors will aid in understanding both PARP specific and PARP–independent cellular effects of PARP inhibitors for development of improved, specific functional targeting.
Highlights.
Mitotic growth arrest by PJ34 is PARP1+2 independent
PJ34 growth arrest occurs in cells from diverse genetic backgrounds
PJ34 causes a G2/M growth arrest, independent of ATM, p53 or Chk1
PJ34 growth arrest is caffeine sensitive and UCN01 insensitive
ATR is necessary for PJ34 growth arrest
PJ34 activates p21 gene expression by both p53 dependent and independent pathways
Supplementary Material
Acknowledgements
This work was supported by grant (NCI K08CA109158) to DLM. We thank Loren Brown and Madeleine Pham for technical assistance and the FACS core facility in the Knight Cancer Center, OHSU for assistance with cell sorting.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conflict of Interest: The authors declare no conflicts of interest.
References
- 1.Schreiber V, Amé JC, Dollé JC, Schultz I, Rinaldi B, Fraulob V, Ménissier-de Murcia J, de Murcia G. Poly(ADP-ribose) polymerase-2 (PARP-2) is required for efficient base excision DNA repair in association with PARP-1 and XRCC1. J. Biol. Chem. 2002;277:23028–23036. doi: 10.1074/jbc.M202390200. [DOI] [PubMed] [Google Scholar]
- 2.Kraus WL. Transcriptional control by PARP-1: chromatin modulation, enhancer binding, co-regulation and insulation. Curr. Opin. Cell Biol. 2008;20:294–302. doi: 10.1016/j.ceb.2008.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rouleau M, Patel A, Hendzel MJ, Kaufmann SH, Poirier GG. PARP inhibition: PARP1 and beyond. Nat. Rev. Cancer. 2010;10:293–300. doi: 10.1038/nrc2812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Farmer H, McCabe N, Lord CJ, Tutt AN, Johnson DA, Richardson TB, Santarosa M, Dillon KJ, Hickson I, Knights C, Martin NM, Jackson SP, Smith GC, Ashworth A. Targeting the DNA repair defect in BRCA mutant cells as a therapeutic strategy. Nature. 2005;434:917–921. doi: 10.1038/nature03445. [DOI] [PubMed] [Google Scholar]
- 5.Bryant H, Schultz N, Thomas HD, Parker KM, Flower D, Lopez E, Kyle S, Meuth M, Curtin NJ, Helleday T. Specific killing of BRCA2-deficient tumours with inhibitors or poly(ADP-ribose) polymerase. Nature. 2005;434:913–917. doi: 10.1038/nature03443. [DOI] [PubMed] [Google Scholar]
- 6.Ratnam K, Low JA. Current development of clinical inhibitors of poly(ADP-Ribose) polymerase in oncology. Clin. Cancer Res. 2007;13:1383–1388. doi: 10.1158/1078-0432.CCR-06-2260. [DOI] [PubMed] [Google Scholar]
- 7.Inbar-Rozensal D, Castiel A, Visochek L, Castel D, Dantzer F, Izraeli S, Cohen-Armon M. A selective eradication of human non-hereditary breast cancer cells by phenanthridine-derived polyADP-ribose polymerase inhibitors. Breast Cancer Res. 2009;11:R78. doi: 10.1186/bcr2445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gaymes TJ, Shall S, McPherson LJ, Twine NA, Lea NC, Farzaneh F, Mufti GJ. Inhibitors of poly ADP-ribose polymerase (PARP) induce apoptosis of myeloid leukemic cells: potential for therapy of myeloid leukemia and myelodysplastic syndromes. Haematologica. 2009;94:638–646. doi: 10.3324/haematol.2008.001933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chevanne M, Zampieri M, Caldini R, Rizzo A, Ciccarone F, Catizone A, D’Angelo C, Gustafierro T, Biroccio A, Reale A, Zupi G, Caiafa P. Inhibition of PARP activity by PJ34 leads to growth impairment and cell death associated with aberrant mitotic pattern and nucleolar actin accumulation in M14 melanoma cell line. J. Cell. Physiol. 2010;222:401–410. doi: 10.1002/jcp.21964. [DOI] [PubMed] [Google Scholar]
- 10.Reinhardt HC, Yaffe MB. Kinases that control the cell cycle in response to DNA damage: Chk1, Chk2 and MK2. Curr. Opin. Cell Biol. 2009;21:245–255. doi: 10.1016/j.ceb.2009.01.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lapenna S, Giordano A. Cell cycle kinases as therapeutic targets for cancer. Nat. Rev. Drug Discov. 2009;8:547–566. doi: 10.1038/nrd2907. [DOI] [PubMed] [Google Scholar]
- 12.Schwartz JK, Lovly CM, Piwnica-Worms H. Regulation of the Chk2 protein kinase by oligomerization-mediated cis- and trans-phosphorylation. Mol. Cancer Res. 2003;1:598–609. [PubMed] [Google Scholar]
- 13.Zhao H, Piwnica-Worms H. ATR-mediated checkpoint pathways regulate phosphorylation and activation of human Chk1. Mol. Cell. Biol. 2001;21:4129–4139. doi: 10.1128/MCB.21.13.4129-4139.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bakkenist CJ, Kastan MB. DNA damage activates ATM through intermolecular autophosphorylation and dimmer dissociation. Nature. 2003;421:499–506. doi: 10.1038/nature01368. [DOI] [PubMed] [Google Scholar]
- 15.Sanchez Y, Wong C, Thoma RS, Richman R, Wu Z, Piwnica-Worms H, Elledge SJ. Conservation of the chk1 checkpoint pathway in mammals: linkage of DNA damage to Cdk regulation through cdc25. Science. 1997;277:1497–1501. doi: 10.1126/science.277.5331.1497. [DOI] [PubMed] [Google Scholar]
- 16.Takai H, Naka K, Okada Y, Watanabe M, Harada N, Saito S, Anderson CW, Appella E, Nakanishi M, Suzuki H, Nagashima K, Sawa H, Ikeda K, Motoyama N. Chk2-deficient mice exhibit radioresistance and defective p53-mediated transcription. EMBO J. 2002;21:5195–5205. doi: 10.1093/emboj/cdf506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yang X, Wood PA, Hrushesky WJM. Mammalain TIMELESS is required for ATM-dependent Chk2 activation and G2/M checkpoint control. J. Biol. Chem. 2010;285:3030–3034. doi: 10.1074/jbc.M109.050237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cliby WA, Roberts CJ, Cimprich KA, Stringer CM, Lamb JR, Schrieber SL, Friend SH. Overexpression of a kinase-inactive ATR protein causes sensitivity to DNA damaging agents and defects in cell cycle checkpoints. EMBO J. 1998;2:159–169. doi: 10.1093/emboj/17.1.159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gatei M, Sloper K, Soresen C, Syljuasen R, Falck J, Hobson K, Savage K, Lukas J, Zhou B-B, Bartek J, Khanna KK. Ataxia-telangectasia-mutated (ATM) and NBS1-dependent phosphorylation of CHk1 an Ser317 in response to Ionizing radiation. J. Biol. Chem. 2003;278:14806–14811. doi: 10.1074/jbc.M210862200. [DOI] [PubMed] [Google Scholar]
- 20.Kedar PS, Stefanick DF, Horton JK, Wilson SH. Interaction between PARP-1 and ATR in mouse fibroblasts is blocked by PARP inhibition. DNA Repair. 2008;7:1787–1798. doi: 10.1016/j.dnarep.2008.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Aguilar-Quesada R, Muñoz-Gámez JA, Martín-Oliva D, Peralta A, Valenzuela MT, Matínez-Romero R, Quiles-Pérez R, Menissier-de Murcia J, de Murcia G, Ruiz de Almodóvar M, Oliver FJ. Interaction between ATM and PARP-1 in response to DNA damage and sensitization of ATM deficient cells through PARP inhibition. BMC Mol. Biol. 2007;8:29. doi: 10.1186/1471-2199-8-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Menisser-de Murcia J, Mark M, Wendling O, Wynshaw-Boris A, de Murcia G. Early embryonic lethality in PARP1 Atm double-mutant mice suggests a functional synergy in cell proliferation during development. Mol. Cell. Biol. 2001;21:1828–1832. doi: 10.1128/MCB.21.5.1828-1832.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fernet M, Megnin-Chanet F, Hall J, Favaudon V. Control of the G2/M checkpoints after exposure to low doses of ionizing radiation: Implications for hyper-radiosensitivity. DNA Repair. 2010;9:48–57. doi: 10.1016/j.dnarep.2009.10.006. [DOI] [PubMed] [Google Scholar]
- 24.Horton JK, Stefanick DF, Kedar PS, Wilson SH. ATR signaling mediates an S-phase checkpoint after inhibition of poly(ADP-ribose) polymerase activity. DNA Repair. 2007;6:742–750. doi: 10.1016/j.dnarep.2006.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Deng C, Zhang P, Harper JW, Elledge SJ, Leder P. Mice lacking p21cip1/waf1 undergo normal development, but are defective in G1 checkpoint control. Cell. 1995;82:675–684. doi: 10.1016/0092-8674(95)90039-x. [DOI] [PubMed] [Google Scholar]
- 26.Bunz F, Dutriaux A, Lengauer C, Waldman T, Zhou S, Brown JP, Sedivy JM, Kinzler KW, Vogelstein B. Requirement for p53 and p21 to sustain G2 arrest after DNA damage. Science. 1998;282:1497–1501. doi: 10.1126/science.282.5393.1497. [DOI] [PubMed] [Google Scholar]
- 27.Abbas T, Dutta A. P21 in cancer: intricate networks and multiple activities. Nat. Rev. Cancer. 2009;9:400–414. doi: 10.1038/nrc2657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Smits VAJ, Klompmaker R, Vallenius T, Rijksen G, Mäkelä T, Medema RH. p21 inhibits Thr161 phosphorylation of Cdc2 to enforce the G2 DNA damage checkpoint. J. Biol. Chem. 2000;275:30638–30643. doi: 10.1074/jbc.M005437200. [DOI] [PubMed] [Google Scholar]
- 29.Lohr K, Moritz C, Contente A, Dobbelstein M. p21/CDKN1A mediates negative regulation of transcription by p53. J. Biol. Chem. 2003;278:32507–32516. doi: 10.1074/jbc.M212517200. [DOI] [PubMed] [Google Scholar]
- 30.Cazzalini O, Dona F, Savio M, Tillhon M, Maccario C, Perucca P, Stivaia LA, Scovassi AI, Prosperi E. p21CDKN1A participates in base excision repair by regulating the activity of poly(ADP-ribose) polymerase-1. DNA Repair (Amst) 2010;9:627–635. doi: 10.1016/j.dnarep.2010.02.011. [DOI] [PubMed] [Google Scholar]
- 31.Wieler S, Gagne JP, Vaziri H, Poirier GG, Benchimol S. Poly(ADP-ribose) polymerase-1 is a positive regulator of the p53-mediated G1 arrest response following ionizing radiation. J. Biol. Chem. 2003;278:18914–18921. doi: 10.1074/jbc.M211641200. [DOI] [PubMed] [Google Scholar]
- 32.Haince J-F, Kozlov S, Dawson VL, Dawson TM, Hendzel MJ, Lavin MF, Poirier GG. Ataxia telangectasia mutated (ATM) signaling network is modulated by a novel poly(ADP-ribose)-dependent pathway in the early response to DNA damage agents. J. Biol. Chem. 2007;282:16441–16453. doi: 10.1074/jbc.M608406200. [DOI] [PubMed] [Google Scholar]
- 33.Madison DL, Lundblad JR. C-terminal binding protein and poly(ADP)ribose polymerase1 contribute to repression of the p21waf1/cip1 promoter. Oncogene. 2010;29:6027–6039. doi: 10.1038/onc.2010.338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Stauffer D, Chang B, Huang J, Dunn A, Thayer M. p300/CREB-binding protein interacts with ATR and is required for the DNA replication checkpoint. J. Biol. Chem. 2007;282:9678–9687. doi: 10.1074/jbc.M609261200. [DOI] [PubMed] [Google Scholar]
- 35.Pfaffl MW. A new mathematical model for relative quantification in real-time PCR. Nucleic Acids Res. 2001;29:e45. doi: 10.1093/nar/29.9.e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Franken NAP, Rodermond HM, Stap J, Haveman J, van Bree C. Clonogenic assay of cells in vitro. Nat. Protocols. 2006;1:2315–2319. doi: 10.1038/nprot.2006.339. [DOI] [PubMed] [Google Scholar]
- 37.Brenner RM, Slayden OD, Rodgers WH, Critchley HOD, Carroll R, Nie XJ, Mah K. Immunocytochemical analysis of mitotic activity with an antibody to phosphorylated histone H3 in the macaque and human endometrium. Hum. Reprod. 2003;6:1185–1193. doi: 10.1093/humrep/deg255. [DOI] [PubMed] [Google Scholar]
- 38.Hollstein M, Rice K, Soussi T, Fuchs R, Sorlie T, Hovig E, Smith-Sorensen B, Montasano R, Harris CC. Database of p53 gene somatic mutations in human tumours and cell lines. Nucl. Acids Res. 1994;22:35551–3555. [PMC free article] [PubMed] [Google Scholar]
- 39.Wu CH, van Riggelen J, Yetil A, Fan AC, Bachireddy P, Fisher DW. Cellular senescence is an important mechanism of tumor regression upon c-Myc inactivation. Proc. Natl. Acad. Sci. USA. 2007;104:13028–13033. doi: 10.1073/pnas.0701953104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Liu M, Casimiro MC, Wang C, Shirley LA, Jiao X, Katiyar S, et al. P21Cip1 attenuates Ras- and c-Myc-dependent breast tumor epithelial mesenchymal transition and cancer stem cell-like gene expression in vivo. Proc. Natl. Acad. Sci. USA. 2009;106:19035–19039. doi: 10.1073/pnas.0910009106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Firzzell KM, Gamble MJ, Berrocal JG, Zhang T, Krishnakumar R, Cen Y, Sauve AA, Kraus WL. Global analysis of transcriptional regulation by poly(ADP-ribose) polymerase-1 and poly(ADP-ribose) glycohydrolase in MCF-7 human breast cancer cells. J. Biol. Chem. 2009;284:33926–33938. doi: 10.1074/jbc.M109.023879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Schlegel R, Pardee AB. Caffeine-induced uncoupling of mitosis from the completion of DNA replication in mammalian cells. Science. 1986;232:1264–1266. doi: 10.1126/science.2422760. [DOI] [PubMed] [Google Scholar]
- 43.Fong PC, Boss DS, Yap TA, Tutt A, Wu P, Mergui-Roelvink M, Mortimer P, Swaisland H, Lau A, O'Connor MJ, Ashworth A, Carmichael J, Kaye SB, Schellens JH, de Bono JS. Inhibition of poly(ADP-Ribose) polymerase in tumors from BRCA mutation carriers. New Engl. J. Med. 2009;361:123–134. doi: 10.1056/NEJMoa0900212. [DOI] [PubMed] [Google Scholar]
- 44.O'Shaughnessy J, Osborne C, Pippen JE, Yoffe M, Patt D, Rocha C, Koo IC, Sherman BM, Bradley C. Iniparib plus chemotherapy in metastatic triple-negative breast cancer. New Engl. J. Med. 2011;364:205–214. doi: 10.1056/NEJMoa1011418. [DOI] [PubMed] [Google Scholar]
- 45.Lu HR, Wang X, Wang Y. A stronger DNA damage-induced G2 checkpoint due to over-activated Chk1 in the absence of PAPR-1. Cell Cycle. 2006;5:2364–2370. doi: 10.4161/cc.5.20.3355. [DOI] [PubMed] [Google Scholar]
- 46.Williamson CT, Muzik H, Turhan AG, Zamo A, O’Conner MJ, Bebb DG, Lees-Miller SP. ATM deficiency sensitizes Mantle Cell lymphoma cells to Poly(ADP-ribose) polymerase-1 inhibitors. Mol. Cancer Ther. 2010;9:347–357. doi: 10.1158/1535-7163.MCT-09-0872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.McCabe N, Turner NC, Lord CJ. Deficiency in the repair of DNA damage by homologous recombination and sensitivity to poly(ADP-ribose) polymerase inhibition. Cancer Res. 2006;66:8109–8115. doi: 10.1158/0008-5472.CAN-06-0140. [DOI] [PubMed] [Google Scholar]
- 48.Li M, Naidu P, Yu Y, Berger NA, Kannan P. Dual regulation of AP-2α transcriptional activation by poly(ADP)-ribose polymerase-1. Biochem. J. 2004;382:323–329. doi: 10.1042/BJ20040593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ju BG, Solum D, Song EJ, Lee KJ, Rose DW, Glass CK, Rosenfeld MG. Activating the PARP-1 sensor component of the groucho/TLE1 corepressor complex mediates a CaMKinaseII-delta dependent neurogenic gene activation pathway. Cell. 2004;119:815–829. doi: 10.1016/j.cell.2004.11.017. [DOI] [PubMed] [Google Scholar]
- 50.Nicolescu AC, Holt A, Kandasamy AD, Pacher P, Schulz R. inhibition of matrix metalloproteinase-2 by PARP inhibitors. Bioch. Biophys. Res. Comm. 2009;387:646–650. doi: 10.1016/j.bbrc.2009.07.080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yang Y, Candelario-Jalil E, Thompson JF, Cuadrado E, Estrada EY, Rosell A, Montaner J, Rosenberg GA. Increased intranuclear matrix metalloproteinase activity in neurons interferes with oxidative DNA repair in focal cerebral ischemia. J. Neurochem. 2010;112:134–149. doi: 10.1111/j.1471-4159.2009.06433.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bryant H, Helleday T. Inhibition of poly(ADP-ribose) polymerase activates ATM which is required for subsequent homologous recombination repair. Nucleic Acids Res. 2006;34:1685–1691. doi: 10.1093/nar/gkl108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ocker M, Schneider-Stock R. Histone Deacetylase inhibitors: signaling towards p21cip1/waf1. Int. J. Biochem. Cell Biol. 2007;39:1367–1374. doi: 10.1016/j.biocel.2007.03.001. [DOI] [PubMed] [Google Scholar]
- 54.el-Deiry W, Tokino T, Velculescu VE, Levy DB, Parsons R, Trent JM, Lin D, Mercer WE, Kinzier KW, Vogelstein B. Waf1, a potential mediator of p53 tumor suppressor. Cell. 1993;75:817–825. doi: 10.1016/0092-8674(93)90500-p. [DOI] [PubMed] [Google Scholar]
- 55.Brugarolas J, Moberg K, Boyd SD, Taya Y, Jacks T, Lees JA. Inhibition of cyclin-dependent kinase 2 by p21 is necessary for retinoblastoma protein-mediated G1 arrest after gamma-irradiation. Proc. Natl. Acad. Sci. USA. 1999;96:1002–1007. doi: 10.1073/pnas.96.3.1002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Gottifredi V, Karni-Schmidt O, Shieh SS, Prives C. p53 down-regulates CHK1 through p21 and the retinoblastoma protein. Mol. Cell. Biol. 2001;21:1066–1076. doi: 10.1128/MCB.21.4.1066-1076.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.