Abstract
The cyclooxygenases (COX-1 and COX-2) are membrane-associated, heme-containing homodimers that generate prostaglandin H2 from arachidonic acid (AA) in the committed step of prostaglandin biogenesis and are the targets for nonsteroidal anti-inflammatory drugs (NSAIDs). N-(2-cyclohexyloxy-4-nitrophenyl) methanesulfonamide (NS-398) was the first in a series of isoform-selective drugs designed to preferentially inhibit COX-2, with the aim of ameliorating many of the toxic gastrointestinal side effects caused by conventional NSAID inhibition. We determined the X-ray crystal structure of murine COX-2 in complex with NS-398 utilizing synchrotron radiation to 3.0A resolution. NS-398 binds in the cyclooxygenase channel in a conformation that is different than that observed for other COX-2-selective inhibitors, such as celecoxib, with no discernible penetration into the side pocket formed in COX-2 by the isoform-specific substitutions of I434V, H513R, and I523V. Instead, the methanesulfonamide moiety of NS-398 interacts with the side chain of Arg-120 at the opening of the cyclooxygenase channel, similar to that observed for acidic, nonselective NSAIDs such as indomethacin and flurbiprofen. Our structure validates inhibitor studies that identified Arg-120 as a molecular determinant for time-dependent inhibition of COX-2 by NS-398.
Keywords: Cyclooxygenase, prostaglandin H2 synthase, NS-398, Nonsteroidal anti-inflammatory drugs, x-ray crystal structure
1. Introduction
Prostaglandin endoperoxide H synthases, also known as cyclooxygenase enzymes (COX-1 and COX-2), catalyze the committed step in the biosynthesis of prostaglandins, prostacyclins, and thromboxanes. Each isoform consists of two separate, but functionally linked active sites, which generate the product prostaglandin H2 (PGH2) from the ω-6 polyunsaturated fatty acid substrate arachidonic acid (AA) in sequential reactions (Rouzer and Marnett, 2009; Smith et al., 2000). COX-1 and COX-2 share a similar three-dimensional fold and the catalytic mechanism is conserved between isoforms (Garavito et al., 2002). The major difference between isoforms is in their observed expression patterns (Tanabe and Tohnai, 2002). In general, COX-1 is constitutively expressed in nearly every tissue, whereas COX-2 expression is tissue-specific and largely induced upon stimulation by inflammatory agents. Additionally, while both COX-1 and COX-2 preferentially oxygenate AA, COX-2 has been shown to selectively utilize eicosapentaenoic acid and a wide spectrum of AA derivatives, such as the endocannabinoids 2-arachidonoyl glycerol and arachidonoyl ethanolamide as substrates (Kozak et al., 2000; Vecchio and Malkowski, 2011; Vecchio et al., 2010; Wada et al., 2007; Yu et al., 1997).
COX-1 and COX-2 are the pharmacological targets of aspirin and other nonsteroidal anti-inflammatory drugs (NSAIDs), utilized for the treatment of pain and inflammation, as well as some cancers (Blobaum and Marnett, 2007). NSAID binding within the cyclooxygenase channel of COX-1 and COX-2 has been extensively studied and many crystal structures of COX-1 and COX-2 in complex with NSAIDs have been elucidated to compliment inhibition studies (Duggan et al., 2010; Harman et al., 2007; Kurumbail et al., 1996; Loll et al., 1995; Luong et al., 1996; Picot et al., 1994; Rowlinson et al., 2003; Selinsky et al., 2001; Wang et al., 2010). Classical nonselective NSAIDs (nsNSAIDs) inhibit both COX isoforms via one of three different modes of inhibition (reviewed in (Blobaum and Marnett, 2007; DeWitt, 1999)): 1) covalent modification (ex. aspirin acetylation of Ser-530); 2) rapid, reversible binding (ex. ibuprofen); and 3) rapid, lower affinity binding followed by time-dependent, high affinity, slowly reversible binding (ex. flurbiprofen). The classic, acidic nsNSAIDs typically bind within the cyclooxygenase channel such that the carboxylic acid group of the inhibitor interacts with the side chain of Arg-120 at the opening of the channel (Kurumbail et al., 1996; Picot et al., 1994). However, diclofenac, which binds with its carboxylic acid group interacting with the side chain of Tyr-385 and Ser-530 at the apex of the channel, is an exception (Rowlinson et al., 2003). COX-2 can also be selectively inhibited in a time-dependent manner by diarylheterocycle-based compounds (termed “coxibs”), which include celecoxib, rofecoxib, valdecoxib, and etoricoxib (Marnett, 2009). The coxibs exploit a side pocket, generated by the substitution Ile-434 and Ile-523 in COX-1 to Val-434 and Val-523 in COX-2, which effectively increases the overall volume of the cyclooxygenase channel of COX-2 by ~20% (Kurumbail et al., 1996; Luong et al., 1996). In addition, the substitution of His-513 in COX-1 to Arg-513 in COX-2 alters the chemical environment of this side pocket by placing a positive charge at the base of the pocket. Indeed, structure-function analyses utilizing diarylheterocyclic COX-2 inhibitors confirm both the insertion of the methylsulfonyl or sulfonamoylphenyl substituent past Val-523 into the side pocket and subsequent interaction with the side chain of Arg-513 (Gierse et al., 1996; Guo et al., 1996; Kurumbail et al., 1996; Wang et al., 2010; Wong et al., 1997).
N-(2-cyclohexyloxy-4-nitrophenyl) methanesulfonamide (NS-398) was one of the earliest time-dependent COX-2-selective compounds identified, with an IC50 value of 0.1μM (Futaki et al., 1993; DeWitt, 1999). Like many nsNSAIDs that preceded it, NS-398 possessed anti-inflammatory and analgesic effects in vivo, but elicited no significant gastrointestinal toxicity (Futaki et al., 1994; Futaki et al., 1993). Despite the presence of a methylsulfonamide moiety and no carboxylic acid group, inhibition studies utilizing NS-398 and R120E and R120Q human (hu) COX-2 mutant constructs revealed a striking dependence on the interaction of the inhibitor with the side chain of Arg-120 as being a determinant for time-dependent inhibition instead of Arg-513 (Greig et al., 1997; Rieke et al., 1999). In addition, NS-398 was ~1000-fold less potent towards the R120E huCOX-2 mutant construct (Greig et al., 1997). Moreover, mutation of Val-523 to isoleucine completely abolished time-dependent inhibition of huCOX-2 by NS-398, but still allowed for rapid, reversible inhibition of this isoform (Gierse et al., 1996; Guo et al., 1996). We report here the X-ray crystal structure of Fe3+-protoporphyrin IX reconstituted murine (mu) COX-2 in complex with NS-398 to 3.0 Å resolution. This study was designed to detail at the molecular level the binding of NS-398 within the cyclooxygenase channel of COX-2 and further investigate the dependence of NS-398 binding with the side chains of Arg-120 and Val-523.
2. Materials and methods
2.1. Crystallization and data collection
Murine COX-2 (muCOX-2) was expressed in baculovirus-infected insect cells and the apo enzyme purified as previously described (Vecchio et al., 2010). Prior to crystallization, the enzyme was concentrated to 3mg/mL, reconstituted with a 2-fold molar excess of Fe3+-protoporphyrin IX and subsequently dialyzed overnight at 4°C against 20mM TRIS, pH 8.0, 100mM NaCl, and 0.6% (w/v) n-octyl β-D-glucopyranoside (βOG). A 5-fold molar excess of NS-398 was then added to the reconstituted enzyme. Crystallization experiments were setup at 296 K using the sitting-drop vapor diffusion method. 3μL protein was combined with 3μL of a drop solution consisting of 23–34% polyacrylic acid 5100, 100mM HEPES, pH 7.5, 20mM MgCl2, and 0.6% (w/v) βOG and equilibrated over a reservoir solution of 23–34% polyacrylic acid 5100, 100mM HEPES, pH 7.5, 20mM MgCl2. Plate-like crystals formed in three days to a week that were brown in color. Prior to data collection, crystals were cryoprotected by soaking in 30% polyacrylic acid 5100, 100mM HEPES, pH 7.5, 20mM MgCl2, and 0.6% (w/v) βOG supplemented with 10% glycerol. Data were collected on beamline A1 at the Cornell High Energy Synchrotron Source (Ithaca, NY) using an Area Detector Systems CCD Quantum-210. Data collection statistics are summarized in Table 1.
Table 1.
Summary of the data collection and refinement statistics.
| Crystallographic Parameter | COX-2:NS398 |
|---|---|
| Space group | I222 |
| No. in Asymmetric Unit | 2 |
| Unit cell length (Å) | |
| a | 120.43 |
| b | 131.21 |
| c | 179.57 |
| α=β=γ(°) | 90° |
| Wavelength (Å) | 0.9777 |
| Resolution (Å) | 20.0-3.00 |
| Highest resolution shell (Å)a | 3.16-3.00 |
| Rmergeb | 12.4 (47.5) |
| Rpim | 7.5 (28.5) |
| Total observations | 112276 (16511) |
| Total uniquec | 28681 (4158) |
| I/σ(I) | 9.8 (3.0) |
| Completeness (%) | 99.6 (100.0) |
| Multiplicity | 3.9 (4.0) |
| Wilson B factor (Å2) | 63.7 |
|
| |
| Number of atoms in refinement | 9237 |
| Rwork | 0.176 (0.252) |
| Rfreed | 0.225 (0.288) |
| Average B factor, protein (Å2) | 34.0 |
| Average B factor, solvent (Å2) | 23.8 |
| Average B factor, inhibitor (Å2): | |
| Monomer A (Å2) | 34.7 |
| Monomer B (Å2) | 41.9 |
| Mean positional error (Å)e | 0.434 |
| RMSD in bond length (Å) | 0.011 |
| RMSD in bond angle (°) | 1.465 |
| Ramachandran Plot (%) | |
| Favored | 97.2 |
| Allowed | 2.8 |
| Disallowed | 0 |
The values in parentheses represent the values in the outermost resolution shell.
RMERGE and RPIM as defined in [Evans, 2006 #27].
Represents reflections with F > 0 σF, which were used in the refinement.
5.1% of the total reflections were used to generate the test set.
Coordinate error as calculated by Luzatti plot.
2.2. Structure solution and refinement
The data were processed using MOSFLM and SCALA in the CCP4 suite of programs (Dodson et al., 1997) in the orthorhombic space group I222. The structure was solved by molecular replacement (MR) using the program PHASER (McCoy et al., 2007) and a truncated search model of muCOX-2 derived from PDB entry 1CVU (Kiefer et al., 2000), with residues 33–144, 320–325, 344–391, 500–553, and all ligands and waters removed. Two monomers constituting the muCOX-2 homodimer were found in the crystallographic asymmetric unit. Phases from MR were then input into ARP/wARP, utilizing the “automated model building starting from experimental phases” option (Langer et al., 2008). Given the moderate resolution of the data, ARP/wARP built only 53% (290 residues) of the model. Iterative cycles of manual model building in COOT (Emsley and Cowtan, 2004), followed by refinement in REFMAC5 (Murshudov et al., 1997) were carried out to fit all the remaining residues, ligands, and waters. Tight NCS restraints were applied to residues 33–582 in each monomer during cycles of refinement. NS-398 was built and a stereochemical dictionary was generated using SKETCHER (Potterton et al., 2003). The final model (Rwork = 17.6% and Rfree = 22.5%) consists of residues 33–582, Fe3+-protoporphyrin IX, carbohydrate moieties linked to Asn-68, Asn-144, and Asn-410, and NS-398 bound in each monomer. A single βOG, glycerol, and 120 water molecules were also modeled into the electron density. Details of the refinement statistics are shown in Table 1.
2.3. Validation
Model validation was carried out in MOLPROBITY (Davis et al., 2007) and PROCHECK (Laskowski et al., 1993). Simulated annealing omit maps were created in CNS (Brunger et al., 1998). Figures, van der Waals, and hydrogen bond interactions were generated using CCP4MG (Potterton et al., 2004). In order to unambiguously determine the correct position of the sulfonamide group of NS-398 within the cyclooxygenase channel, the inhibitor was modeled into the electron density map of the final model in three different orientations, followed by refinement and inspection of 2Fo-Fc and Fo-Fc difference maps (Supplemental Figure S1). Coordinates and structure factors have been deposited in the protein data bank (PDB id 3QMO).
3. Results and Discussion
3.1. Overview of the muCOX-2:NS398 complex
The structure of muCOX-2 in complex with NS-398 (muCOX-2:NS398) was determined to 3.0Å resolution using synchrotron radiation. There are two monomers in the crystallographic asymmetric unit that form the canonical dimer, which is observed in other muCOX-2 substrate and inhibitor crystal structure complexes (Duggan et al., 2010; Kurumbail et al., 1996; Vecchio and Malkowski, 2011; Vecchio et al., 2010). The N-terminal epidermal growth factor-like domain, membrane-binding domain, and the catalytic domain are all well resolved and there is clear electron density for the Fe3+-protoporphyrin IX and N-linked carbohydrate moieties in both monomers. Interpretable electron density is present for NS-398 bound within the cyclooxygenase channel of each monomer. The root mean square deviation (rmsd) between inhibitors is 0.36Å. As such, the conformation of NS-398 in monomer A will be utilized to describe the details associated with cyclooxygenase channel residue interactions and in comparisons to other COX-1 and COX-2 inhibitor complexes. Moreover, as a matter of convention, cyclooxygenase residues are labeled according to the ovine COX-1 numbering scheme.
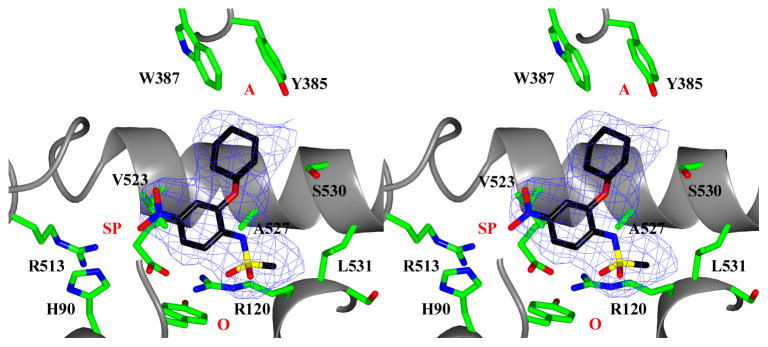
NS-398 binds in the cyclooxygenase channel of muCOX-2 with its methanesulfonamide moiety positioned at the mouth of the channel, near the side chains of Arg-120 and Tyr-355 (Figure 1). The methanesulfonamide moiety is stabilized via an ionic and hydrogen bond, which forms between one oxygen atom of the methanesulfonamide moiety and the Nε and Nη2 atoms of Arg-120 (Supplemental Table S1). The methyl group of the methanesulfonamide moiety makes additional van der Waals contacts to stabilize the inhibitor within the channel; these include interactions with the side chain atoms of Val-116, Arg-120, Ala-527, and three contacts with Leu-531. The nitro group of the nitrophenyl moiety of NS-398 is situated in the vicinity of side pocket residues His-90, Arg-513 and Val-523. One oxygen atom of the nitro group forms a hydrogen bond with the Nε2 of His-90. The phenyl group of the nitrophenyl moiety makes van der Waals contacts with the side chain of Val-523. The side chain atoms of Phe-381, Leu-384, Trp-387, and Phe-518 form a cleft at the apex of the cyclooxygenase channel that has been implicated in the stabilization of the prostaglandin product PGG2 during cyclopentane ring formation (Schneider et al., 2004). The cyclohexane moiety of NS-398 binds near this cleft, making van der Waals contacts with the side chain of Trp-387. In addition, the cyclohexane moiety makes van der Waals contacts with the main-chain atoms of Gly-526 and Ala-527, as well as with the Cβ atom of Ser-530. The nitrogen atom of the methanesulfonamide moiety and the oxygen atom bridging the methanesulfonamide and cyclohexane moieties also form hydrogen bonds with an ordered water molecule located at the base of the cyclooxygenase channel.
Figure 1. NS-398 bound in the cyclooxygenase channel of muCOX-2.
Stereo view of NS-398 bound within the cyclooxygenase channel of monomer A of the muCOX-2:NS398 crystal structure. Fo-Fc simulated annealing omit electron density, contoured at 4.5σ, is shown with the final refined model of NS-398 (black). Residues lining the cyclooxygenase channel, along with the spatial locations of the channel opening (O), channel apex (A), and COX-2-specific side pocket (SP) are labeled accordingly. Carbon atoms of residues lining the channel are colored green, while nitrogen, oxygen, and sulfur atoms are colored blue, red, and yellow, respectively.
3.2. Comparison of NS-398 binding mode with other COX:inhibitor structures
The X-ray crystal structures of COX-2 in complex with indomethacin and celecoxib have been determined previously (Kurumbail et al., 1996; Wang et al., 2010). These structures exemplify how acidic, nsNSAIDs and diarylheterocyclic coxibs bind within the cyclooxygenase channel of COX-2 and provide useful comparison and insight into the unique binding mode observed for NS-398 (see Supplemental Figure S2 for the chemical structures of the inhibitors). NS-398 binds within the cyclooxygenase channel of COX-2 in a similar fashion to indomethacin (Supplemental Figure S3A). The carboxylate of indomethacin and methanesulfonamide moiety of NS-398 both interact with the side chain of Arg-120 at the opening of the channel. The interaction of the carboxylate with the side chain of Arg-120 is a molecular determinant required for inhibition of COX-2 by indomethacin (Kalgutkar et al., 2000; Kurumbail et al., 1996). The carboxylate group of indomethacin also forms a hydrogen bond with the phenolic hydroxyl group of Tyr-355, which coincides with the movement of the side chain of Arg-120 with respect to its position in the muCOX-2:NS398 crystal structure (Supplemental Figure S3A).
Celecoxib binds in the cyclooxygenase channel of COX-2 in a manner indicative of every diaryl heterocyclic coxib studied to date, with its sulfonamide moiety interacting with His-90 and Arg-513 within the side pocket of COX-2 (Wang et al., 2010). As a result, the sulfonamide moiety of celecoxib penetrates the side pocket much deeper than the nitro group of NS-398 (Supplemental Figure S3B). The trifluoromethyl group of celecoxib binds near the mouth of the cyclooxygenase channel, where it forms a hydrogen bond with the side chain of Arg-120. Again, this interaction results in the movement of the side chain of Arg-120 with respect to its positioning the muCOX-2:NS398 crystal structure (Supplemental Figure S3B).
Nimesulide is a close structural analog of NS-398, containing a benzene ring instead of a cyclohexane moiety (Supplemental Figure S2). Although a crystal structure of nimesulide bound to COX-2 has not been elucidated to date, the structure of nimesulide bound within the cyclooxygenase channel of ovine (ov) COX-1 has recently been solved (Sidhu et al., 2010). Interestingly, nimesulide binds in an almost identical orientation to that observed for NS-398, with its methanesulfonamide moiety interacting with the side chain of Arg-120 at the opening of the channel (Supplemental Figure S3C). Additionally, the benzene ring of nimesulide binds in a similar orientation and location to that observed for the cyclohexane of NS-398 despite the more planar character of this moiety. The side chain of Arg-120 is not significantly altered by nimesulide binding within the cyclooxygenase channel of ovCOX-1, despite the classification of nimesulide as a COX-2-selective inhibitor. Instead, the side chains of Glu-524 and Tyr-355 move further apart from the methanesulfonamide moiety of nimesulide and Arg-120. Finally, the side chain of Ile-523 moves to accommodate the nitro group in the nitrophenyl moiety of nimesulide, compared to the position of the Val-523 side chain (Supplemental Figure S3C).
3.3. Functional Significance of Arg-120 and Val-523 in Time-Dependent Inhibition of COX-2 by NS-398
The observed conformation of NS-398 within the cyclooxygenase channel of the muCOX-2:NS398 crystal structure provides insight into previous biochemical data that identified Arg-120 and Val-523 as determinants critical to time-dependent inhibition of COX-2 (Greig et al., 1997; Rieke et al., 1999). Mutation of Arg-120 to glutamine results in the loss of time-dependent inhibition of huCOX-2 by NS-398, although the IC50 value decreased 5-fold compared to wild-type enzyme (Rieke et al., 1999). Based on our analysis of the muCOX-2:NS398 crystal structure, these results suggest that Gln-120 can compensate for arginine and maintain the appropriate interactions with the methanesulfonamide moiety for binding within the channel, but cannot rescue the mechanism by which the compound promotes time-dependent inhibition. Conversely, mutation of Arg-120 to glutamate results in both the loss of time- dependent inhibition by NS-398 and a 1000-fold increase in the IC50 value (Greig et al., 1997). In addition to disrupting the mechanism by which NS-398 facilitates time-dependent inhibition, the presence of a glutamate at position 120 would create repulsive interactions that destabilize NS-398 binding within the cyclooxygenase channel. The V523I huCOX-2 mutant construct is not inhibited in a time-dependent manner by NS-398 (Gierse et al., 1996; Guo et al., 1996). In our structure, the side chain of Val-523 makes four van der Waals contacts with the phenyl group of the nitrophenyl moiety of NS-398. Although the V523I mutation could potentially disrupt optimal binding of the nitrophenyl moiety in the cyclooxygenase channel of COX-2, the ovCOX-1:nimesulide crystal structure suggests that NS-398 could be accommodated within the channel. Indeed, while the ability of NS-398 to inhibit V523I huCOX-2 in a time-dependent manner is lost, rapid reversible inhibition of COX-2 is maintained (Guo et al., 1996). Thus, the side chain of Val-523 plays a role in time-dependent inhibition of COX-2 by NS-398, but our structure does not provide any additional details into the mechanism.
4. Summary and conclusions
NS-398 is a COX-2-selective inhibitor of the sulfonanilide class and as our present results establish, binds uniquely within the cyclooxygenase channel of COX-2. In contrast to that observed for the diarylheterocyclic coxibs, NS-398 does not penetrate the side pocket of COX-2 past the side chain of Val-523 or interact with the side chain of Arg-513 at the base of the pocket. Instead, the methanesulfonamide moiety forms an ionic and hydrogen bond with the side chain of Arg-120 at the opening of the cyclooxygenase channel, similar to that observed for the acidic nsNSAID indomethacin and flurbiprofen. The observed interactions with the side chain of Arg-120 compliment mutational studies that implicate Arg-120 as being a molecular determinant for time-dependent inhibition of COX-2 by NS-398.
Supplementary Material
Acknowledgments
This work was supported by National Institutes of Health grant R01 GM077176 from the National Institute of General Medical Sciences. This work is also based upon research conducted at the Cornell High Energy Synchrotron Source (CHESS), which is supported by the National Science Foundation under NSF award DMR-0225180, using the Macromolecular Diffraction at CHESS (MacCHESS) facility, which is supported by award RR-01646 from the National Institutes of Health, through its National Center for Research Resources.
The abbreviations used are
- COX
cyclooxygenase
- hu
human
- mu
murine
- ov
ovine
- AA
arachidonic acid
- NS-398
N-(2-cyclohexyloxy-4-nitrophenyl) methanesulfonamide
- MR
molecular replacement
- NSAIDs
nonsteroidal anti-inflammatory drugs
- βOG
n-octyl β-D-glucopyranoside
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Blobaum AL, Marnett LJ. Structural and functional basis of cyclooxygenase inhibition. J Med Chem. 2007;50:1425–41. doi: 10.1021/jm0613166. [DOI] [PubMed] [Google Scholar]
- Brunger AT, Adams PD, Clore GM, DeLano WL, Gros P, et al. Crystallography & NMR system: A new software suite for macromolecular structure determination. Acta Crystallogr D Biol Crystallogr. 1998;54:905–21. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- Davis IW, Leaver-Fay A, Chen VB, Block JN, Kapral GJ, et al. MolProbity: all-atom contacts and structure validation for proteins and nucleic acids. Nucleic Acids Res. 2007;35:W375–83. doi: 10.1093/nar/gkm216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeWitt DL. Cox-2-selective inhibitors: the new super aspirins. Mol Pharmacol. 1999;55:625–31. [PubMed] [Google Scholar]
- Dodson EJ, Winn M, Ralph A. Collaborative Computational Project, number 4: providing programs for protein crystallography. Methods Enzymol. 1997;277:620–33. doi: 10.1016/s0076-6879(97)77034-4. [DOI] [PubMed] [Google Scholar]
- Duggan KC, Walters MJ, Musee J, Harp JM, Kiefer JR, et al. Molecular basis for cyclooxygenase inhibition by the non-steroidal anti-inflammatory drug naproxen. J Biol Chem. 2010;285:34950–9. doi: 10.1074/jbc.M110.162982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60:2126–32. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- Futaki N, Takahashi S, Yokoyama M, Arai I, Higuchi S, et al. NS-398, a new anti-inflammatory agent, selectively inhibits prostaglandin G/H synthase/cyclooxygenase (COX-2) activity in vitro. Prostaglandins. 1994;47:55–9. doi: 10.1016/0090-6980(94)90074-4. [DOI] [PubMed] [Google Scholar]
- Futaki N, Yoshikawa K, Hamasaka Y, Arai I, Higuchi S, et al. NS-398, a novel non-steroidal anti-inflammatory drug with potent analgesic and antipyretic effects, which causes minimal stomach lesions. Gen Pharmacol. 1993;24:105–10. doi: 10.1016/0306-3623(93)90018-s. [DOI] [PubMed] [Google Scholar]
- Garavito RM, Malkowski MG, DeWitt DL. The structures of prostaglandin endoperoxide H synthases-1 and -2. Prostaglandins Other Lipid Mediat. 2002;68–69:129–52. doi: 10.1016/s0090-6980(02)00026-6. [DOI] [PubMed] [Google Scholar]
- Gierse JK, McDonald JJ, Hauser SD, Rangwala SH, Koboldt CM, et al. A single amino acid difference between cyclooxygenase-1 (COX-1) and -2 (COX-2) reverses the selectivity of COX-2 specific inhibitors. J Biol Chem. 1996;271:15810–4. doi: 10.1074/jbc.271.26.15810. [DOI] [PubMed] [Google Scholar]
- Greig GM, Francis DA, Falgueyret JP, Ouellet M, Percival MD, et al. The interaction of arginine 106 of human prostaglandin G/H synthase-2 with inhibitors is not a universal component of inhibition mediated by nonsteroidal anti-inflammatory drugs. Mol Pharmacol. 1997;52:829–38. doi: 10.1124/mol.52.5.829. [DOI] [PubMed] [Google Scholar]
- Guo Q, Wang LH, Ruan KH, Kulmacz RJ. Role of Val509 in time-dependent inhibition of human prostaglandin H synthase-2 cyclooxygenase activity by isoform-selective agents. J Biol Chem. 1996;271:19134–9. doi: 10.1074/jbc.271.32.19134. [DOI] [PubMed] [Google Scholar]
- Harman CA, Turman MV, Kozak KR, Marnett LJ, Smith WL, et al. Structural basis of enantioselective inhibition of cyclooxygenase-1 by S-alpha-substituted indomethacin ethanolamides. J Biol Chem. 2007;282:28096–105. doi: 10.1074/jbc.M701335200. [DOI] [PubMed] [Google Scholar]
- Kalgutkar AS, Crews BC, Rowlinson SW, Marnett AB, Kozak KR, et al. Biochemically based design of cyclooxygenase-2 (COX-2) inhibitors: facile conversion of nonsteroidal antiinflammatory drugs to potent and highly selective COX-2 inhibitors. Proc Natl Acad Sci U S A. 2000;97:925–30. doi: 10.1073/pnas.97.2.925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiefer JR, Pawlitz JL, Moreland KT, Stegeman RA, Hood WF, et al. Structural insights into the stereochemistry of the cyclooxygenase reaction. Nature. 2000;405:97–101. doi: 10.1038/35011103. [DOI] [PubMed] [Google Scholar]
- Kozak KR, Rowlinson SW, Marnett LJ. Oxygenation of the endocannabinoid, 2-arachidonylglycerol, to glyceryl prostaglandins by cyclooxygenase-2. J Biol Chem. 2000;275:33744–9. doi: 10.1074/jbc.M007088200. [DOI] [PubMed] [Google Scholar]
- Kurumbail RG, Stevens AM, Gierse JK, McDonald JJ, Stegeman RA, et al. Structural basis for selective inhibition of cyclooxygenase-2 by anti-inflammatory agents. Nature. 1996;384:644–8. doi: 10.1038/384644a0. [DOI] [PubMed] [Google Scholar]
- Langer G, Cohen SX, Lamzin VS, Perrakis A. Automated macromolecular model building for X-ray crystallography using ARP/wARP version 7. Nat Protoc. 2008;3:1171–9. doi: 10.1038/nprot.2008.91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laskowski RA, Moss DS, Thornton JM. Main-chain bond lengths and bond angles in protein structures. J Mol Biol. 1993;231:1049–67. doi: 10.1006/jmbi.1993.1351. [DOI] [PubMed] [Google Scholar]
- Loll PJ, Picot D, Garavito RM. The structural basis of aspirin activity inferred from the crystal structure of inactivated prostaglandin H2 synthase. Nat Struct Biol. 1995;2:637–43. doi: 10.1038/nsb0895-637. [DOI] [PubMed] [Google Scholar]
- Luong C, Miller A, Barnett J, Chow J, Ramesha C, et al. Flexibility of the NSAID binding site in the structure of human cyclooxygenase-2. Nat Struct Biol. 1996;3:927–33. doi: 10.1038/nsb1196-927. [DOI] [PubMed] [Google Scholar]
- Marnett LJ. The COXIB experience: a look in the rearview mirror. Annu Rev Pharmacol Toxicol. 2009;49:265–90. doi: 10.1146/annurev.pharmtox.011008.145638. [DOI] [PubMed] [Google Scholar]
- McCoy AJ, Grosse-Kunstleve RW, Adams PD, Winn MD, Storoni LC, et al. Phaser crystallographic software. J Appl Crystallogr. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murshudov GN, Vagin AA, Dodson EJ. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr D Biol Crystallogr. 1997;53:240–55. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- Picot D, Loll PJ, Garavito RM. The X-ray crystal structure of the membrane protein prostaglandin H2 synthase-1. Nature. 1994;367:243–9. doi: 10.1038/367243a0. [DOI] [PubMed] [Google Scholar]
- Potterton E, Briggs P, Turkenburg M, Dodson E. A graphical user interface to the CCP4 program suite. Acta Crystallogr D Biol Crystallogr. 2003;59:1131–7. doi: 10.1107/s0907444903008126. [DOI] [PubMed] [Google Scholar]
- Potterton L, McNicholas S, Krissinel E, Gruber J, Cowtan K, et al. Developments in the CCP4 molecular-graphics project. Acta Crystallogr D Biol Crystallogr. 2004;60:2288–94. doi: 10.1107/S0907444904023716. [DOI] [PubMed] [Google Scholar]
- Rieke CJ, Mulichak AM, Garavito RM, Smith WL. The role of arginine 120 of human prostaglandin endoperoxide H synthase-2 in the interaction with fatty acid substrates and inhibitors. J Biol Chem. 1999;274:17109–14. doi: 10.1074/jbc.274.24.17109. [DOI] [PubMed] [Google Scholar]
- Rouzer CA, Marnett LJ. Cyclooxygenases: structural and functional insights. J Lipid Res. 2009;50(Suppl):S29–34. doi: 10.1194/jlr.R800042-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rowlinson SW, Kiefer JR, Prusakiewicz JJ, Pawlitz JL, Kozak KR, et al. A novel mechanism of cyclooxygenase-2 inhibition involving interactions with Ser-530 and Tyr-385. J Biol Chem. 2003;278:45763–9. doi: 10.1074/jbc.M305481200. [DOI] [PubMed] [Google Scholar]
- Schneider C, Boeglin WE, Brash AR. Identification of two cyclooxygenase active site residues, Leucine 384 and Glycine 526, that control carbon ring cyclization in prostaglandin biosynthesis. J Biol Chem. 2004;279:4404–14. doi: 10.1074/jbc.M307431200. [DOI] [PubMed] [Google Scholar]
- Selinsky BS, Gupta K, Sharkey CT, Loll PJ. Structural analysis of NSAID binding by prostaglandin H2 synthase: time-dependent and time-independent inhibitors elicit identical enzyme conformations. Biochemistry. 2001;40:5172–80. doi: 10.1021/bi010045s. [DOI] [PubMed] [Google Scholar]
- Sidhu RS, Lee JY, Yuan C, Smith WL. Comparison of cyclooxygenase-1 crystal structures: cross-talk between monomers comprising cyclooxygenase-1 homodimers. Biochemistry. 2010;49:7069–79. doi: 10.1021/bi1003298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith WL, DeWitt DL, Garavito RM. Cyclooxygenases: structural, cellular, and molecular biology. Annu Rev Biochem. 2000;69:145–82. doi: 10.1146/annurev.biochem.69.1.145. [DOI] [PubMed] [Google Scholar]
- Tanabe T, Tohnai N. Cyclooxygenase isozymes and their gene structures and expression. Prostaglandins Other Lipid Mediat. 2002;68–69:95–114. doi: 10.1016/s0090-6980(02)00024-2. [DOI] [PubMed] [Google Scholar]
- Vecchio AJ, Malkowski MG. The structural basis of endocannabinoid oxygenation by cyclooxygenase-2. J Biol Chem. 2011;286:20736–45. doi: 10.1074/jbc.M111.230367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vecchio AJ, Simmons DM, Malkowski MG. Structural basis of fatty acid substrate binding to cyclooxygenase-2. J Biol Chem. 2010;285:22152–63. doi: 10.1074/jbc.M110.119867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wada M, DeLong CJ, Hong YH, Rieke CJ, Song I, et al. Enzymes and receptors of prostaglandin pathways with arachidonic acid-derived versus eicosapentaenoic acid-derived substrates and products. J Biol Chem. 2007;282:22254–66. doi: 10.1074/jbc.M703169200. [DOI] [PubMed] [Google Scholar]
- Wang JL, Carter J, Kiefer JR, Kurumbail RG, Pawlitz JL, et al. The novel benzopyran class of selective cyclooxygenase-2 inhibitors-part I: the first clinical candidate. Bioorg Med Chem Lett. 2010;20:7155–8. doi: 10.1016/j.bmcl.2010.07.053. [DOI] [PubMed] [Google Scholar]
- Wong E, Bayly C, Waterman HL, Riendeau D, Mancini JA. Conversion of prostaglandin G/H synthase-1 into an enzyme sensitive to PGHS-2-selective inhibitors by a double His513 --> Arg and Ile523 --> val mutation. J Biol Chem. 1997;272:9280–6. doi: 10.1074/jbc.272.14.9280. [DOI] [PubMed] [Google Scholar]
- Yu M, Ives D, Ramesha CS. Synthesis of prostaglandin E2 ethanolamide from anandamide by cyclooxygenase-2. J Biol Chem. 1997;272:21181–6. doi: 10.1074/jbc.272.34.21181. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.