Abstract
Amelogenin is cleaved by enamelysin (Mmp-20) soon after its secretion, and the cleavage products accumulate in specific locations during enamel formation, suggesting that parent amelogenin proteolysis is necessary for activating its functions. To investigate the precise roles of Mmp-20 and its influence on the assembly of amelogenin, an in vitro enzymatic digestion process mimicking the initial stages of amelogenin proteolysis was investigated at near-physiological conditions using recombinant porcine amelogenin (rP172) and enamelysin. Hierarchically organized nanorod structures formed during different digestion stages were detected by TEM. At the earliest stage, uniformly dispersed parent amelogenin spherical particles, mixed with some darker stained smaller spheres, and accompanying elongated chain-like nanostructures were observed. Cylindrical nanorods, which appeared to be the result of tight assembly of thin subunit cylindrical discs with thicknesses ranging from ~2.5 nm to ~6.0 nm, were formed after an hour of proteolysis. These subunit building blocks stacked to form nanorods with maximum length of ~100 nm. With the production of more cleavage products, additional morphologies spontaneously evolved from the cylindrical nanorods. Larger ball-like aggregates ultimately formed at the end of proteolysis. The uniform spherical particles, nanorods, morphological patterns evolved from nanorods, and globular aggregated microstructures were successively formed by means of co-assembly of amelogenin and its cleavage products during a comparatively slow proteolysis process. We propose that, following the C-terminal cleavage of amelogenin, co-assembly with its fragments leads to formation of nanorod structures whose properties eventually dictate the super-structural organization of enamel matrix, controlling the elongated growth of enamel apatite crystals.
Keywords: amelogenin, Mmp-20, proteolysis, enamel, self-assembly, nanorods
1. INTRODUCTION
Amelogenesis is one of the most dynamic mineralization processes known, producing the hardest tissue found among vertebrates in which the extracellular matrix proteins are involved in construction, regulation, and control of hydroxyapatite crystals causing them to grow and elongate in parallel (Eastoe 1979). The final product is a tissue that is more than 95% mineral by weight with an extraordinary super-structural organization (Robinson et al. 1995, Boyde 1989). The complementary crystal maturation and protein degradation that leads to mature enamel involve specific protein-protein, protein-proteinase, and protein-crystal interactions (Smith 1989, Moradian-Oldak and Paine 2008). Three major enamel matrix proteins (amelogenin, ameloblastin, and enamelin) are cleaved by proteinases after they are secreted, and their cleavage products accumulate in specific and deep enamel layers, suggesting that protein proteolysis is necessary for these enamel proteins to perform their functions (Bartlett and Simmer 1999). Two major enzymes have been described that carry out these processes and degrade the enamel proteins. The matrix metalloproteinase enamelysin (Mmp-20) is the foremost enamel matrix-processing enzyme, being expressed initially prior to the onset of dentin mineralization and continuing throughout the secretory stage of amelogenesis and into early maturation (Simmer and Hu 2002). Expression of the serine proteinase Kallikrein-4 (KLK4) starts during the transition stage and continues throughout maturation. Defects in MMP-20 cause autosomal-recessive amelogenesis imperfecta in humans (Ozdemir et al 2005). The critical function of Mmp-20 is further evidenced by the loss of prism structure and failure to achieve the full thickness of the enamel in Mmp20-null mice (Caterina et al 2002). Generally, KLK4 protease plays a crucial role in matrix-mediated enamel biomineralization by processing and eventually removing the protein matrix in a specific and timely manner (Simmer et al 2009). In contrast, Mmp20 activity is thought to involve production of specific intermediate fragments of extracellular matrix proteins. One proposed function of Mmp20 is control of the protein self-assembly process to allow the programmed elongated growth of apatite crystals in a hierarchical organization (Sun et al 2005, Bartlett & Simmer 1999)
The importance of amelogenin in controlling mineral phase, elongation of crystal growth, and prism organization of enamel has been recognized. There is general agreement that amelogenin plays a critical role in crystallization of hydroxyapatite, especially in the initial nucleation and protection of amorphous calcium phosphate in vitro (Beniash et al 2005, Weidmann-Bidlack et al 2011, Yang et al 2010). Amelogenin nanospheres can be formed under neutral and slightly alkaline pH conditions and they further assemble to form high aspect ratio nano-structures in solution under a variety of in vitro conditions (Du et al 2005, Moradian-Oldak et al 2006, Wiedemann-Bidlack et al 2007). An interesting transition from nanospheres to fibrous amelogenin (25kDa rH174) assemblies was facilitated under conditions that involved interaction between rH174 and its proteolytic cleavage products (rH163) ( Uskoković et al 2008). It was reported that when both amelogenin cleavage products 25-kDa rH174 and 23-kDa rH163 were mixed, self-assembled nano-strings were formed depending on the pH, and incubation time (He et al 2008). Another group has recently reported that full length porcine amelogenin molecules (rP172) formed tightly connected, elongated, high-aspect ratio assemblies, while amelogenin cleavage products (rP147) appeared as loosely associated spherical particles (Weidmann-Bidlack et al 2007). Additional clues to amelogenin function have been revealed by examination of the protein’s primary and secondary structures. Sequence alignment has revealed that several residues at the N- and C-terminal regions have remained unchanged during 250 million years, indicating their importance for amelogenin function (Sire et al 2005). The main hydrophobic part of amelogenin is rich in proline residues (~30%) that are believed to inhibit the formation of ordered secondary structures such as beta-sheet and alpha-helix (Lakshminarayanan et al 2007). It has been reported that this central PXXP rich region tended to adopt a conformation known as a polyproline II helix. The evolutionary importance of the length of the PPII motif and its association to enamel structure has been also recently reported (Jin et al 2009). Despite these clues, the biological mechanisms by which amelogenins assemble into hierarchically organized microstructures that further build the rods and inter-rods of the enamel matrix structure in vivo remain unclear. Furthermore, the significance of the specific cleavage of amelogenin by enamelysin in the context of enamel formation is still not fully understood.
Previous studies have suggested that proteinases are necessary for “activating” enamel proteins so that the parent proteins and their cleavage products may perform different functions (Bartlett and Simmer 1999). In an attempt to identify and characterize the molecular mechanism of porcine amelogenin proteolysis by porcine enamelysin, investigators have applied both in vitro and in vivo approaches. The dominant porcine amelogenin (P173; Met1-Asp173) has 173 amino acids and an apparent molecular weight of 25 kDa. In developing teeth, Mmp-20 has been shown to have significant proteolytic activity in the enamel extracellular space during the secretory stage (Smith et al 2011). Mmp-20 processes the full-length amelogenin into a group of cleavage products that accumulate and are only slowly degraded further. Most of the secreted P173 is cleaved after Ser148 to generate the 20-kDa amelogenin (P148; Met1-Ser148), which in turn is cleaved after Trp45 to yield the tyrosine-rich amelogenin polypeptide (TRAP; P45; Met1-Trp45) and the 13-kDa amelogenin (Leu46-Ser148) (Nagano et al 2009). Recombinant porcine amelogenin (rP172) and recombinant porcine enamelysin (rpMmp-20) have been studied in in vitro experiments showing that the rP172 is processed at the C- and N-terminal regions, after Pro162, Ser148, His62, Ala63, and Trp45 (Sun et al 2010, Nagano et al 2009). While Mmp-20 cleavage sites on amelogenin have been carefully investigated, these in vitro studies have not focused on the dynamics of assembly events of parent amelogenins during enamelysin digestion and knowledge on the mechanisms by which their truncated products might co-assemble and direct the morphologies of such assemblies is currentely limited (He et al 2008, Uskoković et al 2008).
In the study reported herein, we monitored and investigated the amelogenin assembly process during proteolysis in a relatively slow proteolytic reaction conducted at near physiological pH and temperature, using a recombinant porcine amelogenin rP172 as a substrate and a recombinant porcine enamelysin (rpMmp-20) as a proteinase. This is the first time that the progress of amelogenin assembly alternation following its hydrolysis by Mmp-20 is reported and an association with the changes in the ratio of full-length amelogenin to its proteolytic products is made. The progress of proteolysis was monitored by SDS-PAGE and self-assembly was analyzed by high-resolution transmission electron microscopy (HRTEM). Fine nanorod-like assembled structures with known substrate to cleavage product ratios were successfully produced following increased cleavage of proteins during the early proteolysis stages. In addition, different patterns that evolved from the nanorods were detected at later digestion stages, providing supplemental evidence for the multistep co-assembly process and a possible pathway to hierarchical enamel matrix structure formation in vivo.
2. MATERIALS AND METHODS
2.1. Expression and Purification of Recombinant Amelogenins and rpMmp-20
Full-length recombinant pig amelogenin (rP172) and an engineered mutant form lacking the hydrophilic C-terminal 24 amino acids (rP148) were expressed in e-coli and purified as previously described (Ryu et al 1999, Sun et al 2006). The rP172 and rP148 forms have amino acids 2-172 and 2-149 of porcine amelogenin (P173) respectively (Yamakoshi et al 1994, Sun et al 2006). Briefly, rP172 and rP148 were purified by ammonium sulfate precipitation and reverse phase high performance liquid chromatography (RP-HPLC), using buffer A (0.1% trifluoroacetic acid: TFA) and buffer B (buffer A +60% acetonitrile). The homogeneity of the proteins was verified by analytical chromatography (C4 analytical column, Jupiter, 5 μm, 2 mm x 250 mm). The protein solutions were lyophilized and stored at −20°C. The recombinant enamelysin plasmid (provided by Dr. James P. Simmer) was transformed into competent E. coli XL1-Blue cells (Stratagene, La Jolla, CA, USA). Recombinant pig enamelysin (rpMmp-20) was expressed in 500ml media from the cells and purified following the protocol described previously (Ryu et al 1999, Moradian-Oldak et al 2001). 2001). A His-TRAP chelating nickel affinity column (Pharmacia, Piscataway, NJ,USA) with a three buffer system (Loading buffer A: 6M Guanidine-HCl, 0.01M Tris-HCl, 0.1M NaH2PO4, pH=8.0, binding buffer B: 8M urea, 0.1M NaH2PO4 and 0.01 M Tris-HCl, pH=8.0 and eluting buffer C: 8M urea, 0.1 M NaH2PO4, 0.01M Tris-HCl, 0.25M imidazole, pH= 6.3) was used to isolate the recombinant protein from E-coli products. The enamelysin fraction was desalted against desalting buffer (50mM Tris-HCl. pH 7.4, 150mM NaCl, 0.05% Brij35) using a Centriprep-10 spun filter. Recombinant enamelysin was self-activated by its incubation in APMA (4-aminophenylmercuric acetate) in 10 mM CaCl2 for 24 hrs. Enzyme concentration was quantified by UV adsorption at 220 nm.
2.2. rP172 Proteolysis by rpMmp-20
Proteolysis experiments were repeated three times (N=3). Recombinant porcine amelogenin rP172 was dissolved in 25mM Tris-HCl (pH 8.0) to a final rP172 concentration of 0.2mg/ml, and incubated with rpMmp-20 solution at 37°C with a substrate to enzyme ratio of 200:1 (w:w), N=3. The proteolysis mixtures were incubated without shaking or stirring. The final concentration of calcium in the proteolysis mixture was 50μm. The proteolysis products were collected and analyzed at time points 0.1, 1, 2, 4, 10 and 20 hours as described previously (Sun et al 2010) . The undigested rP172 and digested samples were loaded onto a sodium dodecyl sulfate (SDS)-containing 12% acrylamide gel (Bio-Red, 161-0325), electrophoresed at 125V, and stained with Coomassie Brilliant Blue R 250. To monitor the proteolysis process and quantify each band on the SDS-Gel, peptide/protein band densities were analyzed by Image J 1.43 software. Cleavage sites were analyzed by Tandem Mass Spectrometry (LCMSMS).
2.3. Transmission Electron Microscopy (TEM)
Following proteolytic incubation in Tris-HCl buffer at 37 °C, 300 mesh carbon-coated grids were submerged in 30 μl samples of the proteolysis solution for 0.5 minute, followed by a 0.5 minute immersion in 1% uranyl acetate stain, and air dried. The sample grids were examined under the Jeol 1400 TEM with voltage of 100 kV. The diameters of particles (nanorods) were analyzed and measured with ImageJ software. In control experiments, pure rP172, pure engineered rP148, and mixtures of the two at ratios of 3:1, 1:1, or 1:3 were dissolved in 25mM Tris-HCl (pH 8.0) and incubated at 37 °C. Progress of the proteolytic process was monitored by the same TEM technique described above.
3. RESULTS
3.1. Amelogenin Assembly Evolution Captured by TEM
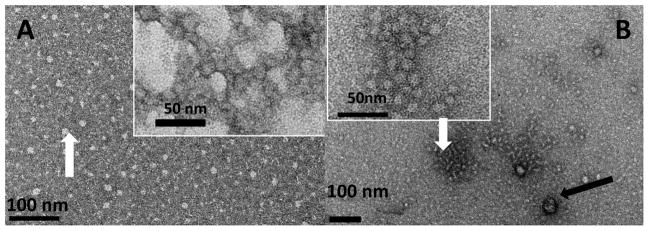
The TEM observations revealed that, in addition to the suspension of spherical particles formed at pH 8 (Fig. 1A, white arrow), full-length amelogenin rP172 occasionally assembled into chain-like structures (inset of Fig. 1A). The average particle size was estimated to be ~19 nm diameter (Table 1). At pH 8, the formation of chain-like structures was favored at 37 °C when compared to room temperature. Under near physiological pH condition (pH 8.0), the truncated rP148 formed spherical particles with a tendency to randomly aggregate to form bigger particles due to the protein’s hydrophobic nature (inset, Fig. 1B), and there was limited tendency to form chain-like structures (Fig. 1B). Note that the chains in A inset are made of more fused particles and have higher aspect ratio as isolated entities, while in B inset the subunits are more separated and isolated chains with high aspect ratios were not observed.
Figure 1.
Typical morphologies of assemblies formed from pure recombinant rP172 and rP148 at pH8.0, in Tris-HCl buffer, at 37 °C. (A) representative uniformly dispersed spherical particles of rP172 (white arrow). Inset shows a typical elongated pearl-like assembly, consisting of assembly particles with ellipsoidal shape. (B) Representative distribution of rP148 spherical particles with some randomly associated aggregates (white arrow), and a small amount of larger aggregates (black arrow), Inset shows the distribution of typical rP148 spherical particles associated to form random morphologies and sizes.
Table 1.
Analysis of composition and morphologies of amelogenin assemblies during Mmp-20 proteolysis. Mass percentage of protein bands corresponding to the cleavage products, stained in SDS-PAGE and estimated by ImageJ software. The dotted line (----) indicates products that were not detected. The main morphology of the assemblies corresponding to specific proteolysis time was accessed by TEM and particle size distribution was estimated by ImageJ software.
| TIME | 0.1 hr | 1 hr | 2 hrs | 4 hrs | 10 hrs | 20 hrs |
|---|---|---|---|---|---|---|
| rP172(*) | 95.38 | 61.11 | 45.98 | 24.62 | 5.66 | 1.28 |
| 2–162 | ---- | 14.45 | 17.24 | 21.78 | 22.64 | 24.3 |
| 2-148 | 4.62 | 24.44 | 36.78 | 49.24 | 56.60 | 53.71 |
| 13k | ---- | ---- | ---- | ---- | 5.66 | 6.39 |
| 11k | ---- | ---- | ---- | ---- | 2.83 | 3.32 |
| Extended TRAP (1-63) | ---- | ---- | ---- | 1.52 | 1.89 | 3.84 |
| TRAP (1-45) | ---- | ---- | ---- | 2.84 | 4.72 | 7.16 |
| rP172: 2-148/2-162 | ~20:1 | ~2:1 | ~1:1 | ~1:3 | ~1:15 | ~1:80 |
| (#) Diameters (nm) | 14.36 | 13.56 | 11.53 | 11.83 | 11.16 | 10.69 |
| Main Morphology | Spherical particles with some pearl-like chains | Spherical particles with longer pearl-like chains | Nanorods with their evolution patterns | Nanorod evolution patterns | Nanorod evolution patterns, some aggregates | Large globular aggregates |
As control measurements, the average diameters were 19.27 nm and 15.16 nm respectively for pure rp172 and pure rp148 incubation within the pH8.0 Tris-Cl buffer at 37 °C, larger aggregation diameters of rp148 are not accounted.
Particle size, as for the chain-like microstructures, the size represents the average diameters of the chains, N=50.
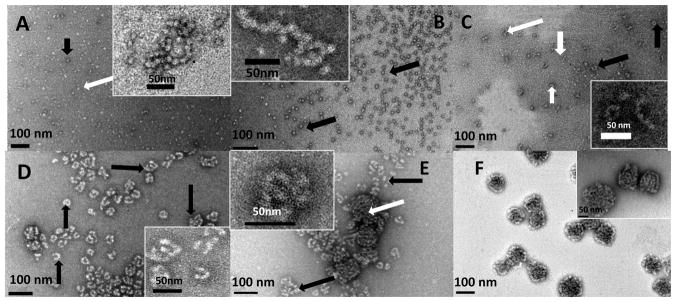
Following the addition of rpMmp-20 to amelogenin rP172 solutions in Tris-HCl (pH 8.0, 37 °C), striking intermediate nanoscale structures started to appear as observed by HRTEM (Fig. 2A-2F). Within approximately ten minutes of digestion, uniform and dispersed spherical particles were observed (Fig. 2A), accompanied by some chain-like nanostructures (Fig. 2A and the inset). Clearly, some particles were surrounded by dense stain (black arrow figure 2A) while some were surrounded by lighter stain (white arrow, Fig. 2A). After one hour of digestion, the uniform and dispersed spherical particles were observed with some longer chain-like nanostructures (Fig. 2B and inset). These chain-like structures were tightly associated (inset of Fig. 2A and 2B), and their building blocks had clear spherical shape. These spherical particles had an average diameter of ~14 nm and were clearly smaller than the dispersed rP172 particles (shown in Fig 1) of ~19 nm diameter (Table 1). Interestingly, about two hours after digestion, nanorods with an average aspect ratio of ~5:1 were formed (white arrows on and inset of Fig. 2C). These high aspect ratio structures had an average diameter of ~11nm, ranging from a minimum of ~6 nm to maximum of ~14 nm. At the same time (2 hours), different morphological patterns that apparently evolved from the nanorods appeared on the grid (black arrows, and inset of Fig. 2C). Remarkably, after four hours digestion, with the production of more hydrophobic products (i.e. 2-148), nanorods evolved into a variety of different morphologies such as donuts and U-shaped structures (black arrows, Fig.2D). The variety of morphologies detected at about four hours included longer nanorods with high aspect ratio (~10:1, perimeter of the ring to diameter of the nanorod) and the nanorods had an average diameter of ~11nm (Table 1). After 10 hours of digestion, new, tightly-aggregated globular structures of about 100 nm in diameter were observed (white arrows, Fig. 2E), and these microstructures grew bigger as digestion continued until they were the dominant particles after 20 hours of proteolysis, by which time the nanorods were completely consumed. The large aggregates were composed of tightly packed chains that had an average diameter of ~10 nm and appeared to be formed as the result of numerous particles aggregating together (Fig. 2F and Table 1).
Figure 2.
Representative TEM images of amelogenin microstructures resulting from co-assembly of rP172 and its proteolytic products during proteolysis in Tris-Cl buffer, pH 8.0, at 37 °C, collected at 0.1 hr (A), 1 hr (B), 2 hrs (C), 4 hrs (D), 10 hrs (E) and 20 hrs (F). (A) The lightly stained rP172 particles (white arrow) are connected with some deeply stained spherical particles (black arrow), forming a beaded-like structure, the inset presenting magnification of typical chains; (B) At 1 hour proteolysis, some tightly associated and elongated chain-like microstructures (inset) are formed within the uniform suspension of spherical particles (black arrows); (C) After 2 hours proteolysis, cylindrical-shaped nanorods (white arrows) formed and some of nanorods curved into O-like or U-shaped microstructures (black arrows). Inset shows typical images of nanorods and their curled microstructures at higher magnification; (D) After 4 hours proteolysis, different morphologies composed of nanorods were formed (black arrows); (E) After 10 hours proteolysis, a new mixture of compact aggregates (white arrows) were detected among the remaining rod-like structures (black arrows). Inset presents a typical morphology at higher magnification; (F) After 20 hours of proteolysis, the tightly compact ball-like aggregates grew larger, concomitant with disappearance of rod-like morphologies. Inset presents a typical morphology at higher magnification. The analyses of all the particles sizes are described in the Table 1.
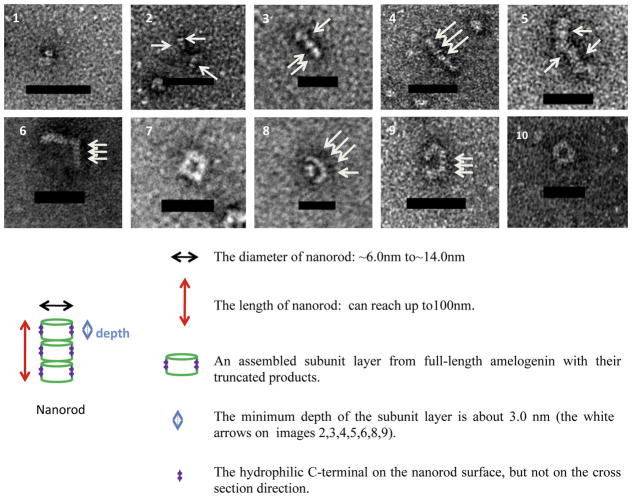
We followed the assembly of nanorods formed between one and two hours of proteolysis and further analyzed the formation of cylindrical-shaped nanorods derived from them. Cylindrical-shaped nanorods (1–5 in Fig. 3) appeared to be the result of tight assembly of thin cylindrical discs (white arrows, Fig. 3). The depth of these subunit layers was estimated from the side view to be ~2.5nm to ~6.0 nm (see 1–5 in Fig. 3). These subunit layers were the basic building blocks which were closely stacked to form longer nanorods of 100nm length. At the same time, these assembled nanorods could further evolve into different morphologies (6–10 in Fig. 3). The average diameter of these nanorods was about 11nm, with a wide range of diameter from a minimum of ~6nm to a maximum of ~14nm.
Figure 3.
Representative nanorod morphologies and sketch of a nanorod structure formed during the first two hours of proteolysis: (1–5) illustrate nanorod growth following accretion of additional subunits; (6–10) illustrate typical morphologies resulting from nanorod evolution patterns, with “⌉”, “□”, “U”, “C” and “O” shapes ( scale bar = 50nm).
To examine the effect of incubation time and amelogenin composition on the formation of nanorods, we monitored the assemblies formed by the mixtures of the full-length amelogenin rP172 and its truncated isoform rp148, combined in certain ratios of rP172:rP148 (0:1 3:1, 1:1, 1:3, 1:0), in 25mM Tris-HCl (pH 8.0), at 37 °C. The TEM observations revealed that, in addition to the suspension of spherical particles formed at pH 8 (Fig. S1: A, D, G, and insets), full-length amelogenin rP172 occasionally assembled into chain-like structures that were favored at 37 °C, and with increasing incubation time (insets of Fig. S1: D, G). The truncated rP148 formed spherical particles with a tendency to randomly aggregate to form bigger particles due to the protein’s hydrophobic nature. The aggregates increased depending on the length of incubation time (Fig.S1 B, E, H, and insets). Representative TEM images of the rp172 and rP148 mixture with 1:1 ratio incubated at 1, 10, and 20 hrs are shown in Fig. S1 C, F, I. This 1:1 ratio is similar to the composition of the proteolytic products after 1-2 hours proteolysis, when nanorods start to form. None of the nanorods structures detected during the proteolysis experiments or other morphologies evolved from them (i.e. Fig’s. 2B-F and Fig. 3) were observed in these control experiments.
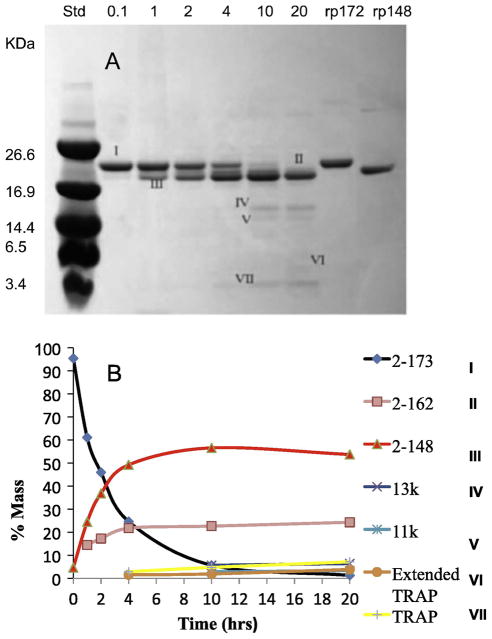
3.2. Progress of Amelogenin Proteolysis by SDS-PAGE
The time course of proteolysis of rP172 was analyzed by SDS-PAGE gels stained with Coomassie Brilliant Blue (Fig. 4A). The proteolysis products were characterized by mass spectroscopy and their percentages were determined based on mass density analysis (Fig. 4B). Based on the full-length porcine amelogenin rP172 (designated as I, amino acids 2-173), the proteolytic products were designated as: II, 2-162; III, 2-148; IV, 46-148 (13k); V, 64-148 (11k), VI, 2-63 (extended TRAP); VII, 2-45 (TRAP). According to the quantitative analysis, about 5% of rP172 was digested within the first 10 min, about 50% of rP172 was digested after 2 hours, and the substrate rP172 was fully cleaved by rpMmp-20 after 20 hours. The 2-148 segment (an analogue to the 20kDa), the main proteolysis product, appeared immediately and was stable against further cleavage by rpMmp-20 even after 20 hours (Fig. 4 and Table 1). Like the 2-148 segment, the segment of 2-162 was an important cleavage product which appeared throughout the proteolysis and reached a stable level after 4 hours. Other products, 13k (46-148), 11k (64-148), extended TRAP (2-63 segment), and TRAP (2-45) became detectable after 4 hours, with a slow increase in their amounts during the progress of digestion. The rP172 to 2-148(2-162) ratios were estimated to be approximately 2:1, 1:1, 1:3, and 1:15 at 1, 2, 4, 10 hours respectively (Table 1).
Figure 4.
Time-course of proteolysis and quantitative analysis of protein cleavage products formed during digestion of rP172 by rpMmp-20. (A) Based on the full-length porcine amelogenin, proteolytic products were designated: I, 2-173 (rP172); II, 2-162; III, 2-148; IV, 46-148 (13k); V, 64-148 (11k), VI, 2-63 (extended TRAP); VII, 2-45 (TRAP); (B) The quantity of proteolytic products corresponding to the stained bands in A is plotted against the proteolytic time in hours as analyzed by Image J.
4. DISCUSSION
To provide further insight into the proposed mechanism of enamelysin function in controlling the assembly of amelogenin, we investigated detailed morphological changes in amelogenin assemblies formed during proteolysis of rP172 by Mmp-20 at near-physiological conditions (pH 8.0, 37.0 °C). Because of difficulties associated with amelogenin solubility and aggregation at pH 7.4, we focused our detailed morphological studies on the experiments conducted at pH 8. We selected conditions under which nano-chain formation by the full-length amelogenin is minimized (i.e. pH 8) and therefore changes in assembly following proteolysis can be followed in more details. We clearly demonstrated that co-assembly between amelogenin and its proteolytic products during a comparatively slow proteolysis process leads to the formation of nanorods and other morphological patterns evolved from them. It has also been reported that full-length human amelogenin and one of its cleavage products have cooperative self-assembly capabilities if incubated for long periods of time (He et al 2008, Uskoković 2008).
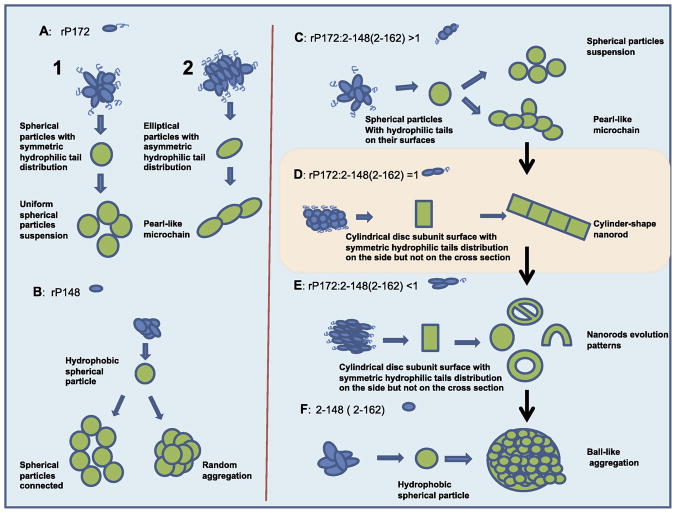
The morphologies that were formed by the rP172 substrate in control experiments, without the presence of rpMmp-20, are schematically presented in Fig. 5 and include: spherical particles of 19 nm diameter (Fig. 5A1) that have isotropically distributed hydrophilic sequence residues on their surfaces and appear as uniformly distributed particles in solution; and ellipsoidal non-isotropic nano-particles (Fig. 5A2) that depending on the incubation time further assemble into chains-like arrays, with defined directionality. Note that the 19 nm diameter particles (9.5 radius) are not the nanosphres originally described (Moradian-Oldak et al 2002, Moradian-Oldak 2007) but oligomeric particles that resulted from the disassembly or re-arrangement of the nanospheres on TEM grids (Bromley et al, in revision). Using in situ atomic force microscopy we recently reported that, although amelogenin exists primarily as ~26 nm in diameter nanospheres in bulk solution at pH 8.0, it behaves dramatically differently upon interacting with charged substrates and with TEM grids (Chen et al, submitted). Our results were further consistent with previous findings that indicated amelogenin-surface interactions could promote disassembly of nanospheres (Tarasevicth et al 2009).
Figure 5.
Proposed mechanism for the assembly of amelogenin hierarchically associated microstructures, formed from co-assembly of rP172 with its cleavage products during in vitro digestion by rpMmp-20. (A) and (B) represent the assembly tendencies of pure rP172 and rP148, respectively, in solution; (C-F) Represent the rP172 co-assembly with its digestion products during the four stages of proteolysis in vitro by rpMmp-20, resulting in the formation of hierarchically associated microstructures. The detailed analysis is described in the text. The scale is arbitrary and does not represent the actual ratios of the morphologies evolved.
4.1. Nanostructures detected at different stages of proteolysis
For clarity, we have divided the in vitro rP172 proteolysis by rpMmp-20 involving amelogenins co-assembly into four distinct stages. These stages are based on the dynamic of proteolysis and development of different morphologies of amelogenin assemblies throughout the time-course of proteolysis. Stage 1 corresponds to the period when less than 50% mass of rP172 substrate has been digested by (the first 5-10 minutes). Stage 2 represents the time when the mass ratio of rP172 to 2-148+2-162 products is about equal (the period between 1–2 hrs). Stage 3 corresponds to the period when more than 50% of rp172 mass has been cleaved by (the period between 2–4 hrs). Stage 4 represents the condition when all of the rP172 substrate has been digested by (beginning at 20 hours). The distinct morphologies resulting from the co-assembly of amelogenin with its proteolytic products during each stage are schematically presented in Fig. 5(C-F).
We propose the following mechanism of co-assembly during proteolysis: At stage 1, the relative low quantity of the hydrophobic cleavage products (2-148 and/or 2-162) are associated with the “amphiphathic” parent rP172 that covers the particle surface (Fig. 5C). These relatively small spherical particles with isotropic distribution of C-termini on their surface lead to the formation of uniform spherical particles. Once some spherical subunits with nonisotropic distribution of C-termini on their surface are formed however, the hydrophobic cores are exposed, giving them a high tendency to interact with other particles. This, in turn, leads to elongated assembly in a specific direction, causing formation of closely packed, elongated, chain-like nanostructures (Fig. 5C). At stage 2, as the quantity of the hydrophobic cleavage products (2-148+2-162) reaches that of the full-length rp172 (1:1 ratio), cylindrical-shaped nanorods with uniform hydrophilic tails exposed on the outer cylinder surfaces are formed. The hydrophobic cross section at the ends of these cylinder-shaped particles has a tendency to grow into elongated nanorods (Fig. 5D). In stage 3, as the quantity of nanorods increases, they curve into different morphologies such as “donuts” or “U-shapes”, which are believed to be developed from the growing nanorods. These organized morphologies occupy the entire solution (Fig. 5E) after the nanorods growth is completed. In stage 4, at the end of digestion, when almost all of the parent rp172 is digested by rpMmp-20, the assemblies completely lose their surface layer of C-termini. Without this protection from further aggregation, they will closely cluster into bigger aggregated microstructures. As 50% of the mass of the full-length rp172 is digested by , the cylindrical-shaped nanorods start to form by close stacking of subunit building blocks (cylindrical discs) (Fig, 3).
We hypothesize that the uniform layer of hydrophilic tails located on the outer surface of the cylindrical particle causes repulsion with other neighboring cylinders, protecting them from further lateral aggregation. On the other hand, the cross section at the ends of the nanorods, which have no hydrophilic tails to protect them, will cause nanorod elongation through hydrophobic interactions. As the C-termini are gradually and progressively cleaved by rpMmp-20, surface tension leads to the curvature of the nanorods to form particles with donut-shaped, U-shaped or other curved morphologies (Fig. 5E). Evidently, the tailored distribution of hydrophilic C-termini on the nanorod surfaces results in unprecedented control over the surface properties and morphology of the assembled particles. It has been reported that assembling of amphiphilic polymers, or designed peptides, by coupling of tailored hydrophilic and hydrophobic branched segments, has resulted in a rich palette of morphologies (Percec et al 2010, Hartgernick et al 2001, Ballister et al 2008). We therefore suggest that nanorod co-assembly of rp172 and 2-148(2-162) is a function of hydrophobic and hydrophilic chemical properties and depends on the precise distribution of the C-termimi (or N-termini) within the assemblies.
4.3 Nanorod formation depends on gradual proteolysis of full-length amelogenin by Mmp-20
Under our experimental pH and temperature conditions, combination of the substrate rP172 and its major proteolytic product rP148 incubated for one hour did not result in the formation of nanorod structures similar to those captured during proteolysis after the same time period (Fig. S1). Our findings lead us to propose that two or more full-length amelogenin molecules self-assemble into one subunit (dimer or oligomer) that cannot tightly assemble into rod-like nano-structures and higher level nanorod architectures unless combined with the truncated cleavage products that formed the subunits. At least in our in vitro experiments, it is clear that this assembly process is a dynamic event that depends upon the action of protease Mmp-20. Although, the exact structure of the assembled subunits is not known, it is clear that the driving factor for the dynamic assembly process that directs the rod-like microstructure formation is the gradual truncation of full -length amelogenin by rpMmp-20. The truncated hydrophobic products play a critical role in nanorods formation, although whether it is due to participation of the hydrophilic cleavaged C-termimi and extended TRAP, and TRAP peptides, or whether the cleavage of 2-162 makes a specific contribution to the ordered nanorod formation, remains unclear.
4.4. The biological significance of nanorod formation
While the composition of amelogenin proteolytic products described in stages 1–4 is considered to be relevant to in vivo composition (Yamakoshi et al 1994, Wen et al 1999), the morphologies developed in E and F do not have in vivo relevance, presumably because of the presence of apatite crystals or other potential amelogenin targets that might prevent the aggregation observed in our in vitro system. Our findings provide new insights into the mechanisms of amelogenin assembly with its proteolytic products. We propose that cylinderical nanorods formed by the co-linear arrangement of full-length amelogenins assembling with their hydrophobic cleavage products (Fig 5D, stage two), represent the elementary building blocks controlling the growth of enamel crystals. It is noteworthy that the scale, and spatial organization of these nanorods are similar to the enamel crystals formed at the early stage of enamel formation in vivo, indicating that the mineral morphology and organization of enamel formation are determined by the “rodlike” assembly of amelogenin molecules through association of their hydrophobic parts. The presence of periodic “filamentous” structures in developing enamel and the relationship of such structures to the c-axial elongated apatite crystals was reported in the early sixties. Rönnholm used decalcified embryonic human enamel to demonstrated the presence of low contrast organic matrix structural patterns he termed “septa” (Ronholm 1962). The 80 Å thick septa, which originated from the “stippled material” (Watson, 1959) were parallel to the row of crystallites. Rönnholm proposed that the crystallites were formed on the two broad surfaces of the longitudinal septa. Travis and Glimcher prepared electron microscopic images of longitudinal tissue sections from developing embryonic bovine enamel to analyze the intra-prismatic organic matrix after staining with uranyl acetate and partial decalcification (Travis and Glimcher 1964). They reported many short, swollen, fragmented filaments described as “dense lines approximately 48 Å wide” that were parallel to each other, with their long axes parallel to the enamel prisms, and were “separated by approximately 120 Å”
Amelogenin nanospheres were proposed to be the basic building blocks of developing enamel (Moradian-Oldak 2007, Fincham et al 1995). The finding of the present study together with our most recent studies on amelogenin oligomeric particles (Bromley et al, in revision), and recent reports on amelogenin-substrate interactions (Chen et al, submitted, Tarasevich et al 2009) , all support the notion that nanospheres are composed of oligomeric subunits particles and disintegrate as soon as they are degraded or interact with surfaces. It is therefore reasonable to argue that nanospheres are transient structures during enamel development. We suggest that the gradual digestion of full-length amelogenin by Mmp-20 in vivo is a critical prerequisite for controlling the continuous alteration of enamel matrix microstructures to allow elongated growth of enamel crystals. The dynamic co-assembly process can prevent uncontrolled random aggregation of the hydrophobic parts that may lead to disorganized crystal growth. On the other hand, full-length amelogenin, with its hydrophilic tails, will lead to loose contact between the subunits. Based on our proteolysis results, we hypothesize that, in vivo, the rod-like enamel matrix structures are composed of cylinder-shaped subunits formed by co-assembly of the hydrophobic parts to form a core, exposing the C-termini through which the nanorods will interact with other contiguous cylinder blocks.
5. CONCLUSIONS
The in vitro experimental data presented here provides evidence that, in addition to spherical particles, full-length amelogenin tends to form an anisometric-shaped assembly under a wide range of conditions. This anisometric shape was a key factor for the formation of pearl-like chains, which were favored under physiological temperature conditions. We have confirmed the crucial roles of C-termini, both in assembled subunit formation and in nanorods formation. Furthermore, the present study emphasizes the dynamics of co-assembly that occurs between the full-length rp172 and its cleavage products during proteolysis by rpMmp-20. It is difficult to relate all aspects of our in vitro results to the ultra-structure of enamel extracellular matrix because of differences between the solution conditions used in our proteolysis system and those in the physiological microenvironment within the enamel extracellular matrix. However, our present study takes a further step towards an in-depth understanding of how protease activity during the earliest secretory stages can lead to the assembly of amelogenin into rod-like microstructures.
Supplementary Material
Acknowledgments
The study was funded by NIH-NIDCR grants DE-015644 and DE-013414. We thank Mr. David Maltby of the Mass Spectrometry Laboratory of the School of Pharmacy at the University of California, San Francisco, for the mass spectrometry analysis (Project # 403). We thank Keith Bromley and Sowmya Lokappa for their valuable discussion. We thank Prof. Ralf Langen’s group (Biochemistry & Molecular Biology Zilkha Neurogenetic Institute Keck School of Medicine of University of Southern California) for providing the transmission electron microscopy instrument.
References
- Beniash E, Simmer JP, Margolis HC. The effect of recombinant mouse amelogenins on the formation and organization of hydroxyapatite crystals in vitro. J Struct Biol. 2005;149:182–90. doi: 10.1016/j.jsb.2004.11.001. [DOI] [PubMed] [Google Scholar]
- Ballister ER, Lai AH, Zuckermann RN, Cheng Y, Mougous JD. In vitro self-assembly of tailorable nanotubes from a simple protein building block. Proc Natl Acad Sci. 2008;105:3733–3738. doi: 10.1073/pnas.0712247105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartlett JD, Simmer JP. Proteinases in developing dental enamel. Crit Rev Oral Biol Med. 1999;10:425–441. doi: 10.1177/10454411990100040101. [DOI] [PubMed] [Google Scholar]
- Boyde A. Enamel. In: Oksche A, Vollrath L, editors. Handbook of Microscopic Anatomy. Springer-Verlag; Berlin: 1989. pp. 309–473. [Google Scholar]
- Bromley KM, Kiss AS, Lokappa SB, Lakshminarayanan R, Fan D, Ndao M, Evans JS, Moradian-Oldak J. Dissecting Amelogenin Nanospheres; Characterization of Metastable Oligomers. J Biol Chem. doi: 10.1074/jbc.M111.250928. In revision. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caterina JJ, Skobe Z, Shi J, Ding Y, Simmer JP, et al. Enamelysin (Matrix Metalloproteinase 20)-deficient Mice Display an Amelogenesis Imperfecta Phenotype. J Biol Chem. 2002;277:49598–49604. doi: 10.1074/jbc.M209100200. [DOI] [PubMed] [Google Scholar]
- Chen C-L, Bromley KM, Moradian-Oldak J, DeYoreo J. In Situ AFM Study of Amelogenin Assembly and Dis-assembly Dynamics on Charged Surfaces provides Insights on Matrix Protein Self-Assembly. doi: 10.1021/ja206849c. Submitted. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du C, Falini G, Fermani S, Abbott C, Moradian-Oldak J. Supramolecular Assembly of Amelogenin Nanospheres into Birefringent Microribbons. Science. 2005;4:1450–1454. doi: 10.1126/science.1105675. Erratum in: Science. 2005 Sep 30;309(5744):2166. [DOI] [PubMed] [Google Scholar]
- Eastoe JE. Enamel protein chemistry--past, present and future. J Dent Res. 1979;58:753–764. doi: 10.1177/00220345790580022701. [DOI] [PubMed] [Google Scholar]
- Fincham AG, Moradian-Oldak J, Diekwisch TG, Lyaruu DM, Wright JTP, et al. Evidence for amelogenin “nanospheres” as functional components of secretory-stage enamel matrix. J Struct Biol. 1995;115:50–9. doi: 10.1006/jsbi.1995.1029. [DOI] [PubMed] [Google Scholar]
- Hartgerink JD, Beniash E, Samuel IS. Self-assembly and mineralization of peptide-amphiphile nanofibers. Science. 2001;23:1684–1688. doi: 10.1126/science.1063187. [DOI] [PubMed] [Google Scholar]
- He XD, Li Wu, Habelitz S. The cooperative self-assembly of 25 and 23 kDa amelogenins. Journal of Structural Biology. 2008;164:314–321. doi: 10.1016/j.jsb.2008.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin T, Ito Y, Luan X, Dangaria S, Walker C, et al. Elongated polyproline motifs facilitate enamel evolution through matrix subunit compaction. PLoS Biol. 2009;7:1–10. doi: 10.1371/journal.pbio.1000262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J, Holtzman DM. Medicine. Prion-like behavior of amyloid-beta. Science. 2010;12:918–919. doi: 10.1126/science.1198314. [DOI] [PubMed] [Google Scholar]
- Lakshminarayanan R, Fan D, Du C, Moradian-Oldak J. The role of secondary structure in the entropically driven amelogenin self-assembly. Biophys J. 2007;93:3664–3674. doi: 10.1529/biophysj.107.113936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyngstadaas SP, Wohlfahrt JC, Brookes SJ, Paine ML, Snead ML, et al. Enamel matrix proteins; old molecules for new applications. Orthod Craniofac Res. 12:243–253. doi: 10.1111/j.1601-6343.2009.01459.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moradian-Oldak J. The emergence of “nanospheres” as basic structural components adopted by amelogenin. J Dent Res. 2007;86:487–90. doi: 10.1177/154405910708600603. [DOI] [PubMed] [Google Scholar]
- Moradian-Oldak J, Paine ML. Mammalian Enamel Formation. In: Sigel Astrid, et al., editors. Metal Ions In Life Sciences. John Wiley & Sons, Ltd; Chichester: 2008. pp. 507–546. [Google Scholar]
- Moradian-Oldak J, Du C, Falini G. On the formation of amelogenin microribbons. Eur J Oral Sci. 2006;114:289–296. doi: 10.1111/j.1600-0722.2006.00285.x. [DOI] [PubMed] [Google Scholar]
- Moradian-Oldak J, Jimenez I, Maltby D, Fincham AG. Controlled proteolysis of amelogenins reveals exposure of both carboxy- and amino-terminal regions. Biopolymers. 2001;58:606–616. doi: 10.1002/1097-0282(200106)58:7<606::AID-BIP1034>3.0.CO;2-8. [DOI] [PubMed] [Google Scholar]
- Moradian-Oldak J, Bouropoulos N, Wang L, Gharakhanian N. Analysis of self-assembly and apatite binding properties of amelogenin proteins lacking the hydrophilic C-terminal. Matrix Biol. 2002;21:197–205. doi: 10.1016/s0945-053x(01)00190-1. [DOI] [PubMed] [Google Scholar]
- Nagano T, Kakegawa A, Yamakosh Y, Tsuchiya S, Hu JC, et al. Mmp-20 and Klk4 cleavage site preferences for amelogenin sequences. J Dent Res. 2009;88:823–828. doi: 10.1177/0022034509342694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozdemir D, Hart PS, Ryu OH, Choi SJ, Ozdemir-Karatas M, et al. amelogenesis imperfecta. J Dent Res. 2005;84:1031–1035. doi: 10.1177/154405910508401112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Percec V, Wilson DA, Leowanawat P, Wilson CJ, Hughes AD, Kaucher MS, et al. Self-assembly of Janus dendrimers into uniform dendrimersomes and other complex architectures. Science. 2010;328:1009–1014. doi: 10.1126/science.1185547. [DOI] [PubMed] [Google Scholar]
- Robinson C, Kirkham J, Shore R. Dental Enamel: Formation to Destruction. CRC Press; Boca Raton, FL: 1995. [Google Scholar]
- Rönnholm E. III. The structure of the organic stroma of human enamel during amelogenesis. J Ultrastruct Res. 1962;6:368–89. doi: 10.1016/s0022-5320(62)80041-0. [DOI] [PubMed] [Google Scholar]
- Ryu OH, Fincham AG, Hu CC, Zhang C, Qian Q, et al. Characterization of recombinant pig enamelysin activity and cleavage of recombinant pig and mouse amelogenins. J Dent Res. 1999;78:743–750. doi: 10.1177/00220345990780030601. [DOI] [PubMed] [Google Scholar]
- Simmer JP, Hu JC. Expression, structure, and function of enamel proteinases. Connect Tissue Res. 2002;43:441–449. doi: 10.1080/03008200290001159. [DOI] [PubMed] [Google Scholar]
- Simmer JP, Hu Y, Lertlam R, Yamakoshi Y, Hu JC. Hypomaturation enamel defects in Klk4 knockout/LacZ knockin mice. J Biol Chem. 2009;284:19110–21. doi: 10.1074/jbc.M109.013623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sire JY, Delgado S, Fromentin D, Girondot M. Amelogenin: lessons from evolution. Arch Oral Biol. 2005;50:205–212. doi: 10.1016/j.archoralbio.2004.09.004. [DOI] [PubMed] [Google Scholar]
- Smith C. Cellular and chemical events during enamel maturation. Crit Rev Oral Biol Med. 1998;9:128–161. doi: 10.1177/10454411980090020101. [DOI] [PubMed] [Google Scholar]
- Smith CE, Richardson AS, Hu Y, Bartlett JD, Hu JC, Simmer JP. Effect of Kallikrein 4 Loss on Enamel Mineralization: Comparison With Mice Lacking Matrix Metalloproteinase 20. J Biol Chem. 2011;286:18149–60. doi: 10.1074/jbc.M110.194258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Z, Ahsan MM, Wang H, Du C, Abbott C, et al. Assembly and processing of an engineered amelogenin proteolytic product (rP148) Eur J Oral Sci. 2006;114:59–63. doi: 10.1111/j.1600-0722.2006.00296.x. [DOI] [PubMed] [Google Scholar]
- Sun Z, Abbott C, Moradian-Oldak J. Control of amelogenin assembly and dis-assembly by enamelysin (MMP-20). The 8th International Conference on Chemistry and Biology of Mineralized Tissue; Banff, Canada: University of Toronto Press Inc; 2005. [Google Scholar]
- Sun Z, Carpiaux W, Fan D, Fan Y, Lakshminarayanan R, et al. Apatite reduces amelogenin proteolysis by MMP-20 and KLK4 in vitro. J Dent Res. 2010;89:344–348. doi: 10.1177/0022034509360660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tarasevich BJ, Lea S, Bernt W, Engelhard MH, Shaw WJ. Changes in the quaternary structure of amelogenin when adsorbed onto surfaces. Biopolymers. 2009;91:103–7. doi: 10.1002/bip.21095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Travis DF, Glimcher MJ. The Structure and Organization of, and the relationship between the organic matrix and the inorganic crystals of embryonic bovine enamel. J Cell Biol. 1964;23:447–97. doi: 10.1083/jcb.23.3.447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uskoković V, Kim MK, Li W, Habelitz S. Enzymatic processing of amelogenin during continous crystallization of apatite. J Mater Res. 2008;23:3184–3195. doi: 10.1557/JMR.2008.0387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watson ML. The extracellular nature of enamel in the rat. J Biophys Biochem Cytol. 1960;7:489–92. doi: 10.1083/jcb.7.3.489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wen HB, Moradian-Oldak J, Leung W, Bringas P, Jr, Fincham AG. Microstructures of an amelogenin gel matrix. J Struct Biol. 1999;126:42–51. doi: 10.1006/jsbi.1999.4086. [DOI] [PubMed] [Google Scholar]
- Wiedemann-Bidlack FB, Beniash E, Yamakoshi Y, Simmer JP, Margolis HC. pH triggered self-assembly of native and recombinant amelogenins under physiological pH and temperature in vitro. Journal of Structural Biology. 2007;160:57–69. doi: 10.1016/j.jsb.2007.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiedemann-Bidlack FB, Kwak SY, Beniash E, Yamakoshi Y, Simmer JP, et al. Effects of phosphorylation on the self-assembly of native full-length porcine amelogenin and its regulation of calcium phosphate formation in vitro. J Struct Biol. 2011;173:250–60. doi: 10.1016/j.jsb.2010.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamakoshi Y, Tanabe T, Fukae M, Shimizu M. Porcine amelogenins Calcif. Tissue Int. 1994;54:69–75. doi: 10.1007/BF00316293. [DOI] [PubMed] [Google Scholar]
- Yang X, Wang L, Qin Y, Sun Z, Henneman ZJ, et al. How Amelogenin Orchestrates the Organization of Hierarchical Elongated Microstructures of Apatite. J Phys Chem B. 2010;114:2293–2300. doi: 10.1021/jp910219s. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.