Abstract
The repair of cis-syn cyclobutane pyrimidine dimers (CPDs) can be initiated via the base excision repair (BER) pathway, utilizing pyrimidine dimer-specific DNA glycosylase/lyase enzymes (pdgs). However, prior to incision at lesion sites, these enzymes bind to non-damaged DNAs through charge-charge interactions. Following initial binding to DNA containing multiple lesions, the enzyme incises at most of these sites prior to dissociation. If a subset of these lesions are in close proximity, clustered breaks may be produced that could lead to decreased cell viability or increased mutagenesis. Based on the co-crystal structures of bacteriophage T4-pdg and homology modeling of a related enzyme from Paramecium bursaria Chlorella virus-1, the structure-function basis for the processive incision activity for both enzymes was investigated using site-directed mutagenesis. An assay was developed that quantitatively measured the rates of incision by these enzymes at clustered apurinic/apyrimidinic (AP) sites. Mathematical modeling of random (distributive) versus processive incisions predicted major differences in the rate and extent of the accumulation of singly nicked DNAs between these two mechanisms. Comparisons of these models with biochemical nicking data revealed significant changes in the damage search mechanisms between wild-type pdgs and most of the mutant enzymes. Several conserved arginine residues were shown to be critical for the processivity of the incision activity, without interfering with catalysis at AP sites. Comparable results were measured for incision at clustered CPD sites in plasmid DNAs. These data reveal that pdgs can be rationally engineered to retain full catalytic activity, while dramatically altering mechanisms of target site location.
Keywords: Abasic sites, base excision repair, structure-function, protein engineering
1. Introduction
Although human cells possess DNA glycosylases to initiate base excision repair (BER) for many forms of base damage, they lack enzymes that initiate BER on the major form of ultraviolet (UV) light-induced damage, cis-syn cyclobutane pyrimidine dimers (CPDs) [1–3]. In most organisms, repair of these lesions occurs via the nucleotide excision repair (NER) pathway. However, subsets of bacteria, bacteriophages, and eukaryotic viruses also use DNA glycosylases with an associated abasic site lysase activity to initiate repair of CPDs via the BER pathway. While the most extensively studied glycosylase with UV-damage specific activity is encoded by the denV gene from the bacteriophage T4 (reviewed in [4]), another closely related enzyme is produced by Paramecium bursaria Chlorella virus-1 (Cv-pdg) [5–9]. Interest in the Cv-pdg enzyme stems from data demonstrating that relative to the T4-pdg, it has broader substrate specificity [8, 9] and an ~ 10-fold increased catalytic efficiency [8]. Both of these enzymes utilize the α-NH2 group of the amino terminal threonine residue as the active site nucleophile to initiate chemistry at the C1′ of an abasic site or a CPD [10–12]. These reactions proceed through an imino intermediate that can be reduced to a stable DNA-protein complex. Structure-function analyses of T4-pdg include the crystal structure of the native enzyme [13] and two co-crystal structures of the protein in complex with CPD- or apurinic/apyrimidinic (AP) site- containing DNA [14, 15]. These structures have revealed that the charge distribution on the surface of the comma-shaped T4-pdg is highly asymmetric with the positively charged amino acids clustered on the concave, DNA-binding surface, while the convex backside surface contains predominantly acidic residues. These structures also reveal the identity of residues that may be critical for binding to non-damaged, as well as CPD- or AP site-containing DNA.
Under physiologically relevant salt concentrations, wild-type T4-pdg or Cv-pdg encounter DNA via a random collision mechanism, followed by association with, and binding to non-damaged DNA, a process that is guided by local charge-charge interactions in the microenvironment of the DNA. This general binding to DNA is maintained over considerable distances such that experimentally, it is observed that these enzymes will incise most, if not all, CPDs in an ~ 5 kb plasmid containing 5–10 CPDs per DNA prior to dissociation [16]. Analyses of the co-crystal structures of a catalytically inactive mutant, T4-pdg (E23Q), with CPD-containing DNA [15] and the covalent, reduced-imine intermediate of wild-type T4-pdg with AP site-containing DNA [14] revealed multiple sites of contact between basic amino acid residues, primarily arginines, and DNA in the local vicinity of the lesion (Figure 1A). Protein sequence alignments of genes with homology to Cv-pdg and T4-pdg revealed significant conservation of these sites (Figure 1B). Molecular modeling of Cv-pdg, using T4-pdg as a template, predicts a similar charge distribution [17]. It was hypothesized that many of these residues are critical for establishing correct pre-catalytic alignment and orientation of the enzyme on non-damaged DNA and thus, facilitate location of sites of damaged DNA. To test this hypothesis, site directed mutants were made at these sites in T4-pdg and Cv-pdg, and analyzed in vitro.
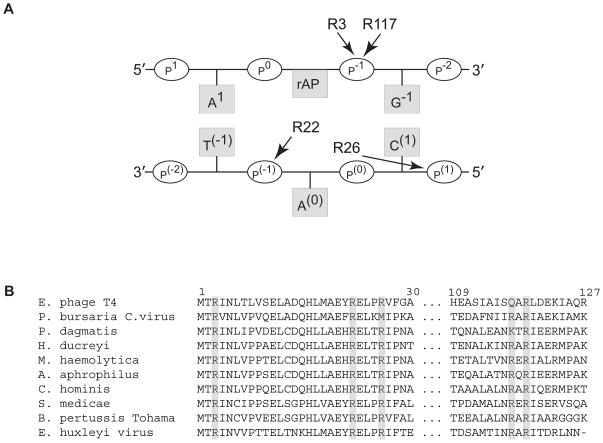
Fig. 1. Arginine residues in pyrimidine dimer glycosylases that may be important for DNA binding and processivity.
(A) Schematic representation of the sites of interaction between T4-pdg and AP site-containing DNA as revealed by a co-crystal structure of a reduced imine covalent complex (derived from [14]). The reduced AP site-containing strand to which the N-terminal threonine of T4-pdg is linked is shown as the upper strand in which both arginine (R) 3 and 117 bind to the p−1 phosphate immediately 3′ to the site of covalent linkage. In the complementary strand (lower) the extrahelical nucleotide (A(0)) is bound by R22 and R26 at p(−1) and p(1), respectively. (B) Alignment of highly conserved arginine residues within phylogenetically related pdgs. Enterobacteria phage T4; Paramecium bursaria Chlorella virus; Pasteurella dagmatis; Haemophilus ducreyi; Mannheimia haemolytica; Aggregatibacter aphrophilus; Cardiobacterium hominis; Sinorhizobium medicae; Bordetella pertussis Tohama; Emiliania huxleyi virus. Biochemical characterization of pdg activity has only been confirmed for Enterobacteria phage T4 and Paramecium bursaria Chlorella virus.
Since human cells are unable to initiate BER at sites containing CPDs, numerous studies have investigated the effects of expressing or introducing T4-pdg or Cv-pdg in mammalian cells [18–27], with an ultimate objective being to use these enzymes in human therapeutics [28]. Collectively, these studies demonstrated that introduction of T4-pdg resulted in: 1) accelerated rates of CPD repair; 2) increased unscheduled DNA repair synthesis; 3) suppression of UV-induced mutagenesis in wild-type cells; 4) increased survival of most NER-deficient cells; and 5) significantly reduced survival of normal repair-proficient cells. In human clinical trials, wild-type T4-pdg was encapsulated into a liposomal delivery vehicle and topically used as a treatment for NER deficient xeroderma pigmentosum (XP) patients [28]. In a one year randomized trial, XP patients who received the T4-pdg-containing lotion showed reduced rates of new actinic keratoses and basal cell carcinomas (68% and 30%, respectively) relative to placebo treated patients. These data are very consistent with the survival studies using XP-derived cells in which activation of BER for CPDs was beneficial [24].
However, since expression of wild-type T4-pdg in repair-proficient mammalian cells resulted in decreased, not enhanced survival, these data suggest that the activity of the enzyme can produce cytotoxic repair intermediates, possibly through the formation of double-strand breaks at clustered CPDs. Similar mechanisms of increased cytotoxicity have been postulated for other glycosylases, NTH1, OGG1 and MAG1 due to their overexpression [29–31]. Insights into a biochemical basis for decreased survival of wild-type mammalian cells expressing T4-pdg come from studies that demonstrated the ability of T4-pdg to catalyze a processive nicking activity in duplex DNA. This activity leads to incision of most, if not all, CPDs in plasmid DNA prior to dissociation of the enzyme from the DNA molecule to which it was initially bound, both in vitro and in vivo [16, 32–34]. The current investigation was designed to test the hypothesis that neutralization of selected positively charged residues in the vicinity of the active site of T4- and Cv-pdg could minimize or eliminate the processive nicking activity of these enzymes without affecting the efficiency of catalysis.
2. Materials and methods
2.1 Site-directed mutagenesis
Site-directed mutagenesis at the designated positions in the T4-pdg and Cv-pdg genes was performed using the bidirectional QuickChange Site-Directed Mutagenesis Kit™ (Stratagene, Santa Clara, CA). Primers for mutagenesis were synthesized by the OHSU MMI DNA Services Core (Portland, OR) (Table 1, complementary sequences not shown). Wild-type and mutant pdg proteins were expressed in frame with a six histidine tag using the pET-22b vector (Novagen, San Diego, CA). The complete sequences of all pdg mutants were confirmed by DNA sequence analyses by the MMI DNA Services Core.
Table 1.
Primers for Site-directed Mutagenesis of T4-pdg and Cv-pdg
| T4-pdg
|
| T2P: 5′-GGAGATATACATATGCCTCGTATCAACCTTAC-3′ |
| R3K: 5′-GATATACATATGACTAAGATCAACCTTACTTTAG-3′ |
| R22Q: 5′-ATGGCTGAATATCAAGAATTGCCGCGT-3′ |
| E23D: 5′-GCTGAATATCGTGACTTGCCGCGTGTTTTTG-3′ |
| R26Q: 5′-CGTGAATTGCCGCAAGTTTTTGGTGCAGTTCG-3′ |
| R117Q: 5′-GCTATATCACAAGCTCAATTAGATGAAAAAA TTGCA-3′
|
| Cv-pdg
|
| R3Q: 5′-GAGGAGATATACATATGACACAAGTGAATCTCG-3′ |
| R22Q: 5′-CATCTCATGGCAGAATTTCAAGAACTTAAGATG-3′ |
| K25Q: 5′-CGTGAACTTCAAATGATTCCGAAGGCA-3′ |
| R22A: 5′-CTCATGGCAGAATTTGCAGAACTTAAGCTG-3′ |
| R117Q: 5′-GCGTTTAATATTATTCAAGCGAGGATTGCCGAA-3′ |
| R119Q: 5′-ATTCGAGCGCAAATTGCCGAAAAAATCGCT-3′ |
2.2 Protein purification
pET-22b plasmids containing point mutations in the genes encoding either T4-pdg or Cv-pdg were transformed into BL21(DE3) cells (Invitrogen, Carlsbad, CA). For each mutant enzyme, bacterial cultures were grown at 37°C of 0.5 in LB medium (250 ml) to an OD600 containing 100 μg/ml ampicillin. The cultures were cooled to 18°C and protein expression was induced by the addition of 0.5 mM IPTG. Cultures were grown at 18°C for 4 hr and the cells harvested by centrifugation at 3,000 × g for 20 min. Cell pellets were resuspended in 30 ml 0°C phosphate-sodium chloride buffer (50 mM Na2HPO4 (pH 7.0), 300 mM NaCl) containing 10 mM imidazole and protease inhibitors (Complete, EDTA-free; Roche Diagnostics, Corp., Indianapolis, IN). Cells were lysed by passage through a French pressure cell at 1,100 psi, and cell lysates were collected in siliconized tubes. Cell debris was cleared from the lysates by centrifugation at 10,000–12,000 × g for 20 min at 4°C. Cleared lysates were mixed with pre-equilibrated Ni-NTA agarose beads (Qiagen, Valencia, CA) and rocked for 2 hr at 4°C. Beads were collected by centrifugation at 700 × g for 5 min and the supernatant decanted. The beads were sequentially washed as follows: twice with 40 ml of phosphate-sodium chloride buffer, twice with 40 ml of phosphate-sodium chloride buffer containing 50 mM imidazole, and once with 40 ml of phosphate-sodium chloride buffer containing 100 mM imidazole. Bound proteins were eluted twice with 5–6 ml of phosphate-sodium chloride buffer containing 200 mM imidazole and stored at 4°C in siliconized tubes. Aliquots of purified proteins were analyzed by electrophoresis through a 0.1% SDS, 15% polyacrylamide gel. Protein concentrations were determined using the Bradford Assay (Bio-Rad, Hercules, CA).
2.3 DNA nicking assay using AP site-containing oligodeoxynucleotides
The oligodeoxynucleotides were synthesized by Integrated DNA Technologies (Coralville, IA). The internally-labeled 46-mer oligodeoxynucleotides containing two uracil (U) bases was constructed as follows. The U-containing 30-mer (5′-CTCAATCTCTUCCGCAATCAGGCCAGATC-3′) was 32P-labeled at the 5′ end with T4 polynucleotide kinase (New England BioLabs, Ipswich, MA) and purified on a Micro-Bio-Spin P-6 chromatography column (Bio-Rad). The labeled U-containing 30-mer and U-containing 16-mer (5′-ACTACGTAC UGTTACG-3′) were annealed to the 41-mer scaffold oligodeoxynucleotide (5′-GGCCTGATTGCGGAAGAGATGGAGCCGTAACAGTACG TAGT-3′) in the presence of 40 mM NaCl, by heating for 5 min at 95°C, followed by slow cooling to 22°C. Ligation was carried out using T4 ligase (400 units) and 100 μM ATP at 12°C overnight. An equal volume of gel loading solution (95% (v/v) formamide, 20 mM EDTA, 0.02% (w/v) bromophenol blue, 0.02% (w/v) xylene cyanol) was added, and following incubation at 95°C for 5 min, DNAs were separated by electrophoresis through a gel (10% polyacrylamide and 8% urea in 90 mM Tris-borate, 2 mM EDTA (TBE)) at 20 W for 3 hr. The labeled single-stranded 46-mer oligodeoxynucleotide was purified from gel slices by extraction in a solution containing 0.5 M ammonium acetate and 10 mM magnesium acetate, followed by dialysis against 10 mM Tris-HCl, pH 7.4, 1 mM EDTA (TE) buffer. The purified 46-mer oligodeoxynucleotide was annealed to a complementary 46-mer oligodeoxynucleotide (5′-GATCTGGCCTGATTGCGGAAGAGATGGAGCCGTAACAGTACGTAGT-3′). The resulting internally-labeled U-containing 46-mer DNA duplexes were incubated for 1 hr at 37°C with uracil DNA glycosylase (New England BioLabs) to form AP sites. The AP site-containing duplex oligodeoxynucleotide was incubated for various times with wild-type and mutant T4-pdg or Cv-pdg in 25 mM K2HPO4 (pH 7.0), 5 mM DTT, 100 μg/ml BSA, 10% glycerol, 25 mM KCl at 37°C. In order to achieve comparable rates of incision, the following final enzyme concentrations were used in the AP lyase nicking reactions: T4-pdg wild-type, 0.7 nM; T4-pdg R3K, 11.6 nM; T4-pdg R22Q, 0.7 nM; T4-pdg R26Q, 5.0 nM; T4-pdg R117Q, 0.8 nM; Cv-pdg wild-type, 0.7 nM; Cv-pdg R3Q, 10.4 nM; Cv-pdg R22Q, 14 nM; Cv-pdg R117Q, 1.2 nM; Cv-pdg R119Q, 6.0 nM. Aliquots (10 μl) were removed at increasing times and the reactions terminated by the addition of 20 mM NaBH4 that rapidly reduces all remaining abasic sites. An equal volume of gel loading buffer (95% (w/v) formamide, 20 mM EDTA, 0.02% (w/v) bromophenol blue, 0.02% (w/v) xylene cyanol) was added to each aliquot. The samples were heated to 90°C for 5 min. The DNAs were loaded on a gel (15% polyacrylamide, 8% urea in TBE), separated by electrophoresis at 20 W for 2 hr, and visualized using a PhosphorImager screen (GE Healthcare, Piscataway, NJ). Quantitative analyses were performed using Image-Quant 5.2 software (GE Healthcare).
2.4 Modeling of sequential AP site nicking
Computer simulation was used to characterize the proportion of the DNA molecule population in each of the three states (0, 1, or 2 nicks). The simulation initially started with 1000 elements in the population, all with zero nicks, and then evolved for 10,000 time steps (10 times population size). At each time step, a random element from the population was selected and either nicked at one of the two sites (determined at random) with probability p, or nicked at both sites with probability 1-p. Simulations were conducted for p=0, 0.20, 0.40, 0.50, 0.60, 0.80, and 1. Each simulation was run for 50 replications and the proportion of the population existing in a specific state was averaged and plotted against time measured relative to the population size. All simulations were run using Octave (v.3.0.0, http://www.octave.org).
2.5 Protein sequence alignments
Pdgs with homology to that of the T4-pdg were identified by querying the non-redundant nucleotide database (NCBI) using BLASTn [35]. Representative samples were chosen from major taxa identified in the query, aligned with ClustalX [36], and manually edited with Seaview [37]. Genbank accession numbers for the aligned sequences are as follows: Paramecium bursaria Chlorella Virus: YP_001498146.1, Enterobacteria phage T4: NP_049733.1, Pasteurella dagmatis: ZP_05920417.1, Haemophilus ducreyi: NP_874115.1, Mannheimia haemolytica: ZP_04977951.1, Aggregatibacter aphrophilus: YP_003008325.1, Cardiobacterium hominis: ZP_05704180.1, Sinorhizobium medicae: YP_001329043.1, Bordetella pertussis Tohama: NP_881306.1, Emiliania huxleyi virus 86: YP_293985.1, Vibrio phage KVP40: NP_899451.1, Aeromonas phage 25: NC_008208.1, Prochlorococcus marinus str.: YP_001017378.1.
3. Results
3.1 Enzyme design and purification
Based on analyses of the co-crystal structures, site directed mutations were introduced into the genes encoding T4-pdg and Cv-pdg at the following sites: T4-pdg R3K, R22Q, R26Q, R117Q and Cv-pdg R3Q, R22Q, R117Q and R119Q. All proteins were expressed with a His tag at the C-terminus to facilitate purification by nickel affinity chromatography. All enzymes were purified to > 95% purity and all preparations were free of nonspecific DNA nicking or exonuclease activities (data not shown).
3.2 Assay design and computer simulations
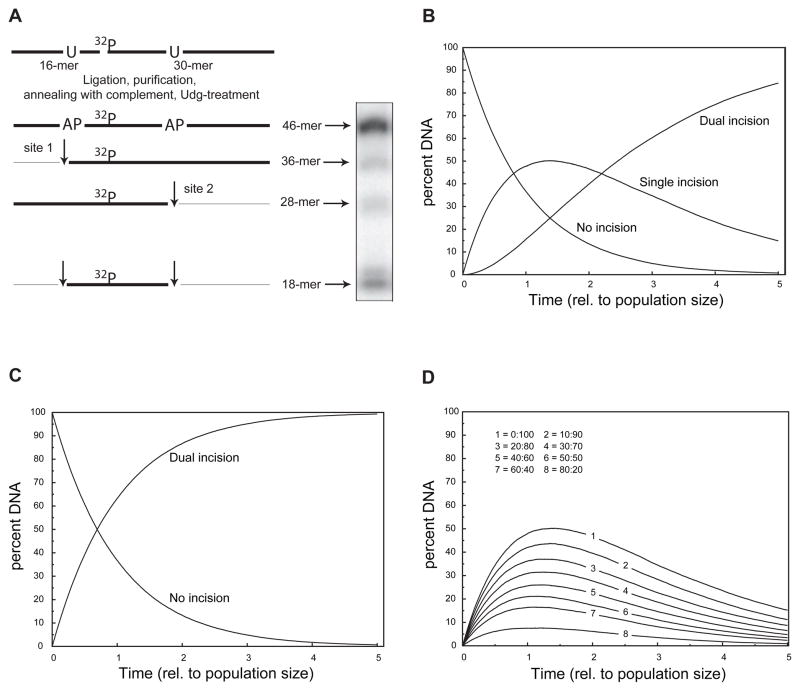
In order to assess which amino acid residues in T4-pdg and Cv-pdg are critical for processive nicking activity, an assay was designed that allowed for the measurement of the efficiency of pdg-catalyzed incisions at AP sites. The active site residues in T4-pdg that are required for the DNA glycosylase activity are identical with those necessary for the lyase activity [14]. Double-stranded oligodeoxynucleotides (46-mers) were generated in which one DNA strand contained two AP sites (at positions 10 and 28) and the 32P isotope (between nucleotides 16 and 17) (Figure 2A). Upon addition of an enzyme with lyase activity, if an incision is only made at the 5′ or 3′ AP site (site 1 or 2, respectively), the radio-label would be contained in either a 36- or 28-mer, respectively. Incision at both AP sites would generate a radio-labeled 18-mer. A sample electrophoretic separation of the DNA substrate and products is shown in Figure 2A.
Fig. 2. Substrate design and computer modeling of processive versus distributive enzyme reactions.
(A) Schematic diagram of the design, construction, and utilization of DNAs containing closely positioned AP sites. The substrate 46-mer was constructed from the ligation of an unlabeled 16-mer containing a uracil (U) at position 10 from the 5′ end with a 32P-labeled 30-mer containing a U at position 11 from the 5′ end of the 30-mer, using a scaffold 41-mer. The resulting 32P-labeled 46-mer was purified and annealed with a 5-fold molar excess of complementary 46-mer. The duplex DNA was digested with uracil DNA glycosylase to create 2 AP sites in the labeled strand in which incision at only the 5′ AP site (site 1) yielded a labeled 36-mer, while incision at the 3′ AP site (site 2) yielded a labeled 28-mer. Dual incisions generated a labeled 18-mer. The appearance of two bands at the 18-mer position is characteristic of T4-pdg catalyzing a β-elimination reaction that is followed by a δ-elimination reaction. (B) Computer simulation of a kinetic nicking reaction in which nicking at either of the two AP sites is completely random (or haphazard). The percentage of DNA molecules with no incisions exponentially decays with a concomitant increase in DNA molecules that contain one incision (at either the 5′ or 3′ AP site, site 1 or 2, respectively). For a completely random incision mechanism, the theoretical maximum percentage of DNA molecules containing only one incision reaches ~50% and begins to decay. The accumulation of DNA molecules containing dual incisions displayed a characteristic lag, followed by a sigmoidal increase as total number of incisions increased. The reaction models a maximally distributive incision reaction. (C) Computer simulation of a kinetic nicking reaction in which every DNA-enzyme encounter results in incision at both AP sites. The exponential decrease in the no incision population is reciprocally matched by an increase in dually-incised DNA molecules. This simulation represents a maximally processive reaction. (D) Computer simulations of the accumulation of single-incised DNA molecules in which the probability of producing dual incisions prior to enzyme-DNA dissociation was varied between 0 to 80% of the encounters. The “no incision” and “dual incision” populations were omitted for clarity. Computer simulation of the reaction mechanism as it shifts from a random (distributive) mechanism toward a more processive incision reveals both that the magnitude of the percent DNA that accumulates with only one incision decreases and the time at which it is maximal occurs earlier in the reaction.
Computer modeling of reactions in which it was assumed that every incision would be introduced randomly through all possible AP sites, showed that the percent of molecules with no hits decreases exponentially with successive incisions, while the appearance of molecules containing one (but not both incisions), transiently increases. Further, in the modeling of the incision reactions, the percent of DNAs containing only one incision decreases due to the accumulation of doubly incised molecules. A kinetic mathematical simulation model of such a reaction is shown in (Figure 2B). If however, at the opposite extreme, incisions were modeled to always occur at both sites within the same DNA molecule prior to enzyme dissociation, the loss of the percentage of molecules not incised is inversely proportional to the dually incised population. The kinetic model of the obligatory dual incision reaction is shown in (Figure 2C).
The kinetic reactions modeled in Figure 2B and 2C illustrate the two extremes of search mechanisms (random versus processive, respectively). In practice, it is likely that wild-type and mutant enzymes will display search mechanisms, as revealed by the nicking kinetics that are intermediate to these cases. In order to predict such data, simulations were run, such that the probability of catalyzing dual incisions prior to dissociation was varied from 0, 10, 20, 30, 40, 50, 60, 70, and 80% (Figure 2D, lines 1–8, respectively). These data capture the sum of the percentage of molecules incised at either the 5′ or 3′ AP site (site 1 or 2, respectively) and reveal that the maximum value of the sum of singly incised molecules reflects the relative processivity of the search mechanism. Thus, application of the values generated through these simulations permit an assessment of the relative changes in processivity that may be conferred by a specific site-directed mutant relative to the wild-type enzyme.
3.3 Modulation of processive nicking activity of T4-pdg at AP sites
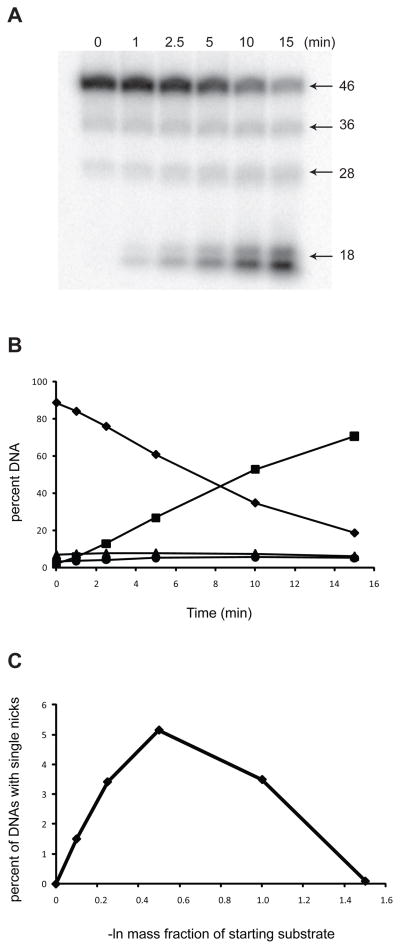
When a limiting concentration of wild-type T4-pdg (0.7 nM) was incubated with oligodeoxynucleotides containing the dual AP sites, concomitant with the decrease in DNAs containing no incisions (46-mer), there were no significant increases in the percentage of DNAs containing only one incision at either the 5′ site 1 (36-mer) or the 3′ site 2 (27-mer) (Figure 3A, B). The starting substrate DNA contained a small percentage of 36-mer and 28-mer DNAs that were formed due to non-enzymatic cleavage at AP sites during the preceding incubation at 37°C with uracil DNA glycosylase. However, the loss of initial, un-incised DNAs corresponded to the appearance of doubly incised (18-mer) fragment. These data match well with the model predicting a highly processive incision mechanism, as previously suggested for the wild-type T4-pdg. Data from four replicate experiments were re-analyzed to normalize for the extent to which the reactions had proceeded by calculating the –ln of the mass fraction of the intact (not incised) population of DNA molecules (the zeroth term of the Poisson distribution) versus the sum of the observed percentage of incisions at site 1 or 2 only (Figure 3C). These data revealed that the sum of singly incised DNAs peaked at ~ 5%. Extrapolation of these data to the computational simulations of nicking reactions indicates that >80% of the encounters of wild-type T4-pdg with this substrate resulted in dually-incised DNAs.
Fig. 3. T4-pdg exhibits processive activity on oligodeoxynucleotides containing AP sites.
T4-pdg was incubated with substrate DNA containing two AP sites for the indicated times. Reactions were quenched with NaBH4 and formamide/dye solution, and analyzed by electrophoresis through denaturing polyacrylamide gels. Bands were visualized by PhosphorImager analyses and the percent DNA (of total DNA in reaction) occurring at each band was quantitated using Image-Quant software. (A) Representative autoradiograph of a denaturing polyacrylamide gel analysis of AP site nicking by wild-type T4-pdg. The 46-mer is the full length substrate. The 36-mer and 28-mer represent the singly incised substrate at either the 5′ site or 3′ site, respectively. The 18-mer is indicative of a doubly incised product. (B) A representative plot of the wild-type T4-pdg nicking on AP site-containing substrate. Symbols: diamond, percent of starting substrate (46-mer); triangle, percent of DNAs nicked at site 1 (36-mer); circle, percent of DNAs nicked at site 2 (28-mer); square, percent of DNAs nicked at both sites (18-mer). (C) Total accumulation of singly incised DNAs. The sum of the percent of site 1 and site 2 nicked molecules minus the percent of site 1 and site 2 nicked molecules at t=0, respectively, as a function of the –ln of the mass fraction of DNAs containing no breaks.
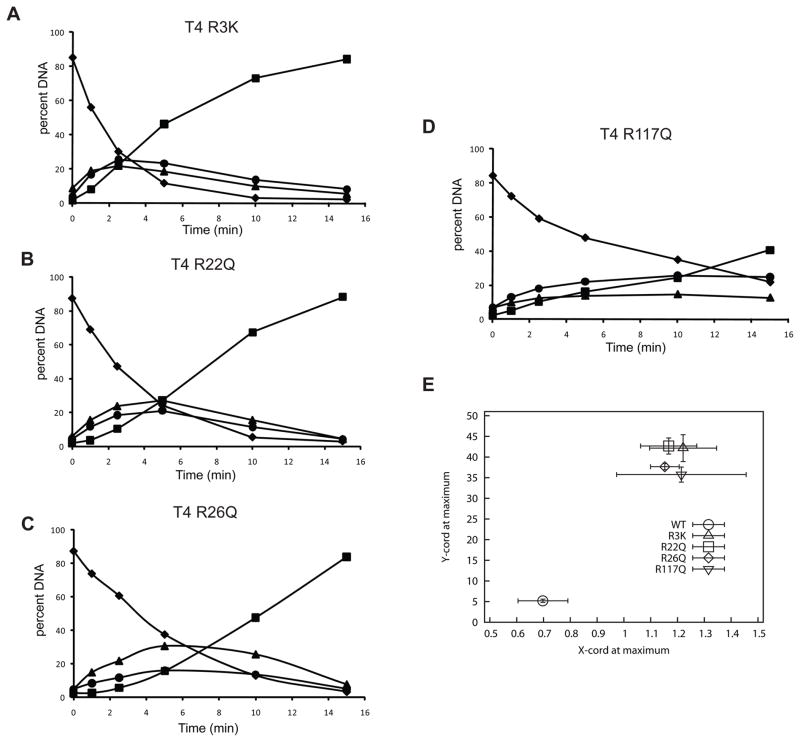
Kinetic analyses of all mutants reveal that they all retain AP lyase activity as evidenced by their nicking activities (Figure 4A–D). However, in contrast to the wild-type enzyme, all of the site-directed mutants of T4-pdg showed significant decreases in their abilities to catalyze dual incisions prior to dissociation from the DNA on which the first incision was made (Figure 4, A–D). All experiments were repeated a minimum of three times with a representative plot shown in panels A–D. The most severely compromised mutant enzymes appear to be T4-pdg R3K (Figure 4A) and R22Q (Figure 4B); with these mutants, the maximal accumulation of singly nicked molecules was observed at either site 1 or 2 at ~ 24% or 20%, respectively. These data suggest that incisions were made both independently and randomly with no significant preference for site 1 or 2. T4-pdg R26Q (Figure 4C) showed a significant decrease in the ability to make dual incisions prior to dissociation, as evidenced by the maximal accumulation of singly nicked molecules at site 1 or 2 reaching ~ 30.7 and 15.6 %, respectively. T4-pdg R117Q (Figure 4D) also displayed reduced processivity in which singly nicked DNAs accumulated maximally at 14.7 and 25.8 % for sites 1 and 2, respectively.
Fig. 4. Altered mechanisms of target site location by T4-pdg arginine mutants.
The site-directed mutants of T4-pdg were incubated with the AP site-containing substrate for the indicated times. Reactions were quenched with NaBH4 and formamide/dye solution, and analyzed by electrophoresis through denaturing polyacrylamide gels. Bands were visualized by PhosphorImager analyses and the percent DNA (of total DNA in reaction) occurring at each band was quantitated using Image-Quant software. Representative plots are shown for each mutant T4-pdg enzyme; (A) T4 R3K; (B) T4 R22Q; (C) T4 R26Q; (D) T4 R117Q. Symbols: diamond, percent of starting substrate (46-mer); triangle, percent of DNAs nicked at site 1 (36-mer); circle, percent of DNAs nicked at site 2 (28-mer); square, percent of DNAs nicked at both sites (18-mer). (E) Relative processivity of incision reactions. Data from all kinetic analyses (minimum of 3 independent experiments) were analyzed to determine the point at which there was a maximum accumulation of DNA molecules with only one incision (sum of site 1 only and site 2 only). The average (± standard deviation) of the percent of singly nicked molecules (y-axis) is plotted as a function of the –ln of the mass fraction (± standard deviation) of unincised molecules at the maximum accumulation of singly nicked molecules. The wild-type T4-pdg (lower left, circle) showed a high degree of processivity, while all mutants clustered in the upper right, indicating a distributive incision reaction.
In order to statistically compare the reproducibility of these assays and quantitate the overall effect on processive nicking at clustered abasic sites, the following analyses were performed (Fig. 4E). The data for all replicates (minimum of 3 independent determinations) of the T4-pdg wild-type and each of the T4-pdg mutants were individually analyzed to determine the point in each reaction at which there was a maximal accumulation of the combined singly nicked DNA molecules (y-axis). Those data points also yielded data on the percentage of DNA molecules containing no nicks, the 0th term of the Poisson distribution and were calculated as the –ln of the mass fraction of un-nicked DNAs (x-axis). These are plotted in Figure 4E ± standard deviations across the independent experiments. As predicted by the computer simulations (Figure 2D), the T4-pdg wild-type enzyme shows a very high degree of processivity, with >80% of the DNA-enzyme encounters resulting in dual incisions (circle). In contrast, all four arginine mutants were severely compromised in their processive nicking activities with T4-pdg R3K (triangle) and R22Q (square) producing dual incisions < 5% of the encounters, while R26Q (diamond) and R117Q (inverted triangle) showed dual incisions produced in ~ 20% of the encounters.
3.4 Modulation of the processive nicking activity of cv-pdg at AP sites
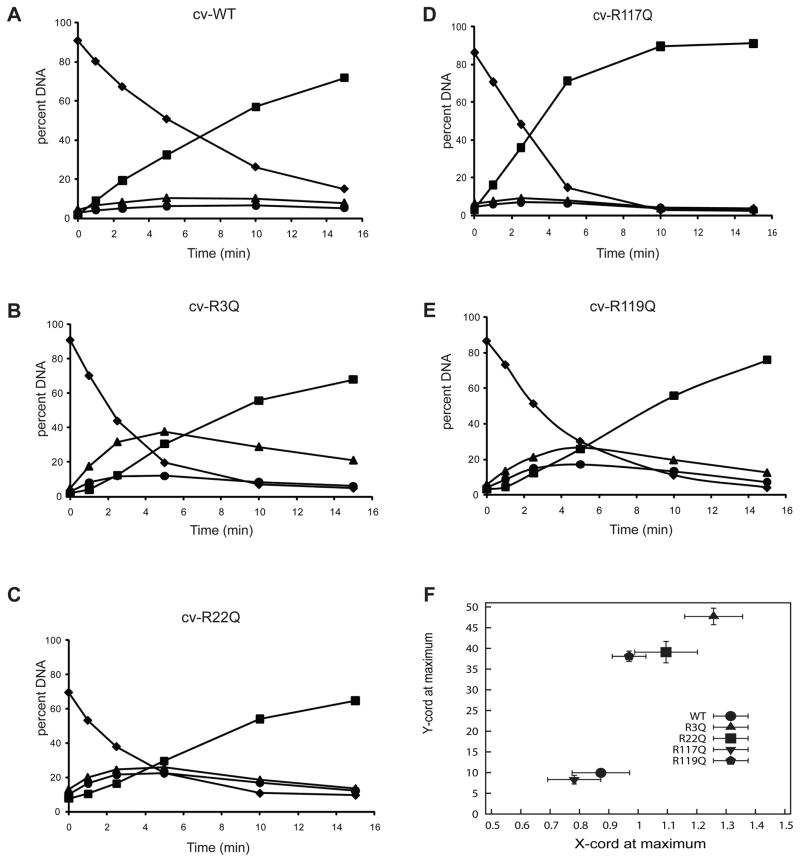
Since both molecular modeling and phylogenetic conservation suggest that Cv-pdg would have conserved DNA binding motifs, comparable analyses were carried out on wild-type Cv-pdg and each of the site-directed mutants (Figure 5A–F). As expected for the wild-type Cv-pdg, it showed a very strong tendency to incise at both AP sites prior to dissociation, with very modest accumulations of either of the singly nicked DNAs (Figure 5A). Three of the Cv-pdg mutants, R3Q, R22Q and R119Q displayed severe reductions in their abilities to catalyze both incisions prior to dissociation, with R3Q being the most severely affected and approaching the theoretical maximum for random incisions, but with a significant preference for site 2 over site 1 (Figure 5B). The Cv-pdg R22Q (Figure 5C) and R119Q (Figure 5E) were impaired to the same extent as the T4-pdg mutants R26Q and R117Q, with each of the singly nicked DNAs accumulating to ~ 20%. In contrast with the other mutant enzymes previously described, Cv-pdg R117Q showed no reduction in its processive incision activity relative to the wild-type Cv-pdg (Figure 5D). These data suggest that R119 in Cv-pdg may be functionally equivalent to R117 in T4-pdg, relative to making critical phosphate backbone contacts.
Fig. 5. Cv-pdg activity on oligodeoxynucleotides containing AP sites and altered mechanisms of target site location by arginine mutants.
Cv-pdg and Cv-pdg mutant enzymes were incubated with the AP site-containing substrate for the indicated times. Reactions were quenched with NaBH4 and formamide/dye solution, and analyzed by electrophoresis through denaturing polyacrylamide gels. Bands were visualized by PhosphorImager analyses and the percent DNA (of total DNA in reaction) occurring at each band was quantitated using Image-Quant software. Bands are as described in Fig. 3. Representative plots are shown for wild-type and each mutant Cv-pdg enzyme; (A) Cv wild-type; (B) Cv R3Q; (C) Cv R22Q; (D) Cv R117Q; (E) Cv R119Q. Symbols: diamond, percent of starting substrate (46-mer); triangle, percent of DNAs nicked at site 1 (36-mer); circle, percent of DNAs nicked at site 2 (28-mer); square, percent of DNAs nicked at both sites (18-mer). (F) Relative processivity of incision reactions. Data from all kinetic analyses (minimum of 3 independent experiments) were analyzed to determine the point at which there was a maximum accumulation of DNA molecules with only one incision (sum of site 1 only and site 2 only). The average (± standard deviation) of the percent of singly nicked molecules (y-axis) is plotted as a function of the –ln of the mass fraction (± standard deviation) of molecules with no incisions at the maximum accumulation of singly nicked molecules. The wild-type Cv-pdg (lower left, circle) and R117Q (inverted triangle) showed a high degree of processivity, while all other mutants were distributive.
All replicates (minimum of 3 independent determinations) of these kinetic experiments for Cv-pdg were analyzed (Figure 5F) as previously described for the T4-pdg enzymes (Figure 4E). The data revealed that both the Cv-pdg wild-type (circle) and R117Q (inverted triangle) show a very high probability of making dual incisions with each encounter (>80% probability), while Cv-pdg R22Q (square) and R119Q (pentagon) have only ~ 20% probabilities to make both incisions per encounter. However, the data for Cv-pdg R3Q reveal that this single amino acid change has disabled this enzyme from producing dual incisions per enzyme-DNA encounter.
In order to determine whether the same biochemical characteristics would also be manifested in the mechanism by which pdgs efficiently locate CPDs within large segments of undamaged DNA, plasmid DNAs were UV irradiated to introduce ~25 CPDs per molecule [32], incubated with limiting concentrations of wild-type and mutant enzymes, and the kinetics of incision assayed.
As shown in Supplemental Figure 1 A–E, addition of wild-type T4-pdg resulted in a loss of form I DNA, with concomitant increases in forms II and III DNAs. Form III DNA accumulates rapidly during the initial phase of the reaction and plateaus following incision of all form I DNA molecules. This increase in form III DNA while form I DNA is still present is a manifestation of the processive nicking activity. However, when identical assays were performed with T4-pdg R22Q or R117Q, there were very significant delays in the appearance of form III DNA, relative to the loss of form I DNA. For T4-pdg R3K and R117Q, no form III DNA was detected, even with prolonged incubations (data not shown). Comparable data were observed for the wild-type and mutant forms of the Cv-pdg enzyme (Supplemental Figure 2 A–E).
4. Discussion and Conclusions
Structure-function analyses of pdgs have been greatly facilitated through the determination of two co-crystal structures of the protein in complex with CPD- or AP site containing DNA [14, 15]. Although these structures have revealed amino acid residues that interact with the DNA phosphate backbone in both pre-catalytic and post-catalytic complexes, the charge-charge interactions with non-damaged DNAs has not been structurally determined, but inferred from these structures. However, it is reasonable to extrapolate the identity of key residues from these structures that may serve to promote non-target DNA binding, since the core structure of T4-pdg changes remarkably little between the native enzyme and the structures observed in the co-crystal structures [14].
The generalized electrostatic interaction between T4-pdg and duplex DNA has been hypothesized to be responsible for the processive nicking activity on defined substrates both in vitro and in vivo [16, 32–34]. The discovery of the processive nicking activity of the T4-pdg was made by incubating limiting concentrations of T4-pdg with supercoiled, covalently-closed circular form I DNA that had been UV irradiated such that ~ 30% of the plasmid molecules contained two CPDs in close proximity on complementary strands. While following the incision kinetics of T4-pdg, it was observed that concomitant with the conversion of form I DNA to a nicked form II DNA via incision at CPD sites, there was a significant and linear accumulation of form III DNA molecules created by a DSB [32]. Velocity sedimentation analyses of the DNA products through alkaline sucrose gradients revealed that there was a bimodal distribution in the molecular weights of the DNAs, such that plasmid DNAs were either fully incised or contained no single-strand breaks. This result not only demonstrated that the kinetics of the accumulation of form III DNA (while starting form I DNA substrate remains in the reaction) is a surrogate measure of the enzyme’s processivity, but also demonstrated that T4-pdg can incise all CPDs within a DNA domain, where the average distance between CPDs varied from 0.27 to 2.7 kb.
In vivo processive nicking activity was shown to occur in cells, using an assay in which the kinetics of complete repair of intracellular plasmids containing 5 and 10 CPDs per molecule was measured [16]. The experimental data showed all the expected parameters of in vivo processive nicking activity. Following UV irradiation, fully repaired plasmid DNAs accumulated linearly and with no time lag throughout the time course of repair, a result that strongly suggested that at physiological salt concentrations, T4-pdg initiated repair at all CPD sites within a subset of plasmids prior to reinitiating repair on additional plasmids. The overall conclusion that can be drawn from these data is that it is very likely that in a cellular context, if CPDs are in close proximity in complementary strands, wild-type T4-pdg has a high probability to convert these into DSBs that could prove to be deleterious to the cell.
The use of DNA repair enzymes in increasing cellular resistance to environmental toxicants and radiation-induced damages has potential applications in preventing, and possibly treating, a number of human diseases. To a limited degree, this concept has been reduced to practice in the treatment of XP patients through a topical delivery of wild-type T4-pdg (reviewed in [28]). In this phase II clinical trial, XP patients showed a reduction of the appearance of new actinic keratoses and basal cell carcinomas through daily administration of the enzyme. These data validated and extended earlier cell-based studies in which delivery of T4-pdg enhanced survival in most XP complementation groups tested (except XPG) [24]. However, in no case, did the survivals come close to achieving UV resistances that are comparable to normal human cells. The reasons for this are likely to be complex and may include the lack of repair of other UV-induced photoproducts, such as 6–4 photoproducts, ring-fragmented guanines, and DNA-protein crosslinks. Additionally, the coordinated hierarchy of transcription-coupled repair and domain-specific repair are not captured through the initiation of BER of CPDs.
However, extrapolation of the use of wild-type T4-pdg in normal human keratinocytes and in normal human populations may face significant challenges, since all attempts to increase cell survival in normal human cells via introduction of T4-pdg has resulted in increased cytotoxicity rather than increased survival. Since the relative survival curves of most XP and normal human cells expressing wild-type-T4-pdg are quite similar, these data suggest that an activity of wild-type T4-pdg creates cytotoxic intermediates which limit the degree of enhanced survival that can be achieved in an XP cell and reduces cell survival in repair-proficient cells. We hypothesize that this cytotoxic intermediate may be the formation of DSBs by the action of T4-pdg cleaving at sites of tightly clustered CPDs in complementary strands.
Thus, the goal of the current investigation was to create a repertoire of T4-pdg and Cv-pdg mutants that retain their catalytic activities as both a DNA glycosylase and AP lyase, but lose or at least minimize, the capacity to incise clustered AP sites and CPDs. Guided by insights gained from analyses of the co-crystal structures of T4-pdg, we were able to predict residues which may contribute to the electrostatic interactions that do not allow dissociation of the enzyme from a DNA domain. It is this affinity that ultimately leads to DSB formation. Our data show that neutralization of one of several residues in fact yielded enzymes with the desired properties and these include T4-pdg R3K, R22Q, R26Q and R117Q and Cv-pdg R3Q, R22Q, and R119Q. These enzymes maintain appropriate glycosylase and AP lyase activities, but fail to incise two AP sites or CPDs in very close proximity. However, the 10–20-fold reduction in catalytic efficiency of T4-pdg R3K and R26Q and Cv-pdg R3Q and R22Q may diminish their efficacy in enhancing overall repair rates. The immediate goal of our future studies will be to express these mutants in both repair-proficient and repair-deficient cells and assay effects on cell survival.
Finally, these data also may provide insight into the mechanisms of target site location. The original descriptions of the processive nicking activity of T4-pdg were described as a “sliding” mechanism. The implicit assumption of that study was that the enzyme remained in contact with the DNA molecule to which it was originally bound and tracked on that molecule until a dissociation event occurred that released the enzyme into bulk solution [32]. Such tracking on DNA has recently been shown using single molecule diffusion data, in which eight proteins of varying size and function were shown to utilize a rotation-coupled sliding mechanism [38].
However, in the case of T4- or Cv-pdg, such analyses have yet to be performed and it is equally plausible that the processive nicking activity could represent a series of hundreds or thousands of dissociations/re-association events, in which the net reaction is manifest as a processive reaction. In this regard, the current study analyzed DNA substrates in which the clustered sites were either in the same strand (the AP nicking assay) or in both strands (the UV irradiated plasmid assay) in which to detect the formation of the double-stranded break, the two CPDs must be in complementary strands.
Analyses of incision data for both assays using the wild-type pdgs reveal that these enzymes readily make dual incisions per binding event, independent of whether the lesions are in the same or different strands. These data favor a mechanism of frequent microscopic dissociation/re-association events such that lesions in either strand are equally well recognized. In contrast, the majority of the mutant enzymes described herein infrequently re-associate with the same DNA molecule, thus producing a random incision pattern.
Supplementary Material
pBR322 plasmid DNA containing ~25 cyclobutane pyrimidine dimers per DNA molecule, was incubated with T4-pdg enzymes and at the indicated time points, the products analyzed by electrophoresis on 1% agarose gels. Gels were stained with ethidium bromide and the intensity of each band was determined using an Alpha Innotech Imaging System, Representative plots are shown for wild-type and each mutant enzyme: (A) T4 wild-type; (B) T4 R3Q; (C) T4 R22Q; (D) T4 R26Q; (E) T4 R117Q. Symbols: Form I, diamonds; Form II, circles; Form III, X.
pBR322 plasmid DNA containing ~25 cyclobutane pyrimidine dimers per DNA molecule, was incubated with Cv-pdg enzymes and at the indicated time points, the products analyzed by electrophoresis on 1% agarose gels. Gels were stained with ethidium bromide and the intensity of each band was determined using an Alpha Innotech Imaging System, Representative plots are shown for wild-type and each mutant enzyme: (A) Cv wild-type; (B) Cv R3Q; (C) Cv R22Q; (D) Cv R117Q; (E) Cv R119Q. Symbols: Form I, diamonds; Form II, circles; Form III, X.
Highlights.
Processivity of DNA glycosylases can cause double-strand breaks at clustered lesions.
We engineered pyrimidine dimer-specific DNA glycosylases with reduced processivity.
We developed an assay to quantitatively measure incisions at clustered abasic sites
Biochemical and computational data reveal residues critical for processive nicking.
Enzymes with reduced processivity but full catalytic function can be engineered.
Acknowledgments
The authors are indebted to Brian Lowell and Lauriel Earley for technical assistance and to Drs. Adam Clore and Anuradha Kumari for help with data analyses. This work was funded by NIH ES04091 (RSL).
Abbreviations
- AP
apurinic/apyrimidinic
- BER
base excision repair
- CPDs
cyclobutane pyrimidine dimers
- NER
nucleotide excision repair
- pdg
pyrimidine dimer DNA glycosylase/AP site lyase
- UV
ultraviolet
Footnotes
Conflict of Interest Statement
The authors declare that there are no conflicts of interest.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Douki T, Reynaud-Angelin A, Cadet J, Sage E. Bipyrimidine photoproducts rather than oxidative lesions are the main type of DNA damage involved in the genotoxic effect of solar UVA radiation. Biochemistry. 2003;42:9221–9226. doi: 10.1021/bi034593c. [DOI] [PubMed] [Google Scholar]
- 2.Galloway AM, Liuzzi M, Paterson MC. Metabolic processing of cyclobutyl pyrimidine dimers and (6–4) photoproducts in UV-treated human cells. Evidence for distinct excision-repair pathways. J Biol Chem. 1994;269:974–980. [PubMed] [Google Scholar]
- 3.Kvam E, Tyrrell RM. Induction of oxidative DNA base damage in human skin cells by UV and near visible radiation. Carcinogenesis. 1997;18:2379–2384. doi: 10.1093/carcin/18.12.2379. [DOI] [PubMed] [Google Scholar]
- 4.Lloyd RS. Investigations of pyrimidine dimer glycosylases--a paradigm for DNA base excision repair enzymology. Mutat Res. 2005;577:77–91. doi: 10.1016/j.mrfmmm.2005.04.003. [DOI] [PubMed] [Google Scholar]
- 5.Furuta M, Schrader JO, Schrader HS, Kokjohn TA, Nyaga S, McCullough AK, Lloyd RS, Burbank DE, Landstein D, Lane L, Van Etten JL. Chlorella virus PBCV-1 encodes a homolog of the bacteriophage T4 UV damage repair gene denV. Appl Environ Microbiol. 1997;63:1551–1556. doi: 10.1128/aem.63.4.1551-1556.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Garvish JF, Lloyd RS. The catalytic mechanism of a pyrimidine dimer-specific glycosylase (pdg)/Abasic lyase, chlorella virus-pdg [In Process Citation] J Biol Chem. 1999;274:9786–9794. doi: 10.1074/jbc.274.14.9786. [DOI] [PubMed] [Google Scholar]
- 7.Garvish JF, Lloyd RS. Active-site determination of a pyrimidine dimer glycosylase. J Mol Biol. 2000;295:479–488. doi: 10.1006/jmbi.1999.3369. [DOI] [PubMed] [Google Scholar]
- 8.McCullough AK, Romberg MT, Nyaga S, Wei Y, Wood TG, Taylor JS, Van Etten JL, Dodson ML, Lloyd RS. Characterization of a novel cis-syn and trans-syn-II pyrimidine dimer glycosylase/AP lyase from a eukaryotic algal virus, Paramecium bursaria chlorella virus-1. J Biol Chem. 1998;273:13136–13142. doi: 10.1074/jbc.273.21.13136. [DOI] [PubMed] [Google Scholar]
- 9.Jaruga P, Jabil R, McCullough AK, Rodriguez H, Dizdaroglu M, Lloyd RS. Chlorella virus pyrimidine dimer glycosylase excises ultraviolet radiation- and hydroxyl radical-induced products 4,6-diamino-5-formamidopyrimidine and 2,6-diamino-4-hydroxy-5-formamidopyrimidine from DNA. Photochem Photobiol. 2002;75:85–91. doi: 10.1562/0031-8655(2002)075<0085:cvpdge>2.0.co;2. [DOI] [PubMed] [Google Scholar]
- 10.Dodson ML, Schrock RDd, Lloyd RS. Evidence for an imino intermediate in the T4 endonuclease V reaction. Biochemistry. 1993;32:8284–8290. doi: 10.1021/bi00083a032. [DOI] [PubMed] [Google Scholar]
- 11.Schrock RDd, Lloyd RS. Reductive methylation of the amino terminus of endonuclease V eradicates catalytic activities. Evidence for an essential role of the amino terminus in the chemical mechanisms of catalysis. J Biol Chem. 1991;266:17631–17639. [PubMed] [Google Scholar]
- 12.Schrock RDd, Lloyd RS. Site-directed mutagenesis of the NH2 terminus of T4 endonuclease V. The position of the alpha NH2 moiety affects catalytic activity. J Biol Chem. 1993;268:880–886. [PubMed] [Google Scholar]
- 13.Morikawa K, Matsumoto O, Tsujimoto M, Katayanagi K, Ariyoshi M, Doi T, Ikehara M, Inaoka T, Ohtsuka E. X-ray structure of T4 endonuclease V: an excision repair enzyme specific for a pyrimidine dimer. Science. 1992;256:523–526. doi: 10.1126/science.1575827. [DOI] [PubMed] [Google Scholar]
- 14.Golan G, Zharkov DO, Grollman AP, Dodson ML, McCullough AK, Lloyd RS, Shoham G. Structure of T4 Pyrimidine Dimer Glycosylase in a Reduced Imine Covalent Complex with Abasic Site-containing DNA. J Mol Biol. 2006;362:241–258. doi: 10.1016/j.jmb.2006.06.059. [DOI] [PubMed] [Google Scholar]
- 15.Vassylyev DG, Kashiwagi T, Mikami Y, Ariyoshi M, Iwai S, Ohtsuka E, Morikawa K. Atomic model of a pyrimidine dimer excision repair enzyme complexed with a DNA substrate: structural basis for damaged DNA recognition. Cell. 1995;83:773–782. doi: 10.1016/0092-8674(95)90190-6. [DOI] [PubMed] [Google Scholar]
- 16.Gruskin EA, Lloyd RS. Molecular analysis of plasmid DNA repair within ultraviolet-irradiated Escherichia coli. I. T4 endonuclease V-initiated excision repair. J Biol Chem. 1988;263:12728–12737. [PubMed] [Google Scholar]
- 17.Soman KV, Schein CH, Zhu H, Braun W. Homology modeling and simulations of nuclease structures. Methods Mol Biol. 2001;160:263–286. doi: 10.1385/1-59259-233-3:263. [DOI] [PubMed] [Google Scholar]
- 18.Tanaka K, Sekiguchi M, Okada Y. Restoration of ultraviolet-induced unscheduled DNA synthesis of xeroderma pigmentosum cells by the concomitant treatment with bacteriophage T4 endonuclease V and HVJ (Sendai virus) Proc Natl Acad Sci U S A. 1975;72:4071–4075. doi: 10.1073/pnas.72.10.4071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kibitel JT, Yee V, Yarosh DB. Enhancement of ultraviolet-DNA repair in denV gene transfectants and T4 endonuclease V-liposome recipients. Photochem Photobiol. 1991;54:753–760. doi: 10.1111/j.1751-1097.1991.tb02086.x. [DOI] [PubMed] [Google Scholar]
- 20.Francis MA, Bagga PS, Athwal RS, Rainbow AJ. Incomplete complementation of the DNA repair defect in cockayne syndrome cells by the denV gene from bacteriophage T4 suggests a deficiency in base excision repair. Mutat Res. 1997;385:59–74. doi: 10.1016/s0921-8777(97)00039-6. [DOI] [PubMed] [Google Scholar]
- 21.Kusewitt DF, Ley RD, Henderson EE. Enhanced pyrimidine dimer removal in repair-proficient murine fibroblasts transformed with the denV gene of bacteriophage T4. Mutat Res. 1991;255:1–9. doi: 10.1016/0921-8777(91)90012-e. [DOI] [PubMed] [Google Scholar]
- 22.Kusewitt DF, Budge CL, Anderson MM, Ryan SL, Ley RD. Frequency of ultraviolet radiation-induced mutation at the hprt locus in repair-proficient murine fibroblasts transfected with the denV gene of bacteriophage T4. Photochem Photobiol. 1993;58:450–454. doi: 10.1111/j.1751-1097.1993.tb09589.x. [DOI] [PubMed] [Google Scholar]
- 23.Kusewitt DF, Budge CL, Ley RD. Enhanced pyrimidine dimer repair in cultured murine epithelial cells transfected with the denV gene of bacteriophage T4. J Invest Dermatol. 1994;102:485–489. doi: 10.1111/1523-1747.ep12373084. [DOI] [PubMed] [Google Scholar]
- 24.Francis MA, Bagga P, Athwal R, Rainbow AJ. Partial complementation of the DNA repair defects in cells from xeroderma pigmentosum groups A, C, D and F but not G by the denV gene from bacteriophage T4. Photochem Photobiol. 2000;72:365–373. doi: 10.1562/0031-8655(2000)072<0365:pcotdr>2.0.co;2. [DOI] [PubMed] [Google Scholar]
- 25.Kusewitt DF, Dyble J, Sherburn TE, Ryan SL, Ji JY. Altered UV resistance and UV mutational spectrum in repair-proficient murine fibroblasts expressing endonuclease V. Mutat Res. 1998;407:157–168. doi: 10.1016/s0921-8777(98)00004-4. [DOI] [PubMed] [Google Scholar]
- 26.Yamaizumi M, Inaoka T, Uchida T, Ohtsuka E. Microinjection of T4 endonuclease V produced by a synthetic denV gene stimulates unscheduled DNA synthesis in both xeroderma pigmentosum and normal cells. Mutat Res. 1989;217:135–140. doi: 10.1016/0921-8777(89)90065-7. [DOI] [PubMed] [Google Scholar]
- 27.Johnson JL, Lowell BC, Ryabinina OP, Lloyd RS, McCullough AK. TAT-Mediated Delivery of a DNA Repair Enzyme to Skin Cells Rapidly Initiates Repair of UV-Induced DNA Damage. J Invest Dermatol. 2011 doi: 10.1038/jid.2010.300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yarosh D, Klein J, O’Connor A, Hawk J, Rafal E, Wolf P. Effect of topically applied T4 endonuclease V in liposomes on skin cancer in xeroderma pigmentosum: a randomised study. Xeroderma Pigmentosum Study Group. Lancet. 2001;357:926–929. doi: 10.1016/s0140-6736(00)04214-8. [DOI] [PubMed] [Google Scholar]
- 29.Hofseth LJ, Khan MA, Ambrose M, Nikolayeva O, Xu-Welliver M, Kartalou M, Hussain SP, Roth RB, Zhou X, Mechanic LE, Zurer I, Rotter V, Samson LD, Harris CC. The adaptive imbalance in base excision-repair enzymes generates microsatellite instability in chronic inflammation. J Clin Invest. 2003;112:1887–1894. doi: 10.1172/JCI19757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sobol RW, Kartalou M, Almeida KH, Joyce DF, Engelward BP, Horton JK, Prasad R, Samson LD, Wilson SH. Base excision repair intermediates induce p53-independent cytotoxic and genotoxic responses. J Biol Chem. 2003;278:39951–39959. doi: 10.1074/jbc.M306592200. [DOI] [PubMed] [Google Scholar]
- 31.Yang N, Galick H, Wallace SS. Attempted base excision repair of ionizing radiation damage in human lymphoblastoid cells produces lethal and mutagenic double strand breaks. DNA Repair (Amst) 2004;3:1323–1334. doi: 10.1016/j.dnarep.2004.04.014. [DOI] [PubMed] [Google Scholar]
- 32.Lloyd RS, Hanawalt PC, Dodson ML. Processive action of T4 endonuclease V on ultraviolet-irradiated DNA. Nucleic Acids Res. 1980;8:5113–5127. doi: 10.1093/nar/8.21.5113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ganesan AK, Seawell PC, Lewis RJ, Hanawalt PC. Processivity of T4 endonuclease V is sensitive to NaCl concentration. Biochemistry. 1986;25:5751–5755. doi: 10.1021/bi00367a060. [DOI] [PubMed] [Google Scholar]
- 34.Gruskin EA, Lloyd RS. The DNA scanning mechanism of T4 endonuclease V. Effect of NaCl concentration on processive nicking activity. J Biol Chem. 1986;261:9607–9613. [PubMed] [Google Scholar]
- 35.Zhang Z, Schwartz S, Wagner L, Miller W. A greedy algorithm for aligning DNA sequences. J Comput Biol. 2000;7:203–214. doi: 10.1089/10665270050081478. [DOI] [PubMed] [Google Scholar]
- 36.Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, Thompson JD, Gibson TJ, Higgins DG. Clustal W and Clustal X version 2.0. Bioinformatics. 2007;23:2947–2948. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- 37.Galtier N, Gouy M, Gautier C. SEAVIEW and PHYLO_WIN: two graphic tools for sequence alignment and molecular phylogeny. Comput Appl Biosci. 1996;12:543–548. doi: 10.1093/bioinformatics/12.6.543. [DOI] [PubMed] [Google Scholar]
- 38.Blainey PC, Luo G, Kou SC, Mangel WF, Verdine GL, Bagchi B, Xie XS. Nonspecifically bound proteins spin while diffusing along DNA. Nat Struct Mol Biol. 2009;16:1224–1229. doi: 10.1038/nsmb.1716. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
pBR322 plasmid DNA containing ~25 cyclobutane pyrimidine dimers per DNA molecule, was incubated with T4-pdg enzymes and at the indicated time points, the products analyzed by electrophoresis on 1% agarose gels. Gels were stained with ethidium bromide and the intensity of each band was determined using an Alpha Innotech Imaging System, Representative plots are shown for wild-type and each mutant enzyme: (A) T4 wild-type; (B) T4 R3Q; (C) T4 R22Q; (D) T4 R26Q; (E) T4 R117Q. Symbols: Form I, diamonds; Form II, circles; Form III, X.
pBR322 plasmid DNA containing ~25 cyclobutane pyrimidine dimers per DNA molecule, was incubated with Cv-pdg enzymes and at the indicated time points, the products analyzed by electrophoresis on 1% agarose gels. Gels were stained with ethidium bromide and the intensity of each band was determined using an Alpha Innotech Imaging System, Representative plots are shown for wild-type and each mutant enzyme: (A) Cv wild-type; (B) Cv R3Q; (C) Cv R22Q; (D) Cv R117Q; (E) Cv R119Q. Symbols: Form I, diamonds; Form II, circles; Form III, X.