Abstract
Background
Abiraterone acetate (AA) is an androgen biosynthesis inhibitor shown to prolong life in patients with castration-resistant prostate cancer (CRPC) already treated with chemotherapy. AA treatment results in dramatic declines in prostate-specific antigen (PSA) in some patients and no declines in others, suggesting the presence of molecular determinants of sensitivity in tumors.
Objective
To study the role of transmembrane protease, serine 2 (TMPRSS2)–v-ets erythroblastosis virus E26 oncogene homolog (ERG) fusion, an androgen-dependent growth factor, in circulating tumor cells (CTCs) as a biomarker of sensitivity to AA.
Design, setting, and participants
The predictive value of TMPRSS2-ERG status was studied in 41 of 48 men with postchemotherapy-treated CRPC enrolled in sequential phase 2 AA trials.
Intervention
Patients received AA 1000 mg daily and continuously.
Measurements
TMPRSS2-ERG status was characterized by a sensitive, analytically valid reverse transcription polymerase chain reaction assay in CTCs enriched from ethylene-diaminetetraacetic acid anticoagulated blood obtained prior to AA treatment. Outcomes were measured by PSA Working Group 1 criteria.
Results and limitations
Standard procedures for specimen acquisition, processing, and testing using the validated TMPRSS2-ERG assay on a multiplex platform gave intra-assay and interassay coefficients of variation <7%. TMPRSS2-ERG fusion was present in 15 of 41 patients (37%), who had a median baseline CTC count of 17 (interquartile range: 7–103 cells per 7.5 ml). A PSA decline ≥50% was observed in 7 of 15 patients (47%) with the fusion and in 10 of 26 patients (38%) without the fusion. Although limited by the low number of patients, a posttherapy CTC count of less than five per 7.5 ml was prognostic for longer survival relative to a CTC count five or more. TMPRSS2-ERG status did not predict a decline in PSA or other clinical outcomes.
Conclusions
Molecular profiles of CTCs with an analytically valid assay identified the presence of the prostate cancer–specific TMPRSS2-ERG fusion but did not predict for response to AA treatment. This finding demonstrates the role of CTCs as surrogate tissue that can be obtained in a routine practice setting.
Keywords: Abiraterone, Biomarker, Circulating tumor cells, Prostate cancer, Prostate-specific antigen, TMPRSS2-ERG fusion
1. Introduction
Abiraterone acetate (AA) is a 17α-hydroxylase/C17,20-lyase (CYP17) inhibitor that blocks androgen synthesis in the testes and adrenal glands and in prostate cancers. AA has been shown to prolong survival in men with castration-resistant prostate cancer (CRPC) who have progressed after chemotherapy with docetaxel, and AA was recently approved by the US Food and Drug Administration (FDA) for this indication [1]. The results validate earlier profiling studies showing that overexpression of androgen biosynthesis pathway enzymes and increased intratumoral androgen levels were frequent alterations in CRPC. Notable in the trials was the pattern of posttreatment prostate-specific antigen (PSA) changes, which ranged from dramatic declines in some patients to continued elevations in others. This wide variation in PSA-decline rates suggested the presence of predictive molecular markers of sensitivity [2–6], with a number of biomarkers being available for evaluation. Analogous to the sequence of trials used to evaluate new therapeutic approaches is the need for defined metrics to determine which biomarkers warrant testing in large-scale phase 3 trials.
Ttransmembrane protease, serine 2 (TMPRSS2)–v-ets erythroblastosis virus E26 oncogene homolog (ERG) fusion, first identified in 2005, is created by the translocation of the androgen-driven 5′ TMRPSS2 chromosomal region to the ETS transcription factor family member ERG(21q22.2) [7]. This fusion, present in approximately 50% (range: 30–70%) of newly diagnosed prostate cancers [7–10], represents >90% of the identified ETS translocations. In experimental models, the TMPRSS2-ERG fusion has shown a limited role in prostate tumorigenesis [9]. In clinical studies, presence of the fusion was associated with low-grade disease [11] but not with higher risk of biochemical recurrence, metastases, or death [9,10]. A role in androgen-dependent tumor growth has been postulated [12], and a relationship between TMPRSS2-ERG status and degree of PSA decline was shown in chemotherapy-naive CRPC patients treated with AA, suggesting a role as a predictive biomarker of AA sensitivity [13]. For prostate cancer in particular, establishing the clinical significance of a molecular determinant is hindered by the difficulty of obtaining representative tumor samples for analysis in a routine clinical practice setting. Circulating tumor cells (CTCs) isolated from peripheral blood have the potential to fulfill this unmet need. Before clinical utility can be established, it is essential that a robust assay that gives consistent results be available [14]; this process is termed analytic validation and is outlined in the FDA Critical Path [15].
This report describes the development and analytic validation of a sensitive polymerase chain reaction (PCR)–based assay to detect TMPRSS2-ERG fusions in CTC and explores the relationship between fusion detection and clinical outcome of patients treated in phase 2 AA trials at Memorial Sloan-Kettering Cancer Center (MSKCC).
2. Patients and methods
2.1. Patients
Samples were obtained from 48 men with progressive metastatic CRPC following docetaxel-based chemotherapy who were treated at MSKCC in two sequential phase 2 trials with AA 1000 mg plus prednisone 10 mg daily and continuously until clinical or radiologic progression [2,4], according to PSA Working Group 1 criteria [16]. Declines in PSA were illustrated by waterfall plots [17]. Informed consent was obtained prior to the initiation of institutional review board–approved protocols.
2.2. Laboratory methods
CTCs were enumerated from 7.5 ml of blood collected in CellSave tubes (Veridex, Raritan, NJ, USA) before treatment and every 4 wk while on treatment using the FDA-cleared CellSearch system (Veridex), as previously described [18,19].
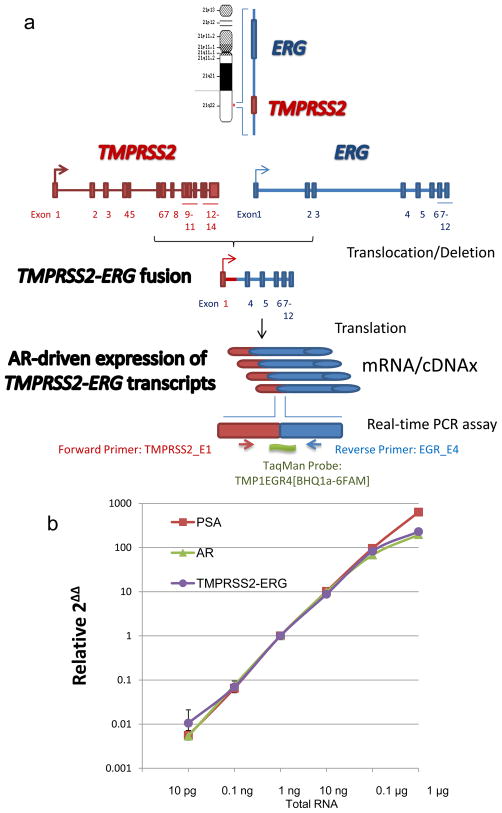
A reverse transcription PCR (RT-PCR) assay was used for TMPRSS2-ERG transcripts in CTCs, as described in Figure 1a, from cell pellets enriched from blood collected into bipotassium ethylene-diaminetetraacetic acid (EDTA)–containing Vacutainers (BD, Franklin Lakes, NJ, USA) using an epithelial cellular adhesion molecule–based immunomagnetic isolation according to the CTC Profiling Kit (Veridex) [18–20]. CTC-enriched pellets were immediately lysed in 500 μl TRIzol (Invitrogen, Carlsbad, CA, USA) and stored at −80°C for batched processing for total RNA extraction. Primer-directed reverse transcription was performed using the CellDirect One Step (Invitrogen) method, followed by multiplex real-time PCR onto 48 × 48 dynamic array chips (Fluidigm, South San Francisco, CA, USA). TaqMan assays (Applied Biosystems, Carlsbad, CA, USA) for each gene were used. TMPRSS2 exon 1 and ERG exon 4 (TMPRSS2-ERG) fusion product was detected using for forward primer TMPRSS2_E1-TAGGCGCGAGCTAAGCAG, reverse primer EGR_E4-GTCCATAGTCGCTGGAGGAG, and TMP1EGR4 probe-[BHQ1a-6FAM]TGGAGCGCGGCAGGAAGCCTTA. Analytic validation was performed according to our standard operating procedures.
Fig. 1.
TMPRSS2-ERG fusion reverse transcription polymerase chain reaction (RT-PCR) assay: (a) schematic of TMPRSS2-ERG fusion and design of TaqMan probes for the RT-PCR assay; (b) dynamic range of the assay was determined for TMPRSS2-ERG, glyceraldehyde-3-phosphate dehydrogenase, androgen receptor (AR), and prostate-specific antigen (PSA) genes.
To assess TMPRSS2-ERG status in the primary tumor by fluorescence in situ hybridization (FISH), we used archived pathology slides from the diagnostic prostate biopsy or from the radical prostatectomy specimen, if available, as previously described [9,10].
2.3. Statistical methods
The Kaplan-Meier method was used to estimate overall survival. The relationship between TMPRSS2-ERG status and PSA decline was assessed using the Fisher exact test. We assessed the number of patient samples needed to produce sufficient power to detect a relationship between TMPRSS2-ERG fusion status and the probability of remaining alive for at least 1 yr from the start of treatment.
3. Results
3.1. Analytic validation of TMPRSS2-ERG reverse transcription polymerase chain reaction assay
3.1.1. Standard procedures for specimen collection and handling
Blood samples were collected prior to drug administration, first in CellSave tubes for CTC enumeration (n = 48) and second in an EDTA tube for RNA profiling (n = 41). Samples were delivered and processed in the clinical chemistry laboratory within 4 h.
3.1.2. Assay performance characteristics
The intra-assay coefficient of variation (CV) was 0.34% (n = 1152). Repeat experiments performed in triplicate by different analysts at different time points showed an interassay Pearson correlation coefficient of 0.994, while the interassay CV across a six-log dilution was <7%. Using VCaP prostate cancer cells, TMPRSS2-ERG was detected in the multiplex panel with a robust CV <5% down to a single cell. The dynamic linearity of the assay between the range of dilutions of 400 ng to 10 pg had a calculated linear correlation of r2 = 0.992 for TMPRSS2-ERG, r2 = 0.963 for glyceraldehyde-3-phosphate dehydrogenase, r2 = 0.935 for androgen receptor (AR), and r2 = 0.97 for PSA (Fig. 1b). The concordance between a standard PCR platform (Eppendorf Realplex, Eppendorf, Hauppauge, NY, USA) and the chip was r2 = 0.94 testing five serial dilutions.
3.1.3. Reporting
CTC counts were reported as the number of cells meeting CellSearch-defined criteria per 7.5 ml of blood. Counts were considered favorable if less than five and unfavorable if five or more. TMPRSS2-ERG fusion status was reported as present or absent.
3.2. Patient data
The clinical characteristics of the 48 patients treated at MSKCC are detailed in Table 1.
Table 1.
Patient baseline demographics and clinical characteristics
| Overall, n = 48 | TMPRSS2-ERG fusion | ||
|---|---|---|---|
| Present, n = 15 | Absent, n = 26 | ||
| Age, yr, median (IQR) | 70 (63–79) | 71 (64–78) | 68 (63–80) |
| Primary therapy | |||
| Radical prostatectomy, no. (%) | 16 (33) | 6 (40) | 6 (23) |
| Radiation therapy to the prostate, no. (%) | 18 (38) | 4 (27) | 14 (54) |
| No primary treatment, no. (%) | 14 (29) | 5 (33) | 6 (23) |
| Prior systemic therapy | |||
| Androgen depletion, no. | 48 | 15 | 26 |
| Three or more hormonal therapies, no. (%) | 21 (44) | 8 (53) | 8 (31) |
| Chemotherapy, no. | 48 | 15 | 26 |
| One prior regimen, no. (%) | 26 (54) | 9 (60) | 14 (54) |
| Two prior regimens, no. (%) | 22 (46) | 6 (40) | 12 (46) |
| Sites of disease | |||
| Visceral, no. (%) | 18 (38) | 5 (33) | 13 (50) |
| Soft tissue and no bone disease, no. (%) | 2 (4) | 0 (0) | 1 (4) |
| Bone and soft tissue, no. (%) | 18 (38) | 6 (40) | 8 (31) |
| Bone only, no. (%) | 11 (23) | 4 (27) | 4 (15) |
| Baseline PSA, ng/ml, median (IQR) | 116 (37–343) | 185 (50–563) | 66 (33–329) |
| Baseline CTCs per 7.5 ml blood, median (IQR) | 16 (4–56) | 17 (7–103) | 12 (2–38) |
| Patients with ≥5 CTCs, no. (IQR of CTC counts) | 35 (13–99) | 12 (17–99) | 18 (12–49) |
| Patients with <5 CTCs, no. (IQR of CTC counts) | 13 (0–2) | 3 (0–4) | 8 (0–4) |
| Baseline LDH, U/l, median (IQR) | 239 (200–357) | 239 (223–351) | 235 (194–276) |
| Baseline hemoglobin, g/dl, median (IQR) | 12 (10.4–12.6) | 11 (10.4–12.3) | 12.0 (11.2–12.6) |
| Baseline albumin, g/dl, median (IQR) | 4 (4–4.4) | 4.2 (4–4.35) | 4.2 (4–4.4) |
| Baseline alkaline phosphatase, U/l, median (IQR) | 130 (73–233) | 152 (71–310) | 117 (73–232) |
IQR = interquartile range; PSA = prostate-specific antigen; CTC = circulating tumor cell; LDH = lactase dehydrogenase.
3.2.1. Circulating tumor cell enumeration
Overall, 35 of 48 patients (73%; 95% confidence interval [CI], 59–83%) had unfavorable CTC counts at baseline, and 13 of those (37%; 95% CI, 23–54%) had favorable CTC counts after 4 wk of therapy. Of the 13 patients with favorable baseline CTC counts, 2 showed an increase to unfavorable during therapy.
3.2.2. TMPRSS2-ERG fusion by reverse transcription polymerase chain reaction from circulating tumor cells
Total RNA was extracted from CTCs enriched from baseline blood samples in 41 of 48 enrolled patients (85%). As expected, the total RNA yield, ranging from 520 to 4636 ng, correlated poorly with CTC counts (Pearson r = 0.15) because of the variable contamination with peripheral blood mononuclear cells. Baseline RNA samples were not obtained from seven patients (15%) who did not have blood drawn in EDTA tubes.
TMPRSS2-ERG fusions were detected in CTCs from 15 of 41 patients (37%; 95% CI, 24–52%), with a median baseline CTC count of 17 cells per 7.5 ml blood (interquartile range (IQR): 7–103 cells). The fusion was not detected in CTCs from 26 of 41 patients. Baseline clinical characteristics of each group are described in Table 1.
3.2.3. TMPRSS2-ERG fusion by fluorescence in situ hybridization from primary tumor tissue
Of the 41 patients with TMPRSS2-ERG status assessed in CTCs, 23 patients had tissue samples available for FISH analysis, including 9 prostatectomy specimens, 13 diagnostic prostate biopsies, and 1 transurethral resection. Table 2 shows the comparison of TMPRSS2-ERG status by RT-PCR in CTCs and FISH analysis. The overall rate of detection of TMPRSS2-ERG fusion by FISH in primary tumors was 39% (9 of 23), similar to the 37% observed in CTC. However, the concordance between the RT-PCR assay in CTCs and FISH assay in primary tumors was only 15 of 23 patients (65%). Of the seven patients with positive TMPRSS2-ERG fusion by RT-PCR in CTCs, four showed concordant FISH results in primary tumor tissue, but three patients had negative FISH.
Table 2.
Comparison of TMPRSS2-ERG status in circulating tumor cell and primary prostate cancer tissue*
| Patient | TMPRSS2-ERG status | |
|---|---|---|
| CTCs/RT-PCR | Primary tissue/FISH | |
| 1 | + | + |
| 2 | + | + |
| 3 | + | + |
| 4 | + | + |
| 5 | + | − |
| 6 | + | − |
| 7 | + | − |
| 8 | − | − |
| 9 | − | − |
| 10 | − | − |
| 11 | − | − |
| 12 | − | − |
| 13 | − | − |
| 14 | − | − |
| 15 | − | − |
| 16 | − | − |
| 17 | − | − |
| 18 | − | − |
| 19 | − | + |
| 20 | − | + |
| 21 | − | + |
| 22 | − | + |
| 23 | − | + |
CTC = circulating tumor cell; FISH = fluorescent in situ hybridization; RT-PCR = reverse transcription polymerase chain reaction.
For 23 patients, TMPRSS2-ERG fusion presence or absence was detected in parallel by RT-PCR in enriched CTC tissue and by FISH analysis in tissue available from prostatectomy specimens obtained from 9 patients, biopsy tissue obtained from 13 patients, and chips from transurethral resection of the prostate obtained from 1 patient.
3.2.4. Associations with clinical outcomes
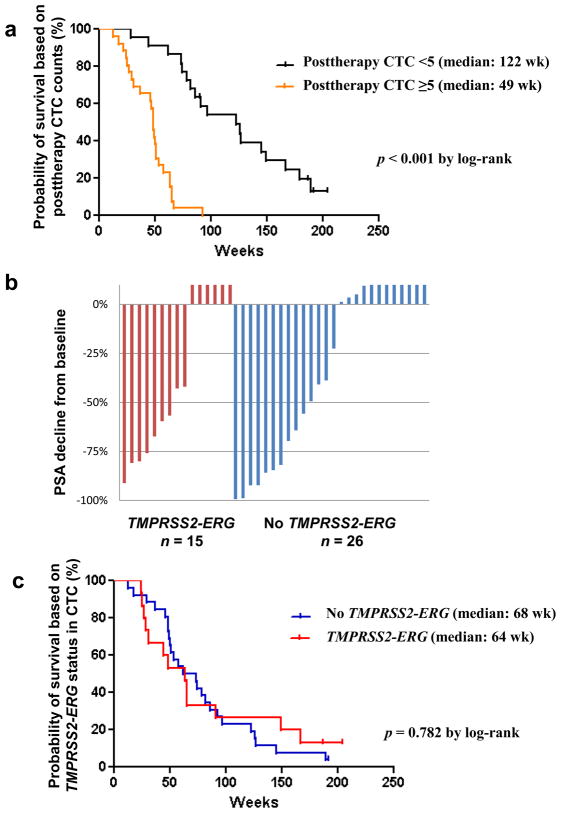
Based on CTC counts after 4 wk of treatment, patients with posttreatment favorable CTC counts of less than five (n = 24) had longer overall survival (median: 122 wk) relative to patients with unfavorable CTC counts of five or more (n = 24; median: 49 wk) by log-rank analysis (p < 0.001; Fig. 2a). There was no difference in survival between patients with favorable CTC counts throughout the study and patients who converted to favorable counts following treatment (p = 0.47).
Fig. 2.
(a) Patients with circulating tumor cell (CTC) counts of five or more at 4 wk after therapy showed significantly shorter overall survival compared with patients with CTC counts of less than five (p < 0.001 by log-rank test); (b) waterfall plots showing prostate-specific antigen (PSA) decline from baseline at 12 wk and TMPRSS2-ERG fusion measured by reverse transcription polymerase chain reaction in CTCs from patients with castration-resistant prostate cancer treated with abiraterone acetate (41 of 48 patients); (c) overall survival by TMPRSS2-ERG fusion status in CTCs (p = 0.782 by log-rank test).
Figure 2b shows the posttherapy PSA change waterfall plot at 12 wk by TMPRSS2-ERG fusion status in CTCs. In fusion-positive patients (n = 15) versus fusion-negative patients (n = 26), PSA declines of ≥30%, ≥50%, and ≥90% were seen in 9 (60%) versus 12 (46%), 7 (47%) versus 10 (38%), and 1 (7%) versus 4 (15%), respectively. These differences were not statistically significant with the Fisher exact test.
There was also no difference in survival between the fusion-positive and fusion-negative groups in overall survival (p = 0.782; Fig. 2c), even after adjusting for posttreatment CTC counts (p = 0.661). A subgroup analysis of patients with baseline CTC count of five or more (n = 30) showed no significant differences in overall survival based on TMPRSS2-ERG status (p = 0.38). The probability of remaining alive for at least 1 yr from the start of treatment was 0.53 for TMPRSS2-ERG fusion–positive patients (n = 15) and 0.62 for fusion-negative patients (n = 26). Based on this estimate, the sample size needed to detect a difference in the 1-yr survival probability with 0.80 power and a two-sided type 1 error rate equal to 0.05 would be a minimum of 990 patients. These calculations were based on the raw proportions of patients remaining alive for 1 yr. No patients were censored prior to the 1-yr follow-up mark.
4. Discussion
The revolution in cancer genomics is rapidly changing the field of cancer diagnostics and therapeutics, bringing us to an era of personalized medicine in which treatment selection is based on the molecular characteristics of an individual patient’s tumor. Achieving this objective in clinical practice requires the identification of predictive biomarkers of sensitivity, the validation of assays to measure the biomarker, and, separately, prospective clinical trials designed to qualify the biomarker for the specific context of use. Our group has established a formal collaboration with the FDA to qualify CTC number as a component of an efficacy-response surrogate biomarker panel for survival. In this paper, we describe a study of the androgen-driven TMPRSS2-ERG fusion in CTCs as a predictive biomarker of sensitivity to the androgen biosynthesis inhibitor AA.
The first step was to analytically validate a sensitive RT-PCR assay to detect the TMPRSS2-ERG fusion in CTCs before studying the association between this fusion and clinical outcomes [2,4]. The frequency of detection of the fusion in CTCs from patients with metastatic CRPC was 37% (95% CI, 24–52%), consistent with prior studies from tumor samples [7–10]. In relation to patient outcome following treatment with abiraterone, fusion status in CTCs was not predictive of PSA-decline rates or survival. Significant discordance between TMPRSS2-ERG status in the primary tumor by FISH and in CTCs by RT-PCR has also been reported by others and may reflect in part the multifocal nature of primary prostate cancers [21,22] and the inability to sample divergent clones in the primary tumor [23]. Of the 12 patients with TMPRSS2-ERG fusion detected in either CTCs or primary tumor tissue, only 4 patients were positive in both analyses, supporting the importance of analyzing metastatic tumor samples when treatment is being considered.
The results of this analysis contrast with a report that showed an association between the 90% PSA-decline rate from baseline and the presence of TMPRSS2-ERG rearrangements in CTCs assayed by FISH [13]. However, on critically analyzing both studies, it is apparent that while the two studies are evaluating the same gene target, they are not using the same assay or reporting the same biomarker. The patient populations are different (one included both prechemotherapy- and postchemotherapy-treated patients and the other only postchemotherapy-treated patients). An additional question is whether any posttherapy change in PSA is the relevant clinical outcome to report for a drug that can modify PSA independent of tumor cell growth. In our study, posttreatment CTC number, a biomarker not modulated by AR, was associated with overall survival.
Qualification of a predictive biomarker requires prospective testing in multiple phase 3 trials in which the biomarker question is embedded. The randomized phase 3 trial of AA plus prednisone (COU-AA-301), which included the study of CTC enumeration as an efficacy-response surrogate biomarker for survival, met the primary end point of an improvement in survival in postchemotherapy-treated CRPC and led to the FDA approval of the drug for this indication [1]. Exploratory studies are ongoing to prioritize biomarkers for evaluation in the context of this trial. The results reported in this paper for TMPRSS2-ERG status in CTCs measured with a validated assay and the sample size needed to detect a difference in survival argue against further study of this biomarker alone in this patient group. Whether TMPRSS2-ERG status in combination with other biomarkers such as phosphoinositide 3-kinase signaling status would be predictive remains open [24–27].
5. Conclusions
A significant proportion of patients have the prostate cancer–specific TMPRSS2-ERG fusion product present in CTCs. These results demonstrate the feasibility of CTCs as an easily obtained tissue for molecular analysis such as the detection of kinase mutations in other tumor types [28,29]. The results also illustrate the importance of developing standards for biomarker development that include establishing the performance of the assay itself, followed by a prospectively planned sequence of clinical investigations that prioritize biomarkers for further study in large-scale trials. TMPRSS2-ERG fusion status by itself has a limited role as a predictive biomarker of sensitivity to AA in postchemotherapy-treated CRPC.
Acknowledgments
Funding/Support and role of the sponsor: The design and conduct of this research was funded by the National Cancer Institute SPORE in Prostate Cancer (P50 CA92629); the Prostate Cancer Foundation; Mr. William H. and Mrs. Alice Goodwin and the Commonwealth Foundation for Cancer Research and the Experimental Therapeutics Center of Memorial Sloan-Kettering Cancer Center; the Department of Defense Prostate Cancer Clinical Trials Consortium; and the ASCO Young Investigator Award and Department of Defense Prostate Cancer Research Program Physician Research Award W81XWH-09–1-0307 to D.C. Danila.
Footnotes
Trial registration: ClinicalTrials.gov: NCT00474383 (COU-AA-003), NCT00485303 (COU-AA-004).
Author contributions: Howard I. Scher had full access to all the data in the study and takes responsibility for the integrity of the data and the accuracy of the data analysis.
Study concept and design: Danila, Fleisher, Scher.
Acquisition of data: Danila, Anand, Sung, Leversha, Cao, Scher.
Analysis and interpretation of data: Danila, Anand, Heller, Leversha, Lilja, Fleisher, Scher.
Drafting of the manuscript: Danila, Lilja, Fleisher, Scher.
Critical revision of the manuscript for important intellectual content: Danila, Heller, Lilja, Molina, Sung, Fleisher, Scher.
Statistical analysis: Danila, Heller.
Obtaining funding: Danila, Scher.
Administrative, technical, or material support: Danila, Fleisher, Scher.
Supervision: Danila, Scher.
Other (specify): None.
Financial disclosures: I certify that all conflicts of interest, including specific financial interests and relationships and affiliations relevant to the subject matter or materials discussed in the manuscript (eg, employment/affiliation, grants or funding, consultancies, honoraria, stock ownership or options, expert testimony, royalties, or patents filed, received, or pending), are the following: Arturo Molina is employed by Ortho Biotech Oncology Research and Development, a unit of Cougar Biotechnology. Howard I. Scher receives research funding from Ortho Biotech Oncology Research and Development, a unit of Cougar Biotechnology and Veridex, and had stock in Johnson and Johnson.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.de Bono JS, Logothetis CJ, Molina A, et al. Abiraterone and increased survival in metastatic prostate cancer. N Engl J Med. 2011;364:1995–2005. doi: 10.1056/NEJMoa1014618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Danila DC, Morris MJ, de Bono JS, et al. Phase II multicenter study of abiraterone acetate plus prednisone therapy in patients with docetaxel-treated castration-resistant prostate cancer. J Clin Oncol. 2010;28:1496–501. doi: 10.1200/JCO.2009.25.9259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Holzbeierlein J, Lal P, LaTulippe E, et al. Gene expression analysis of human prostate carcinoma during hormonal therapy identifies androgen-responsive genes and mechanisms of therapy resistance. Am J Pathol. 2004;164:217–27. doi: 10.1016/S0002-9440(10)63112-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Reid AH, Attard G, Danila DC, et al. Significant and sustained antitumor activity in post-docetaxel, castration-resistant prostate cancer with the CYP17 inhibitor abiraterone acetate. J Clin Oncol. 2010;28:1489–95. doi: 10.1200/JCO.2009.24.6819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Scher HI, Sawyers C. Biology of progressive castration resistant prostate cancer: directed therapies targeting the androgen receptor signaling axis. J Clin Oncol. 2005;23:8253–61. doi: 10.1200/JCO.2005.03.4777. [DOI] [PubMed] [Google Scholar]
- 6.Stanbrough M, Bubley GJ, Ross K, et al. Increased expression of genes converting adrenal androgens to testosterone in androgen-independent prostate cancer. Cancer Res. 2006;66:2815–25. doi: 10.1158/0008-5472.CAN-05-4000. [DOI] [PubMed] [Google Scholar]
- 7.Tomlins SA, Rhodes DR, Perner S, et al. Recurrent fusion of TMPRSS2 and ETS transcription factor genes in prostate cancer. Science. 2005;310:644–8. doi: 10.1126/science.1117679. [DOI] [PubMed] [Google Scholar]
- 8.Lapointe J, Kim YH, Miller MA, et al. A variant TMPRSS2 isoform and ERG fusion product in prostate cancer with implications for molecular diagnosis. Mod Pathol. 2007;20:467–73. doi: 10.1038/modpathol.3800759. [DOI] [PubMed] [Google Scholar]
- 9.Carver BS, Tran J, Chen Z, et al. ETS rearrangements and prostate cancer initiation. Nature. 2009;457:E1. doi: 10.1038/nature07738. discussion E2–E3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gopalan A, Leversha MA, Satagopan JM, et al. TMPRSS2-ERG gene fusion is not associated with outcome in patients treated by prostatectomy. Cancer Res. 2009;69:1400–6. doi: 10.1158/0008-5472.CAN-08-2467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fine SW, Gopalan A, Leversha MA, et al. TMPRSS2-ERG gene fusion is associated with low Gleason scores and not with high-grade morphological features. Mod Pathol. 2010;23:1325–33. doi: 10.1038/modpathol.2010.120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yu J, Mani RS, Cao Q, et al. An integrated network of androgen receptor, polycomb, and TMPRSS2-ERG gene fusions in prostate cancer progression. Cancer Cell. 2010;17:443–54. doi: 10.1016/j.ccr.2010.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Attard G, Swennenhuis JF, Olmos D, et al. Characterization of ERG, AR and PTEN gene status in circulating tumor cells from patients with castration-resistant prostate cancer. Cancer Res. 2009;69:2912–8. doi: 10.1158/0008-5472.CAN-08-3667. [DOI] [PubMed] [Google Scholar]
- 14.McShane LM, Altman DG, Sauerbrei W, Taube SE, Gion M, Clark GM. Reporting recommendations for tumor marker prognostic studies (REMARK) J Natl Cancer Inst. 2005;97:1180–4. doi: 10.1093/jnci/dji237. [DOI] [PubMed] [Google Scholar]
- 15.Altar CA. The Biomarkers Consortium: on the critical path of drug discovery. Clin Pharmacol Ther. 2008;83:361–4. doi: 10.1038/sj.clpt.6100471. [DOI] [PubMed] [Google Scholar]
- 16.Bubley GJ, Carducci M, Dahut W, et al. Eligibility and response guidelines for phase II clinical trials in androgen-independent prostate cancer: recommendations from the PSA Working Group. J Clin Oncol. 1999;17:3461–7. doi: 10.1200/JCO.1999.17.11.3461. [DOI] [PubMed] [Google Scholar]
- 17.Scher HI, Halabi S, Tannock I, et al. Design and end points of clinical trials for patients with progressive prostate cancer and castrate levels of testosterone: recommendations of the Prostate Cancer Clinical Trials Working Group. J Clin Oncol. 2008;26:1148–59. doi: 10.1200/JCO.2007.12.4487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Allard WJ, Matera J, Miller MC, et al. Tumor cells circulate in the peripheral blood of all major carcinomas but not in healthy subjects or patients with nonmalignant diseases. Clin Cancer Res. 2004;10:6897–904. doi: 10.1158/1078-0432.CCR-04-0378. [DOI] [PubMed] [Google Scholar]
- 19.Shaffer DR, Leversha MA, Danila DC, et al. Circulating tumor cell analysis in patients with progressive castration-resistant prostate cancer. Clin Cancer Res. 2007;13:2023–9. doi: 10.1158/1078-0432.CCR-06-2701. [DOI] [PubMed] [Google Scholar]
- 20.Danila DC, Heller G, Gignac GA, et al. Circulating tumor cell number and prognosis in progressive castration-resistant prostate cancer. Clin Cancer Res. 2007;13:7053–8. doi: 10.1158/1078-0432.CCR-07-1506. [DOI] [PubMed] [Google Scholar]
- 21.Attard G, de Bono JS, Clark J, Cooper CS. Studies of TMPRSS2-ERG gene fusions in diagnostic trans-rectal prostate biopsies. Clin Cancer Res. 2010;16:1340. doi: 10.1158/1078-0432.CCR-09-2253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Stott SL, Lee RJ, Nagrath S, et al. Isolation and characterization of circulating tumor cells from patients with localized and metastatic prostate cancer. Sci Transl Med. 2010;2:25ra23. doi: 10.1126/scitranslmed.3000403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Liu W, Laitinen S, Khan S, et al. Copy number analysis indicates monoclonal origin of lethal metastatic prostate cancer. Nat Med. 2009;15:559–65. doi: 10.1038/nm.1944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.King JC, Xu J, Wongvipat J, et al. Cooperativity of TMPRSS2-ERG with PI3-kinase pathway activation in prostate oncogenesis. Nat Genet. 2009;41:524–6. doi: 10.1038/ng.371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Clark JP, Cooper CS. ETS gene fusions in prostate cancer. Nat Rev Urol. 2009;6:429–39. doi: 10.1038/nrurol.2009.127. [DOI] [PubMed] [Google Scholar]
- 26.Berger MF, Lawrence MS, Demichelis F, et al. The genomic complexity of primary human prostate cancer. Nature. 2011;470:214–20. doi: 10.1038/nature09744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Taylor BS, Schultz N, Hieronymus H, et al. Integrative genomic profiling of human prostate cancer. Cancer Cell. 2010;18:11–22. doi: 10.1016/j.ccr.2010.05.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lankiewicz S, Rother E, Zimmermann S, Hollmann C, Korangy F, Greten TF. Tumour-associated transcripts and EGFR deletion variants in colorectal cancer in primary tumour, metastases and circulating tumour cells. Cell Oncol. 2008;30:463–71. doi: 10.3233/CLO-2008-0432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Maheswaran S, Sequist LV, Nagrath S, et al. Detection of mutations in EGFR in circulating lung-cancer cells. N Engl J Med. 2008;359:366–77. doi: 10.1056/NEJMoa0800668. [DOI] [PMC free article] [PubMed] [Google Scholar]