Summary
The release of Ca2+ ions from the sarcoplasmic reticulum through ryanodine receptor calcium release channels represents the critical step linking electrical excitation to muscular contraction in the heart and skeletal muscle (excitation-contraction coupling). Two small Ca2+ binding proteins, S100A1 and calmodulin, have been demonstrated to bind and regulate ryanodine receptor activity in vitro. This review focuses on recent work that has revealed new information about the endogenous roles of S100A1 and calmodulin in regulating skeletal muscle excitation-contraction coupling. S100A1 and calmodulin bind to an overlapping domain on the ryanodine receptor type 1 to tune the Ca2+ release process, and thereby regulate skeletal muscle function. We also discuss past, current and future work surrounding the regulation of ryanodine receptors by calmodulin and S100A1 in both cardiac and skeletal muscle, and the implications for excitation-contraction coupling.
1. Introduction
Excitation-contraction (EC) coupling is the process by which membrane depolarization triggers sarcoplasmic reticulum (SR) Ca2+ release and subsequent muscle contraction. In skeletal muscle, membrane depolarization alters the molecular interaction between dihydropyridine receptors (DHPRs) in transverse (t) tubule membranes and the ryanodine receptor Ca2+ release channels (RyR1) in the adjacent junctional SR membranes, triggering rapid RyR1 activation and Ca2+ release into the cytosol [1–5]. Both activation and deactivation of Ca2+ release in skeletal muscle are under tight control of the voltage-dependent conformational changes in the DHPR, and the resulting change in molecular interaction between DHPRs and RyR1s. EC coupling in cardiac muscle uses different isoforms of the DHPR and RyR, which are coupled by Ca2+ ions that enter the myocyte via DHPR Ca2+ channels and activate nearby RyR2 Ca2+ release channels through Ca2+-induced Ca2+ release (CICR) (Reviewed in [6]).
Despite the tight inter-molecular regulation of skeletal muscle SR Ca2+ release, numerous intracellular modulators interact directly or indirectly with RyR1 to fine tune Ca2+ release. RyR1 is a massive 560 kD homotetramer, with approximately 4/5ths of the channel exposed to the cytoplasm and the remaining 1/5th located in the transmembrane or luminal SR [7]. This large cytoplasmic face is the target of several small molecule regulators, such as ATP, Ca2+, and Mg2+ [8], and is the putative DHPR interacting site [9, 10]. Multiple endogenous proteins have also been proposed to regulate RyR1 function at the cytoplasmic face, including FKBP12 [11], calmodulin (CaM) [12], and S100A1 [13]. In cardiac muscle the same regulators serve to modulate RyR2 activity [14–16]. In recent years, work from our group and collaborators has provided significant insight into the regulation of RyR1 by CaM and S100A1, primarily through the use of genetically engineered mouse models. This review will focus on the emerging picture of how CaM and S100A1 compete for binding RyR1 to regulate Ca2+ release in skeletal muscle. We will also discuss the possibilities and implications for similar interactions between CaM and S100A1 with RyR2 in cardiac muscle.
2. Calmodulin and S100A1: small Ca2+ binding proteins that bind and regulate RyR1
Calmodulin is a small (~17 kD), ubiquitous, highly conserved EF-hand containing Ca2+ binding protein. Roughly 50% of CaM localizes to cellular membranes, while the other half resides in the cytosol and nucleus [17]. CaM binds four Ca2+ ions cooperatively, and undergoes a Ca2+-dependent conformation change that increases its affinity for target proteins [17]. CaM binds directly to RyR1 and is well documented to regulate activity of isolated RyR1 channels in vitro. EM and FRET based studies show CaM binds to the large cytoplasmic face of RyR1 ([18, 19], and single channel measurements demonstrated that at low [Ca2+] (100nM) Ca2+ free (ie, apo-CaM) increases the activity of RyR1, while at higher Ca2+ concentrations (≥1µM) Ca-CaM inhibits channel activity several fold [20]. Further studies complemented these findings, leading to the classification of apo-CaM as a weak agonist of RyR1 and Ca-CaM as a stronger inhibitor of the channel [21, 22]. CaM binds to one site per RyR1 subunit (4 per tetramer), and protects RyR1 from proteolytic cleavage at amino acid residues 3630 and 3637. Combined with sequence analysis for CaM binding motifs, the CaM binding domain (CaM BD) has been identified as residues 3614–3643 of rabbit RyR1 [23] on the cytoplasmic face of each RyR1 subunit [24]. Rabbit RyR1 3614–3643 corresponds to mouse RyR1 3615–3644; in the rabbit sequence residue numbers are shifted one residue lower for RyR1 and one residue higher for RyR2 compared to the mouse sequence, which should be kept in mind when comparing reports in the literature.
More recently, CaM has been shown to interact with RyR1 in a more complex manner. The C-terminal lobe of CaM binds to the previously classified CaM BD of RyR1, while the N-terminal lobe may interact with a second site non-contiguous with the primary sequence [25]. The N-terminal lobe has been proposed to bind between residues 1975 and 1999 of the RyR1 monomer adjacent to the C-terminal binding site. Disulfide bonds between these sites supports proximity between the two domains, which could explain CaM binding non-contiguously to this site of intersubunit interaction [26]. Upon binding Ca2+, CaM has been shown to shift in its binding site [18, 19]. Zhang et al. (2003) proposed that this shift mediates the switch between apo-CaM activating RyR1 to Ca-CaM inhibiting the channel. Furthermore, Rodney et al. (2005) proposed that apo-CaM binding the CaM BD may sensitize RyR1 to activation by disrupting intersubunit interactions, while Ca-CaM may produce an inhibitory effect by stabilizing intersubunit interactions and promoting a closed configuration of the channel.
Although CaM binding to RyR1 and modulation of RyR1 activity in vitro has been thoroughly demonstrated [20, 22, 27], in situ analysis of CaM’s role in EC coupling with functionally coupled DHPR-RyR1 has been limited. O’Connell et al. (2002) showed that myotubes expressing RyR1 with mutations to the CaM BD showed minimal alterations in EC coupling [28]. However, until recently no studies had evaluated endogenous CaM’s in vivo role in adult skeletal muscle, leaving a void between in vitro analysis of CaM’s effect on RyR1 and an analysis of its physiologic contribution to EC coupling.
2.1. S100A1
The S100 family of proteins are Ca2+ binding proteins that appeared relatively late in evolution and are only expressed in vertebrates. One of the first of the now more than 20 family members characterized was S100A1, originally referred to as S100α [29, 30]. S100A1 is a small (21kD), dimeric Ca2+ binding protein that, like CaM, contains 4 EF-hand Ca2+ binding domains in its dimerized form [31, 32]. The first EF hand is a “pseudo” EF hand, as it binds Ca2+ with lower affinity (100–500 µM) than the second, canonical EF hand, which binds Ca2+ in the standard range (1–50 µM) [31, 32]. This relatively low Ca2+ affinity of S100A1 can be greatly strengthened by glutathionylation [33] as well as the presence of a target interacting protein [34, 35], allowing S100A1 to sense nanomolar intracellular Ca2+ concentrations in vivo. Ca2+ affinity is a key determinant in the function of S100A1, since like CaM it has no intrinsic enzymatic activity, but instead interacts with target proteins though a Ca2+-dependent mechanism to elicit biological responses [29, 36]. Upon binding Ca2+, two of the EF-hands undergo dramatic conformational changes, exposing a hydrophobic binding pocket that is thought to be the target protein interacting region [32, 37].
Although S100 family members share considerable sequence homology, they are expressed in tissue specific patterns. S100A1 is the most highly expressed family member in striated muscle, and also exhibits expression in other organ systems such as the brain, kidney, and spleen [30, 38, 39]. In striated muscle, S100A1 demonstrates highest expression in cardiac muscle, followed by slow twitch and fast twitch skeletal muscle, respectively [40, 41]. In fast twitch skeletal muscle, estimates of S100A1 protein concentration range from 0.5 – 15 µM [40, 42, 43]. The effective concentration of S100A1 locally available for the modulation of RyR1 is likely higher, however, as S100A1 has been shown, both at an ultrastructural level and with immunofluorescence, to localize to A band/I band junctions and SR membranes [44–46], where RyR1 Ca2+ release occurs in skeletal muscle.
While a considerable body of knowledge has amassed about S100A1’s role in cardiac muscle (see below and review by [47]), research on this protein in skeletal muscle has lagged behind. Early experiments showed that S100A1 enhanced RyR1 Ca2+ release in SR terminal cisternae preparations [48]. These authors presciently noted that despite belonging to the same superfamily of EF-hand Ca2+ binding proteins, CaM and S100A1 appeared to have contrasting effects on the activation of RyR1-mediated Ca2+ release, a notion we have explored in detail in recent years and will focus on in this review. S100A1 has also been shown to bind purified RyR1 by affinity chromatography (with mid-nanomolar affinity, similar to CaM), and to potentiate the open probability of RyR1 reconstituted in a lipid bilayer [13]. Furthermore, exogenous S100A1 increases caffeine-evoked force transients in skinned skeletal muscle fibers [41]. These in vitro studies laid mechanistic groundwork for our understanding of S100A1 regulation of RyR1 Ca2+ release; however, they did not explore physiologic voltage-gated Ca2+ release in a functionally intact skeletal muscle system.
2.2. Calmodulin and S100A1 bind to an overlapping conserved region of the ryanodine receptor Ca2+ release channel
It is not uncommon for S100 proteins to bind similar structural motifs as CaM [49, 50]. Sequence scanning of mouse RyR1 showed that residues 3617–3628, which begin 2 residues into the N-terminus of the CaM BD, very closely match the S100 consensus binding sequence (Fig. 1), closer than any other sequence in the entirety of RyR1. S100A1 and CaM were shown to bind with similar affinities to a peptide generated from this region of RyR1 [46]. The solution structure of S100A1 bound to this peptide of RyR1 has since been solved using NMR spectroscopy [35], and the crystal structure of CaM bound to the CaM BD is also available [51]. Of note, the S100A1 binding domain of RyR1 (mouse residues 3617–3628) is perfectly conserved in the corresponding sequence of RyR2 (mouse residues 3582–3593; Fig. 1; see below). Furthermore, there are only three residue differences between RyR1 and RyR2 over the entire CaM binding domain.
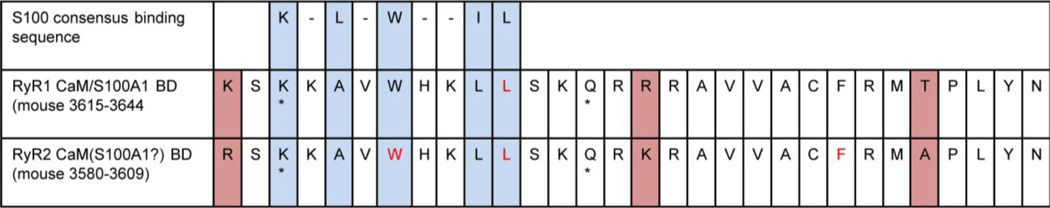
Fig. 1. S100A1 and calmodulin compete for binding to an overlapping region of RyR1.
The S100 consensus binding sequence is identified in RyR1 residues 3617–3625. This sequence lies in the N-terminus of the previously characterized calmodulin binding domain (CaM BD) of RyR1 (3615–3644). A 12 amino acid peptide containing this sequence (residues within the asterisks) is perfectly conserved between RyR1 and RyR2 and has a similar affinity for S100A1 and Ca-CaM [46]. Mutation of L3625D (red) abrogates both CaM and S100A1 binding to RyR1. This mutation impairs S100A1 activation of SR Ca2+ release and Ca-CaM inactivation of SR Ca2+ release [53]. We therefore refer to this region as the CaM/S100A1 binding domain. The CaM BD is highly conserved between RyR1 and RyR2 (red shading marks differing residues). Mutations of W3586A/L3590D/F3602A (green) abrogate CaM from binding and inactivating RyR2 in heart cells [73]. It is currently unknown whether S100A1 regulates Ca2+ release in the heart through interaction with this domain. The residue numbers used here correspond to mouse RyR1 and RyR2. Note that for the rabbit, residue numbers are shifted one residue lower for RyR1 and one residue higher for RyR2 compared to the mouse sequence.
Competition assays using isolated SR vesicle preparations were used to evaluate whether CaM and S100A1 compete for binding the CaM BD of the full length RyR1. CaM-linked beads were mixed with intact SR vesicles expressing full length RyR1, allowing CaM to bind to the CaM BD of RyR1 in the SR vesicles. Addition of S100A1 displaced the RyR1-containing vesicles from the CaM beads in a dose dependent fashion [46]. Importantly, S100A1 could displace the RyR1-containing vesicles from the CaM-linked beads at 100 nM Ca2+, suggesting that S100A1 binds RyR1 at [Ca2+]i inherent to resting conditions in skeletal muscle fibers. The reverse experiment using S100A1-linked beads showed that addition of CaM also fully displaced S100A1 from RyR1 [35]. As both CaM and S100A1 fully displace one another from the CaM BD, CaM and S100A1 likely share one major, high affinity binding site on each RyR1 monomer (or 4 per tetrameric RyR1 channel), although there are additional sites of interaction [13, 20, 52]. In addition, the N-terminal CaM BD peptide also fully displaced the SR vesicles from S100A1-linked beads, further suggesting that S100A1 interaction with RyR1 occurs through this domain [35]. We will thus refer to this site of presumed competitive binding of CaM or S100A1 to RyR1 as the CaM/S100A1 BD for the remainder of this review.
3. S100A1−/− and RyR1D/D mice: studying the endogenous regulation of RyR1 by S100A1 and CaM
As detailed above, there is considerable evidence that exogenous S100A1 and CaM both bind RyR1 and regulate channel activity in vitro. To complement and advance these studies, our group has evaluated the in situ roles of endogenous S100A1 and CaM through the use of two genetic mouse models: 1) S100A1−/− mice [46]; 2) transgenic mice expressing a specific point mutation to the CaM/S100A1 BD of RyR1 (RyR1-L3625D, RyR1D/D mice) [53]. This mutation has previously been demonstrated to impair CaM binding and regulation of RyR1 in vitro [22], and we recently showed that the mutation also abolishes the in vitro binding of S100A1 to RyR1 [53] (Fig. 1). Single channel studies performed in isolated membrane fractions from RyR1D/D mice demonstrated that this point mutation impairs apo-CaM activation of RyR1 and Ca-CaM inhibition of the channel. Importantly, the large activating effect S100A1 had on single channels isolated from control mice was fully eliminated in channels isolated from RyR1D/D mice [53]. This finding provides strong support that S100A1 primarily regulates RyR1 through the CaM/S100A1 BD. RyR1D/D mice thus provide the unique advantage of eliminating non-specific effects that arise from global protein ablation, allowing a focused investigation on regulation of the CaM/S100A1 BD of RyR1. Additionally, when analyzed in parallel to results from S100A1−/− mice, the study of RyR1D/D mice has allowed us to begin to isolate the specific roles of CaM and S100A1 in the regulation of RyR1 Ca2+ release.
3.1. S100A1 potentiates SR Ca2+ release and twitch force in skeletal muscle
To study the endogenous roles of CaM and S100A1 we studied EC coupling in enzymatically dissociated flexor digitorum brevis (FDB) skeletal muscle fibers [54] isolated from S100A1−/− and RyR1D/D mice. When stimulated by a brief field stimulus to initiate a single action potential (AP) [55], S100A1−/− fibers demonstrate a ~30% reduction in the amplitude of cytosolic Ca2+ transients when compared to fibers from wild-type (WT) littermates [46]. Similarly, Ca2+ transients are suppressed at all test membrane potentials in voltage-clamped S100A1−/− fibers [56]. This reduction in Ca2+ transient amplitude is independent of changes in either resting cytosolic [Ca2+] [46] or releasable SR Ca2+ stores [55]. Additionally, there is no change in the waveform of the propagated AP (Fig. 2A) [55], or the RyR1-activating component of DHPR charge movement (Qβ) in S100A1−/− fibers (Fig. 2B) [57], suggesting a direct effect on RyR1 (Fig. 2). When S100A1 protein expression is restored in S100A1−/− fibers by adenoviral delivery of S100A1, Ca2+ transient amplitude is restored back to the WT level, suggesting that the depressed Ca2+ transients in S100A1−/− fibers did not result from a compensatory response to genetic manipulation [46]. A Ca2+ removal model [58] was used to calculate the time course of SR Ca2+ release flux underlying the AP-evoked Ca2+ transients in S100A1−/− fibers. Using this model the peak Ca2+ release flux during a single AP was found to be reduced ~30% in S100A1−/− fibers. This depressed Ca2+ release on a single cell level translates to weaker force production of the whole muscle in vivo; the isometric force generated by the tibialis anterior (TA) muscles of anesthetized S100A1−/− animals is reduced ~25% when stimulated with a single AP [55].
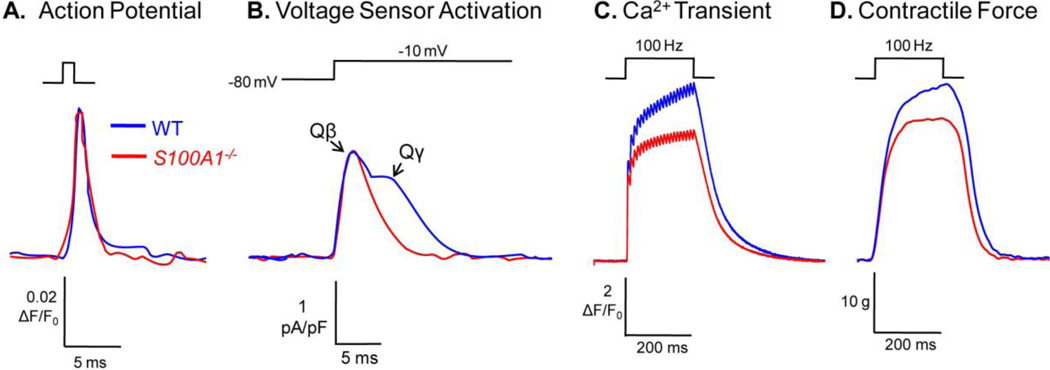
Fig. 2. S100A1 modulation of skeletal muscle EC coupling.
A) Di-8 ANEPPS recordings of action potentials elicited by field stimulation of WT (blue) and S100A1−/− (red) fibers. Genetic ablation of S100A1 has no effect on the propagated AP in the t-system of skeletal muscle fibers [55]. B) DHPR intramembrane charge movement currents elicited by a voltage clamp depolarization to −10 mV of WT and S100A1−/− fibers. Ablation of S100A1 has no effect on the RyR1-activating component of DHPR charge movement (Qβ), but does blunt a secondary component of DHPR charge movement (Qγ) that is a consequence of optimal SR Ca2+ release [57]. C) Fluo-4 recordings of Ca2+ transients elicited by 100 Hz field stimulation of WT and S100A1−/− fibers. The Ca2+ transient is depressed in S100A1−/− fibers, and demonstrates less summation during the train of stimuli [46, 55]. D) Tetanic force generated by the tibialis anterior of anesthetized WT and S100A1−/− animals in response to 100 Hz stimulation. Maximal force is suppressed in S100A1−/− muscle [55].
SR Ca2+ release is also suppressed in RyR1D/D fibers when compared to littermate controls. Both indo-1 and fluo-4 recordings of Ca2+ transients are similarly reduced in amplitude in RyR1D/D and S100A1−/− fibers stimulated by a single AP [46, 53]. Furthermore, there is a 41% average decrease in peak SR Ca2+ release flux during voltage clamp depolarization of RyR1D/D fibers [53], a remarkably similar suppression to the 42% decrease seen in S100A1−/− fibers under identical conditions [56]. As the elimination of S100A1 (S100A1−/−) and mutation to the CaM/S100A1 BD (RyR1D/D) cause a virtually identical suppression of Ca2+ release, we hypothesize that the elimination of the S100A1 activating effects on RyR1 may fully account for the suppressed Ca2+ release in RyR1D/D fibers stimulated with a single AP (Fig. 3A). Consistent with this proposal, in vivo TA twitch force elicited by a single AP is reduced 25% in RyR1D/D muscle, the same degree of suppression seen in S100A1−/− muscle [53].
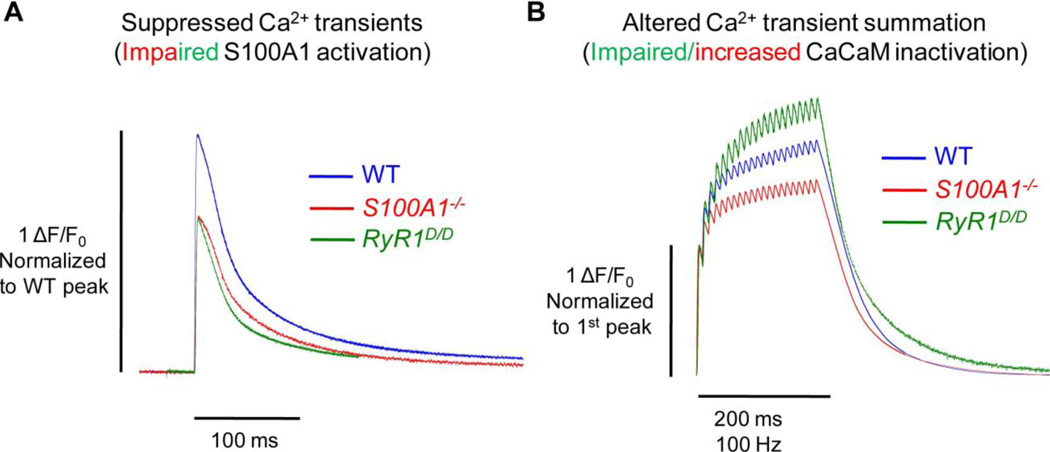
Fig. 3. S100A1 and CaM differentially regulate RyR1 Ca2+ release.
A) In response to a single action potential, the peak amplitude of the Ca2+ transient is similarly suppressed in the absence of S100A1 (S100A1−/−, red) and in fibers with a mutated CaM/S100A1 binding domain of RyR1 (RyR1D/D, green). This suggests that under resting conditions, S100A1 may predominantly occupy the CaM/S100A1 BD and potentiate Ca2+ release upon muscle activation. B) Fibers with a mutated CaM/S100A1 BD demonstrate greater relative summation of Ca2+ transients during repetitive activation. Conversely, fibers lacking S100A1 show less summation. This suggests that upon a rise in [Ca2+]i during prolonged muscle activation, Ca-CaM may predominantly occupy the CaM/S100A1 BD and inactivate some portion of Ca2+ release. This inactivation is impaired in RyR1D/D fibers, and enhanced in S100A1−/− fibers, as they lack CaM’s endogenous competitor. All data traces represent average data from [46, 53, 55].
3.2. CaM contributes to the inactivation of SR Ca2+ release during repetitive skeletal muscle stimulation
Based on the above rationale, disrupting just CaM binding to the CaM/S100A1 BD is hypothesized to have little effect on Ca2+ release elicited by a single AP from rest. However, during high frequency (100Hz) trains of APs (a more physiologic firing pattern of motor units in small mammals [59]), a clear role for CaM regulation of RyR1 Ca2+ release becomes apparent. Although the amplitude of the Ca2+ transient in response to a single AP is depressed in RyR1D/D fibers, during a 100Hz train of stimuli the Ca2+ transient amplitude (relative to the amplitude due to the initial pulse) increases more and more with each stimulus when compared to WT counterparts (i.e., there is a greater relative “summation” of Ca2+ transients in RyR1D/D fibers, as seen in Fig. 3B [53]). The time course of SR Ca2+ release flux during repetitive stimulation suggests that the greater summation of Ca2+ transients is a result of slowed inactivation of release flux in RyR1D/D fibers. This is in stark contrast to S100A1−/− fibers, where there is less summation of the Ca2+ transients and greater inactivation of Ca2+ release flux during repetitive stimulation (Fig. 3B; [55]). These finding are supported by voltage clamp studies, where a prolonged “shoulder” of Ca2+ release flux is evident during sustained depolarization of RyR1D/D fibers, again suggesting slowed inactivation of release flux [53]. Importantly, this shoulder is absent in S100A1−/− fibers [56].
Taken together these results suggest that as cytosolic [Ca2+] increases during repetitive or prolonged fiber depolarization, endogenous Ca-CaM contributes to the inactivation of Ca2+ release. This is consistent with earlier in vitro studies (see above). Impaired Ca-CaM inactivation accounts for the greater relative summation of Ca2+ transients and slowed inactivation of release flux in fibers with a mutated CaM/S100A1 BD (RyR1D/D). As S100A1−/− fibers demonstrate the opposite effects (reduced summation, greater inactivation), this may be attributed to increased Ca-CaM-dependent inactivation in these fibers, as Ca-CaM is free to bind and inactivate RyR1 more readily in the absence of its endogenous competitor, S100A1 (Fig. 3B). Consistent with this model, TA muscles of anesthetized RyR1D/D mice demonstrate greater summation of force during tetanic stimulation (attributable to impaired Ca-CaM inactivation of Ca2+ release). S100A1−/− muscle, on the other hand, shows reduced force summation (greater Ca-CaM-dependent inactivation), and during prolonged tetanic stimulation S100A1−/− muscle fatigues more rapidly than WT muscle. This suggests that un-inhibited Ca-CaM inactivation of Ca2+ release is negatively affecting muscular performance in these animals [55]. One possible reservation concerning the preceding interpretation is that the two genetically engineered mouse lines that we have examined, S100A1−/− and RyR1D/D are derived from slightly different mouse strains. Whether this influences the observed effects of these genetic manipulations remains to be determined.
3.3. Reaction schemes for activation and inactivation of WT, S100A1−/− and RyR1D/D skeletal muscle fibers
Figure 4 presents our hypothesized reaction schemes for SR Ca2+ channel gating in skeletal muscle from wild type (WT; Fig. 4A), S100A1−/− (Fig. 4B) and RyR1D/D (Fig. 4C) mice. In these schemes, the TT voltage sensor, RyR1, S100A1 and CaM are represented by the symbols V, R, S and C, respectively, and V* represents the voltage sensor in its voltage activated state. In WT fibers (Fig. 4A) in the resting state (left), the CaM/S100A1 BD is assumed to be either unoccupied (left foreground) or to bind S100A1 (left, background) at the CaM/S100A1 BD. Since S100A1−/− has the same effect as RyR1D/D on initial Ca2+ release (see above), Ca2+-free calmodulin (i.e., apo-CaM) is assumed not to bind significantly to RyR1 in resting muscle fibers. Because interaction of RyR1 with the TT voltage sensor in resting mammalian fibers suppresses Ca2+ spark activity [60–62], we represent both the resting and activated state of the voltage sensor as interacting with RyR1, but this interaction has opposite effects on RyR1 depending on whether the voltage sensor is in its resting (V, blue) or activated (V*, red) state.
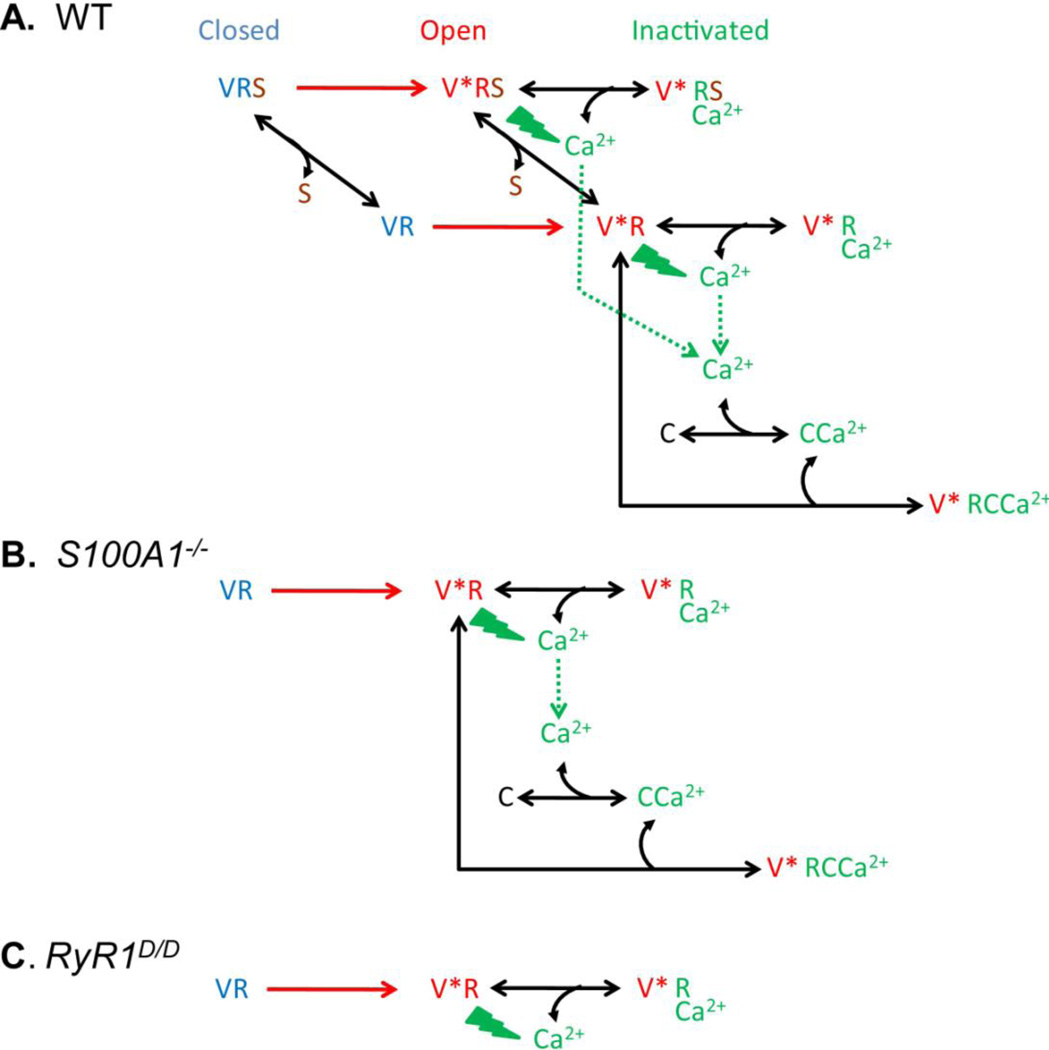
Fig.4. Reaction schemes for activation and inactivation of RyR1 Ca2+ release channels in skeletal muscle fibers from wild type (A), S100A1−/− (B) and RyR1D/D (C) mice.
V, R, S and C represent the TT voltage sensor, the SR ryanodine receptor/Ca2+ release channel, S100A1 and calmodulin, with V* representing the voltage activated state of V. Channels move sequentially left to right from the resting (blue) to the activated (red) to the inactivated state (green) due to voltage activation of V (red) to Ca2+-dependent inactivated states (green). In A, S100A1 occupied states are depicted in the background, and S100A1 free states are in the foreground. See text (3.3, 3.4) for further details.
Muscle fiber depolarization (red) during an action potential or by voltage clamp activates the TT voltage sensor, which in turn opens the SR Ca2+ release channel (red) with or without bound S100A1 (middle; background or foreground, respectively), but the release is less in the absence of S100A1, as shown by the comparison of release in fibers from WT and S100A1−/− mice (above). For simplicity, the likely multi-state sequence for voltage sensor activation [58, 63] is here represented by a single voltage dependent transition, either with or without bound S100A1. The activated RyR1 channels release Ca2+ ions into the myoplasm (the elevation in myoplasmic [Ca2+] is indicated by green bolts), resulting in Ca2+ dependent inactivation of the RyR1 channels by two different binding mechanisms (middle, right; green). First, Ca2+ ions can bind directly to Ca2+ regulatory inhibitory sites on RyR1 and inactivate the channel independently of whether or not S100A1 is bound at the CaM/S100A1 BD (right; background or foreground, respectively). Alternatively, Ca2+ ions can bind to calmodulin, and Ca-CaM can then bind to the CaM/S100A1 BD of RyR1 and also contribute to channel inactivation providing that S100A1 is not occupying that site (right, foreground). The scheme in Fig 4A assumes that inactivation by direct binding of Ca2+ ions to RyR1 is mediated by locally elevated [Ca2+] in the immediate vicinity of open channels, whereas inactivation by CaCaM binding to RyR1 is mediated by global pool cytosolic Ca2+ from both channel types driving Ca2+ binding to CaM followed by Ca-CaM binding to RyR1. Alternative scenarios for effects of global and local Ca2+ may also be possible.
In S100A1−/− fibers (Fig. 4B), there is no S100A1 present so any states in the WT reaction scheme with S100A1 bound (background of Scheme A) cannot be attained and are thus missing from Scheme B. In this case there is only a single resting state of RyR1, and after voltage dependent activation, all channels can undergo inactivation by either Ca2+ binding directly to RyR1 (upper right) or by Ca-CaM binding to the otherwise unoccupied CaM/S100A1 BD (lower right). In the RyR1D/D fibers (Fig. 4C), neither S100A1 nor Ca-CaM can bind to the CaM/S100A1 BD, so there is only a single voltage activated state and a single inactivated state in which Ca2+ binds directly to RyR1.
3.4. Limitations of our interpretations
There are several caveats regarding Fig. 4. First, Fig. 4 represents a state diagram, with no indication of molecular mechanism for the functional properties of any of the indicated states. Second, it is highly simplified. For example, each RyR1 channel is a homo tetramer, with one binding site per monomer or four sites per tetrameric channel, yet we represent binding only to a single site. This works for independent binding, but would require modification for cooperative binding or if the functional effect is not proportional to fractional occupancy. Third, RyR1 is also modulated by binding of a number of other ligands (e.g., ATP, Mg2+, FKBP12, homer protein and others). However, if the contributions of each of these ligands is the same in WT, S100A1−/− and RyR1D/D fibers, then they can simply be considered as constant contributors to the overall effectiveness of voltage dependent channel activation, and thus not require explicit inclusion in the reaction schemes comparing WT, S100A1−/− and RyR1D/D fibers. Finally, the lack of apo-CaM binding to the resting channels was deduced assuming that all effects on SR Ca2+ release seen in S100A1−/− fibers were due solely to elimination of S100A1 binding to the CaM/S100A1 BD, which we concluded from the rationale presented in preceding sections.
4. Regulation of cardiac muscle by CaM and S100A1
Activation of cardiac myocytes involves voltage dependent activation of DHPR voltage sensors and opening of SR RyR Ca2+ release channels, but the isoforms of both molecules are different in cardiac (CaV1.2 or α1c; RyR2) and skeletal (CaV1.1 or α1s; RyR1) muscle. Whereas DHPR-RyR1 communication in skeletal muscle is via direct molecular communication (above), in cardiac myocytes voltage sensor activation causes opening of L-type calcium channels in the sarcolemma and transverse tubules, and the resulting Ca2+ influx triggers SR Ca2+ release via RyR2 Ca2+ release channels, which in cardiac muscle are activated by CICR [64].
Intensive research over the last decade has provided clues about the mechanisms involved in control of cardiac contractility by CaM and S100A1. A multitude of studies have shown that both CaM and S100A1 can interact with dozens of different proteins, enzymes and structural proteins critical for cardiac performance. CaM regulates RyR2, CaV1.2, SERCA2a-phospholamban complex, IP3R2 (type-2 inositol trisphosphate Ca2+ release channel), and phosphorylase kinase (reviewed in : [65, 66]. S100A1 also modulates RyR2, CaV1.2, SERCA2a-phospholamban complex, the mitochondrial ATP synthase, and binds to the myofilament protein titin (reviewed in [47]). These CaM and S100A1 target proteins represent important regulators of cardiac EC coupling, Ca2+ and energy homeostasis. Here, we will focus on the actions of CaM and S100A1 on the RyR2-mediated Ca2+ release process (for reviews regarding molecular, structural, and disease related details of RyR2s and their modulation by CaM, S100A1 and other regulators see: [47, 65, 67–70]).
4.1. CaM modulates cardiac RyR2
When studied using isolated SR vesicles or purified RyRs incorporated into planar lipid bilayers, modulation of RyR2 activity by CaM is unique when compared to modulation of RyR1 and RyR3. At nanomolar free [Ca2+], apo-CaM has an inhibitory effect on RyR2 channel activity, whereas apo-CaM potentiates RyR1 and RyR3 channel activity. At micromolar free [Ca2+], Ca-CaM inhibits all RyR isoforms in isolated preparations [67].
Mutagenesis studies showed that apo-CaM and Ca-CaM inhibit RyR2 via preferential binding to a high-affinity binding domain comprised of amino acid residues 3580–3609 of mouse RyR2 that is conserved among the RyR isoforms (Figure 1; [15]). CaM shifts the Ca2+-dependence of RyR2 activation to higher Ca2+ concentrations and hence decreases RyR2 opening at all Ca2+ concentrations [15, 71]. It has been proposed that CaM promotes RyR2 channel closing following SR Ca2+ release during EC coupling [72]. In support of this hypothesis, neonatal cardiomyocytes isolated from mice engineered with a disrupted RyR2 CaM BD (RyR2-W3586A/L3590D/F3602A) show impaired CaM regulation of RyR2 and have prolonged and abnormal SR Ca2+ release, which leads to cardiac hypertrophy and early death [73]. Our results in skeletal muscle fibers (above) with a disrupted RyR1 CaM/S100A1 BD indicate that Ca-CaM has a less pronounced inhibitory effect on RyR1, which manifests during repetitive AP stimulation (above), whereas Ca-CaM has a stronger inhibitory effect on RyR2 which is evident during a single AP [73]. These differences between RyR1 and RyR2 inhibition by Ca-CaM might arise from different mechanisms (i.e., differential Ca-CaM binding affinities at the CaM BD, variable CaM-CaM BD interaction dwell times, fluctuations of local [CaM], etc.). It is also noteworthy that three point mutations to the CaM BD were required to block CaM inhibition of RyR2 while only a single mutation was needed for RyR1. It is not teleologically surprising that Ca-CaM inhibition may play a more dramatic role in cardiac EC coupling, as the lack of tight voltage sensor control of Ca2+ release in heart cells may allow greater regulation of CICR by modulatory proteins such as CaM. Regardless of the mechanisms at play, these contrasting differences between RyR1 and RyR2 inhibition by CaM demand further research.
4.2. S100A1 effects on cardiac RyR2
S100A1 is predominantly expressed in the heart [38]. In ventricular cardiomyocytes, as in the case of skeletal FDB fibers, S100A1 mostly displays a striated-like pattern and is also localized at both the junctional and longitudinal SR, myofilaments, intercalated discs, and mitochondria [74, 75].
Several studies have identified the functional impact of S100A1 on intracellular Ca2+ homeostasis and have found that S100A1 increases diastolic and systolic performance via augmented SR Ca2+ reuptake and by increasing the gain of CICR, respectively [16, 74, 76–79]. S100A1 reduces [3H]-ryanodine binding to RyR2-enriched SR vesicles at low (nanomolar) [Ca2+] [74, 80], however, at high (micromolar) [Ca2+], S100A1 enhances [3H]-ryanodine binding to RyR2 [74]. Other evidence in support of the inhibitory role of S100A1 on RyR2 at low [Ca2+]i comes from a study showing that S100A1 can reduce Ca2+ spark frequency in quiescent cardiomyocytes [78] and SR leak in cardiac SR vesicle preparations [74, 79], supporting the idea that S100A1 promotes diastolic RyR2 closure during cardiomyocyte relaxation. On the other hand, experiments in voltage-clamped cardiomyocytes and SR vesicle preparations [8, 77, 79, 80] demonstrated that S100A1 increased SR Ca2+ release, suggesting that S100A1 can augment RyR2 opening during systole. Interestingly, the intracellular actions of S100A1 were independent of sarcolemmal Ca2+ fluxes (i.e., S100A1 did not affect Ca2+ entry via L-type Ca2+ channels or via the Na-Ca2+ exchanger [80]. However, other studies have shown that exogenous S100A1 enhances L-type Ca2+ current in both neonatal ventricular myocytes [81] and in neuronal cultures [82], but in contrast, chronic ablation of S100A1 leads to enhanced L-type Ca2+ influx in adult myocytes [83]. These contrasting findings, coupled with the multitude of roles proposed for S100A1 in cardiac EC coupling, has created the need for further experimentation to define the precise contribution of S100A1 to EC coupling in the young, mature, and failing heart.
4.3. Do S100A1 and CaM also compete for binding to the CaM BD of RyR2 in heart cells?
As with many of its other binding partners, S100A1 binding to RyR2 is Ca2+ dependent, and amino acid residues 75–94 within the α-helical S100A1 C-terminal domain are a crucial interface for interaction [41]. Recent collaborative studies of our group revealed that S100A1 binds to and competes with CaM at the CaM/S100A1 BD of RyR1 in skeletal muscle [35, 46]. Given that this region is highly conserved between RyR1 and RyR2 [15] (Fig. 1), we hypothesize that S100A1 and CaM may compete for binding to the CaM BD of RyR2 as they do for RyR1 in skeletal muscle. However, the S100A1 interaction site(s) as well as binding stoichiometries with the macromolecular RyR2 complex remain to be characterized.
5. Do CaM and/or S100A1 bind to more than one site at RyRs?
The preceding sections provide evidence that both CaM and S100A1 are ubiquitous calcium sensors that can fine-tune the Ca2+ sensitivity of RyRs by binding to an overlapping region exposed to the cytoplasm. This modulation is RyR isoform specific and depends on the concentration of free cytosolic Ca2+. The differential effects of CaM and S100A1 binding on RyR activity could also be explained by binding interactions at more than one CaM/S100A1 binding site. While we have presented evidence above suggesting one major, high affinity, overlapping binding site shared by CaM and S100A1, S100A1 ligand overlays on a panel of RyR1 fusion proteins have identified at least three distinct S100A1 binding domains with different binding affinities for the cytoplasmic portion of RyR1 [13]. Recent observations employing isothermal titration calorimetry has determined the thermodynamic parameters of CaM binding to three distinct regions in skeletal (RyR1) and cardiac (RyR2) channels [52]. It is clear that one goal of future research will be to establish the functional consequences of CaM and/or S100A1 binding to these distinct regions in RyR1 and RyR2 channels.
6. What other factors affect CaM or S100A1 binding to RyRs?
Recently, reduced affinity for CaM binding to RyR2 with PKA phosphorylation was found in a catecholaminergic polymorphic ventricular tachycardia (CPVT)-associated mouse model (Arg2474Ser), resulting in spontaneous local Ca2+ release events leading to lethal arrythmias [84]. Modified CaM binding affinity for RyR1 or RyR2 may occur when the RyR channel or CaM itself is modulated by other endogenous regulators (i.e., FKBP12, homer protein, PKA, others). As S100A1’s affinity to bind both Ca2+ and target protein is regulated by diverse processes [33, 34], similar effects are expected for S100A1 binding to both RyR1 and RyR2 upon convergent modulation. This scenario of complex RyR meta-modulation by CaM/S100A1 and other important signaling molecules is largely unexplored and warrants further research, particularly in the context of altered protein expression profiles and activity in heart disease and muscular fatigue.
7. Concluding remarks
Our results are consistent with a simplified model of S100A1 and CaM regulation of RyR1 Ca2+ release in skeletal muscle. Under resting conditions, the CaM/S100A1 binding domain of RyR1 is predominantly regulated by S100A1, which potentiates SR Ca2+ release and force production when the muscle is stimulated (Fig. 3A). As [Ca2+]i becomes elevated during maintained stimulation, Ca-CaM binding becomes more dominant, displacing S100A1 from RyR1 and promoting channel inactivation (Fig. 3B). In this manner CaM and S100A1 compete to fine tune SR Ca2+ release during normal muscle activation. Disrupting this system leads to decreased force production and impaired muscular performance. However, many questions remain. How is this competitive balance affected by other cellular regulatory mechanisms, or during muscular exercise, fatigue and disease? For instance, does muscle activity or muscle chronic paralysis regulate the expression levels of CaM/S100A1? What are the molecular details of CaM/S100A1 interaction with RyR1 and RyR2, and how do these interactions exert their effect on channel activity? Does the differential expression of S100A1 and CaM among fast and slow fiber types confer some specificity to Ca2+ release to meet the varied demands of these muscle types? Clearly, more work is needed to address these important issues.
Acknowledgements
The writing of this review was supported by NIH grant R01AR055099. We are grateful to Drs. Nate Wright, Richard Lovering, Danna Zimmer, David Weber, Werner Melzer and Gerhard Meissner for their participation and insights during the recent collaborative experimental work by our groups in this area, which forms much of the basis for the present review.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Schneider MF, Chandler WK. Voltage-dependent charge movement in skeletal muscle: a possible step in excitation-contraction coupling. Nature. 1973;242:244–247. doi: 10.1038/242244a0. [DOI] [PubMed] [Google Scholar]
- 2.Horowicz P, Schneider MF. Membrane charge moved at contraction theresholds in skeletal muscle fibres. Journal of Physiology. 1981;314:595–633. doi: 10.1113/jphysiol.1981.sp013726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rios E, Brum G. Involvement of dihydropyridine receptors in excitation-contraction coupling in skeletal muscle. Nature. 1987;325:717–720. doi: 10.1038/325717a0. [DOI] [PubMed] [Google Scholar]
- 4.Block BA, Imagawa T, Campbell KP, Franzini-Armstrong C. Structural evidence for direct interaction between the molecular components of the transverse tubule/sarcoplasmic reticulum junction in skeletal muscle. J Cell Biol. 1988;107:2587–2600. doi: 10.1083/jcb.107.6.2587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fahrenbach WH. Sarcoplasmic Reticulum: Ultrastructure of the Triadic Junction. Science. 1965;147:1308–1309. doi: 10.1126/science.147.3663.1308. [DOI] [PubMed] [Google Scholar]
- 6.Cheng H, Lederer WJ. Calcium sparks. Physiol Rev. 2008;88:1491–1545. doi: 10.1152/physrev.00030.2007. [DOI] [PubMed] [Google Scholar]
- 7.Wolf M, Eberhart A, Glossmann H, Striessnig J, Grigorieff N. Visualization of the domain structure of an L-type Ca2+ channel using electron cryo-microscopy. J Mol Biol. 2003;332:171–182. doi: 10.1016/s0022-2836(03)00899-4. [DOI] [PubMed] [Google Scholar]
- 8.Meissner G. Ryanodine receptor/Ca2+ release channels and their regulation by endogenous effectors. Annu Rev Physiol. 1994;56:485–508. doi: 10.1146/annurev.ph.56.030194.002413. [DOI] [PubMed] [Google Scholar]
- 9.Tanabe T, Beam KG, Adams BA, Niidome T, Numa S. Regions of the skeletal muscle dihydropyridine receptor critical for excitation-contraction coupling. Nature. 1990;346:567–569. doi: 10.1038/346567a0. [DOI] [PubMed] [Google Scholar]
- 10.el Hayek R, Antoniu B, Wang J, Hamilton SL, Ikemoto N. Identification of calcium release-triggering and blocking regions of the II–III loop of the skeletal muscle dihydropyridine receptor. Journal of Biological Chemistry. 1995;270:22116–22118. doi: 10.1074/jbc.270.38.22116. [DOI] [PubMed] [Google Scholar]
- 11.Jayaraman T, Brillantes AM, Timerman AP, Fleischer S, Erdjument-Bromage H, Tempst P, Marks AR. FK506 binding protein associated with the calcium release channel (ryanodine receptor) Journal of Biological Chemistry. 1992;267:9474–9477. [PubMed] [Google Scholar]
- 12.Meissner G. Evidence of a role for calmodulin in the regulation of calcium release from skeletal muscle sarcoplasmic reticulum. Biochemistry. 1986;25:244–251. doi: 10.1021/bi00349a034. [DOI] [PubMed] [Google Scholar]
- 13.Treves S, Scutari E, Robert M, Groh S, Ottolia M, Prestipino G, Ronjat M, Zorzato F. Interaction of S100A1 with the Ca 2+ release channel (ryanodine receptor) of skeletal muscle. Biochemistry. 1997;36:11496–11503. doi: 10.1021/bi970160w. [DOI] [PubMed] [Google Scholar]
- 14.Marx SO, Reiken S, Hisamatsu Y, Jayaraman T, Burkhoff D, Rosemblit N, Marks AR. PKA phosphorylation dissociates FKBP12.6 from the calcium release channel (ryanodine receptor): defective regulation in failing hearts. Cell. 2000;101:365–376. doi: 10.1016/s0092-8674(00)80847-8. [DOI] [PubMed] [Google Scholar]
- 15.Yamaguchi N, Xu L, Pasek DA, Evans KE, Meissner G. Molecular basis of calmodulin binding to cardiac muscle Ca(2+) release channel (ryanodine receptor) Journal of Biological Chemistry. 2003;278:23480–23486. doi: 10.1074/jbc.M301125200. [DOI] [PubMed] [Google Scholar]
- 16.Most P, Bernotat J, Ehlermann P, Pleger ST, Reppel M, Borries M, Niroomand F, Pieske B, Janssen PM, Eschenhagen T, Karczewski P, Smith GL, Koch WJ, Katus HA, Remppis A. S100A1: a regulator of myocardial contractility. Proc.Natl.Acad.Sci.U.S.A. 2001;%20(98):13889–13894. doi: 10.1073/pnas.241393598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Maier LS, Bers DM. Calcium, calmodulin, and calcium-calmodulin kinase II: heartbeat to heartbeat and beyond. J.Mol.Cell Cardiol. 2002;34:919–939. doi: 10.1006/jmcc.2002.2038. [DOI] [PubMed] [Google Scholar]
- 18.Wagenknecht T, Berkowitz J, Grassucci R, Timerman AP, Fleischer S. Localization of calmodulin binding sites on the ryanodine receptor from skeletal muscle by electron microscopy. Biophysical Journal. 1994;67:2286–2295. doi: 10.1016/S0006-3495(94)80714-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cornea RL, Nitu F, Gruber S, Kohler K, Satzer M, Thomas DD, Fruen BR. FRET-based mapping of calmodulin bound to the RyR1 Ca2+ release channel. Proc Natl Acad Sci U S A. 2009;106:6128–6133. doi: 10.1073/pnas.0813010106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tripathy A, Xu L, Mann G, Meissner G. Calmodulin activation and inhibition of skeletal muscle Ca 2+ release channel (ryanodine receptor) Biophysical Journal. 1995;69:106–119. doi: 10.1016/S0006-3495(95)79880-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rodney GG, Williams BY, Strasburg GM, Beckingham K, Hamilton SL. Regulation of RYR1 activity by Ca(2+) and calmodulin. Biochemistry. 2000;39:7807–7812. doi: 10.1021/bi0005660. [DOI] [PubMed] [Google Scholar]
- 22.Yamaguchi N, Xin C, Meissner G. Identification of apocalmodulin and Ca2+-calmodulin regulatory domain in skeletal muscle Ca2+ release channel, ryanodine receptor. Journal of Biological Chemistry. 2001;276:22579–22585. doi: 10.1074/jbc.M102729200. [DOI] [PubMed] [Google Scholar]
- 23.Moore CP, Rodney G, Zhang JZ, Santacruz-Toloza L, Strasburg G, Hamilton SL. Apocalmodulin and Ca2+ calmodulin bind to the same region on the skeletal muscle Ca2+ release channel. Biochemistry. 1999;38:8532–8537. doi: 10.1021/bi9907431. [DOI] [PubMed] [Google Scholar]
- 24.Meissner G. Regulation of mammalian ryanodine receptors. Front Biosci. 2002;7(d2072–80):d2072–d2080. doi: 10.2741/A899. [DOI] [PubMed] [Google Scholar]
- 25.Xiong LW, Newman RA, Rodney GG, Thomas O, Zhang JZ, Persechini A, Shea MA, Hamilton SL. Lobe-dependent regulation of ryanodine receptor type 1 by calmodulin. Journal of Biological Chemistry. 2002;277:40862–40870. doi: 10.1074/jbc.M206763200. [DOI] [PubMed] [Google Scholar]
- 26.Zhang H, Zhang JZ, Danila CI, Hamilton SL. A noncontiguous, intersubunit binding site for calmodulin on the skeletal muscle Ca2+ release channel. Journal of Biological Chemistry. 2003;278:8348–8355. doi: 10.1074/jbc.M209565200. [DOI] [PubMed] [Google Scholar]
- 27.Rodney GG. Calmodulin in Adult Mammalian Skeletal Muscle: Localization and Effect on Sarcoplasmic Reticulum Ca2+ Release. Am.J Physiol Cell Physiol. 2008 doi: 10.1152/ajpcell.00033.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.O'Connell KM, Yamaguchi N, Meissner G, Dirksen RT. Calmodulin binding to the 3614–3643 region of RyR1 is not essential for excitation-contraction coupling in skeletal myotubes. J.Gen.Physiol. 2002;120:337–347. doi: 10.1085/jgp.20028617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Moore BW. A soluble protein characteristic of the nervous system. Biochem Biophys Res Commun. 1965;19:739–744. doi: 10.1016/0006-291x(65)90320-7. [DOI] [PubMed] [Google Scholar]
- 30.Zimmer DB, Cornwall EH, Landar A, Song W. The S100 protein family: history, function, and expression. Brain Res Bull. 1995;37:417–429. doi: 10.1016/0361-9230(95)00040-2. [DOI] [PubMed] [Google Scholar]
- 31.Rustandi RR, Baldisseri DM, Inman KG, Nizner P, Hamilton SM, Landar A, Zimmer DB, Weber DJ. Three-dimensional solution structure of the calcium-signaling protein apo-S100A1 as determined by NMR. Biochemistry. 2002;41:788–796. doi: 10.1021/bi0118308. [DOI] [PubMed] [Google Scholar]
- 32.Wright NT, Varney KM, Ellis KC, Markowitz J, Gitti RK, Zimmer DB, Weber DJ. The three-dimensional solution structure of Ca(2+)-bound S100A1 as determined by NMR spectroscopy. J Mol Biol. 2005;353:410–426. doi: 10.1016/j.jmb.2005.08.027. [DOI] [PubMed] [Google Scholar]
- 33.Goch G, Vdovenko S, Kozlowska H, Bierzynski A. Affinity of S100A1 protein for calcium increases dramatically upon glutathionylation. FEBS J. 2005;272:2557–2565. doi: 10.1111/j.1742-4658.2005.04680.x. [DOI] [PubMed] [Google Scholar]
- 34.Baudier J, Mochly-Rosen D, Newton A, Lee SH, Koshland DE, Jr, Cole RD. Comparison of S100b protein with calmodulin: interactions with melittin and microtubule-associated tau proteins and inhibition of phosphorylation of tau proteins by protein kinase C. Biochemistry. 1987;26:2886–2893. doi: 10.1021/bi00384a033. [DOI] [PubMed] [Google Scholar]
- 35.Wright NT, Prosser BL, Varney KM, Zimmer DB, Schneider MF, Weber DJ. S100A1 and calmodulin compete for the same binding site on ryanodine receptor. Journal of Biological Chemistry. 2008;283:26676–26683. doi: 10.1074/jbc.M804432200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Landar A, Rustandi RR, Weber DJ, Zimmer DB. S100A1 utilizes different mechanisms for interacting with calcium-dependent and calcium-independent target proteins. Biochemistry. 1998;37:17429–17438. doi: 10.1021/bi9817921. [DOI] [PubMed] [Google Scholar]
- 37.Inman KG, Yang R, Rustandi RR, Miller KE, Baldisseri DM, Weber DJ. Solution NMR structure of S100B bound to the high-affinity target peptide TRTK-12. J Mol Biol. 2002;324:1003–1014. doi: 10.1016/s0022-2836(02)01152-x. [DOI] [PubMed] [Google Scholar]
- 38.Kato K, Kimura S. S100ao (alpha alpha) protein is mainly located in the heart and striated muscles. Biochim Biophys Acta. 1985;842:146–150. doi: 10.1016/0304-4165(85)90196-5. [DOI] [PubMed] [Google Scholar]
- 39.Zimmer DB, Landar A. Analysis of S100A1 expression during skeletal muscle and neuronal cell differentiation. J Neurochem. 1995;64:2727–2736. doi: 10.1046/j.1471-4159.1995.64062727.x. [DOI] [PubMed] [Google Scholar]
- 40.Haimoto H, Kato K. S100a0 (alpha alpha) protein, a calcium-binding protein, is localized in the slow-twitch muscle fiber. J Neurochem. 1987;48:917–923. doi: 10.1111/j.1471-4159.1987.tb05604.x. [DOI] [PubMed] [Google Scholar]
- 41.Most P, Remppis A, Weber C, Bernotat J, Ehlermann P, Pleger ST, Kirsch W, Weber M, Uttenweiler D, Smith GL, Katus HA, Fink RH. The C terminus (amino acids 75–94) and the linker region (amino acids 42–54) of the Ca2+-binding protein S100A1 differentially enhance sarcoplasmic Ca2+ release in murine skinned skeletal muscle fibers. Journal of Biological Chemistry. 2003;278:26356–26364. doi: 10.1074/jbc.M303338200. [DOI] [PubMed] [Google Scholar]
- 42.Zimmer DB, Song W, Zimmer WE. Isolation of a rat S100 alpha cDNA and distribution of its mRNA in rat tissues. Brain Res Bull. 1991;27:157–162. doi: 10.1016/0361-9230(91)90061-n. [DOI] [PubMed] [Google Scholar]
- 43.Adhikari BB, Wang K. S100A1 modulates skeletal muscle contraction by desensitizing calcium activation of isometric tension, stiffness and ATPase. FEBS Lett. 2001;497:95–98. doi: 10.1016/s0014-5793(01)02444-9. [DOI] [PubMed] [Google Scholar]
- 44.Maco B, Brezova A, Schafer BW, Uhrik B, Heizmann CW. Localization of the Ca(2+)-binding S100A1 protein in slow and fast skeletal muscles of the rat. Gen Physiol Biophys. 1997;16:373–377. [PubMed] [Google Scholar]
- 45.Arcuri C, Giambanco I, Bianchi R, Donato R. Annexin V, annexin VI, S100A1 and S100B in developing and adult avian skeletal muscles. Neuroscience. 2002;109:371–388. doi: 10.1016/s0306-4522(01)00330-x. [DOI] [PubMed] [Google Scholar]
- 46.Prosser BL, Wright NT, Hernandez-Ochoa EO, Varney KM, Liu Y, Olojo RO, Zimmer DB, Weber DJ, Schneider MF. S100A1 binds to the calmodulin-binding site of ryanodine receptor and modulates skeletal muscle excitation-contraction coupling. Journal of Biological Chemistry. 2008;283:5046–5057. doi: 10.1074/jbc.M709231200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Most P, Remppis A, Pleger ST, Katus HA, Koch WJ. S100A1: a novel inotropic regulator of cardiac performance. Transition from molecular physiology to pathophysiological relevance. Am.J.Physiol Regul.Integr.Comp Physiol. 2007;293:R568–R577. doi: 10.1152/ajpregu.00075.2007. [DOI] [PubMed] [Google Scholar]
- 48.Fano G, Marsili V, Angelella P, Aisa MC, Giambanco I, Donato R. S-100a0 protein stimulates Ca2+-induced Ca2+ release from isolated sarcoplasmic reticulum vesicles. FEBS Lett. 1989;255:381–384. doi: 10.1016/0014-5793(89)81127-5. [DOI] [PubMed] [Google Scholar]
- 49.Rhoads AR, Friedberg F. Sequence motifs for calmodulin recognition. FASEB J. 1997;11:331–340. doi: 10.1096/fasebj.11.5.9141499. [DOI] [PubMed] [Google Scholar]
- 50.Wilder PT, Lin J, Bair CL, Charpentier TH, Yang D, Liriano M, Varney KM, Lee A, Oppenheim AB, Adhya S, Carrier F, Weber DJ. Recognition of the tumor suppressor protein p53 and other protein targets by the calcium-binding protein S100B. Biochim Biophys Acta. 2006;1763:1284–1297. doi: 10.1016/j.bbamcr.2006.08.024. [DOI] [PubMed] [Google Scholar]
- 51.Maximciuc AA, Putkey JA, Shamoo Y, Mackenzie KR. Complex of calmodulin with a ryanodine receptor target reveals a novel, flexible binding mode. Structure. 2006;14:1547–1556. doi: 10.1016/j.str.2006.08.011. [DOI] [PubMed] [Google Scholar]
- 52.Lau K, Chan MY, van Petegem F. Studies in the Binding of Calmodulin to Skeletal and Cardiac Ryanodine Receptors. Biophys J. 2011;100:188a. [Google Scholar]
- 53.Yamaguchi N, Prosser BL, Ghassemi F, Xu L, Pasek DA, Eu JP, Hernandez-Ochoa EO, Cannon BR, Wilder PT, Lovering RM, Weber D, Melzer W, Schneider MF, Meissner G. Modulation of sarcoplasmic reticulum Ca2+ release in skeletal muscle expressing ryanodine receptor impaired in regulation by calmodulin and S100A1. Am J Physiol Cell Physiol. 2011;300:C998–C1012. doi: 10.1152/ajpcell.00370.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Liu YW, Carroll SL, Klein MG, Schneider MF. Calcium transients and calcium homeostasis in adult mouse fast- twitch skeletal muscle fibers in culture. American Journal of Physiology: Cell Physiology. 1997;272:C1919–C1927. doi: 10.1152/ajpcell.1997.272.6.C1919. [DOI] [PubMed] [Google Scholar]
- 55.Prosser BL, Hernandez-Ochoa EO, Lovering RM, Andronache Z, Zimmer DB, Melzer W, Schneider MF. S100A1 promotes action potential-initiated calcium release flux and force production in skeletal muscle. Am J Physiol Cell Physiol. 2010;299:C891–C902. doi: 10.1152/ajpcell.00180.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Prosser BL, Hernandez-Ochoa EO, Zimmer DB, Schneider MF. Simultaneous recording of intramembrane charge movement components and calcium release in wild-type and S100A1−/− muscle fibres. J.Physiol. 2009;587:4543–4559. doi: 10.1113/jphysiol.2009.177246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Prosser BL, Hernandez-Ochoa EO, Zimmer DB, Schneider MF. The Qgamma component of intra-membrane charge movement is present in mammalian muscle fibres, but suppressed in the absence of S100A1. J.Physiol. 2009;587:4523–4541. doi: 10.1113/jphysiol.2009.177238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Melzer W, Rios E, Schneider MF. A general procedure for determining the rate of calcium release from the sarcoplasmic reticulum in skeletal muscle fibers. Biophysical Journal. 1987;51:849–863. doi: 10.1016/S0006-3495(87)83413-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Hennig R, Lomo T. Firing patterns of motor units in normal rats. Nature. 1985;314:164–166. doi: 10.1038/314164a0. [DOI] [PubMed] [Google Scholar]
- 60.Zhou J, Yi J, Royer L, Launikonis BS, Gonzalez AA, Garcia J, Rios E. A probable role of dihydropyridine receptors in repression of Ca2+ sparks, demonstrated in cultured mammalian muscle. Am.J.Physiol Cell Physiol. 2005 doi: 10.1152/ajpcell.00592.2004. [DOI] [PubMed] [Google Scholar]
- 61.Brown LD, Rodney GG, Hernandez-Ochoa E, Ward CW, Schneider MF. Ca2+ sparks and T tubule reorganization in dedifferentiating adult mouse skeletal muscle fibers. Am.J.Physiol Cell Physiol. 2007;292:C1156–C1166. doi: 10.1152/ajpcell.00397.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Chun LG, Ward CW, Schneider MF. Ca2+ sparks are initiated by Ca2+ entry in embryonic mouse skeletal muscle and decrease in frequency postnatally. Am J Physiol Cell Physiol. 2003;285:C686–C697. doi: 10.1152/ajpcell.00072.2003. [DOI] [PubMed] [Google Scholar]
- 63.Horowicz P, Schneider MF. Membrane charge movement in contracting and non-contracting skeletal muscle fibres. J Physiol. 1981;314:565–593. doi: 10.1113/jphysiol.1981.sp013725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Bers DM. Cardiac excitation-contraction coupling. Nature. 2002;415:198–205. doi: 10.1038/415198a. [DOI] [PubMed] [Google Scholar]
- 65.Tang W, Sencer S, Hamilton SL. Calmodulin modulation of proteins involved in excitation-contraction coupling. Front Biosci. 2002;7:d1583–d1589. doi: 10.2741/tang. [DOI] [PubMed] [Google Scholar]
- 66.Schaub MC, Hefti MA, Zaugg M. Integration of calcium with the signaling network in cardiac myocytes. J Mol Cell Cardiol. 2006;41:183–214. doi: 10.1016/j.yjmcc.2006.04.005. [DOI] [PubMed] [Google Scholar]
- 67.Meissner G. Molecular regulation of cardiac ryanodine receptor ion channel. Cell Calcium. 2004;35:621–628. doi: 10.1016/j.ceca.2004.01.015. [DOI] [PubMed] [Google Scholar]
- 68.Kushnir A, Marks AR. The ryanodine receptor in cardiac physiology and disease. Adv Pharmacol. 2010;59:1–30. doi: 10.1016/S1054-3589(10)59001-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Kraus C, Rohde D, Weidenhammer C, Qiu G, Pleger ST, Voelkers M, Boerries M, Remppis A, Katus HA, Most P. S100A1 in cardiovascular health and disease: closing the gap between basic science and clinical therapy. J Mol Cell Cardiol. 2009;47:445–455. doi: 10.1016/j.yjmcc.2009.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Lanner JT, Georgiou DK, Joshi AD, Hamilton SL. Ryanodine receptors: structure, expression, molecular details, and function in calcium release. Cold Spring Harb Perspect Biol. 2010;2 doi: 10.1101/cshperspect.a003996. a003996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Balshaw DM, Xu L, Yamaguchi N, Pasek DA, Meissner G. Calmodulin binding and inhibition of cardiac muscle calcium release channel (ryanodine receptor) Journal of Biological Chemistry. 2001;276:20144–20153. doi: 10.1074/jbc.M010771200. [DOI] [PubMed] [Google Scholar]
- 72.Xu L, Meissner G. Mechanism of calmodulin inhibition of cardiac sarcoplasmic reticulum Ca2+ release channel (ryanodine receptor) Biophysical Journal. 2004;86:797–804. doi: 10.1016/S0006-3495(04)74155-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Yamaguchi N, Takahashi N, Xu L, Smithies O, Meissner G. Early cardiac hypertrophy in mice with impaired calmodulin regulation of cardiac muscle Ca release channel. J.Clin.Invest. 2007;117:1344–1353. doi: 10.1172/JCI29515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Most P, Pleger ST, Volkers M, Heidt B, Boerries M, Weichenhan D, Loffler E, Janssen PM, Eckhart AD, Martini J, Williams ML, Katus HA, Remppis A, Koch WJ. Cardiac adenoviral S100A1 gene delivery rescues failing myocardium. J.Clin.Invest. 2004;114:1550–1563. doi: 10.1172/JCI21454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Brezova A, Heizmann CW, Uhrik B. Immunocytochemical localization of S100A1 in mitochondria on cryosections of the rat heart. Gen Physiol Biophys. 2007;26:143–149. [PubMed] [Google Scholar]
- 76.Most P, Boerries M, Eicher C, Schweda C, Volkers M, Wedel T, Sollner S, Katus HA, Remppis A, Aebi U, Koch WJ, Schoenenberger CA. Distinct subcellular location of the Ca2+-binding protein S100A1 differentially modulates Ca2+-cycling in ventricular rat cardiomyocytes. J.Cell Sci. 2005;118:421–431. doi: 10.1242/jcs.01614. [DOI] [PubMed] [Google Scholar]
- 77.Most P, Remppis A, Pleger ST, Loffler E, Ehlermann P, Bernotat J, Kleuss C, Heierhorst J, Ruiz P, Witt H, Karczewski P, Mao L, Rockman HA, Duncan SJ, Katus HA, Koch WJ. Transgenic overexpression of the Ca2+-binding protein S100A1 in the heart leads to increased in vivo myocardial contractile performance. Journal of Biological Chemistry. 2003;278:33809–33817. doi: 10.1074/jbc.M301788200. [DOI] [PubMed] [Google Scholar]
- 78.Volkers M, Loughrey CM, Macquaide N, Remppis A, DeGeorge BR, Jr, Wegner FV, Friedrich O, Fink RH, Koch WJ, Smith GL, Most P. S100A1 decreases calcium spark frequency and alters their spatial characteristics in permeabilized adult ventricular cardiomyocytes. Cell Calcium. 2007;41:135–143. doi: 10.1016/j.ceca.2006.06.001. [DOI] [PubMed] [Google Scholar]
- 79.Most P, Seifert H, Gao E, Funakoshi H, Volkers M, Heierhorst J, Remppis A, Pleger ST, DeGeorge BR, Jr, Eckhart AD, Feldman AM, Koch WJ. Cardiac S100A1 protein levels determine contractile performance and propensity toward heart failure after myocardial infarction. Circulation. 2006;114:1258–1268. doi: 10.1161/CIRCULATIONAHA.106.622415. [DOI] [PubMed] [Google Scholar]
- 80.Kettlewell S, Most P, Currie S, Koch WJ, Smith GL. S100A1 increases the gain of excitation-contraction coupling in isolated rabbit ventricular cardiomyocytes. J Mol Cell Cardiol. 2005;39:900–910. doi: 10.1016/j.yjmcc.2005.06.018. [DOI] [PubMed] [Google Scholar]
- 81.Reppel M, Sasse P, Piekorz R, Tang M, Roell W, Duan Y, Kletke A, Hescheler J, Nurnberg B, Fleischmann BK. S100A1 enhances the L-type Ca2+ current in embryonic mouse and neonatal rat ventricular cardiomyocytes. Journal of Biological Chemistry. 2005;280:36019–36028. doi: 10.1074/jbc.M504750200. [DOI] [PubMed] [Google Scholar]
- 82.Hernandez-Ochoa EO, Prosser BL, Wright NT, Contreras M, Weber DJ, Schneider MF. Augmentation of Cav1 channel current and action potential duration after uptake of S100A1 in sympathetic ganglion neurons. Am.J.Physiol Cell Physiol. 2009;297:C955–C970. doi: 10.1152/ajpcell.00140.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Gusev K, Ackermann GE, Heizmann CW, Niggli E. Ca2+ signaling in mouse cardiomyocytes with ablated S100A1 protein. Gen Physiol Biophys. 2009;28:371–383. doi: 10.4149/gpb_2009_04_371. [DOI] [PubMed] [Google Scholar]
- 84.Xu X, Yano M, Uchinoumi H, Hino A, Suetomi T, Ono M, Tateishi H, Oda T, Okuda S, Doi M, Kobayashi S, Yamamoto T, Ikeda Y, Ikemoto N, Matsuzaki M. Defective calmodulin binding to the cardiac ryanodine receptor plays a key role in CPVT-associated channel dysfunction. Biochem Biophys Res Commun. 2010;394:660–666. doi: 10.1016/j.bbrc.2010.03.046. [DOI] [PMC free article] [PubMed] [Google Scholar]