Abstract
The abundance of genome polymorphism and divergence data has provided unprecedented insight into how mutation, drift and natural selection shape genome evolution. Application of the McDonald-Kreitman test to such data indicates a pervasive influence of positive selection, particularly in Drosophila species. However, evidence for positive selection in other species ranging from yeast to humans is often weak or absent. While evidence for positive selection may be obscured in some species, there is also reason to believe that the frequency of adaptive substitutions may be overestimated due to epistatic fitness effects or hitchhiking of deleterious mutations. Based on these considerations I argue that the common assumption of independence among sites must be relaxed before abandoning the neutral theory of molecular evolution.
Keywords: neutral theory, positive selection, McDonald-Kreitman test, epistasis, hitchhiking
Estimating the frequency of positive selection using the McDonald-Kreitman test
The extent to which molecular evolution is driven by positive selection has long been debated. The neutral theory holds that the vast majority of DNA sequence differences between species are neutral [1] or nearly neutral [2] with respect to fitness. However, models that assume natural selection plays a dominant role in driving molecular evolution can also explain many features of DNA polymorphism and divergence data [3]. As a consequence, great efforts have been made to identify patterns of variation that distinguish neutral and selective models [1, 3–5]. Because of these efforts, many examples of genes evolving under positive selection have been documented. However, genome-wide estimates of the fraction of substitutions driven by positive selection are required to test the neutral theory of molecular evolution.
One test for positive selection that has been extensively applied to genome-wide polymorphism and divergence data is the McDonald-Kreitman (MK) test [6]. The MK test and related methods can be used to estimate the fraction of substitutions driven by positive selection by comparing observed rates of divergence to those expected based on polymorphism data (Box 1). Application of the MK test to a number of species has provided evidence that positive selection has driven a large portion of interspecific differences, bringing the neutral theory into question [7–11]. However, other species show little or no evidence that positive selection has driven a large portion of interspecific differences. The disparate results of the MK test necessitate a careful consideration of what factors can explain these results, and particularly whether estimates of the rate of adaptive evolution are robust to the assumptions of the test. Here, I discuss one of the assumptions of the MK test that has received relatively little attention: sites evolve independently of one another. Given both theoretical and empirical evidence that this assumption is not valid, I argue that estimates of the frequency of adaptive substitution are too unreliable to reject the neutral theory of molecular evolution.
Box 1. The McDonald-Kreitman test.
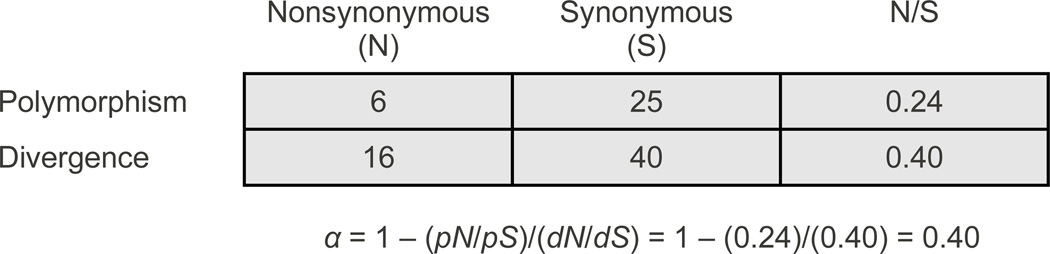
The McDonald-Kreitman (MK) test compares rates of DNA polymorphism within species to divergence between species [6]. If all nonsynonymous and synonymous variation is neutral, the ratio of the nonsynonymous to synonymous substitution rate between species (dN/dS) is expected to be equal to the ratio of nonsynonymous to synonymous polymorphism within species (pN/pS). However, positive selection can increase dN/dS above pN/pS since adaptive mutations spread quickly through a population but have a cumulative effect on divergence. In its original formulation, the MK test determines whether the ratio of nonsynonymous to synonymous fixed differences is significantly different from that of polymorphic sites in a single gene. Subsequent studies used the combined data from multiple genes across the genome to test whether dN/dS is significantly greater than pN/pS and estimate the fraction of nonsynonymous substitutions driven by positive selection, α, from the degree to which dN/dS is elevated over pN/pS, or more specifically: α = 1 - (pN/pS)/(dN/dS) [74, 75]. An example illustrating how α is estimated is shown in Figure I. A similar logic can be used to estimate the fraction of noncoding substitutions driven by positive selection [76].
A significant obstacle in estimating the frequency of positive selection is accounting for deleterious mutations. Deleterious polymorphism can increase pN/pS above dN/dS, even in the presence of positive selection [77]. The effect of deleterious polymorphism on pN/pS can be enhanced by a recent reduction in population size [78]. A number of methods have been developed to account for segregating deleterious polymorphism in order to more accurately estimate the frequency of positive selection [16, 19, 22, 65, 75]. However, deleterious mutations can also become fixed during historic periods of reduced population size, leading to an increase in dN/dS and spurious signals of positive selection [40, 75].
Evidence for positive selection based on the McDonald-Kreitman test
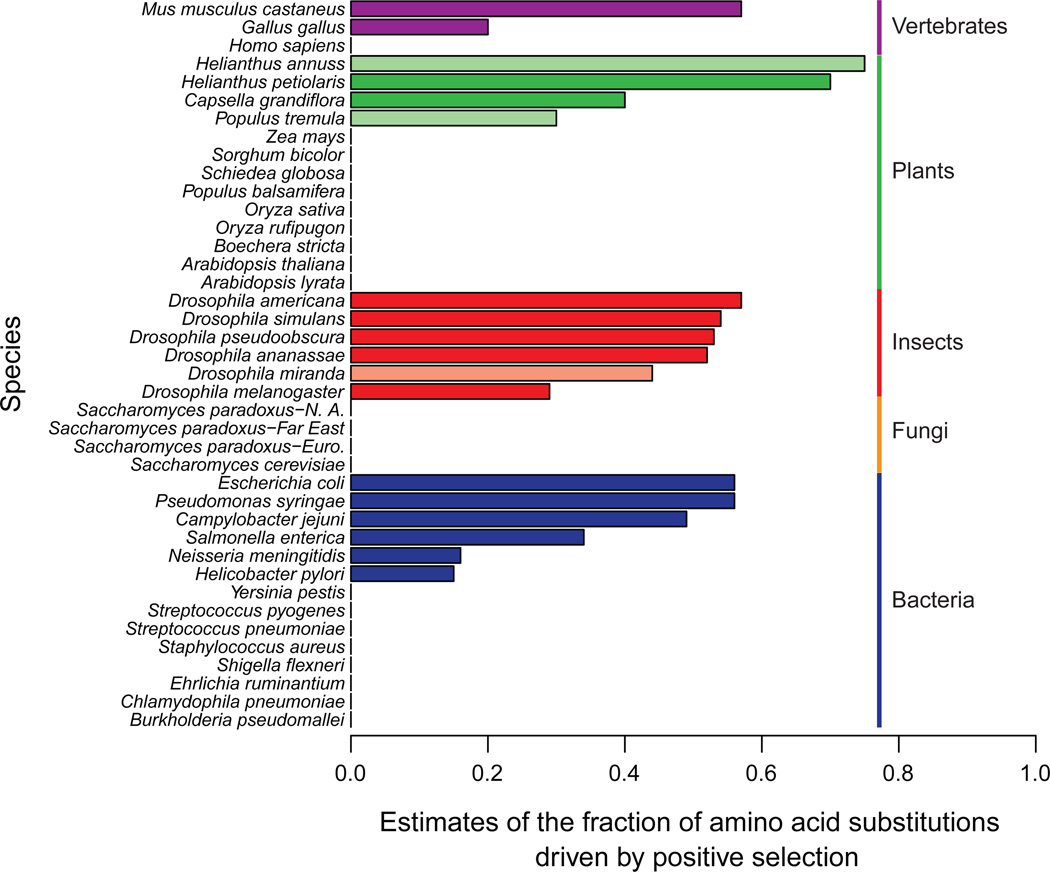
Out of 38 species for which polymorphism has been compared to divergence across multiple genes, 47% show evidence that positive selection has significantly increased the nonsynonymous substitution rate based on the McDonald-Kreitman (MK) test or related tests (Table 1 and Table S1). For those species that do show significant evidence of positive selection, the estimated fraction of nonsynonymous substitutions driven by positive selection is quite high, often around 40% (Figure 1).
Table 1.
Reported evidence of positive selection based on the McDonald-Kreitman test.
| Group | Species | Evidence of positive selectiona (%) |
References |
|---|---|---|---|
| Fungi | 2 | 0 (0%) | [33] |
| Plantsb | 13 | 4 (31%) | [25–27, 30, 79] |
| Bacteria | 14 | 6 (43%) | [23, 24] |
| Vertebrates | 3 | 2 (67%) | [17, 18, 22] |
| Insectsb | 6 | 6 (100%) | [13, 15, 37, 80–82] |
| All | 38 | 18 (47%) |
Evidence for positive selection is based on a significant excess of nonsynonymous divergence based on the MK test.
There are both positive and negative reports of positive selection for some species.
Figure 1.
Estimates of the fraction of nonsynonymous substitutions driven by positive selection (α) grouped by taxa. Estimates of α are from published reports (Table S1) using a variety of different methods based on the MK test. Estimates of α are only shown for species with significant MK test results with the exception of 12 bacterial species [24] for which α was not estimated in the original publication and for which the equation in Box 1 was used. Species with conflicting reports of positive selection are shown in lighter shades. Separate results are shown for subpopulations of Saccharomyces paradoxus since they are monophyletic and show evidence of reproductive isolation [83]. Together, these results indicate that there is substantial variation in evidence for positive selection based on the MK test.
Evidence for positive selection varies among taxonomic groups. The strongest evidence comes from Drosophila, where all six species that have been examined show evidence of positive selection. One of the six species, Drosophila miranda, has conflicting reports of positive selection, which may be a consequence of low levels of polymorphism or a dependency on which genes were studied [12–16]. In vertebrates, plants, bacteria and fungi, some species show strong evidence of positive selection and others show weak or no evidence. In vertebrates, there is evidence of positive selection in chickens [17] and mice [18], but evidence in humans is weak or absent [19–22]. The lack of evidence in humans could be a consequence of an obscured signal due to a recent reduction in effective population size [22](Box 1). In bacteria, Escherichia coli and Salmonella enterica show strong evidence of positive selection [23]. Yet, the ratio of the nonsynonymous to synonymous substitution rate (dN/dS) is greater than that of the rate of polymorphism (pN/pS) for only four out of twelve other bacterial species [24]. Note that the authors of the latter study interpreted their results as more likely a consequence of purifying selection. In plants, there is evidence of positive selection in four out of thirteen species. Interestingly, the species showing evidence of positive selection are also those estimated to have the largest effective population size [25–27], a trend that is also reflected in other taxa [8]. Under some but not all models of molecular evolution [28], species with large effective population sizes are expected to have higher rates of adaptive substitution and signals of positive selection are less likely to be obscured by deleterious polymorphism [29]. A valuable feature of the plant research is that evidence of positive selection does not appear to be related to the mating system since both the predominantly selfing species, Arabidopsis thaliana, and its outcrossing relative, Arabidopsis lyrata, show no evidence of positive selection [30]. Finally, two yeast species show no evidence of positive selection [31–33].
The neutralist and selectionist interpretations
As is often the case, there are both neutral and selective interpretations of the mixed evidence for positive selection based on the MK test. The selectionist view is that positive selection is indeed pervasive, at least in some species, and that the absence of evidence in other species is a consequence of an obscured signal of selection or a lower rate of adaptive evolution. In humans and multiple plant species, the absence of a strong signal of selection could be a consequence of their small effective population size [7, 8, 10, 11, 18, 22, 25, 26, 29]. Yet, the absence of evidence in other species, particularly bacteria and yeast species, is not easy to explain since they have large effective population sizes [34, 35]. One potential explanation for yeast and bacteria species is that they may have a greater propensity for local adaptation, in which case pN/pS may be inflated above dN/dS even when positive selection has had a significant impact on dN/dS [8].
The neutralist view is that positive selection is rare and that factors other than positive selection generate significant MK test results. A reduction in population size can lead to fixation of deleterious mutations and spurious evidence of positive selection (Box 1). While changes in population size may be responsible for spurious signals of positive selection in some species, it seems unlikely that all of the Drosophila species experienced a demographic history that would incur a similar false signal of selection. Positive selection may also be overestimated due to statistical biases related to the assumption that synonymous and nonsynonymous sites share the same genealogical history [36] and the summation over sparse contingency tables [37, 38]. However, neither of these statistical considerations appear to account for the Drosophila data. Thus, positive selection is often [7–11] but not always [24, 39] the preferred interpretation for significant MK test results.
In weighing the evidence for positive selection, it is important to consider the many assumptions of the MK test [6, 40]. One assumption which has received much less attention than others is that sites evolve independently of one another. While non-independence among sites may not always generate false signals of selection, there are two scenarios discussed below where non-independence may lead to overestimates of the frequency of positive selection. First, epistasis can result in changes in selective constraint. Second, positive selection can increase the substitution rate of linked deleterious mutations through hitchhiking.
Epistasis can result in changes in selective constraint
The MK test assumes that the selective constraints on a sequence remain constant over time. The selective constraint on a site is determined by the degree to which fitness is reduced when mutated and can be measured by the reduction in the substitution rate relative to a neutral site, e.g. by dN relative to dS. In the absence of any fitness effect, a site is unconstrained and will evolve neutrally. Changes in effective population size influence selective constraints since populations have trouble removing deleterious mutations with small fitness effects [2, 40]. However, the selective constraint on a site may also depend on epistatic interactions with other sites in the genome.
Epistasis is relevant to the evolution of a site whenever the fitness effect of a mutation depends on other genetic or environmental variation. In the case of genetic dependencies, epistasis is defined by fitness effects at one site that depend on the genotype present at other sites. Epistatic fitness effects can result from non-additive relationships between genotype and phenotype, or phenotype and fitness. Epistatic fitness effects may be common as they are expected to occur as a general consequence of any non-linear phenotype-fitness function, even under an additive genetic model [41] (Box 2). A number of models of molecular evolution include epistatic fitness effects in order to account for the possibility that selective constraints are not static but can change over time [42, 43]. While models that incorporate epistasis have been investigated in the context of substitution rate heterogeneity [44], their predictions in the context of the MK test are at present unknown. In addition to changes in selective constraint, epistatic interactions between a mutation and its genetic background can also result in a mutation having positive effects in some individuals but negative effects in others (Box 2). This complicates the classification of evolutionary models with epistasis into the neutral regime since it seems unlikely that mutations with epistatic fitness effects would only cause changes in selective constraint without also being able to cause previously deleterious or neutral mutations to become advantageous. Interestingly, rapid temporal fluctuations between positive and negative selection can result in significant MK test results since the dynamics of sites under fluctuating selection are determined by their effects on the variance in fitness rather than the mean, which is assumed to be zero [45, 46].
Box 2. Changes in selective constraint generated by epistasis.
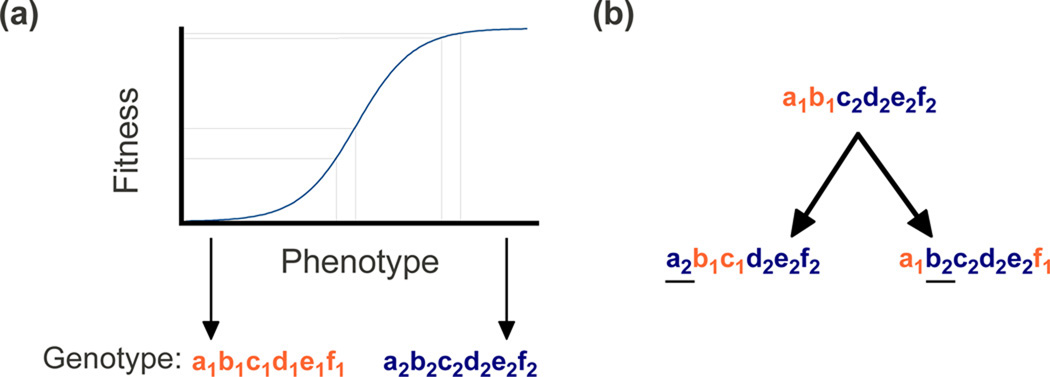
Given a non-linear phenotype-fitness function, an allele can have a positive, negative or no fitness effect depending on its genetic background. Figure I shows a simple example of a phenotype-fitness function where a mutation that causes a slight increase in an individual's phenotype (e.g. height) is expected to increase fitness within individuals who have intermediate phenotypes but to have little or no effect on fitness within individuals who have extreme phenotypes. In this context, maximizing population fitness results in a loss of constraint and evolution of neutrality [42]. Note that if the phenotype-fitness function is bell-shaped a mutation can have positive or negative effects depending on the phenotype of the individual in which it occurs. In models that allow for epistatic fitness effects [68], species can become fixed for alternate genotypes without a change in fitness, resulting in divergence between species at sites under negative selection within species. Figure I shows an example where four out of six sites have diverged between species despite the presence of purifying selection on four of the six sites within species.
Empirical evidence of temporal changes in selective constraint
There is an abundance of evidence that the effects of a mutation depend on its genetic background. Empirical evidence of epistasis is frequently encountered in quantitative trait mapping [47], experimental evolution [48, 49], reconstruction of substitutions that have occurred during evolution [50, 51], and patterns of molecular evolution [52, 53]. In many cases, large epistatic effects can be inferred from the strongly deleterious or even lethal effects of a mutation in one genetic background relative to the absence of any noticeable effects in another background [54, 55]. However, the mere presence of epistasis does not imply that the frequency of positive selection has been overestimated by the MK test.
In order for epistasis to cause spurious signals of positive selection dN/dS must be consistently greater than pN/pS across the genome, such that a large number of sites are constrained within a species but were less constrained or unconstrained in the past. Yet, without an explicit model for how epistasis generates changes in constraint over time, it is hard to know whether there are also a large number of neutral sites within a species that were constrained during divergence between species.
Two observations suggest that selective constraint may systematically change over time. First, duplicated genes exhibit low levels of constraint immediately after gene duplication followed by increased levels of constraint over time, as measured by a decrease in dN/dS over time [56]. Based on this empirical pattern, pN/pS may be consistently lower than dN/dS for duplicated genes since the high rate of divergence following gene duplication is transient and so would contribute to divergence between species but not polymorphism within species. This pattern of temporal changes in selective constraint may occur as a consequence of subfunctionalization of duplicate genes [57], but may also occur as a consequence of stabilizing selection on other types of redundant genetic elements such as transcription factor binding sites within enhancers [58].
A second observation relevant to temporal changes in selective constraint is that in both bacteria and vertebrates, dN/dS of all genes decreases as a function of the time-period over which it is measured [59, 60]. Explanations for this pattern include fixation of deleterious polymorphism in ancestral populations [61], and statistical biases of the methods used to estimate dN/dS [60]. Regardless of the cause, the pattern implies that dN/dS or estimates of dN/dS are higher for closely related species compared to distantly related species. Interestingly, there is no evidence of positive selection for many of the species with high rates of divergence to their outgroup, e.g. bacterial and yeast species (Table S1). Yet, without knowing why dN/dS changes over time, it is hard to know how pN/pS would be affected relative to dN/dS.
In summary, even though epistasis can result in changes in selective constraint, its impact on the MK test has yet to be determined. Gene duplication provides a concrete example of how epistatic fitness effects can result in changes in constraint over time. Although the genome-wide changes in dN/dS may not be related to epistasis [60], they suggests that the elevation of dN/dS over pN/pS may be an unreliable estimator of the frequency of positive selection.
Hitchhiking of deleterious mutations
The MK test assumes that any increase in dN/dS above pN/pS is due to positive selection. While weakly deleterious mutations can become polymorphic in a population and reach fixation, they are expected to increase pN/pS more than dN/dS [40, 62], assuming selective constraints have not changed and that there are no effects of selection at linked sites. However, when the effects of a deleterious mutation do not outweigh the effects of an advantageous mutation, deleterious mutations can be rapidly fixed due to hitchhiking with a linked advantageous mutation. As a consequence, positive selection can result in a dramatic increase in the deleterious substitution rate [63, 64]. Although hitchhiking of deleterious mutations requires the presence of positive selection, a single strong hitchhiking event could result in the fixation of multiple slightly deleterious mutations. Without accounting for this possibility, it is hard to know the degree to which hitchhiking of deleterious mutations causes overestimation of the frequency of positive selection based on the MK test.
Weighing evidence for adaptation based on the McDonald-Kreitman test
Application of the MK test to DNA polymorphism and divergence data has revealed two features regarding evidence of positive selection: heterogeneity across species [7, 8, 10] and uniformity among genes within a species [23, 65, 66]. Thus far, neither neutral nor selective models have provided compelling explanations for these observations. While deleterious polymorphism may obscure evidence of positive selection in species with small population sizes, this cannot explain the absence of evidence for selection in species with large effective population sizes. Furthermore, it seems unlikely that positive selection would have a uniform impact across genes, unless positive selection is driven by the continual fixation of deleterious mutations [67]. In comparison, neutral models that invoke population bottlenecks may explain results from some species but don't provide a compelling explanation for the Drosophila data. Given the vexing patterns of heterogeneous MK test results, I argue that other factors known to affect the MK test must be considered when estimating the fraction of substitutions driven by positive selection.
One of the factors that may have a considerable influence on the MK test is the assumption that sites evolve independently of one another. Given both theoretical considerations and empirical support that sites do not evolve independently of one another [42, 43, 43, 44, 47–55, 57, 58, 63, 64, 68], I suggest that estimates of positive selection based on the MK test are unreliable. Epistatic fitness effects is one mechanism that can lead to non-independence among sites and can result in changes in selective constraint over time. Any case where a site is constrained within a species but was free to diverge between species will produce spurious evidence of positive selection based on the MK test. Purifying selection on redundant genetic elements, such as duplicated genes or transcription factor binding sites, provides a potential but as yet unexplored explanation for positive MK test results. Furthermore, the observation that constraint or measurement of constraint consistently changes as a function of the time interval over which it is measured [59, 60] raises questions over the reliability of using the elevation of dN/dS over pN/pS to estimate the frequency of positive selection. In addition to epistasis, hitchhiking also causes non-independence among sites. Hitchhiking may lead to overestimation of the frequency of positive selection due to the fixation of weakly deleterious mutations linked to a strongly advantageous mutation. While fixation of deleterious mutations creates the opportunity for further adaptive substitutions, it makes it difficult to reliably estimate the frequency of positive selection based on the MK test.
Despite the concerns regarding the interpretation of the MK test results, many other patterns of molecular evolution are relevant to understanding how positive selection shapes genome evolution and they often point to a pervasive influence of positive selection [7, 9, 10]. The reduced levels of synonymous polymorphism surrounding recently fixed nonsynonymous substitutions is just one pattern that supports a dominant influence of positive selection [13, 27, 69–72]. While this pattern is also expected to occur due to background selection, it is best explained by positive selection due to a number of patterns that are not consistent with a background selection model [13, 27, 69–72]. However, disentangling the effects of positive and negative selection, especially in the context of hitchhiking of deleterious mutations [71], makes it difficult to estimate the fraction of substitutions driven by positive selection.
Concluding remarks
Even if non-independence among sites limits our ability to distinguish positive and negative selection based on significant MK test results, it is worth investigating the theoretical consequences and empirical evidence for more realistic models of molecular evolution that include both positive and negative selection and that do not assume sites evolve independently of one another. In this context, it may be less relevant to distinguish between models of positive and negative selection and more relevant to distinguish models of purifying selection from those of adaptive evolution, where there is a positive fitness flux [73], or where there is a change in phenotype. Regardless of the model of selection, non-independence among sites must be investigated before we conclude that positive selection is a pervasive feature of molecular evolution and dismiss the neutral theory of molecular evolution.
Supplementary Material
Box 1 Figure I.
An estimate of the fraction of nonsynonymous substitutions driven by positive selection (α) using the McDonald-Kreitman test. The MK test determines whether the ratio of nonsynonymous to synonymous polymorphisms (N/S) is significantly different from the same ratio obtained from divergence data using Fisher's Exact test or a comparable test statistic. In this example, N/S of divergence is twice as great as that of polymorphism and leads to an estimate that 40% of nonsynonymous substitutions were driven by positive selection. Note that α can be estimate from the number of changes or rates of change since rates are measured by the number of changes divided by the number of sites surveyed, which must be the same for polymorphism and divergence data.
Box 2 Figure I.
Epistatic fitness effects under a simple evolutionary model. (a) Schematic of a phenotype-fitness function that generates epistatic fitness effects. Lines in the graph show how the expected fitness effect of a mutation depends on an individual's phenotype. An individual's phenotype is assumed be an additive function of multiple loci, six are shown for illustrative purposes. Genotypes at six loci (a–f) are shown for two extreme phenotypes below the graph. (b) A representation of species divergence that can arise as a consequence of the model shown in (a). As species diverge, their genotypes can diverge via drift through neutral phenotype-space. This results in sites (underlined) that have undergone neutral divergence between species but that are subject to purifying selection within species.
Acknowledgements
I would like to acknowledge participants of the Kavli Institute for Theoretic Physics conference in Population Genetics and Genomics for discussions, members of the Fay lab for comments, and multiple anonymous reviewers whose comments on this and a previous version of this work were extremely helpful in formulating my views in relation to the views of others in the field. This work was supported by the National Institute of General Medical Sciences (GM080669 and GM086412).
Glossary
- Neutral theory
a theory that the vast majority of DNA substitutions between species is the result of neutral mutations and random drift rather than selectively driven substitutions. The neutral theory does not assert that all mutations are neutral or that there is no adaptive evolution, but rather that deleterious mutations are eliminated from a population and that positively selected mutations make only a small contribution to divergence between species.
- Effective population size
The size of a randomly mating population of constant size that would effectively recapitulate patterns and levels of variation observed in a real population. A population's effective size rather than its actual size is used to account for the fact that most real populations are not constant in size and mating is not entirely random. A species' effective population size is relevant to estimates of α since weakly deleterious and advantageous mutations are effectively neutral when their fitness effects are less than the reciprocal of a population's effective size.
- Alpha (α)
Alpha is the estimated fraction of substitutions that were driven by positive selection and for nonsynonymous substitutions can be estimated by 1-(pN/pS)/(dN/dS).
- Selective constraint
The selective constraint on a site, or the average constraint across multiple sites, is a function of the degree to which fitness is reduced when mutated. In the absence of positive selection, pN/pS and dN/dS provide measures of the selective constraint on a site since they measure the extent to which the rate of nonsynonymous variation is reduced by negative selection relative to synonymous variation (assuming synonymous variation is neutral).
- Epistasis
While epistasis is used in many different contexts, here it is used to refer to mutations with effects that depend on genotypes at other loci.
- Hitchhiking
Hitchhiking is the process by which the frequency of mutations linked to an advantageous mutation are influenced by the spread of the advantageous mutation through a population.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Kimura M. The neutral theory of molecular evolution. Cambridge University Press; 1983. [Google Scholar]
- 2.Ohta T. Slightly deleterious mutant substitutions in evolution. Nature. 1973;246:96–98. doi: 10.1038/246096a0. [DOI] [PubMed] [Google Scholar]
- 3.Gillespie JH. The causes of molecular evolution. Oxford University Press; 1991. [Google Scholar]
- 4.Kreitman M, Akashi H. Molecular evidence for natural selection. Annu Rev Ecol Syst. 1995;26:403–422. [Google Scholar]
- 5.Fay J, Wu C-I. Sequence divergence, functional constraint, and selection in protein evolution. Annu Rev Genomics Hum Genet. 2003;4:213–235. doi: 10.1146/annurev.genom.4.020303.162528. [DOI] [PubMed] [Google Scholar]
- 6.McDonald J, Kreitman M. Adaptive protein evolution at the Adh locus in Drosophila. Nature. 1991;351:652–654. doi: 10.1038/351652a0. [DOI] [PubMed] [Google Scholar]
- 7.Sella G, et al. Pervasive natural selection in the Drosophila genome? PLoS Genet. 2009;5:e1000495. doi: 10.1371/journal.pgen.1000495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Siol M, et al. The population genomics of plant adaptation. New Phytol. 2010;188:313–332. doi: 10.1111/j.1469-8137.2010.03401.x. [DOI] [PubMed] [Google Scholar]
- 9.Hahn MW. Toward a selection theory of molecular evolution. Evolution. 2008;62:255–265. doi: 10.1111/j.1558-5646.2007.00308.x. [DOI] [PubMed] [Google Scholar]
- 10.Wright S, Andolfatto P. The impact of natural selection on the genome: Emerging patterns in Drosophila and Arabidopsis. Annu Rev Ecol Syst. 2008;39:193–213. [Google Scholar]
- 11.Eyre-Walker A. The genomic rate of adaptive evolution. Trends Ecol. Evol. 2006;21:569–575. doi: 10.1016/j.tree.2006.06.015. [DOI] [PubMed] [Google Scholar]
- 12.Bachtrog D, Andolfatto P. Selection, recombination and demographic history in Drosophila miranda. Genetics. 2006;174:2045–2059. doi: 10.1534/genetics.106.062760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bachtrog D. Similar rates of protein adaptation in Drosophila miranda and D. melanogaster, two species with different current effective population sizes. BMC Evol. Biol. 2008;8:334. doi: 10.1186/1471-2148-8-334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bartolomé C, et al. Patterns of selection on synonymous and nonsynonymous variants in Drosophila miranda. Genetics. 2005;169:1495–1507. doi: 10.1534/genetics.104.033068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Haddrill PR, et al. Estimating the parameters of selection on nonsynonymous mutations in Drosophila pseudoobscura and D. miranda. Genetics. 2010;185:1381–1396. doi: 10.1534/genetics.110.117614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Loewe L, Charlesworth B. Inferring the distribution of mutational effects on fitness in Drosophila. Biol. Lett. 2006;2:426–430. doi: 10.1098/rsbl.2006.0481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Axelsson E, Ellegren H. Quantification of adaptive evolution of genes expressed in avian brain and the population size effect on the efficacy of selection. Mol. Biol. Evol. 2009;26:1073–1079. doi: 10.1093/molbev/msp019. [DOI] [PubMed] [Google Scholar]
- 18.Halligan DL, et al. Evidence for pervasive adaptive protein evolution in wild mice. PLoS Genet. 2010;6:e1000825. doi: 10.1371/journal.pgen.1000825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Boyko AR, et al. Assessing the evolutionary impact of amino acid mutations in the human genome. PLoS Genet. 2008;4:e1000083. doi: 10.1371/journal.pgen.1000083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gojobori J, et al. Adaptive evolution in humans revealed by the negative correlation between the polymorphism and fixation phases of evolution. Proc. Natl. Acad. Sci. U.S.A. 2007;104:3907–3912. doi: 10.1073/pnas.0605565104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhang L, Li WH. Human SNPs reveal no evidence of frequent positive selection. Mol. Biol. Evol. 2005;22:2504–2507. doi: 10.1093/molbev/msi240. [DOI] [PubMed] [Google Scholar]
- 22.Eyre-Walker A, Keightley PD. Estimating the rate of adaptive molecular evolution in the presence of slightly deleterious mutations and population size change. Mol. Biol. Evol. 2009;26:2097–2108. doi: 10.1093/molbev/msp119. [DOI] [PubMed] [Google Scholar]
- 23.Charlesworth J, Eyre-Walker A. The rate of adaptive evolution in enteric bacteria. Mol. Biol. Evol. 2006;23:1348–1356. doi: 10.1093/molbev/msk025. [DOI] [PubMed] [Google Scholar]
- 24.Hughes AL, et al. Synonymous and nonsynonymous polymorphisms versus divergences in bacterial genomes. Mol. Biol. Evol. 2008;25:2199–2209. doi: 10.1093/molbev/msn166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gossmann TI, et al. Genome wide analyses reveal little evidence for adaptive evolution in many plant species. Mol. Biol. Evol. 2010;27:1822–1832. doi: 10.1093/molbev/msq079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Slotte T, et al. Genome-wide evidence for efficient positive and purifying selection in Capsella grandiflora, a plant species with a large effective population size. Mol. Biol. Evol. 2010;27:1813–1821. doi: 10.1093/molbev/msq062. [DOI] [PubMed] [Google Scholar]
- 27.Ingvarsson PK. Natural selection on synonymous and nonsynonymous mutations shapes patterns of polymorphism in Populus tremula. Mol. Biol. Evol. 2010;27:650–660. doi: 10.1093/molbev/msp255. [DOI] [PubMed] [Google Scholar]
- 28.Gillespie JH. Is the population size of a species relevant to its evolution? Evolution. 2001;55:2161–2169. doi: 10.1111/j.0014-3820.2001.tb00732.x. [DOI] [PubMed] [Google Scholar]
- 29.Ellegren H. A selection model of molecular evolution incorporating the effective population size. Evolution. 2009;63:301–305. doi: 10.1111/j.1558-5646.2008.00560.x. [DOI] [PubMed] [Google Scholar]
- 30.Foxe JP, et al. Selection on amino acid substitutions in Arabidopsis. Mol. Biol. Evol. 2008;25:1375–1383. doi: 10.1093/molbev/msn079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Doniger SW, et al. A catalog of neutral and deleterious polymorphism in yeast. PLoS Genet. 2008;4:e1000183. doi: 10.1371/journal.pgen.1000183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Liti G, et al. Population genomics of domestic and wild yeasts. Nature. 2009;458:337–341. doi: 10.1038/nature07743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Elyashiv E, et al. Shifts in the intensity of purifying selection: an analysis of genome-wide polymorphism data from two closely related yeast species. Genome Res. 2010;20:1558–1573. doi: 10.1101/gr.108993.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lynch M, Conery JS. The origins of genome complexity. Science. 2003;302:1401–1404. doi: 10.1126/science.1089370. [DOI] [PubMed] [Google Scholar]
- 35.Tsai IJ, et al. Population genomics of the wild yeast Saccharomyces paradoxus: Quantifying the life cycle. Proc. Natl. Acad. Sci. U.S.A. 2008;105:4957–4962. doi: 10.1073/pnas.0707314105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Andolfatto P. Controlling type-I error of the McDonald-Kreitman test in genomewide scans for selection on noncoding DNA. Genetics. 2008;180:1767–1771. doi: 10.1534/genetics.108.091850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Shapiro JA, et al. Adaptive genic evolution in the Drosophila genomes. Proc. Natl. Acad. Sci. U.S.A. 2007;104:2271–2276. doi: 10.1073/pnas.0610385104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Stoletzki N, Eyre-Walker A. Estimation of the neutrality index. Mol. Biol. Evol. 2011;28:63–70. doi: 10.1093/molbev/msq249. [DOI] [PubMed] [Google Scholar]
- 39.Nei M, et al. The neutral theory of molecular evolution in the genomic era. Annu Rev Genomics Hum Genet. 2010;11:265–289. doi: 10.1146/annurev-genom-082908-150129. [DOI] [PubMed] [Google Scholar]
- 40.Fay J, Wu C-I. The neutral theory in the genomic era. Curr. Opin. Genet. Dev. 2001;11:642–646. doi: 10.1016/s0959-437x(00)00247-1. [DOI] [PubMed] [Google Scholar]
- 41.Brodie E., III . Why evolutionary genetics does not always add up. In: Wolf J, Brodie E III, Wade M, editors. Epistasis and the evolutionary process. Oxford University Press; 2000. pp. 3–19. [Google Scholar]
- 42.Hartl DL, et al. Limits of adaptation: the evolution of selective neutrality. Genetics. 1985;111:655–674. doi: 10.1093/genetics/111.3.655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Takahata N. Statistical models of the overdispersed molecular clock. Theor Popul Biol. 1991;39:329–344. doi: 10.1016/0040-5809(91)90027-d. [DOI] [PubMed] [Google Scholar]
- 44.Cutler DJ. Understanding the overdispersed molecular clock. Genetics. 2000;154:1403–1417. doi: 10.1093/genetics/154.3.1403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Huerta-Sanchez E, et al. Population genetics of polymorphism and divergence under fluctuating selection. Genetics. 2008;178:325–337. doi: 10.1534/genetics.107.073361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mustonen V, Lässig M. Adaptations to fluctuating selection in Drosophila. Proc. Natl. Acad. Sci. U.S.A. 2007;104:2277–2282. doi: 10.1073/pnas.0607105104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Malmberg RL, Mauricio R. QTL-based evidence for the role of epistasis in evolution. Genet. Res. 2005;86:89–95. doi: 10.1017/S0016672305007780. [DOI] [PubMed] [Google Scholar]
- 48.Sanjuán R, et al. The contribution of epistasis to the architecture of fitness in an RNA virus. Proc. Natl. Acad. Sci. U.S.A. 2004;101:15376–15379. doi: 10.1073/pnas.0404125101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Pepin KM, Wichman HA. Variable epistatic effects between mutations at host recognition sites in phiX174 bacteriophage. Evolution. 2007;61:1710–1724. doi: 10.1111/j.1558-5646.2007.00143.x. [DOI] [PubMed] [Google Scholar]
- 50.Weinreich DM, et al. Darwinian evolution can follow only very few mutational paths to fitter proteins. Science. 2006;312:111–114. doi: 10.1126/science.1123539. [DOI] [PubMed] [Google Scholar]
- 51.Lunzer M, et al. Pervasive cryptic epistasis in molecular evolution. PLoS Genet. 2010;6:e1001162. doi: 10.1371/journal.pgen.1001162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Huelsenbeck J. Testing a covariotide model of DNA substitution. Mol. Biol. Evol. 2002;19(5):698–707. doi: 10.1093/oxfordjournals.molbev.a004128. [DOI] [PubMed] [Google Scholar]
- 53.Callahan B, et al. Correlated evolution of nearby residues in Drosophilid proteins. PLoS Genet. 2011;7:e1001315. doi: 10.1371/journal.pgen.1001315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kondrashov A, et al. Dobzhansky-Muller incompatibilities in protein evolution. Proc. Natl. Acad. Sci. U.S.A. 2002;99:14878–14883. doi: 10.1073/pnas.232565499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Dowell RD, et al. Genotype to phenotype: a complex problem. Science. 2010;328:469. doi: 10.1126/science.1189015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lynch M, Conery JS. The evolutionary fate and consequences of duplicate genes. Science. 2000;290:1151–1155. doi: 10.1126/science.290.5494.1151. [DOI] [PubMed] [Google Scholar]
- 57.Force A, et al. Preservation of duplicate genes by complementary, degenerative mutations. Genetics. 1999;151:1531–1545. doi: 10.1093/genetics/151.4.1531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Bullaughey K. Changes in selective effects over time facilitate turnover of enhancer sequences. Genetics. 2011;187:567–582. doi: 10.1534/genetics.110.121590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Rocha EPC, et al. Comparisons of dN/dS are time dependent for closely related bacterial genomes. J. Theor. Biol. 2006;239:226–235. doi: 10.1016/j.jtbi.2005.08.037. [DOI] [PubMed] [Google Scholar]
- 60.Wolf JBW, et al. Nonlinear dynamics of nonsynonymous (dN) and synonymous (dS) substitution rates affects inference of selection. Genome Biol Evol. 2009;1:308–319. doi: 10.1093/gbe/evp030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Peterson GI, Masel J. Quantitative prediction of molecular clock and ka/ks at short timescales. Mol. Biol. Evol. 2009;26:2595–2603. doi: 10.1093/molbev/msp175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Fay J, et al. Positive and negative selection on the human genome. Genetics. 2001;158:1227–1234. doi: 10.1093/genetics/158.3.1227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Bachtrog D, Gordo I. Adaptive evolution of asexual populations under Muller’s ratchet. Evolution. 2004;58:1403–1413. doi: 10.1111/j.0014-3820.2004.tb01722.x. [DOI] [PubMed] [Google Scholar]
- 64.Hartfield M, Otto S. Recombination and hitchhiking of deleterious alleles. Evolution. 2011 doi: 10.1111/j.1558-5646.2011.01311.x. in press and available online. [DOI] [PubMed] [Google Scholar]
- 65.Bierne N, Eyre-Walker A. The genomic rate of adaptive amino acid substitution in Drosophila. Mol. Biol. Evol. 2004;21:1350–1360. doi: 10.1093/molbev/msh134. [DOI] [PubMed] [Google Scholar]
- 66.Welch JJ. Estimating the genomewide rate of adaptive protein evolution in Drosophila. Genetics. 2006;173:821–837. doi: 10.1534/genetics.106.056911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Charlesworth J, Eyre-Walker A. The other side of the nearly neutral theory, evidence of slightly advantageous back-mutations. Proc. Natl. Acad. Sci. U.S.A. 2007;104:16992–16997. doi: 10.1073/pnas.0705456104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Gavrilets S. Evolution and speciation on holey adaptive landscapes. Trends Ecol. Evol. 1997;12:307–312. doi: 10.1016/S0169-5347(97)01098-7. [DOI] [PubMed] [Google Scholar]
- 69.Sattath S, et al. Pervasive adaptive protein evolution apparent in diversity patterns around amino acid substitutions in Drosophila simulans. PLoS Genet. 2011;7:e1001302. doi: 10.1371/journal.pgen.1001302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Macpherson JM, et al. Genomewide spatial correspondence between nonsynonymous divergence and neutral polymorphism reveals extensive adaptation in Drosophila. Genetics. 2007;177:2083–2099. doi: 10.1534/genetics.107.080226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Andolfatto P. Hitchhiking effects of recurrent beneficial amino acid substitutions in the Drosophila melanogaster genome. Genome Res. 2007;17:1755–1762. doi: 10.1101/gr.6691007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Cai JJ, et al. Pervasive hitchhiking at coding and regulatory sites in humans. PLoS Genet. 2009;5:e1000336. doi: 10.1371/journal.pgen.1000336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Mustonen V, Lässig M. From fitness landscapes to seascapes: non-equilibrium dynamics of selection and adaptation. Trends Genet. 2009;25:111–119. doi: 10.1016/j.tig.2009.01.002. [DOI] [PubMed] [Google Scholar]
- 74.Smith N, Eyre-Walker A. Adaptive protein evolution in Drosophila. Nature. 2002;415:1022–1024. doi: 10.1038/4151022a. [DOI] [PubMed] [Google Scholar]
- 75.Fay J, et al. Testing the neutral theory of molecular evolution with genomic data from Drosophila. Nature. 2002;415:1024–1026. doi: 10.1038/4151024a. [DOI] [PubMed] [Google Scholar]
- 76.Andolfatto P. Adaptive evolution of non-coding DNA in Drosophila. Nature. 2005;437:1149–1152. doi: 10.1038/nature04107. [DOI] [PubMed] [Google Scholar]
- 77.Charlesworth J, Eyre-Walker A. The McDonald-Kreitman test and slightly deleterious mutations. Mol. Biol. Evol. 2008;25:1007–1015. doi: 10.1093/molbev/msn005. [DOI] [PubMed] [Google Scholar]
- 78.Eyre-Walker A. Changing effective population size and the McDonald-Kreitman test. Genetics. 2002;162:2017–2024. doi: 10.1093/genetics/162.4.2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Begun DJ, et al. Population genomics: whole-genome analysis of polymorphism and divergence in Drosophila simulans. PLoS Biol. 2007;5:e310. doi: 10.1371/journal.pbio.0050310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Maside X, Charlesworth B. Patterns of molecular variation and evolution in Drosophila americana and its relatives. Genetics. 2007;176:2293–2305. doi: 10.1534/genetics.107.071191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Grath S, et al. Molecular evolution of sex-biased genes in the Drosophila ananassae subgroup. BMC Evol. Biol. 2009;9:291. doi: 10.1186/1471-2148-9-291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Sniegowski P, et al. Saccharomyces cerevisiae and Saccharomyces paradoxus coexist in a natural woodland site in North America and display different levels of reproductive isolation from European conspecifics. FEM Yeast Res. 2002;1:299–306. doi: 10.1111/j.1567-1364.2002.tb00048.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.