Abstract
A combination of biophysical, biochemical, and computational techniques was used to delineate mechanistic differences between the platinum–acridine hybrid agent [PtCl(en)(L)](NO3)2 (complex 1, en = ethane-1,2-diamine, L = 1-[2-(acridin-9-ylamino)ethyl]-1,3- dimethylthiourea) and a considerably more potent second-generation analogue containing L′ = N-[2-(acridin-9-ylamino)ethyl]-Nmethylpropionamidine (complex 2). Calculations at the density functional theory level provide a rationale for the binding preference of both complexes for guanine-N7 and the relatively high level of adenine adducts observed for compound 1. A significant rate enhancement is observed for binding of the amidine-based complex 2 with DNA compared with the thiourea-based prototype 1. Studies conducted with chemical probes and on the bending and unwinding of model duplex DNA suggest that adducts of complex 2 perturb B-form DNA more severely than complex 1, however, without denaturing the double strand and significantly less than cisplatin. Circular and linear dichroism spectroscopies and viscosity measurements suggest that subtle differences exist between the intercalation modes and adduct geometries of the two complexes. The adducts formed by complex 2 most efficiently inhibit transcription of the damaged DNA by RNA polymerase II. Not only do complexes 1 and 2 cause less distortion to DNA than cisplatin, they also do not compromise the thermodynamic stability of the modified duplex. This leads to a decreased or negligible affinity of HMG domain proteins for the adducts formed by either Pt-acridine complex. In a DNA repair synthesis assay the lesions formed by complex 2 were repaired less efficiently than those formed by complex 1. These significant differences in DNA adduct formation, structure, and recognition between the two acridine complexes and cisplatin help to elucidate why compound 2 is highly active in cisplatin-resistant, repair proficient cancer cell lines.
Keywords: platinum drug, antitumor, DNA binding, conformational distortion, DNA repair
Introduction
Cisplatin (Figure 1) and several of its second-generation analogues enjoy the status of the World’s best-selling anticancer drugs. Unfortunately, like most chemotherapies, platinum antitumor drugs suffer from major drawbacks, including intrinsic and acquired resistance and severe systemic toxicity.1 To overcome these limitations, structurally and functionally, unique metallodrugs have been designed. These include trans-configured complexes,2 inert platinum(IV) compounds,3 polynuclear complexes,4 and photoactivatable complexes,5 as well as agents containing alternative metals, such as ruthenium, osmium, or iridium.6–8 The mechanism of action of cisplatin involves several critical events, such as cellular uptake and transport of the drug to the nucleus, formation of DNA adducts in chromatin, and recognition by DNA-binding proteins and DNA-processing enzymes.9 Subsequent activation of signal transduction pathways, as a consequence of the DNA damage and its interference with nuclear proteins, results in cell-cycle arrest and, if the lesions are not repaired, in apoptosis or necrosis.10
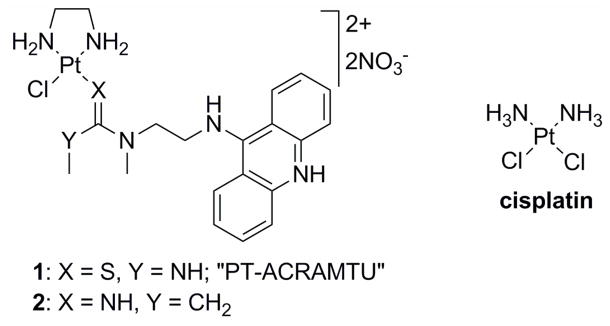
Figure 1.
Structures of the platinum complexes studied.
Recent insights into the events triggering cell death have led to the hypothesis that platinum and other transition-metal based drugs that damage DNA in a fundamentally different manner than cisplatin may show unique biological properties, including an altered spectrum of antitumor activity.11 To test this hypothesis conjugates of metal complexes with organic intercalators have been reported previously.12–16 Platinum–acridine complexes represented by the prototype [PtCl(en)(ACRAMTU)](NO3)2 (1, “PT-ACRAMTU”) (en = ethane-1,2-diamine, ACRAMTU = 1-[2-(acridin-9-ylamino)ethyl]-1,3-dimethylthiourea) (Figure 1) are a new class of hybrid agents also designed towards this goal. These PtII compounds have demonstrated promising activity in a wide range of human cancer cell lines including those resistant to cisplatin.17 The DNA binding mechanism of 1 differs in many ways from that of clinical platinum drugs. Cisplatin and its analogues form bifunctional adducts with neighboring purine bases (mainly 1,2-GG and AG intrastrand cross-links).9 These cross-links unwind the DNA duplex locally and bend it toward the major groove.18 This severe distortion is recognized by nuclear proteins,19 including those involved in repair processes and proteins containing high-mobility group (HMG) domains.9 By contrast, the DNA binding mode of 1 involves a combination of intercalation and monofunctional coordination. Complex 1 forms unique monoadducts with the N7 donor site of G residues (80%) in the sequence 5′-CG and with N7 (major), N3, and N1 of A residues (20%) at 5′-TA and 5′-GA sites.20 In these adducts, the acridine moiety intercalates into the 5′ base-pair step adjacent to the platinated nucleobase.20–25
Recently, a derivative of PT-ACRAMTU (1), [PtCl(en)(L)](NO3)2 (2) (L = N-[2-(acridin- 9-ylamino)ethyl]-N-methylpropionamidine) (Figure 1) was synthesized,26 in which the thiourea donor group was replaced with an amidine group. The substitution of thiourea sulfur with amidine nitrogen was done as part of a structure–activity relationship study to explore the possibility of modulating the reactivity of the hybrid agent with biological nucleophiles by altering the donor set in the metal’s coordination sphere. This simple modification led to a significant increase in the rate of platinum adduct formation with DNA, which translated into greatly enhanced cytotoxicity in non-small-cell lung cancer (NSCLC) and tumor growth inhibition in the corresponding mouse xenograft model.26 These observations suggest that the biological activity of platinum–acridines can be tuned at the DNA adduct level. The type of DNA conformational changes caused by a specific adduct and its recognition and repair are critical determinants of a platinum drug’s efficacy.9 In the current study, we aimed to delineate differences in the DNA interactions between complexes 1 and 2 and compare the effects of the hybrid adducts with the damage produced by cisplatin. A combination of biophysical, biochemical, and computational techniques were used to establish mechanistic differences between the two compounds with respect to the DNA adducts they produce, their effect on the structure and stability of the damaged biopolymer, as well as their interactions with critical DNA-binding proteins and DNA-processing enzymes. The latter aspect focused on high-mobility-group (HMG) proteins9 and human RNA polymerase II,27 which are critical mediators of the antitumor effect of clinical platinum drugs. The repair of DNA adducts formed by 1 and 2 was also studied since repair efficiency is a potential biomarker for the sensitivity of a tumor to platinum-based therapy.28
Materials and Methods
Chemicals
Cisplatin (purity was ≥99.9% based on elemental and ICP trace analysis) was obtained from Sigma (Prague, Czech Republic). The platinum–acridine complexes 1 and 2 were synthesized according to published procedures.21, 26 Stock solutions of the platinum complexes [5 × 10−4 M in NaClO4 (10 mM)] were stored in the dark at 277 K. Calf thymus (CT) DNA (42% G + C, mean molecular mass ca. 20 000 kDa) was prepared and characterized as described previously.29, 30 The plasmids, pUC19 (2686 bp) and pBR322 (4361 bp), were isolated according to standard procedures. Restriction endonucleases, plasmid DNA pCMV-GLuc (5764 bp), T4 DNA ligase and T4 polynucleotide kinase were purchased from New England Biolabs (Beverly, MA). The synthetic oligodeoxyribonucleotides were purchased from VBC-GENOMICS (Vienna, Austria) or DNA Technology (Aarhus, Denmark). The purity of the oligonucleotides was verified by either high-pressure liquid chromatography (HPLC) or gel electrophoresis. Expression and purification of the domains A and B (residues 1–84 and 85–180, respectively) of the HMGB1 protein (HMGB1a and HMGB1b, respectively) were carried out as described previously31, 32 (in this study the HMGB1b protein also contained the A/B linker (residues 85–91)33). HeLaScribe®Nuclear Extract in vitro Transcription system kit was from Promega (Mannheim, Germany). Deoxyribonucleotide triphosphates were from Roche Diagnostics, GmbH (Mannheim, Germany). Agarose was from FMC BioProducts (Rockland, ME). Acrylamide, bis(acrylamide), ethidium bromide (EtBr), NaCN and urea were from Merck KgaA (Darmstadt, Germany). Dimethyl sulfate (DMS), KMnO4, diethylpyrocarbonate (DEPC), KBr, and KHSO5 were from Sigma (Prague, Czech Republic). Sodium dodecyl sulfate (SDS) was from Serva (Heidelberg, Germany). Proteinase K and ATP were from Boehringer (Mannheim, Germany). Radioactive products were from Amersham (Arlington Heights, IL, USA). The cell-free extract (CFE) was prepared from the repair-proficient HeLa S3 cell line as reported previously.34
Platination Reactions
If not stated otherwise, CT or plasmid DNAs were incubated with the platinum complex in 10 mM NaClO4 at 310 K in the dark. After 24 h, the samples were exhaustively dialyzed against the medium required for subsequent biochemical or biophysical analysis. An aliquot of these samples was used to determine rb values (the number of molecules of the platinum complex bound per nucleotide residue) by flameless atomic absorption spectrometry (FAAS), or by differential pulse polarography (DPP).35 The duplexes containing single, site-specific adducts of the platinum compounds were prepared as follows: the single-stranded oligonucleotides (the top strands of the duplexes used in the present work) were reacted with 1, 2 or cisplatin in the dark. The platinated oligonucleotide was repurified by HPLC. FAAS and optical density measurements were used to verify that the modified oligonucleotides contained one molecule of platinum complex per one strand. By using Maxam-Gilbert (DMS) footprinting of platinum on DNA,36 we also verified that the N7 position of the guanine residue in the platinated top strands was not accessible for reaction with DMS. The platinated top strand was allowed to anneal with the complementary strand in NaClO4 (0.1 M) and incubated with platinum at 310 K for 24 h.
Computational Studies
Models were built using the Gaussview program (Semichem Inc., Shawnee Mission, KS, 2009). The crystal structure coordinates of compound 226 and the 2′-deoxyguanosine adduct of compound 137 served as starting geometries where applicable. Calculations were performed with truncated models lacking the N-methylacridin-9-amine moieties. These truncated models were modified in Gaussview to produce PT-AMD-AQ, PTAMD- A, PT-AMD-G, PT-TU-AQ, PT-TU-A, and PT-TU-G. 9-Methylguanine and 9- methyladenine were used to mimic binding of platinum to the N7 positions of the DNA nucleobases. Transition states were built using a trigonal bipyramidal platinum template and the appropriate crystal structure fragments in Discovery Studio (Accelrys, San Diego, CA, version 2.1, 2008). To sample the conformational space of both the transition states and platinum adducts, the torsional angles about the Pt–nucleobase-N7 bonds were systematically varied to allow the models to relax into (hydrogen-bonded) optimized geometries. Optimizations and single-point energy calculations were performed with the Gaussian 03 (G03) software package.38 All geometries were fully optimized in the gas phase at the gradient-corrected DFT level using the restricted B3LYP functional.39, 40 The LANL2DZ valence basis set,41 which includes relativistic core potentials, and the D95V basis set42 were used to describe the elements Na–Bi and H–Ne, respectively. Vibrational frequencies were used to confirm that the optimized structures had converged to their local minima (or maxima in the case of the transition states). Equilibrium structures contained no imaginary frequencies, whereas transition state structures contained one imaginary frequency. Single-point energy calculations on the gas-phase structures were performed using the self-consistent reaction field (SCRF) approach,43 which employed a constant dielectric (ε = 78.3553) to mimic solvation in water. Cartesian coordinates and vibrational frequencies for the optimized structures have been submitted as Supporting Information (Tables S1 and S2).
Circular and Linear Dichroism Spectroscopy
Isothermal circular dichroism (CD) spectra of CT DNA at the concentration of 0.032 mg mL−1 (1 × 10−4 M in nucleotides) modified by 1 or 2 at rb in the range of 0.01–0.1 were recorded at 298 K in 10 mM NaClO4 with 10 mM Tris·HCl, pH 7.0 using a Jasco J-720 spectropolarimeter equipped with a thermoelectrically controlled cell holder. The cell path length was 10 mm. CD spectra were recorded in the range of 200–500 nm in 0.2 nm increments with an averaging time of 0.5 s. Flow linear dichroism (LD) spectra were collected using a flow Couette cell in a Jasco J-720 spectropolarimeter adapted for LD measurements. The flow cell consisted of a fixed outer cylinder and a rotating solid quartz inner cylinder, separated by a gap of 0.5 mm, giving a total path length of 1 mm.44, 45 LD spectra of CT DNA at a concentration of 0.032 mg mL−1 (1 × 10−4 M in terms of monomeric nucleotide content) modified with 1 or 2 were recorded at 298 K in 10 mM NaClO4 with 10 mM sodium cacodylate, pH 7.0.
Viscometry
The relative viscosity of the solutions of CT DNA (150 μg mL−1), unmodified or modified with 1 or 2, was measured in 0.01 M NaClO4 at 310 K by microviscometry (AMVn Automated Micro Viscometer, Anton Paar GmbH, Austria) in a 1.6-mm capillary tube. The densities of the solutions were measured using a Density Meter DMA4500 instrument (Anton Paar GmbH, Austria).
Differential Scanning Calorimetry (DSC)
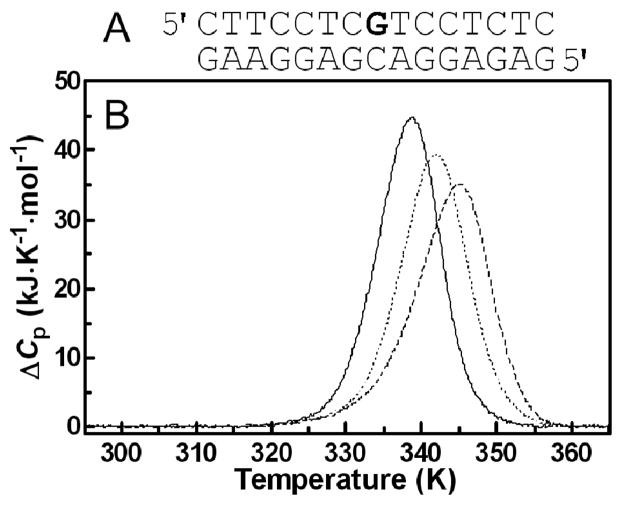
Excess heat capacity (ΔCp) versus temperature profiles for the thermally induced transitions of the 15-bp duplex (see Figure 11A for its sequence) were measured using a VP-DSC calorimeter (Microcal, Northampton, MA). In the DSC experiments the concentration of duplex was 30 μM, the heating rate was 60 K h−1, and the maximum temperature was 368 K. After reaching the maximum temperature the samples were cooled at the same rate to the starting temperature of 298 K. In this study ΔCp is defined as the excess heat capacity, which is baseline-subtracted and concentration-normalized.46 The reference scans were subtracted from the sample scans to obtain ΔCp versus temperature profiles. The enthalpies (ΔHcal) and entropies (ΔS) of duplex melting were calculated from the areas under the experimental ΔCp versus T and the derived ΔCp/T versus T curves, respectively, using ORIGIN (version 5.0, Microcal, Studio City, CA). The free energy of duplex dissociation at 298 K (ΔG0 298) was calculated from the standard thermodynamic relationship and the corresponding ΔHcal and ΔS values:
| (1) |
Figure 11.
Differential scanning calorimetry of the unmodified 15-bp duplex and duplexes containing a single, monofunctional adduct of 1 or 2. A. Nucleotide sequence of the 15-bp duplex; the bold letter (central G residue) in the top strand indicates the location of the monofunctional adduct of the platinum complex. B. DSC thermograms; lines: solid, unmodified duplex; dashed, duplex containing the adduct of 1; dotted, duplex containing the adduct of 2. The duplex concentration was 30 μM, and the buffer conditions were 10 mM phosphate buffer (pH 7) plus 150 mM NaCl. For other details, see the text.
The duplexes were dissolved in buffer, at pH 7.0, containing sodium phosphate (NaH2PO4/Na2HPO4, 10 mM) and NaCl (150 mM). It was also verified, as described previously,47, 48 that the melting transitions of both the platinated and unmodified duplexes were fully reversible.
Chemical Probing of DNA Conformation
The reaction of the platinated oligonucleotide duplexes with KMnO4, DEPC and KBr/KHSO5 were performed as described previously.49 The top or bottom strands of the oligonucleotide duplexes were 5′-end-labeled with [γ-32P]ATP and T4 polynucleotide kinase. In the case of the platinated oligonucleotides, platinum was removed after reaction of the DNA with the probe by incubation with NaCN (0.2 M, pH 11) at 318 K for 16 h in the dark.
Ligation and Electrophoresis of Oligonucleotides
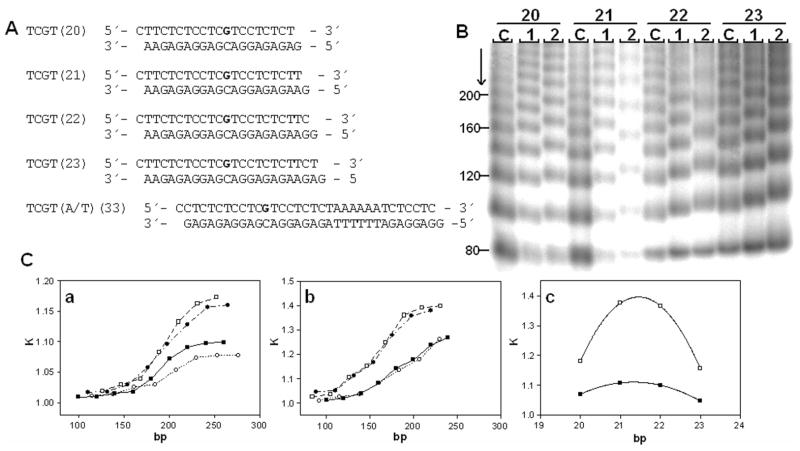
Unplatinated and monoadducted 20- 23-bp duplexes (shown in Figure 10) were 5′-end-labeled with [γ-32P]ATP by using T4 polynucleotide kinase. The duplexes were allowed to react with T4 DNA ligase. The resulting samples were subsequently examined on 8% native PAA [mono:bis(acrylamide) ratio 29:1] electrophoresis gels. Other details of these experiments were as described in previous papers50, 51 or are described in the text.
Figure 10.
Ligation and electrophoresis of platinated model oligodeoxyribonucleotides. A. Sequences of the synthetic oligodeoxyribonucleotides used in this study with their abbreviations. The top and bottom strands of each pair are designated ‘top’ and ‘bottom’, respectively, in the text. The bold letters in the top strand indicate the location of the adduct after the modification of the oligonucleotides by 1 or 2 as described in the text. B. Autoradiogram of the ligation products of double-stranded oligonucleotides (20–23); platinum-free duplexes (lanes C) and duplexes containing a unique adduct of 1 or 2 separated on an 8% polyacrylamide gel (lanes 1 and 2, respectively). C(a,b). Plots showing the relative mobility K (defined as the ratios of the calculated and the actual lengths) versus sequence length for the 20–23-bp oligomers containing the adducts of 1 (a) or 2 (b), (■), 20- mer; (□), 21-mer; (●), 22-mer; (○), and 23-mer. C(c). The plot showing the relative mobility K versus interadduct distance in bp for the 20–23-bp oligomers containing the adducts of 1(■) or 2(□) with total length of 190 bp. The experimental points represent the average of three independent electrophoresis experiments, and the curves represent the best fit of the data to the equation K = ad2 + bd + c.50
Electrophoretic mobility shift assays with HMGB1 domain proteins
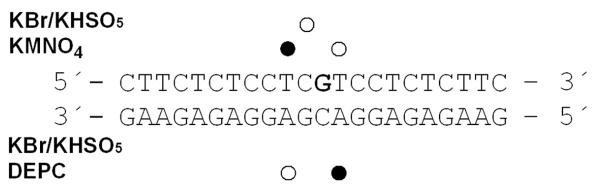
Radioactively labeled 22-bp DNA probes with blunt ends (their sequence is shown in Figure 9A) were titrated with HMGB1a or HMGB1b proteins. The duplexes (1 pmol) were incubated with the proteins in 10-μL sample volumes in a buffer composed of HEPES (10 mM, pH 7.5), MgCl2 (10 mM), LiCl (50 mM), NaCl (0.1 M), spermidine (1 mM), bovine serum albumin (0.2 mg.mL−1), and Nonidet P40 (0.05% v/v). For all gel mobility shift experiments, samples were incubated on ice for 1 h and made 7% in sucrose and 0.017% in xylene cyanol before loading on running, pre-cooled (277 K), pre-run (300 V, 1–2 h) 5% native PAA gels (29:1 acrylamide:bisacrylamide, 0.5× Tris-borate-Na2H2EDTA buffer, Tris-HCl (45 mM), boric acid (45 mM), and Na2H2EDTA (1 mM, pH 8.3). Gels were electrophoresed at 277 K and 300 V for ~1.5 h, dried, exposed to a molecular imaging plate, and analyzed on a Fujifilm bioimaging analyzer. The radioactivities associated with the bands were quantitated with the AIDA image analyzer software. Other details have been published previously.33, 52
Figure 9.
Chemical probes of DNA conformation. Summary of the reactivity of chemical probes with the 22-bp duplex containing a single, site-specific adduct of 1 and 2. The platinated nucleobase is highlighted in bold. Closed and open circles designate strong and weak reactivity, respectively.
Transcription by RNA Polymerase II In Vitro
In vitro transcription was performed using HeLaScribe®Nuclear Extract in vitro Transcription system kit. The system contains all necessary components for in vitro transcription from a CMV promoter of plasmid DNA pCMV-GLuc. Plasmid pCMV-GLuc linearized by XbaI was incubated with platinum complexes in 10 mM NaClO4 for 24 h at 310 K at an rb of 5 × 10−4 to 5 × 10−3. The DNA was precipitated with ethanol to remove unbound platinum. In vitro transcription was performed using the HeLa nuclear extract supplied with the kit following the manufacturer’s protocol with small modifications. Briefly, 100 ng of platinated and unmodified, linearized pCMVGLuc DNA was incubated in the transcription buffer supplemented with 4 mM MgCl2, 0.4 mM rATP, 0.4 mM UTP, 0.4 mM rGTP, 16 μM rCTP, 10 mCi [α-32P]CTP (3000 Ci/mmol), 20 U RNase inhibitor and nuclear extract (8 U) in a final reaction volume of 25 μL at 303 K for 60 min. The reaction was terminated by adding 175 μL HeLa Extract Stop Solution followed by extraction with phenol and chloroform. The transcripts were precipitated with ethanol and the pellet was washed, dried and resuspended in a loading buffer containing 90% formamide, 10 mM Na2H2EDTA, 0.1% xylene cyanol and 0.1% bromophenol blue. The samples were separated on a 6% denaturing PAA gel, and the radioactivity associated with the bands corresponding to full-length run-off transcription products was quantified.
Repair DNA Synthesis by Human Cell-Free Extract
Repair DNA synthesis was assayed using platinated pUC19 plasmid and cell-free extracts (CFEs). Each reaction of 50 μL contained 600 ng of unmodified pBR322 and 600 ng of unmodified or platinated pUC19, 2 mM ATP, 30 mM KCl, 0.05 mg mL−1 creatine phosphokinase (rabbit muscle), 20 mM each dGTP, dATP, and dTTP, 8 mM dCTP, 74 kBq of [α-32P]dCTP in the buffer composed of 40 mM HEPES–KOH, pH 7.5, 5 mM MgCl2, 0.5 mM dithiothreitol, 22 mM creatine phosphate, 1.4 mg of bovine serum albumin/mL, and 20 mg of CFE from the HeLa S3 cells. Reactions were incubated for 3 h at 303 K and terminated by incubating for 20 min with 20 mM Na2H2EDTA, 0.6% SDS, and 250 mg mL−1 proteinase K. The products were extracted once with phenol/chloroform (1:1) and precipitated by adding 3 M sodium acetate and ethanol. After 30 min of incubation at 253 K and centrifugation at 12,000 × g for 30 min at 277 K, the pellet was washed with 0.2 mL of 80% ethanol and dried in a vacuum centrifuge. The DNA was linearized before electrophoresis on a 1% agarose gel. Gels were stained with EtBr for photodocumentation. Experiments were performed in quadruplicate.
Other Physical Methods
Absorption spectra were measured with a Beckman 7400 DU spectrophotometer equipped with a thermoelectrically controlled cell holder. HPLC purification of oligonucleotides was carried out on a Waters HPLC system consisting of a Waters 262 pump, Waters 2487 UV detector, and Waters 600S controller with MonoQ HR 5/50 GL column. The FAAS measurements were carried out on a Varian AA240Z Zeeman atomic absorption spectrometer equipped with a GTA 120 graphite tube atomizer. For FAAS analyses, DNA was precipitated with ethanol and dissolved in HCl (0.1 M). DPP was performed with an EG&G Princeton Applied Research Corporation Model 384B Polarographic Analyzer. The gels were visualized on a BAS 2500 FUJIFILM bioimaging analyzer, and the radioactivity associated with bands was quantified with the AIDA image analyzer software (Raytest, Germany).
Results
Platination of DNA
Prior to the synthesis of the site-specific adducts in this study, we investigated the reactions of 1 and 2 with CT DNA in cell-free media and compared their binding properties with those of cisplatin. For this purpose, solutions of double-helical CT DNA were incubated with 1 and 2 under well-defined conditions (see section Materials and Methods). At various time intervals, suitable aliquots were withdrawn from the reaction mixture and assayed by DPP for unbound platinum. The amount of platinum bound to DNA increased with time, and the time at which the binding of 1 or 2 reached 50% (t50) was 12±1 or 6±1 min, respectively. Both complexes were quantitatively bound after ~6 h. These results are in agreement with a recent kinetic study, which showed that replacement of a thiourea sulfur with an amidine nitrogen increased the rate of platinum binding by approximately three- to four-fold.53 On the other hand, intercalator-driven platination of DNA by 1 and 2 proved to be considerably faster than formation of adducts by cisplatin.54
Circular and Linear Dichroism (CD and LD) Studies
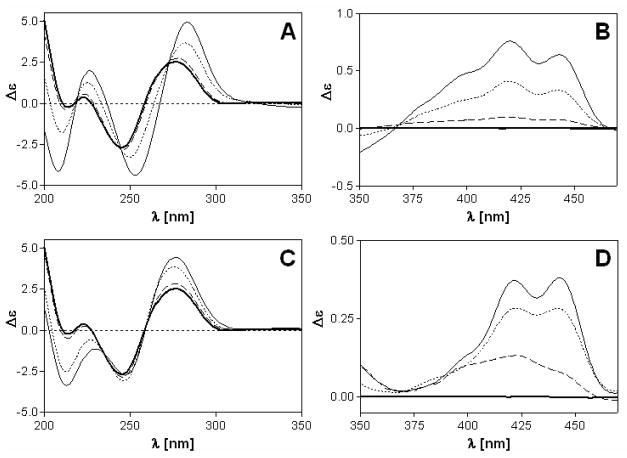
CD spectra of CT DNA modified with complexes 1 and 2 at varying rb values were recorded to study the binding modes and global conformational changes produced by the conjugates (Figure 2). Complexes 1 and 2 are achiral molecules and therefore do not show intrinsic CD signals. The CD signal above 320 nm (ligand region), can be attributed to the interaction of the complexes with the chiral DNA, resulting in induced circular dichroism (ICD). The ICD signatures of complexes 1 and 2 bound to CT DNA comprise broad positive features in the 380–460 nm region, which essentially mimic the vibronically coupled absorption spectrum of the intercalated 9- aminoacridine moiety.24, 55 The band can be assigned to the π– π* transition polarized along the short axis of the intercalated chromophore (Figures 2A,B).56, 57 The same feature has been observed previously for a double-stranded octamer modified with a single adduct of compound 1.24, 55 Its positive sign is consistent with an intercalation mode in which the long axes of the acridines in 1 and 2 are aligned with the long dimension of the adjacent base pairs.56 Subtle differences, however, seem to exist between the intercalation geometries of the two compounds since the long-wavelength ICD signal for compound 1 is more intense than the corresponding feature for compound 2.
Figure 2.
Circular dichroism (CD) spectra of calf thymus DNA modified by 1 (A,B) and 2 (C,D). CD spectra were recorded for DNA at the concentration of 32 μg mL−1 in 10 mM NaClO4 with 10 mM Tris.HCl, pH 7.0. Curves (A, B): bold solid line, control (unplatinated) DNA; dashed line, rb = 0.01; dotted line, rb = 0.05; solid line, rb = 0.1.
While compound 1 shows a weak negative ICD band at 340 nm (n-π* transition57), the corresponding feature in the spectrum of derivative 2 shows a positive sign. The negative ICD in the former adduct suggests that the acridine moiety deviates significantly from coplanarity with the nucleobases of the intercalation pocket, possibly indicating partial intercalation or significant distortions within the adjacent base pairs. Below 320 nm (DNA region), changes in the CD spectrum upon modification of the DNA with the platinum–intercalators can be attributed to structural perturbations induced in the DNA. However, since complexes 1 and 2 absorb below 300 nm, ligand-based ICD bands may also contribute to the observed changes in this region, which complicates their interpretation. Overall, the CD data suggest that the DNA adducts formed by compound 1 produce a less ideal geometry for classical intercalation than adducts formed by the new derivative, 2.
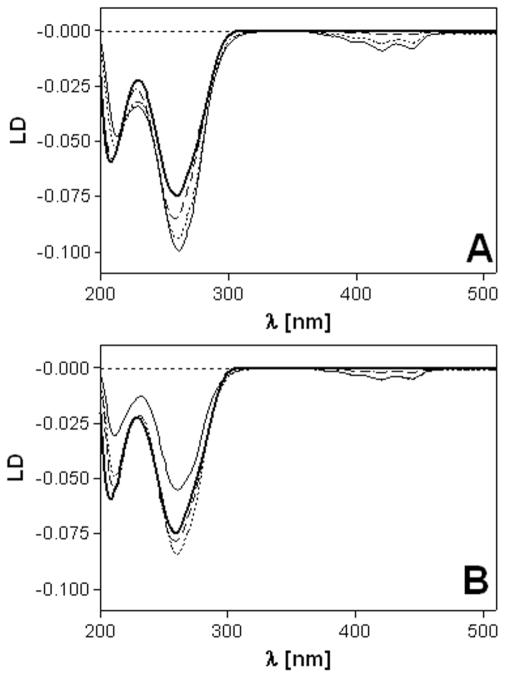
LD was also used to characterize the DNA binding mode of complexes 1 and 2. Long molecules such as DNA (minimum length ~250 base pairs) can be oriented through viscous drag produced in a Couette flow cell.58 Small unbound molecules and molecules bound randomly to CT DNA show no LD signal. However, molecules bound in a specific orientation with respect to the biopolymer give rise to LD. Thus, the LD signals in the ligand regions of calf thymus DNA modified with complexes 1 and 2 arise from the intercalative monoadducts. The LD spectra show a ligand-based negative signal in the 370–460 nm range and negative bands in the DNA region (220–300 nm) (Figure 3). The latter feature confirms that the DNA modified with complexes 1 and 2 remains in a B-type conformation. The short-axis transition within the ligand chromophore gives rise to a signal exhibiting the same sign as the DNA bands. This observation is in agreement with a coplanar arrangement of the DNA base pairs and the acridine moiety, which is highly suggestive of classical intercalative binding.57
Figure 3.
Linear dichroism (LD) spectra of calf thymus DNA modified by 1 (A) and 2 (B). LD spectra were recorded for DNA in 10 mM NaClO4 with 10 mM sodium cacodylate, pH 7.0 at 25 °C. The concentration of DNA was 32 μg mL−1. Curves (A, B): bold solid line, control (unplatinated) DNA; dashed line, rb = 0.01; dotted line, rb = 0.05; solid line, rb = 0.08.
Viscometry Studies of Platinated DNA
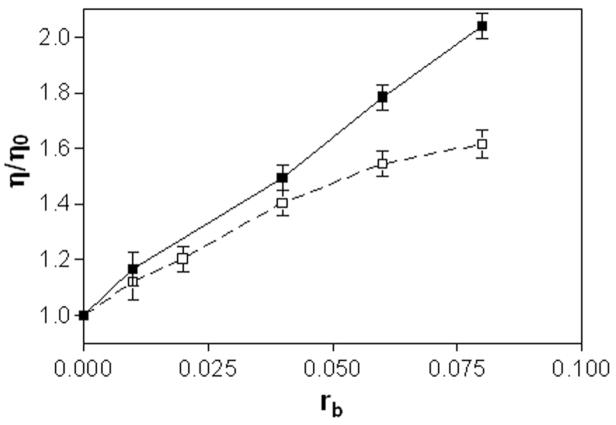
The effects of complexes 1 and 2 on the viscosity of calf thymus DNA were also studied (Figure 4). On increasing the amounts of 1 or 2 bound to DNA (in the range of rb values of 0.01 — 0.08), the relative viscosity of the DNA increased steadily. The linear increase in viscosity as a function of platinum content observed for compound 1 across the entire rb range is in agreement with the classical intercalation model. For compound 2, a significant deviation from linearity is observed at rb values greater than 0.05. This suggests that formation of monofunctional adducts by 2, in which the acridine chromophore is able to intercalate into the DNA base stack, becomes unfavorable at higher adduct levels. The saturation behavior observed for 2 is consistent with the change in base selectivity that results from the substitution of the thiourea linker in 1 with an amidine group in 2 (see section Discussion).
Figure 4.
Viscometry of calf thymus DNA modified by 1(■) and 2(□). Dependence of relative viscosity of calf thymus DNA on rb. The data were recorded for DNA concentration of 0.15 mg mL−1 in 10 mM NaClO4 at 310 K.
Recognition of Adducts by HMG Domain Proteins
An important feature of the mechanism of action of cisplatin is that the altered structures produced by the bending of the helical axis induced in DNA by 1,2-intrastrand or 1,2-interstrand cross-links of cisplatin attract HMG domain proteins and other proteins.19, 59, 60 This binding of HMG domain proteins to cisplatin-modified DNA has been postulated to mediate or enhance the drug’s antitumor properties.19 Full-length HMGB1 or HMGB2 proteins and the domains A and B of HMGB1 protein (HMGB1a and HMGB1b, respectively) bind to 1,2-GG intrastrand crosslinks of cisplatin. Interestingly, the full-length HMGB1 protein and its domain B that contains the N-terminal lysine-rich region (seven amino acid residues) of the A/B linker specifically recognize the interstrand cross-link.33, 60
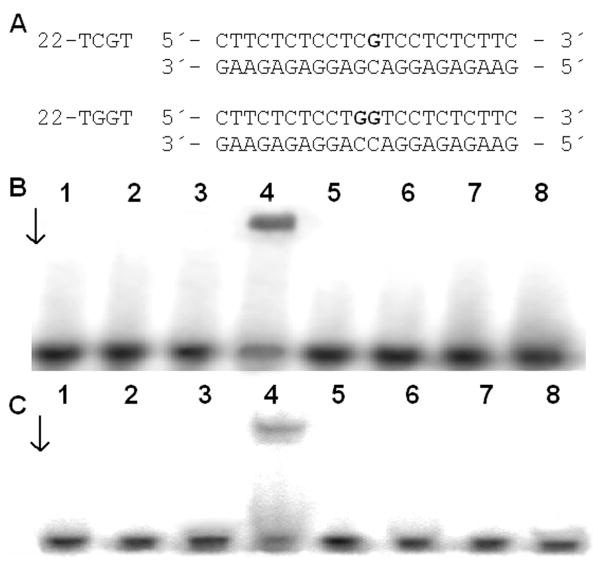
Since the DNA conformational changes caused by 1 and 2 are very different from those caused by cisplatin, experiments were performed to determine if differences exist in the recognition of the monofunctional-intercalative and bifunctional adducts by HMG domain proteins. The interactions of the domains A and B of HMGB1, which is considered the prototype of this family of proteins, with the adducts of 1 or 2 were investigated using a gel mobility shift assay.33, 61 In these experiments, the 22-bp duplex with blunt ends (see Figure 5A for its sequence) was modified so that it contained a single, site-specific, monofunctional adduct formed by 1 or 2; or for comparative purposes, the 22-bp duplex was also used which was identical to that containing the monofunctional adduct of 1 or 2 except that its central sequence was TGGT/ACCA at which a single, site-specific 1,2-GG intrastrand cross-link of cisplatin was formed (Figure 5A). The binding of the domains HMGB1a or HMGB1b to these DNA probes was detected as a band of reduced electrophoretic mobility on the gels.33, 61 These proteins exhibited negligible binding to the unmodified 22-bp duplexes, whereas both HMGB1a and HMGB1b recognized and bound to the duplex containing the 1,2-GG intrastrand cross-link of cisplatin. The results of the incubation of the duplexes modified with 1 or 2 with HMGB1a or HMGB1b indicate that neither of these proteins bound the probes under conditions where the HMGB1a or HMGB1b proteins associated with the duplex containing the 1,2-GG intrastrand cross-link (Figures 5B,C). Hence, the monofunctional adducts of 1 or 2 are either not recognized at all by HMG domain proteins, or the affinity of these proteins is markedly lower for the monofunctional adducts than for the 1,2-GG intrastrand cross-link.
Figure 5.
Gel mobility shift assay analysis of the interaction of a 22 bp duplex containing the 1,2-GG intrastrand cross-link of cisplatin (22-TGGT) or adducts of 1 or 2 (22-TCGT) with HMGB1a and HMGB1b. Radioactively labeled oligodeoxyribonucleotide duplexes (1 pmol) were incubated with 10 ng of HMGB1a or 40 ng of HMGB1b. A. Sequences of the synthetic oligodeoxyribonucleotides used in this study. B, C. Autoradiograms of the gel mobility shift assay analysis of the interaction with HMGB1a (B) and HMGB1b (C). Lanes: 1,2 - control (unplatinated) duplex; 3,4 – 1,2-GG intrastrand cross-link of cisplatin (22-TGGT); 5,6 – 22-TCGT duplex modified by 1; 7,8 – 22-TCGT duplex modified by 2. Lanes 1,3,5,7 – no protein; lanes 2,4,6, 8 – 10 ng of HMGB1a (B) or 40 ng of HMGB1b (C).
RNA Polymerase II Transcription and DNA Repair Synthesis in Human Cell-Free Extracts (CFE)
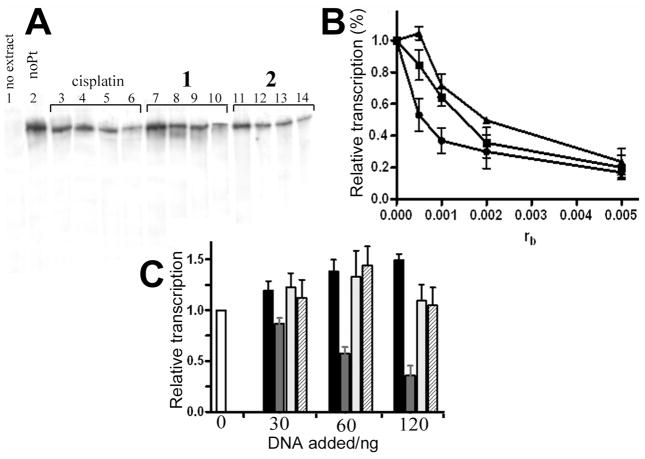
To investigate how the adducts formed by complexes 1 and 2 affect DNAprocessing enzymes, we used an assay to test the ability of RNA polymerase II (RNA pol II) in human CFE to transcribe DNA modified with 1 and 2. In addition, we used an in vitro system to study repair of platinum lesions by CFE. The effect of 1 and 2 on the transcription activity of human RNA pol II was tested using a commercially available in vitro transcription system kit. Using a previously described procedure,62 the RNA pol II transcription template pCMV-Gluc modified with 1, 2 or cisplatin and an unmodified control were incubated with the HeLa nuclear extract supplied with this kit. This extract can support accurate transcription initiation by RNA pol II and exhibits both basal and regulated patterns of RNA transcription.63 This nuclear extract also is the source for a variety of transcription factors, DNA binding proteins and the enzymatic machinery involved in the process of RNA synthesis. Specific transcription from the CMV promoter results in a run-off transcript 688 nucleotides in length. The newly synthesized full-length transcripts can be subsequently detected by gel electrophoresis. As seen in Figure 10A, in the absence of platinum complex a high level of transcript was observed. In contrast, a significant decrease in the amount of full-length transcript was observed as a result of increasing template modification by the three platinum complexes tested in this experiment. The relative amount of transcript generated in each reaction was quantified and plotted as a function of the level of platination (rb) (Figure 6B). Under the specific conditions of this assay, RNA pol II transcription was affected by very low levels of platinum adducts. The lesions produced by 2 were considerably more effective in inhibiting RNA pol II transcription than those produced by 1 or cisplatin, in particular at low adduct levels.
Figure 6.
Inhibition of RNA polymerase II transcription by DNA adducts of complexes 1, 2 and cisplatin. (A) Autoradiogram of the 8% PAA/8 M urea denaturing gel. Lanes: 1, unmodified substrate, no extract added; 2, control, unplatinated pCMV-Gluc substrate; 3, 4, 5 and 6, pCMV-Gluc substrate modified with cisplatin at rb = 5×10−4, 1×10−3, 2×10−3 and 5×10−3, respectively; 7, 8, 9 and 10, pCMV-Gluc substrate modified by complex 1 at rb = 5×10−4, 1×10−3, 2×10−3 and 5×10−3, respectively; 11, 12, 13 and 14, pCMV-Gluc substrate modified by complex 2 at rb = 5×10−4, 1×10−3, 2×10−3 and 5×10−3, respectively. B. Quantitative assessment. The relative transcription was assed as follows: the amount of full length transcript at each rb was quantified (in % of total radioactivity in the lane) and calculated as the percentage of that generated by the control, undamaged template. Data represent results of two independent experiments and are expressed as mean percentages ±SEM. (■) cisplatin; (▲) 1; (●) 2. C. Inhibition of RNA polymerase II transcription by the addition of increasing amount of exogenously platinated pUC19 DNA. The amount of full-length transcript in each lane is expressed as a mean fraction (±SEM) of that generated in the absence of exogenously added DNA (white bar). Black bars: transcription of undamaged DNA; gray bars: transcription of cisplatin modified DNA; light gray bars: transcription of DNA modified with 1; hatched bars: transcription of DNA modified with 2.
To determine if the catalytic activity of RNA pol II was inhibited as a consequence of erroneous recruitment of factors essential for RNA pol II transcription initiation62 to the DNA adducts of 1 or 2, the following competition experiments were performed: RNA pol II transcription of undamaged template pCMV-Gluc was examined in the presence of increasing levels of exogenous pUC19 plasmid containing multiple lesions caused by 1, 2, or cisplatin. As shown in Figure 10C, the initial addition of undamaged exogenous plasmid resulted in an overall increase in the amount of transcript generated by pCMV-Gluc. Additional unmodified plasmid only led to a minor increase in transcript levels. RNA pol II transcription of pCMVGluc template was significantly reduced by the addition of cisplatin modified exogenous plasmid. In contrast, a negligible inhibition effect was observed when the transcription assay was performed in the presence of exogenous plasmid containing the adducts of 1 or 2 (Figure 6C).
DNA repair synthesis by human cell extract
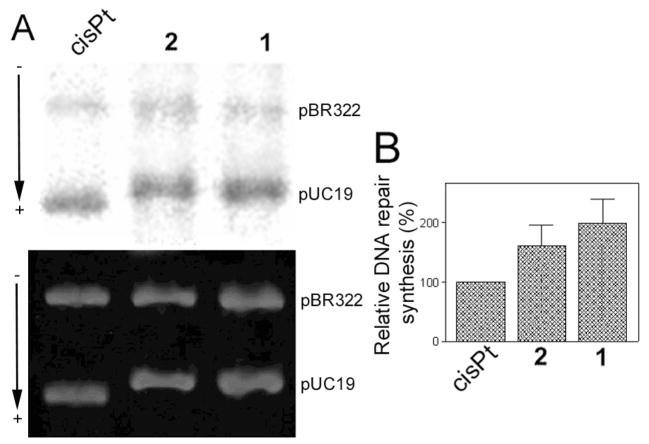
Finally, pUC19 plasmid (2686 bp) randomly modified with 1, 2 or cisplatin at rb = 0.03 was used to study the repair of each type of adduct by the CFE from repair-proficient HeLa cells. The repair activity was monitored by measuring the amount of incorporated radiolabeled nucleotide. The incorporation of radioactive precursor was corrected for the relative DNA content in each band. As illustrated in Figure 7, damage-induced DNA repair synthesis detected in the plasmid modified with 2 was considerably lower than that found for 1 at the same level of modification, although significantly higher than that found for cisplatin.
Figure 7.
In vitro DNA repair synthesis assay. Repair mediated by the extract prepared from the repair-proficient HeLa cell line was monitored using unmodified pBR322 plasmid and pUC19 plasmid, unmodified or modified at rb = 0.03 with cisplatin, 1 or 2. A. Results of a typical experiment. Top panel, autoradiogram of the gel showing the incorporation of [α-32P]dCTP; bottom panel, a photograph of the EtBr stained gel. Lanes: cisplatin, unmodified pBR322 plus pUC19 modified with cisplatin; 1, unmodified pBR322 plus pUC19 modified with 1, 2, unmodified pBR322 plus pUC19 modified with 2. (B) Incorporation of [α-32P]dCTP into unmodified or platinated pUC19 plasmid. For all quantifications representing the mean values of three separate experiments, incorporation of radioactive material is corrected for the relative DNA content in each band. The radioactivity associated with incorporation of [α-32P]dCTP into DNA modified with cisplatin was arbitrarily set to 100%. Values shown in the graph are the means (±SEM) of three separate experiments, each conducted in quadruplicate.
Computational Studies of DNA Adduct Formation
Critical differences exist between the adducts produced by the platinum–acridines and cisplatin-type cross-links with respect to their base selectivity and rate of formation. While guanine-N7 is the major target of cisplatin and the platinum–acridines, one striking feature of thiourea-based 1 is its ability to produce a high percentage (approximately 20% in native DNA) of previously unknown adenine monoadducts. The majority of these adducts are formed with N7 of the nucleobase. Replacement of the thiourea donor with an amidine group in compound 2 has a major impact on the DNA damage profile and the kinetics of platination. Compound 2 reacts with the model nucleotide 2′-deoxyguanosine and double-stranded DNA significantly more rapidly and shows a less pronounced affinity for adenine-containing base-pair steps than complex 1. Attempts to detect adenine adducts in enzymatic digests of DNA treated with the amidinelinked conjugate have been unsuccessful (unpublished results).
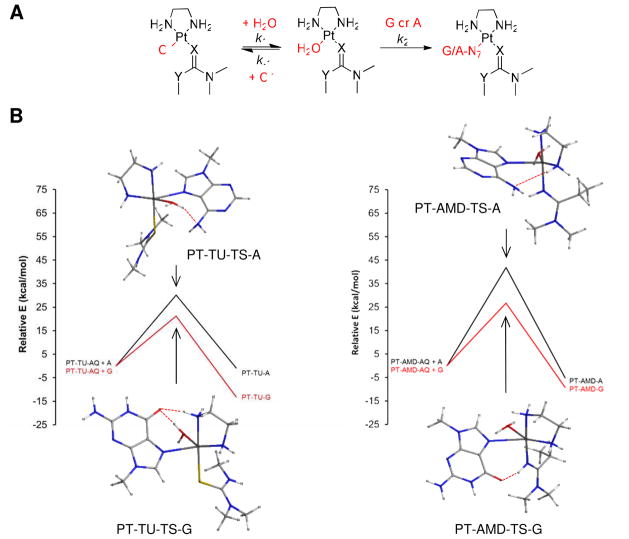
To shed light on the reactivity and nucleobase selectivity of complexes 1 and 2, we performed density functional theory (DFT) calculations on the reaction sequence leading to adduct formation with guanine and adenine bases. Previous model studies performed with compounds 1 and 2 have shown that simple mononucleo(t)sides, while unable to mimic the sequence context and donor site selectivity of intercalator-driven adduct formation in doublestranded DNA, are extremely useful for studying the relative nucleobase affinities of newly designed derivatives.26,64 The proposed reaction pathway involves aquation of the chloro complexes in an equilibrium (k1/k−1) that lies far to the left, followed by substitution of the aqua ligand by the nucleobase nitrogen (k2) (Figure 8A). The binding processes were modeled as associative mechanisms proceeding through pentacoordinate, trigonal-bipyramidal transition states. The reaction profiles along with the lowest-energy optimized transition states for each reaction are shown in Figure 8B, and important geometric parameters and energies are summarized in Tables 1 and 2. A table of absolute energies and views of the optimized geometries of the aqua intermediates and final nucleobase adducts are available as Supporting Information (Table S3).
Figure 8.
Results of the DFT computations. (A) General reaction sequence modeled for compounds 1 (X = S, Y = NH; “TU”) and 2 (X = NH, Y = CH2; “AMD”) using truncated N-dimethyl carrier ligands and 9-methylpurine bases (G, A). (B) Computed reaction pathways for nucleobase binding to the aquated forms of compounds 1 and 2 showing the lowest-energy optimized transition state geometries. For the optimized structures of the aqua complexes and the final nucleobase adducts, see Supporting Information (Figure S1).
Table 1.
Selected Geometrical Parameters in Minimized Models of Platinum Complexes and Transition States (TS).
| Lowest- Energy Structure | Selected Bond Distances | H-Bonding Interactionsa | ||
|---|---|---|---|---|
| Pt-O (Å) | Pt-GN7/AN7 (Å) | Donor-Acceptor Pair | D…A (Å) | |
| PT-AMD-AQb | 2.116 | -- | -- | -- |
| PT-TU-AQ | 2.087 | -- | OHw-Ntuc | 2.659 |
|
| ||||
| PT-AMD-TS-A | 2.459 | 2.442 | NHamd-AN2 | 3.306 (weak) |
| PT-AMD-TS-G | 2.428 | 2.553 | NHamd-GO6 | 2.821 |
| PT-TU-TS-A | 2.365 | 2.570 | OHw-AN2 | 2.647 |
| PT-TU-TS-G | 2.434 | 2.485 | NHen-GO6 | 2.889 |
| OHw-GO6 | 2.681 | |||
|
| ||||
| PT-AMD-A | --- | 2.042 | NHamd-AN2 | 3.277 (weak) |
| PT-AMD-G | --- | 2.049 | NHamd-GO6 | 2.867 |
| PT-TU-A | --- | 2.051 | NHen-AN2 | 2.942 |
| PT-TU-G | --- | 2.065 | NHen-GO6 | 2.684 |
Donor–acceptor distances less than 3.40 Å considered.
For optimized structures of the aqua complexes and the final nucleobase adducts of 1 and 2, see Supporting Information (Figure S1).
Abbreviations: w, aqua ligand; tu, thiourea; amd, amidine; en, ethylenediamine.
Table 2.
Summary of Free Activation Energies and Binding Free Energies.a
| Reaction | ΔG‡ (kJ mol−1) | ΔGrxn (kJ mol−1) |
|---|---|---|
| PT-AMD-AQ + A | 175.06 | −21.77 |
| PT-AMD-AQ + G | 111.96 | −38.77 |
| PT-TU-AQ + A | 126.41 | −3.64 |
| PT-TU-AQ + G | 88.68 | −55.18 |
Solvation-corrected energies based on from single-point calculations of the gasphase structures performed with the self-consistent reaction field (SCRF) approach.
The first reaction modeled is the substitution of chloride in 1 and 2 by water, leading to the aquated species, PT-TU-AQ and PT-AMD-AQ. Based on the total energies calculated in a dielectric mimicking water, formation of the latter amidine-substituted aqua complex is favored by 25 kJ mol−1 over aquation of the analogus thiourea-based complex, suggesting that compound 2 aquates to a greater extent than compound 1. A difference in ΔG‡ of the same order of magnitude would translate into a ~103-fold enhanced rate of aquation. Unfortunately, attempts to generate optimized transition state geometries for the aquation reactions were unsuccessful. Loss of chloride in a hydrolytic pre-equilibrium is the rate-determining step in platinum drug–DNA adduct formation. Thus, the observation that complex 2 is significantly more reactive with DNA than compound 153 is in agreement with a greater rate and extent of aquation of the amidine derivative, which is supported by the computational results in this study.
To mimic DNA adduct formation, substitution of the aqua ligands in PT-TU-AQ and PTAMD- AQ by N7 of guanine (G) or adenine (A) (Figure 8B) was modeled. Based on the binding free energies (ΔGrxn) and free energies of activation (ΔG‡) calculated for the four reactions (Table 2), G-N7 is the thermodynamically and kinetically preferred target of the platinum–acridines, which is in agreement with experimental findings.20–25 The pentacoordinate transition states leading to the adducts show distinct hydrogen-bonding interactions (Figure 8B, Table 1) involving G-O6 and A-N6 as acceptors and the aqua-OH, en-NH, or amidine-NH as H-bond donors. An important observation is that, while the thiourea sulfur is unable to participate in hydrogen bonding, the amidine NH group forms a strong hydrogen bond with G-O6, both in the transition state (PT-AMD-TS-G, Figure 8B) and in the final adduct (PT-AMD-G, see Figure S1 in the Supporting Information). Another striking feature of the modeled reaction pathways is that the reaction of A-N7 with PT-AMDAQ is kinetically disfavored by ΔΔG‡ ≈ 50 kJ mol−1 compared to the analogous reaction with PT-TU-AQ. This difference in transition state free energies would explain why compound 1, but not compound 2, forms a greater percentage of DNA adducts with adenine. Likewise, a comparison of the ΔG‡ values for the substitution reactions involving G-N7 suggests that adduct formation with PT-TU-AQ is kinetically favored over PT-AMD-AQ, which seemingly contradicts the observation that amidine-based compound 2 reacts more rapidly with G-N7 in 2′-deoxyguanosine and double-stranded DNA than the thiourea-based prototype, 1. However, the opposite reactivity is observed for the formation of the aqua species (see above), which is in agreement with experimental data because rapid aquation can be considered a rateenhancing event in guanine adduct formation.
Adduct Characterization Using Chemical Probes
To further characterize the distortion induced in DNA by the monofunctional adducts formed by 1 and 2, the platinum-modified 22-bp oligonucleotide duplex in Figure 9 was treated with reagents that are commonly used as tools for monitoring conformational changes in B-DNA. The single, site-specific adducts of 1 or 2 were generated at the central 5′-CG/CG base-pair step. The choice of the central sequence at which the site-specific adducts of 1 or 2 were formed is substantiated as follows. Using DNA polymerase stop assays we previously demonstrated26 that compound 2 shows a slightly altered damage profile compared to compound 1. Similar to compound 1, compound 2 produced adducts in the sequences 5′-CTG, 5′-CGATG, and 5′-CGG, but not in poly-dG, consistent with the notion that this derivative targets 5′-pyrimidine–guanine steps.26 Unlike compound 1, however, which produces a high percentage of adenine adducts, the only platinum-containing DNA fragment isolated from enzymatic digests of CT DNA treated with compound 2 was 2′-deoxyguanosine (unpublished data). Thus, sequences containing platinated central 5′-CG damage sites common to both derivatives were chosen as models in this study.
The reagents included KMnO4, bromine, and DEPC, which are used as probes for structural changes in the vicinity of thymine, cytosine, and adenine/guanine residues, respectively.49, 65, 66 These probes react, under certain conditions, with nucleobases in single-stranded DNA and distorted double-stranded DNA, but not with nucleobases in intact, double-stranded DNA. For this analysis, we used exactly the same methodology as in our recent studies of DNA adducts of various antitumor platinum drugs.49, 65, 66 The results (Figure S2 in the Supplemental Information) are schematically summarized in Figure 9.
The pattern and degree of reactivity toward the chemical probes shows that the conformational distortion induced by monofunctional adducts of 1 and 2 is delocalized, extending over at least four base pairs around the adduct. The distortion induced by the adduct formed by 2 appears to be somewhat more pronounced than that of 1 (Figure S1 in the Supporting Information). Interestingly, the most pronounced structural perturbations in these adducts are observed at T and A bases adjacent to the 5′-CG/CG base-pair step, the site of intercalation of the acridine chromophore. The fact that the cytosine bases at the intercalation pocket are relatively insensitive to bromination is in agreement with the non-denaturing nature of the hybrid adducts formed by these agents.
Bending and Unwinding in Site-Specifically Modified Duplexes
In this work we performed studies on the bending and unwinding induced by single, site-specific, monofunctional adducts of 1 or 2 formed in oligodeoxyribonucleotide duplexes at guanine residues. As in the previous studies,33, 60, 67, 68 we used variations in electrophoretic mobility as a quantitative measure of the extent of DNA curvature to analyze the bending and unwinding induced by the single, site-specific adduct formed by 1 or 2. The oligodeoxyribonucleotide duplexes whose sequences are shown in Figure 10A were used in these studies. The ligation products of these unplatinated duplexes and duplexes containing a single, site-specific adduct of 1 or 2 were analyzed on native polyacrylamide electrophoresis (PAGE) gels. Details of this assay have been discussed in previous reports.36, 49 A representative experiment showing the mobility of the ligation products for 1 or 2 and the graphical analysis of the gel are shown in Figure 10B,C. The data demonstrate that the monofunctional adducts formed by 1 and 2, in the sequence 5′-TCGT, bend the model duplex by 7° and 13° toward the major groove and concomitantly unwind it by 11° and 16°, respectively. For comparison, using a similar setup, the 1,2-GG intrastrand cross-link has been demonstrated to kink the DNA by 32–34° and unwind it by 13°.50 The direction of the bend was determined using the duplex TCGT(A/T)(33) (Figure 10A), which contained, besides a single monofunctional adduct, an (AT)6 tract located “in phase” with the adduct, separating the platinated base pairs and the center of the A tract by 11 bp.66, 69, 70 In summary, the hybrid adducts cause local unwinding of the model duplexes similar to the 1,2-intrastrand cross-link but cause significantly less severe major-groove directed bending than the adducts formed by the clinical agent.
Effect of Monofunctional Adducts on Duplex Stability
A calorimetric technique was used to study the effect of the monofunctional adducts formed by 1 or 2 on the thermal stability and energetics of a site-specifically platinated 15-base-pair (bp) DNA duplex (Figure 11A). The thermodynamic stability of the drug-damaged DNA has been shown to play an important role in the cellular response to and biological activity of platinum antitumor drugs.47, 48, 67, 71–74 Here, we studied duplexes containing unique monofunctional adducts formed by 1 or 2 at guanine residues in the central sequence 5′-CGT. Figure 11B shows DSC melting profiles (ΔCp versus T) for the parent unmodified 15-bp duplex (solid curve) and the same duplexes containing a single monofunctional adduct of 1 or 2. Each transition showed negligible changes in the heat capacities between the initial and final states, and denaturation (heating) and renaturation (cooling) curves for the unmodified and platinated duplexes were superposable (not shown), which is consistent with the reversibility of the melting equilibrium. The calorimetric data were interpreted based on the assumption that the thermodynamic parameters for the melting of the unmodified and platinated duplexes can be ascribed to differences in the initial duplex states. This implies that the final single-stranded states should be thermodynamically equivalent at the elevated temperatures at which they are formed. This assumption was verified (not shown) similarly to earlier reports by recording identical circular dichroic spectra for samples of unplatinated and platinated duplexes heated at high temperatures (363 K).47, 48, 71, 72
DSC melting profiles (Figure 11B) were analyzed as described in the section Materials and Methods and the results are listed in Table 3. All thermodynamic parameters discussed in this work refer to the duplex dissociation process. Differences in the thermodynamics of strand dissociation due to the presence of a platinum adduct are presented as “ΔΔ” parameters. These parameters are computed by subtracting the appropriate value measured for the control, the unmodified duplex, from the value measured for the duplex containing the single, site-specific platinum adduct and are reported in Table 3 in parentheses. Inspection of these thermodynamic parameters reveals a number of interesting features. First, the formation of monofunctional adducts by 1 and 2 increased the duplex thermal stability by 6.4 and 3.4 K, respectively. Interestingly, the formation of monofunctional adducts by 1 and 2 resulted in a large decrease in the enthalpy of duplex dissociation (Table 3). In other words, the monofunctional adducts of these platinum complexes enthalpically destabilized the duplex relative to their unmodified counterpart. On the other hand, the formation of monofunctional adducts by 1 and 2 resulted in a substantial decrease in the duplex dissociation entropy (Table 3). Thus, the net result of these enthalpic and entropic effects was that the formation of monofunctional adducts by 1 and 2 induced only a very small increase (0.7 kJ mol−1) in the free energy of duplex dissociation at 25 °C (ΔG0298; Table 3). Shape analysis of the experimental DSC curves allows model-dependent ΔHvH enthalpies to be calculated.75 We found that ΔHvH values were similar to ΔHcal ratios for all duplexes and platinum compounds tested in this work; the ratios of ΔHvH/ΔHcal were in the range of 1.01–1.03 (Table 3). The compensatory effects of the melting enthalpies and entropies, which lead to a virtually unchanged free energy of duplex dissociation, have been previously observed in the van’t Hoff analysis of a double-stranded dodecamer modified with 1.55 Based on this data, the energetic consequences of the intercalative hybrid adducts are strikingly different from those observed for the major 1,2-intrastrand cross-link of cisplatin, which significantly reduces the thermodynamic stability of the modified duplex.47
Table 3.
Calorimetry-Derived Thermodynamic Parameters for the Dissociation (Melting) of the Unmodified and Platinum-Modified 15-bp Duplexes.
| Duplex | Tma (K) | ΔHcala(kJ mol−1) | ΔS a (kJ K−1 mol−1) | ΔG0 298 a (kJ mol−1) | ΔHvH (kJ mol−1) | ΔHvH /Δ Hcal |
|---|---|---|---|---|---|---|
| Control | 338.8 | 488.1 | 1.445 | 57.3 | 499.3 | 1.02 |
| 1 | 345.2 (6.4) | 441.4 (−46.7) | 1.286 (−0.159) | 58.0 (0.7) | 447.4 | 1.01 |
| 2 | 342.2 (3.4) | 457.8 (−30.3) | 1.341 (−0.104) | 58.0 (0.7) | 470.0 | 1.03 |
The ΔHcal and ΔS values are averages derived from three independent experiments.
The experimental uncertainties of the parameters are as follows: Tm ± 0.5 K, ΔHcal ± 2%), ΔS ± 3%, ΔG0298 ± 3%). The “ΔΔ” parameters are given in parentheses (these parameters are computed by subtracting the appropriate value measured for the control, the unmodified duplex, from the value measured for the duplex containing the single, site-specific platinum adduct.
Discussion
The goal of the current project was to delineate mechanistic differences between two members of a novel class of platinum–acridine antitumor agents and compare their DNA damage mechanisms with that of cisplatin. Specifically, the experiments aimed to elucidate the consequences of changing a thiourea into an amidine donor group for the molecular mechanism of the hybrid agents at the DNA level. Using model studies in cell-free systems, the two central questions addressed were: (i) what specific DNA binding features might contribute to the enhanced cytotoxicity of the amidine-substituted derivatives(s), and (ii) what distinguishes the mechanism of the hybrids from that of clinical platinum drugs. Based on the data acquired in this study, a subtle structural modification made to the platinum–acridines has a major effect on the rates and selectivity of DNA binding, as well as on the DNA structural impact, recognition, and processing of the adducts formed.
The DNA-binding experiments performed in this study confirm that compound 2 has a major advantage over compound 1 with respect to the kinetics of DNA adduct formation. The DFT calculations suggest that this is due to a larger extent of aquation of the former complex. In the computational study, critical differences also emerged that suggest that the formation of adenine adducts is kinetically more favorable for compound 1 than for compound 2. This is in agreement with the broader array of adducts formed by the thiourea derivative, of which adenine adducts have been demonstrated to contribute significantly to the cytotoxic effect of compound 1.76 Although compound 2 does not show this degree of chemical promiscuity in its interactions with DNA, it forms guanine adducts more rapidly. Based on the relationship between DNA adduct levels in a set of platinum–acridine derivatives and their biological activities in H460 cell line,53 the DNA binding rate appears to be more critical than the nucleobase and sequence specificity of adduct formation in rapidly proliferating, repair proficient NSCLC cells. Amidine-NH–G-O6 hydrogen-bonding, as observed in the models of the transition states and the final adduct, may contribute to the high G affinity of compound 2 and produce structural effects at the adduct site not feasible for thiourea-based complex 1. High-resolution studies are underway to support this notion.
The biophysical and biochemical experiments performed with randomly (Figures 2–4,6 and 7) and site-specifically modified (Figures 5,9–11) DNA also demonstrate that substitution of the thiourea donor group (X = S, Y = NH, Figure 1) with an amidine donor group (X = NH, Y = CH2, Figure 1) has consequences for the local DNA adduct structure and global DNA conformation beyond the adduct sites. One striking difference between the geometries of complexes 1 and 2 is the bond angle Pt–X–C, which is 108.3(4) Å64 and 129.6(4) Å26 for 1 and 2, respectively. This structural alteration, along with differences in drug–DNA hydrogen bonding patterns, may lead to an altered geometry of intercalation. The combined physicochemical and biochemical data suggest that the DNA conformational changes produced by the adducts formed by compound 2 are more pronounced than the effects caused by compound 1. In particular, the higher degree of duplex unwinding (Figure 10) and the CD signatures (Figure 2) observed for compound 2 are in agreement with a geometry more favorable for classical intercalation of the acridine moiety in adducts of this derivative. Despite these structural differences, the adducts formed by both derivatives do not significantly affect the thermodynamic stability of the modified DNA due to complete enthalpy–entropy compensation (Figure 11, Table 3).
An important consequence of the small extent of bending in DNA modified with compounds 1 and 2 is the lack of recognition of the damage by HMG domain proteins (Figure 5). HMG domain proteins have been implicated in the cytotoxicity of cisplatin9 although these proteins have been shown to play no significant role in the mechanism of cisplatin-induced cytotoxicity in mouse embryonic cells.77 The role of HMG domain proteins as mediators or enhancers of cisplatin-triggered cytotoxicity is because HMG protein binding protects the 1,2 intrastrand cross-link from cellular repair.9 This suggests that binding of proteins that have a high affinity for the widened minor groove of severely bent DNA to the monofunctional– intercalative adducts does not play a role in the mechanism of action of the hybrid agents. This notion is further corroborated by the observation that transcription factors in the cell-free extracts used in this study are “hijacked” to cisplatin-induced cross-links but not to the sites of DNA damage produced by complexes 1 and 2 (Figure 6). These important findings may also apply to other nuclear proteins known to recognize severely distorted DNA, such as DNA damage recognition proteins belonging to the nucleotide excision repair (NER) complex. Recent clinical studies suggest that high levels of expression of proteins associated with NER of cisplatin–DNA adducts result in tumor resistance and, ultimately, are responsible for the low efficacy of classical platinum-based regimens.78, 79 NER most efficiently recognizes and removes irreversible DNA adducts that are bulky in nature or severely distort and thermodynamically destabilize double-stranded DNA. Adducts that cause local unstacking of nucleobases, disruption of Watson–Crick hydrogen bonding, or bending of the DNA helix are ideal substrates for the NER complex.80 Thus, based on the outcome of our experiments in cell-free systems, the monofunctional adducts produced by complexes 1 and 2 should be poor substrates for NER repair. The relative resistance to DNA repair would explain why complexes 1 and 2 show major pharmacological advantages over cisplatin in NSCLC, a cancer-type notorious for NER-related drug resistance. The repair synthesis assay in randomly modified plasmid performed in this study (Figure 7) demonstrates that the (presumably most cytotoxic) adducts formed by complex 2 are repaired less efficiently than the damage caused by derivative 1, but to a higher extent than cisplatin-type adducts. Because this assay measures the cumulative effects of all cellular repair pathways, additional experiments will have to be designed to assess the contribution of nucleotide excision to the global repair activity.
Another critical difference between complexes 1 and 2 emerged with respect to their ability to inhibit transcription of DNA by stalling RNA pol II. Complex 2 proves to be a significantly more potent inhibitor of RNA synthesis than either complex 1 or cisplatin (Figure 6). Inhibition of DNA transcription is considered to be a major mediator of the cell kill effect of cisplatin. The observation that monofunctional–intercalative adducts of complex 2 are able to efficiently stall RNA pol II suggests that transcription inhibition may contribute to the high cytotoxicity levels observed for the second-generation platinum–acridine pharmacophore. The ability of monofunctional adducts to interfere with RNA pol II catalyzed transcription in a structurally unique manner has recently been demonstrated for the complex diammine(pyridine)chloroplatinum(II) (“pyriplatin”).81, 82 Pyriplatin and its analogues form non-intercalative adducts different from the hybrid adducts investigated in this study. Thus, the mechanisms by which these two types of agents interfere with RNA synthesis may show critical differences, which may contribute to their distinctly different cytotoxic potencies.
In conclusion, we have undertaken a comparative study of the structural effects and recognition of the DNA damage produced by the prototype of a class platinum–acridine agents, complex 1, and its more potent second-generation derivative, complex 2. The data acquired in this study will help establish structure–activity relationships in this class of compounds, with the ultimate goal of providing novel therapies exhibiting a unique mechanism of action. A unique mode of DNA binding distinct from that of cisplatin appears to be critical for overcoming resistance in chemoresistant cancers.11
Supplementary Material
Acknowledgments
This work was supported by the Czech Science Foundation (Grant P205/11/0856), the student project of the Palacky University Olomouc (Grant PrF 2011 024) (to T. M.), and by the US National Institutes of Health, grant CA101880 (to U. B.). Computations were performed on the Wake Forest University DEAC Cluster, a centrally managed resource with support provided in part by the University. We thank Dr. Akbar Salam (Wake Forest University) for providing computational facilities and technical support.
Footnotes
Supporting Information. Tables of total energies, frequencies, atomic coordinates, and views of the optimized computational models. Figure demonstrating reactivity of chemical probes with the 22-bp duplexes containing single, site-specific adducts of 1 and 2. This material is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Kelland L. The resurgence of platinum-based cancer chemotherapy. Nature Rev Cancer. 2007;7:573–584. doi: 10.1038/nrc2167. [DOI] [PubMed] [Google Scholar]
- 2.Kalinowska-Lis U, Ochocki J, Matlawska-Wasowska K. Trans geometry in platinum antitumor complexes. Coord Chem Rev. 2008;252:1328–1345. [Google Scholar]
- 3.Hall MD, Dolman RC, Hambley TW. Platinum(IV) anticancer complexes. In: Sigel A, Sigel H, editors. Metal Ions in Biological Systems. Vol. 42. Marcel Dekker, Inc; New York, Basel: 2004. pp. 297–322. [PubMed] [Google Scholar]
- 4.Mangrum JB, Farrell NP. Excursions in polynuclear platinum DNA binding. Chem Commun. 2010;46:6640–6650. doi: 10.1039/c0cc01254h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bednarski PJ, Mackay FS, Sadler P. Photoactivatable platinum complexes. Anti- Cancer Agents Med Chem. 2007;7:75–93. doi: 10.2174/187152007779314053. [DOI] [PubMed] [Google Scholar]
- 6.Kostova I. Ruthenium complexes as anticancer agents. Curr Med Chem. 2006;13:1085–1107. doi: 10.2174/092986706776360941. [DOI] [PubMed] [Google Scholar]
- 7.Peacock AFA, Habtemariam A, Fernandez R, Walland V, Fabbiani FPA, Parsons S, Aird RE, Jodrell DI, Sadler PJ. Tuning the reactivity of osmium(II) and ruthenium(II) arene complexes under physiological conditions. J Am Chem Soc. 2006;128:1739–1748. doi: 10.1021/ja055886r. [DOI] [PubMed] [Google Scholar]
- 8.Liu Z, Habtemariam A, Pizarro AM, Fletcher SA, Kisova A, Vrana O, Salassa L, Bruijnincx PCA, Clarkson GJ, Brabec V, Sadler PJ. Organometallic half-sandwich iridium anticancer complexes. J Med Chem. 2011;54:3011–3026. doi: 10.1021/jm2000932. [DOI] [PubMed] [Google Scholar]
- 9.Jung Y, Lippard SJ. Direct cellular responses to platinum-induced DNA damage. Chem Rev. 2007;107:1387–1407. doi: 10.1021/cr068207j. [DOI] [PubMed] [Google Scholar]
- 10.Cepeda V, Fuertes M, Castilla J, Alonso C, Quevedo C, Perez JM. Biochemical mechanisms of cisplatin cytotoxicity. Anti-Cancer Agents Med Chem. 2007;7:3–18. doi: 10.2174/187152007779314044. [DOI] [PubMed] [Google Scholar]
- 11.Vrana O, Brabec V, Kleinwächter V. Polarographic studies on the conformation of some platinum complexes: relations to anti-tumour activity. Anti-Cancer Drug Des. 1986;1:95–109. [PubMed] [Google Scholar]
- 12.Bowler BE, Lippard SJ. Modulation of platinum antitumor drug binding to DNA by linked and free intercalators. Biochemistry. 1986;25:3031–3038. doi: 10.1021/bi00358a044. [DOI] [PubMed] [Google Scholar]
- 13.Petitjean A, Barton JK. Tuning the DNA reactivity of cis-platinum: Conjugation to a mismatch-specific metallointercalator. J Am Chem Soc. 2004;126:14728–14729. doi: 10.1021/ja047235l. [DOI] [PubMed] [Google Scholar]
- 14.Baruah H, Barry CG, Bierbach U. Platinum-intercalator conjugates: From DNAtargeted cisplatin derivatives to adenine binding complexes as potential modulators of gene regulation. Curr Topics Med Chem. 2004;4:1537–1549. doi: 10.2174/1568026043387313. [DOI] [PubMed] [Google Scholar]
- 15.Liu HK, Parkinson JA, Bella J, Wang F, Sadler PJ. Penetrative DNA intercalation and G-base selectivity of an organometallic tetrahydroanthracene RuII anticancer complex. Chem Sci. 2010;1:258–270. [Google Scholar]
- 16.Liu HK, Sadler PJ. Metal complexes as DNA intercalators. Acc Chem Res. 2011;44:349–359. doi: 10.1021/ar100140e. [DOI] [PubMed] [Google Scholar]
- 17.Guddneppanavar R, Bierbach U. Adenine-N3 in the DNA minor groove - An emerging target for platinum containing anticancer pharmacophores. Anti-Cancer Agents Med Chem. 2007;7:125–138. doi: 10.2174/187152007779313991. [DOI] [PubMed] [Google Scholar]
- 18.Takahara PM, Rosenzweig AC, Frederick CA, Lippard SJ. Crystal structure of double-stranded DNA containing the major adduct of the anticancer drug cisplatin. Nature. 1995;377:649–652. doi: 10.1038/377649a0. [DOI] [PubMed] [Google Scholar]
- 19.Kartalou M, Essigmann JM. Recognition of cisplatin adducts by cellular proteins. Mutation Res. 2001;478:1–21. doi: 10.1016/s0027-5107(01)00142-7. [DOI] [PubMed] [Google Scholar]
- 20.Barry CG, Baruah H, Bierbach U. Unprecedented monofunctional metalation of adenine nucleobase in guanine- and thymine-containing dinucleotide sequences by a cytotoxic platinum-acridine hybrid agent. J Am Chem Soc. 2003;125:9629–9637. doi: 10.1021/ja0351443. [DOI] [PubMed] [Google Scholar]
- 21.Martins ET, Baruah H, Kramarczyk J, Saluta G, Day CS, Kucera GL, Bierbach U. Design, synthesis, and biological activity of a novel non-cisplatin-type platinum-acridine pharmacophore. J Med Chem. 2001;44:4492–4496. doi: 10.1021/jm010293m. [DOI] [PubMed] [Google Scholar]
- 22.Baruah H, Rector CL, Monnier SM, Bierbach U. Mechanism of action of noncisplatin type DNA-targeted platinum anticancer agents: DNA interactions of novel acridinylthioureas and their platinum conjugates. Biochem Pharmacol. 2002;64:191–200. doi: 10.1016/s0006-2952(02)01107-3. [DOI] [PubMed] [Google Scholar]
- 23.Barry CG, Day CS, Bierbach U. Duplex-promoted platination of adenine-N3 in the minor groove of DNA: Challenging a longstanding bioinorganic paradigm. J Am Chem Soc. 2005;127:1160–1169. doi: 10.1021/ja0451620. [DOI] [PubMed] [Google Scholar]
- 24.Baruah H, Wright MW, Bierbach U. Solution structural study of a DNA duplex containing the guanine-N7 adduct formed by a cytotoxic platinum-acridine hybrid agent. Biochemistry. 2005;44:6059–6070. doi: 10.1021/bi050021b. [DOI] [PubMed] [Google Scholar]
- 25.Budiman ME, Alexander RW, Bierbach U. Unique base-step recognition by a platinum-acridinylthiourea conjugate leads to a DNA damage profile complementary to that of the anticancer drug cisplatin. Biochemistry. 2004;43:8560–8567. doi: 10.1021/bi049415d. [DOI] [PubMed] [Google Scholar]
- 26.Ma Z, Choudhury JR, Wright MW, Day CS, Saluta G, Kucera GL, Bierbach U. A non-cross-linking platinum-acridine agent with potent activity in nonsmall- cell lung cancer. J Med Chem. 2008;51:7574–7580. doi: 10.1021/jm800900g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Damsma GE, Alt A, Brueckner F, Carell T, Cramer P. Mechanism of transcriptional stalling at cisplatin-damaged DNA. Nature Struct Mol Biol. 2007;14:1127–1133. doi: 10.1038/nsmb1314. [DOI] [PubMed] [Google Scholar]
- 28.Brabec V, Kasparkova J. Role of DNA repair in antitumor effects of platinum drugs. In: Hadjiliadis N, Sletten E, editors. Metal Complex - DNA Interactions. Wiley; Chichester, UK: 2009. pp. 175–208. [Google Scholar]
- 29.Brabec V, Palecek E. Interaction of nucleic acids with electrically charged surfaces. II. Conformational changes in double-helical polynucleotides. Biophys Chem. 1976;4:76–92. doi: 10.1016/0301-4622(76)80009-9. [DOI] [PubMed] [Google Scholar]
- 30.Brabec V, Palecek E. The influence of salts and pH on polarographic currents produced by denatured DNA. Biophysik. 1970;6:290–300. doi: 10.1007/BF01575324. [DOI] [PubMed] [Google Scholar]
- 31.Stros M. DNA bending by the chromosomal protein HMG1 and its high mobility group box domains. Effect of flanking sequences. J Biol Chem. 1998;273:10355–10361. [PubMed] [Google Scholar]
- 32.Stros M. Two mutations of basic residues within the N-terminus of HMG-1 B domain with different effects on DNA supercoiling and binding to bent DNA. Biochemistry. 2001;40:4769–4779. doi: 10.1021/bi002741i. [DOI] [PubMed] [Google Scholar]
- 33.Kasparkova J, Delalande O, Stros M, Elizondo-Riojas MA, Vojtiskova M, Kozelka J, Brabec V. Recognition of DNA interstrand cross-link of antitumor cisplatin by HMGB1 protein. Biochemistry. 2003;42:1234–1244. doi: 10.1021/bi026695t. [DOI] [PubMed] [Google Scholar]
- 34.Reardon JT, Vaisman A, Chaney SG, Sancar A. Efficient nucleotide excision repair of cisplatin, oxaliplatin, and bis-aceto-ammine-dichloro-cyclohexylamineplatinum( IV) (JM216) platinum intrastrand DNA diadducts. Cancer Res. 1999;59:3968–3971. [PubMed] [Google Scholar]
- 35.Kim SD, Vrana O, Kleinwächter V, Niki K, Brabec V. Polarographic determination of subnanogram quantities of free platinum in reaction mixture with DNA. Anal Lett. 1990;23:1505–1518. [Google Scholar]
- 36.Kasparkova J, Mellish KJ, Qu Y, Brabec V, Farrell N. Site-specific d(GpG) intrastrand cross-links formed by dinuclear platinum complexes. Bending and NMR studies. Biochemistry. 1996;35:16705–16713. doi: 10.1021/bi961160j. [DOI] [PubMed] [Google Scholar]
- 37.Baruah H, Day CS, Wright MW, Bierbach U. Metal-intercalator-mediated selfassociation and one-dimensional aggregation in the structure of the excised major DNA adduct of a platinum-acridine agent. J Am Chem Soc. 2004;126:4492–4493. doi: 10.1021/ja038592j. [DOI] [PubMed] [Google Scholar]
- 38.Frisch MJ, Trucks GW, Schlegel HB, Scuseria GE, Robb MA, Cheeseman JR, Montgomery JA, Jr, Vreven T, Kudin KN, Burant JC, Millam JM, Iyengar SS, Tomasi J, Barone V, Mennucci B, Cossi M, Scalmani G, Rega N, Petersson GA, Nakatsuji H, Hada M, Ehara M, Toyota K, Fukuda R, Hasegawa J, Ishida M, Nakajima T, Honda Y, Kitao O, Nakai H, Klene M, Li X, Knox JE, Hratchian HP, Cross JB, Adamo C, Jaramillo J, Gomperts R, Stratmann RE, Yazyev O, Austin AJ, Cammi R, Pomelli C, Ochterski JW, Ayala PY, Morokuma K, Voth GA, Salvador P, Dannenberg JJ, Zakrzewski VG, Dapprich S, Daniels AD, Strain MC, Farkas O, Malick DK, Rabuck AD, Raghavachari K, Foresman JB, Ortiz JV, Cui Q, Baboul AG, Clifford S, Cioslowski J, Stefanov BB, Liu G, Liashenko A, Piskorz P, Komaromi I, Martin RL, Fox DJ, Keith T, Al-Laham MA, Peng CY, Nanayakkara A, Challacombe M, Gill PMW, Johnson B, Chen W, Wong MW, Gonzalez C, Pople JA. Gaussian 03, Revision A.1. Gaussian, Inc; Pittsburgh, PA: 2003. [Google Scholar]
- 39.Becke AD. Density-functional thermochemistry. 3. The role of exact exchange. J Chem Phys. 1993;98:5648–5652. [Google Scholar]
- 40.Lee C, Yang W, Parr RG. Development of the Colle-Salvetti correlation-energy formula into a functional of the electron density. Phys Rev B. 1988;37:785–789. doi: 10.1103/physrevb.37.785. [DOI] [PubMed] [Google Scholar]
- 41.Hay PJ, Wadt WR. Ab initio effective core potentials for molecular calculations. Potentials for K to Au including the outermost core orbitals. J Chem Phys. 1985;82:299–310. [Google Scholar]
- 42.Dunning THJ, Hay PJ. Gaussian basis sets for molecular calculations. Modern Theor Chem. 1977;3:1–27. [Google Scholar]
- 43.Miertus S, Scrocco E, Tomasi J. Electrostatic interaction of a solute with a continuum. A direct utilizaion of AB initio molecular potentials for the prevision of solvent effects. Chem Phys. 1981;55:117–129. [Google Scholar]
- 44.Rodger A. Linear dichroism. Methods Enzymol. 1993;226:232–258. doi: 10.1016/0076-6879(93)26012-x. [DOI] [PubMed] [Google Scholar]
- 45.Rodger A, Norden B. Circular Dichroism and Linear Dichroism. Oxford University Press; Oxford, New York, Tokyo: 1997. [Google Scholar]
- 46.Leharne SA, Chowdhry BZ. Thermodynamic background to differential scanning calorimetry. In: Ladbury JE, Chowdhry BZ, editors. Biocalorimetry: Applications of Calorimetry in the Biological Sciences. J. Wiley & Sons; Chichester, UK: 1998. pp. 157–182. [Google Scholar]
- 47.Hofr C, Farrell N, Brabec V. Thermodynamic properties of duplex DNA containing a site-specific d(GpG) intrastrand crosslink formed by an antitumor dinuclear platinum complex. Nucleic Acids Res. 2001;29:2034–2040. doi: 10.1093/nar/29.10.2034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hofr C, Brabec V. Thermal and thermodynamic properties of duplex DNA containing site-specific interstrand cross-link of antitumor cisplatin or its clinically ineffective trans isomer. J Biol Chem. 2001;276:9655–9661. doi: 10.1074/jbc.M010205200. [DOI] [PubMed] [Google Scholar]
- 49.Brabec V, Sip M, Leng M. DNA conformational distortion produced by site-specific interstrand cross-link of trans-diamminedichloroplatinum(II) Biochemistry. 1993;32:11676–11681. doi: 10.1021/bi00094a025. [DOI] [PubMed] [Google Scholar]
- 50.Bellon SF, Coleman JH, Lippard SJ. DNA unwinding produced by site-specific intrastrand cross- links of the antitumor drug cis-diamminedichloroplatinum(II) Biochemistry. 1991;30:8026–8035. doi: 10.1021/bi00246a021. [DOI] [PubMed] [Google Scholar]
- 51.Kasparkova J, Farrell N, Brabec V. Sequence specificity, conformation, and recognition by HMG1 protein of major DNA interstrand cross-links of antitumor dinuclear platinum complexes. J Biol Chem. 2000;275:15789–15798. doi: 10.1074/jbc.M000777200. [DOI] [PubMed] [Google Scholar]
- 52.Malina J, Kasparkova J, Natile G, Brabec V. Recognition of major DNA adducts of enantiomeric cisplatin analogs by HMG box proteins and nucleotide excision repair of these adducts. Chem Biol. 2002;9:629–638. doi: 10.1016/s1074-5521(02)00134-5. [DOI] [PubMed] [Google Scholar]
- 53.Choudhury J, Rao L, Bierbach U. Rates of intercalator-driven platination of DNA determined by a restriction enzyme cleavage inhibition assay. J Biol Inorg Chem. 2011;16:373–380. doi: 10.1007/s00775-010-0733-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kostrhunova H, Vrana O, Suchankova T, Gibson D, Kasparkova J, Brabec V. Different features of the DNA binding mode of antitumor cis-amminedichlorido(cyclohexylamine)platinum(II) (JM118) and cisplatin in vitro. Chem Res Toxicol. 2010;23:1833–1842. doi: 10.1021/tx1002904. [DOI] [PubMed] [Google Scholar]
- 55.Baruah H, Bierbach U. Biophysical characterization and molecular modeling of the coordinative-intercalative DNA monoadduct of a platinum-acridinylthiourea agent in a site-specifically modified dodecamer. J Biol Inorg Chem. 2004;9:335 – 344. doi: 10.1007/s00775-004-0534-3. [DOI] [PubMed] [Google Scholar]
- 56.Schipper PE, Norden B, Tjerneld F. Determination of binding geometry of DNAadduct systems through induced circular-dichroism. Chem Phys Lett. 1980;70:17–21. [Google Scholar]
- 57.Wirth M, Buchardt O, Koch T, Nielsen PE, Norden B. Interactions between DNA and mono(aminoacridines), bis(aminoacridines), tris(aminoacridines), tetrakis(aminoacridines), and hexakis(aminoacridines) - a linear and circular-dichroism, electric orientation relaxation, viscometry, and equilibrium study. J Am Chem Soc. 1988;110:932–939. [Google Scholar]
- 58.Rodger A, Marington R, Geeves MA, Hicks M, de Alwis L, Halsall DJ, Dafforn TR. Looking at long molecules in solution: what happens when they are subjected to Couette flow? Phys Chem Chem Phys. 2006;8:3161–3171. doi: 10.1039/b604810m. [DOI] [PubMed] [Google Scholar]
- 59.Brabec V. DNA modifications by antitumor platinum and ruthenium compounds: their recognition and repair. Prog Nucleic Acid Res Mol Biol. 2002;71:1–68. doi: 10.1016/s0079-6603(02)71040-4. [DOI] [PubMed] [Google Scholar]
- 60.Kasparkova J, Brabec V. Recognition of DNA interstrand cross-links of cis-diamminedichloroplatinum( II) and its trans isomer by DNA- binding proteins. Biochemistry. 1995;34:12379–12387. doi: 10.1021/bi00038a035. [DOI] [PubMed] [Google Scholar]
- 61.Kasparkova J, Novakova O, Vrana O, Intini F, Natile G, Brabec V. Molecular aspects of antitumor effects of a new platinum(IV) drug. Mol Pharmacol. 2006;70:1708–1719. doi: 10.1124/mol.106.027730. [DOI] [PubMed] [Google Scholar]
- 62.Cullinane C, Mazur SJ, Essigmann JM, Phillips DR, Bohr VA. Inhibition of RNA polymerase II transcription in human cell extracts by cisplatin DNA damage. Biochemistry. 1999;38:6204–6212. doi: 10.1021/bi982685+. [DOI] [PubMed] [Google Scholar]
- 63.Bagchi MK, Tsai SY, Weigel NL, Tsai MJ, O’Malley BW. Regulation of in vitro transcription by progesterone receptor. Characterization and kinetic studies. J Biol Chem. 1990;265:5129–5134. [PubMed] [Google Scholar]
- 64.Ackley MC, Barry CG, Mounce AM, Farmer MC, Springer BE, Day CS, Wright MW, Berners-Price SJ, Hess SH, Bierbach U. Structure–activity relationships in platinum–acridinylthiourea conjugates: effect of the thiourea nonleaving group on drug stability, nucleobase affinity, and in vitro cytotoxicity. J Biol Inorg Chem. 2004;9:453 – 461. doi: 10.1007/s00775-004-0541-4. [DOI] [PubMed] [Google Scholar]
- 65.Zehnulova J, Kasparkova J, Farrell N, Brabec V. Conformation, recognition by high mobility group domain proteins, and nucleotide excision repair of DNA intrastrand cross-links of novel antitumor trinuclear platinum complex BBR3464. J Biol Chem. 2001;276:22191–22199. doi: 10.1074/jbc.M103118200. [DOI] [PubMed] [Google Scholar]
- 66.Kasparkova J, Zehnulova J, Farrell N, Brabec V. DNA interstrand cross-links of the novel antitumor trinuclear platinum complex BBR3464. Conformation, recognition by high mobility group domain proteins, and nucleotide excision repair. J Biol Chem. 2002;277:48076–48086. doi: 10.1074/jbc.M208016200. [DOI] [PubMed] [Google Scholar]
- 67.Malina J, Hofr C, Maresca L, Natile G, Brabec V. DNA interactions of antitumor cisplatin analogs containing enantiomeric amine ligands. Biophys J. 2000;78:2008–2021. doi: 10.1016/S0006-3495(00)76748-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Brabec V. Chemistry and structural biology of 1,2-interstrand adducts of cisplatin. In: Kelland LR, Farrell NP, editors. Platinum-based Drugs in Cancer Therapy. Humana Press Inc; Totowa/NJ: 2000. pp. 37–61. [Google Scholar]
- 69.Kasparkova J, Novakova O, Farrell N, Brabec V. DNA binding by antitumor trans- [PtCl2(NH3)(thiazole)]. Protein recognition and nucleotide excision repair of monofunctional adducts. Biochemistry. 2003;42:792–800. doi: 10.1021/bi026614t. [DOI] [PubMed] [Google Scholar]
- 70.Loskotova H, Brabec V. DNA interactions of cisplatin tethered to the DNA minor groove binder distamycin. Eur J Biochem. 1999;266:392–402. doi: 10.1046/j.1432-1327.1999.00866.x. [DOI] [PubMed] [Google Scholar]
- 71.Pilch DS, Dunham SU, Jamieson ER, Lippard SJ, Breslauer KJ. DNA sequence context modulates the impact of a cisplatin 1,2-d(GpG) intrastrand cross-link an the conformational and thermodynamic properties of duplex DNA. J Mol Biol. 2000;296:803–812. doi: 10.1006/jmbi.2000.3496. [DOI] [PubMed] [Google Scholar]
- 72.Hofr C, Brabec V. Thermal stability and energetics of 15-mer DNA duplex interstrand cross-linked by trans-diamminedichloroplatinum(II) Biopolymers. 2005;77:222–229. doi: 10.1002/bip.20216. [DOI] [PubMed] [Google Scholar]
- 73.Malina J, Novakova O, Vojtiskova M, Natile G, Brabec V. Conformation of DNA GG intrastrand cross-link of antitumor oxaliplatin and its enantiomeric analog. Biophys J. 2007;93:3950–3962. doi: 10.1529/biophysj.107.116996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Bursova V, Kasparkova J, Hofr C, Brabec V. Effects of monofunctional adducts of platinum(II) complexes on thermodynamic stability and energetics of DNA duplexes. Biophys J. 2005;88:1207–1214. doi: 10.1529/biophysj.104.051771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Marky LA, Breslauer KJ. Calculating thermodynamic data for transitions of any molecularity from equilibrium melting curves. Biopolymers. 1987;26:1601–1620. doi: 10.1002/bip.360260911. [DOI] [PubMed] [Google Scholar]
- 76.Rao L, West TK, Saluta G, Kucera GL, Bierbach U. Probing platinumadenine- N3 adduct formation with DNA minor-groove binding agents. Chem Res Toxicol. 2010;23:1148–1150. doi: 10.1021/tx100170p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Wei M, Burenkova O, Lippard SJ. Cisplatin sensitivity in Hmgb1(−/−) and Hmgb1 mouse cells. J Biol Chem. 2003;278:1769–1773. doi: 10.1074/jbc.M210562200. [DOI] [PubMed] [Google Scholar]
- 78.Weaver D, Crawford E, Warner K, Elkhairi F, Khuder S, Willey J. ABCC5, ERCC2, XPA and XRCC1 transcript abundance levels correlate with cisplatin chemoresistance in non-small cell lung cancer cell lines. Mol Cancer. 2005;4:18. doi: 10.1186/1476-4598-4-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Fujii T, Toyooka S, Ichimura K, Fujiwara Y, Hotta K, Soh J, Suehisa H, Kobayashi N, Aoe M, Yoshino T, Kiura K, Date H. ERCC1 protein expression predicts the response of cisplatin-based neoadjuvant chemotherapy in non-small-cell lung cancer. Lung Cancer. 2008;59:377–384. doi: 10.1016/j.lungcan.2007.08.025. [DOI] [PubMed] [Google Scholar]
- 80.Missura M, Buterin T, Hindges R, Hubscher U, Kasparkova J, Brabec V, Naegeli H. Double-check probing of DNA bending and unwinding by XPA-RPA: an architectural function in DNA repair. EMBO J. 2001;20:3554–3564. doi: 10.1093/emboj/20.13.3554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Lovejoy KS, Todd RC, Zhang SZ, McCormick MS, D’Aquino JA, Reardon JT, Sancar A, Giacomini KM, Lippard SJ. cisdiammine( pyridine)chloroplatinum(II), a monofunctional platinum(II) antitumor agent: Uptake, structure, function, and prospects. Proc Natl Acad Sci USA. 2008;105:8902–8907. doi: 10.1073/pnas.0803441105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Wang D, Zhu G, Huang X, Lippard SJ. X-ray structure and mechanism of RNA polymerase II stalled at an antineoplastic monofunctional platinum-DNA adduct. Proc Natl Acad Sci USA. 2010;107:9584–9589. doi: 10.1073/pnas.1002565107. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.