Abstract
The acridinone derivatives with antitumor activity and ability with respect to noncovalent DNA binding were investigated for their quantitative structure–activity relationships (QSAR). Multiple regression analysis was used to model relationships between molecular descriptors and antileukemia activity, or between molecular descriptors and DNA-duplexes stabilization. Studies were performed on molecular modeling using HyperChem and Dragon computer programs, and molecular geometry optimization using MM+ molecular mechanics and semi-empirical AM1 method. Two multiple regression equations were derived and characterized as good and with statistically significant correlations, R = 0.9384 and R = 0.8388, for quantitative structure–antitumor activity relationships and quantitative structure–ability to DNA-duplexes stabilization relationships, respectively. Moreover, hydrophobic and total molecular symmetry properties are important for antitumor activity of acridinone derivatives, and electronic and topological properties are important for physicochemical (noncovalent) DNA-duplexes stabilization of these compounds. The obtained equations can be used for prediction of acridinone derivatives’ activity and their ability to noncovalent interaction with DNA which, as it was shown earlier, play important role in the antitumor mechanism of action of these compounds.
Keywords: Quantitative structure–activity relationships (QSAR), Multiple linear regression (MLR) analysis, Acridinones, Antitumor activity, Physicochemical interaction with DNA
Introduction
The acridinones represented by imidazo- and triazoloacridinones are a new group of potent antitumor compounds (Cholody et al., 1990, 1992, 1996) from which one of the most active derivatives called as C-1305 has been selected for extended preclinical trials and the other one called C-1311 (for review see Mazerska et al., 1998) is currently undergoing phase II clinical trials as drug SymadexTM (Burger et al., 1996; Den Brok et al., 2005a, b, c). Imidazoacridinones exhibit in vivo activity against various tumor cell lines as leukemia, melanoma, colon adenocarcinoma, and colon carcinoma. Triazoloacridinones exhibit in vivo activity against leukemia, murine carcinoma, lung carcinoma, breast carcinoma, and colon carcinoma (Cholody et al., 1990, 1992, 1996; Kusnierczyk et al., 1994; Burger et al., 1996a, b; Lamb and Wheatley, 1996; Calabrese et al., 1998, 1999; Alami et al., 2007; De Marco et al., 2007; Bram et al., 2007). As was previously shown (Składanowski et al., 1999; Lemke et al., 2004; Augustin et al., 2004, 2006; Wesierska-Gadek et al., 2004; Koba and Konopa, 2007; Koba et al., 2009), cellular DNA is important target for the triazoloacridinone drugs, and hence interactions with DNA are naturally the crucial point in view of the biological activity of these compounds. In previous article (Składanowski et al., 1999; Lemke et al., 2004), it was indicated that triazoloacridinones inhibit cleavable complexes of topoisomerase II with DNA. They inhibit also nucleic acid or protein synthesis induced by G2 block of cell cycle followed by apoptosis (Augustin et al., 2004, 2006; Wesierska-Gadek et al., 2004), intercalating to DNA and binding in minor groove (Koba and Konopa, 2007; Koba et al., 2009) and/or forming of interstrand DNA crosslinks (Koba and Konopa, 2007). In addition, it was shown that intercalation to DNA takes place preferentially in guanine triplet regions inducing changes in DNA structures (Lemke et al., 2005). For imidazoacridinones, it was demonstrated that intercalation to DNA undergoes at physiological condition with parallel stabilization of double-stranded DNA and unwinding of supercoiled DNA (Burger et al., 1999; Dziegielewski et al., 2002). The intercalative binding mode of acridinone derivatives was also confirmed with the use of molecular-modeling studies (Mazerski and Muchniewicz, 2000). Similar to other DNA-binding agents, treatment of tumor cells with imidazoacridinones induces topoisomerase II-associated DNA strand breaks (Składanowski et al., 1996), arrests cells in G2 phase, and stimulates apoptosis (Zaffaroni et al., 2001; Skwarska et al., 2007) or mitotic catastrophe (Hyzy et al., 2005; Skwarska et al., 2007). However, after testing imidazoacridinone and triazoloacridinone derivatives, it has been concluded that although the intercalative binding to DNA seems to be necessary for their biological activity (the most active compounds have usually the highest binding affinity), it is not sufficient (some inactive analogs also bind strongly with DNA) (Dziegielewski et al., 2002; Koba and Konopa, 2007). Moreover, acridinones undergo enzymatic oxidation, and this reaction is important for their biological activity as intercalation to DNA and covalent adducts formation (Dziegielewski and Konopa, 1996; Mazerska et al., 1999, 2003). In this context, noncovalent interaction of acridinones may help position drug molecules on DNA for the covalent reaction.
In this article, physicochemical interactions of acridinones with DNA were evaluated in view of quantitative structure–activity relationships (QSAR). Multiple regression analysis was used to model relationships between molecular structural descriptors and biological (antitumor) activity of molecules, or between molecular structural descriptors and physicochemical interaction of acridinone derivatives with DNA. The biological data expressed as anti-leukemia P388 activity and parameter describing ability to physicochemical (noncovalent) interaction with DNA as value of DNA-duplexes stabilization were applied in this study.
Materials and methods
Antitumor and physicochemical DNA-binding activity data of acridinones
The acridinone derivatives examined in this study have been selected to collect analogue compounds differing in chemical structures as well as anticancer activities (Table 1). The data of acridinones’ antitumor activity against P388 leukemia in mice in vivo and expressed as the percentage of increase in survival time of the treated to that of the control mice with P388 leukemia at optimal dose (ILS) were taken from the literature (Table 1) (Cholody et al., 1990, 1992; Koba and Konopa, 2007; Mazerska et al., 1996). The data of physicochemical binding of acridinones to DNA (as values of DNA-duplexes stabilization), which were expressed as an increase in DNA melting temperature in centigrade degrees of ctDNA at drug to DNA base pairs 0.25 M ratio were taken from the literature (Table 1) (Koba and Konopa, 2007; Dziegielewski et al., 2002).
Table 1.
Chemical structures of acridinones studied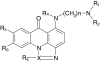
| Compound | X | n | R 1 | R 2 | R 3 | R 4 | R 5 | R 6 | ILSa | ΔT m b |
|---|---|---|---|---|---|---|---|---|---|---|
| C-1310 | C | 2 | CH2CH3 | CH2CH3 | OH | H | CH3 | H | 185 | 15.3 |
| C-1311 | C | 2 | CH2CH3 | CH2CH3 | OH | H | H | H | 93 | 13.7 |
| C-1330 | C | 2 | CH2CH3 | CH2CH3 | OCH3 | H | H | H | 96 | 11.5 |
| C-1415 | C | 2 | CH2CH3 | CH2CH3 | H | H | H | H | 55 | 7.2 |
| C-1419 | C | 2 | CH2CH3 | CH2CH3 | H | H | H | OH | 27 | 8.3 |
| C-1558 | C | 2 | CH2CH3 | CH2CH3 | C(CH3)3 | H | H | H | 0 | 2.4 |
| C-1176 | C | 2 | CH3 | CH3 | H | H | H | H | 90 | 9.5 |
| C-1263 | C | 2 | CH3 | CH3 | OH | H | H | H | 110 | 12.3 |
| C-1212 | C | 3 | CH3 | CH3 | H | H | H | H | 25 | 11.5 |
| C-1371 | C | 3 | CH3 | CH3 | OH | H | H | H | 120 | 3.5 |
| C-1554 | C | 5 | CH2CH3 | CH2CH3 | CH3 | H | H | H | 20 | 10.5 |
| C-1266 | C | 5 | CH3 | CH3 | H | H | H | H | 10 | 9.9 |
| C-1492 | C | 5 | CH3 | CH3 | OH | H | H | H | 85 | 13.1 |
| C-1233 | N | 2 | CH3 | CH3 | H | H | – | H | 77 | 9.1 |
| C-1303 | N | 2 | CH3 | CH3 | OH | H | – | H | 102 | 13.1 |
| C-1533 | N | 2 | CH3 | CH3 | OH | CH3 | – | H | 10 | 8.1 |
| C-1567 | N | 2 | CH3 | CH3 | C(CH3)3 | H | – | H | 0 | 6.8 |
| C-1410 | N | 2 | H | CH2CH3 | OH | H | – | H | 78 | 7.1 |
| C-1296 | N | 3 | CH3 | CH3 | CH3 | H | – | H | 18 | 11.5 |
| C-1305 | N | 3 | CH3 | CH3 | OH | H | – | H | 165 | 15.1 |
aThe percentage of increase in survival time of treated to control mice with P388 leukemia at optimal dose
bThe increase in DNA melting temperature (expressed in centigrade degrees) at drug to DNA base pairs 0.25 M ratio
Structural parameters
The structure of the tested compounds was studied by molecular modeling using HyperChem 7.5 Release software (Kaliszan et al., 1995; Ivanciuc, 1996) and Dragon software (Todeschini et al., 2000). The structures of the compounds were first pre-optimized with the Molecular Mechanics Force Field (MM+) procedure included in the Hyperchem 6.03 (Hypercube) http://www.hyper.com, and the resulting geometries were further refined by means of the Semi-Empirical Molecular Orbitals Method AM1 using the Polak-Ribiere algorithm and a gradient norm limit of 0.01 kcal Å−1.
The following molecular descriptors taken from HyperChem software were considered among quantum and chemical indices: total energy (TE), binding energy (BE), isolated atomic energy (IAE), electron energy (EE), core–core energy (CCE), heat flow (HF), energy of the highest occupied molecular orbitals (E_HOMO), energy of the lowest unoccupied molecular orbitals (E_LUMO), and difference between HOMO and LUMO energies referred to as EG = energy gap; ionization energy (potential) (IE) and electron affinity (EA) were calculated as a difference between the HF of positive molecular ion and electrically neutral molecule, and electronegativity (EN) calculated as average arithmetic potential of ionization and EA. In addition, other parameters were used as the value of electron density of atom orbitals from the lowest to the highest (ED_MIN and ED_MAX, respectively), the highest positive electron charge on the atoms (MAX_POS), and the highest negative electron charge on the atoms (MAX_NEG), the difference between the highest positive and negative charge (DELTA_Q), distribution of dipolar moment along x, y, and z axes (X_DM, Y_DM, and Z_DM, respectively), total dipolar moment (TDM), mean polarizability of molecules (in atom units) MP (Mean Polarizability), energy equal to the length of the wave with the greatest long-wave transfer of electrons, for which the value of oscillator force was different from zero (EL)—the value of wave figures were converted to eV—and the value of the most intensive electron transfer (EMAX—the maximum value of oscillator force calculated with the use of AM1 method—as well as oscillator maximum force used for the transfer (OS_EMAX). Moreover, additional parameters were calculated with the use of QSAR Properties Module of HyperChem. They include the following descriptors: surface area of the molecule available for solvent (SA), molecule volume (V), hydration energy (HE), the calculated distribution coefficient logarithm (logP), refraction (R), and polarizability (P).
On the other hand, using Dragon software, over 1,300 molecular descriptors were calculated and considered for QSAR analysis. They include molecular parameters from different group and class of descriptors as constitutional, topological, walk and path counts, connectivity indices, information indices, 2D autocorrelations, edge adjacency indices, topological charge indices, eigenvalue-based indices, geometrical, 3D-MoRSE, WHIM, GETAWAY, functional group counts, atom-centred fragments, charge, molecular properties and other group of descriptors, and describing some properties of compound as geometry, symmetry, topology, electronic, steric or thermodynamic and other properties. The definitions of these descriptors are reviewed by Todeschini (Todeschini et al., 2000).
Statistical analysis
The statistical QSAR analysis was performed with the use of stepwise regression analysis that used Statistica 8.0 software (StatSoft, Tulsa, OK, USA). The objective of stepwise regression is to construct a multivariate regression model (QSAR equation) for a certain property, y, based on several selected explanatory variables. In stepwise regression, the first selected explanatory variable has the highest correlation with dependent variable, y. Then, explanatory (independent) variables are consecutively added to the model in a forward selection procedure. A new variable is added to the model if a significant change in residuals of the model can be observed. The significance is evaluated using a statistical test, usually F-test (the value of the F-test of significance, F). In addition, the multiple correlation coefficients (R), the standard error of estimate (S), and the significance levels of each term and of whole equation (p) are calculated for the derived QSAR equations. Whenever a new variable is included into a model, a backward elimination step follows in which an F-test detects the earlier selected variables, which can be removed from the model without any significant change on the level of the residuals. The variable selection procedure stops when no additional variable significantly improves the model. Stepwise regression is very much popular in QSAR studies, since the stepwise procedure is simple and based on the classical multiple linear regression (MLR) approach. Moreover, it is implemented in almost all the statistical software packages. One of the drawbacks of the method is the fact that no optimal variable selection is guaranteed, since the new variables are found based on the previously included variables into the model (Put et al., 2006).
During model building, the model fit can be improved proportional to the model complexity. Therefore, the more the factors are included into the model, the better the model fits the training data. Usually, the model fit is evaluated by the root mean-squared error (RMSE), computed for the training data.
The determination of the optimal complexity of the model requires an estimation of its predictive ability, to prevent overfitting to the calibration data. After all, the main goal of QSAR models is to obtain a reasonable prediction of the retention for future samples. To evaluate the prediction by means of an internal validation procedures, cross validation can be used. The predictive ability of a model is characterized by the cross-validated root mean-squared error (RMSECV); test values were calculated with the Matlab software (MathWorks, Natick, MA, USA). The RMSECV as values, which quantify the predictive power of the QSAR model, were calculated by the leave-one-out method and leave-ten-out method.
Results and discussion
The chemical structures of the 20 compounds considered for this study and their antitumor and noncovalent DNA-binding activities are presented in Table 1. In this study, QSAR analysis using multiple regression method was performed for biological activity data (ILS) or physicochemical interaction with DNA (ΔT m), and non-empirical parameters (molecular descriptors). Owing to a large number of non-empirical parameters (over 1,300), treated as independent variables and according to QSAR strategy and multi-parameter regression rule in derived multi-parameter regression equation, the number of independent variables must be 5–6 times less than the number of cases considered in this study. In practice, for obtaining statistically significant equation, one independent variable (in our case structural descriptor) falls, generally out of five to maximum six cases considered, in dependent-variable activity (in our case, activity of acridinones). In the research done, the data set of 20 acridinone derivatives (dependent variables) was taken to QSAR analysis, and for this reason the derived QSAR equations were maximally limited to four statistically significant independent variables (structural descriptors).
Moreover, correlations were limited to the value of regression coefficient R ≥ 0.8 and an additional criterion, considered as relevant to particular independent variables, was established at the significance level p ≤ 0.05.
The calculated equations are presented in Table 2 and characterized by four statistically significant independent variables with a good value of regression coefficient R ≥ 0.8 (R = 0.9384 and R = 0.8388 for quantitative structure–antitumor activity relationships and quantitative structure–ability to DNA-duplexes stabilization relationships, respectively). Moreover, all the regression coefficients are highly statistically significant (p < 0.05) as is the whole equation (p < 7 × 10−4 for quantitative structure–antitumor activity relationships and p < 9 × 10−7 for quantitative structure–ability to DNA-duplexes stabilization relationships, respectively). The values of the multiple correlation coefficient, R; the standard error of the estimate, s; and the value of the F-test of significance, F, are also statistically significant.
Table 2.
Multiple regression QSAR equation (dependent variable = k 0 + k 1 A + k 2 B + k 3 C + k 4 D)
| Dependent variable | Coefficients and statistically significant molecular descriptors | Statistical parameters | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| k 0 | k 1 | A | k 2 | B | k 3 | C | k 4 | D | R (R 2)a | S b | F c | p d | |
| ΔT m | 97.44 ± 55.09 | −6.59 ± 1.50 | GATS7e | 3.03 ± 0.88 | μi | 0.64 ± 0.30 | H-047 | −147.44 ± 83.58 | Mp | 0.8388 (0.7036) | 2.15 | 8.90 | 7 × 10−4 |
| p = 1 × 10−2 | p = 5 × 10−4 | p = 4 × 10−3 | p = 5 × 10−3 | p = 1 × 10−2 | |||||||||
| ILS | 88.80 ± 153.44 | 8914.33 ± 1225.69 | G3m | −36.31 ± 6.02 | logP | −4691.69 ± 1227.99 | G2p | −4744.01 ± 1451.51 | G3p | 0.9384 (0.8806) | 21.03 | 27.65 | 9 × 10−7 |
| p = 1 × 10−2 | p = 3 × 10−6 | p = 2 × 10−5 | p = 2 × 10−3 | p = 5 × 10−3 | |||||||||
aMultiple correlation coefficient (determination coefficient)
bStandard error of estimate
cValue of the F-test of significance
dSignificance level
Statistically significant parameters used in QSAR analysis (values of molecular descriptors are presented in the Table 3) as logarithm of theoretically calculated n-octanol–water partition coefficient (logP) from the class of hydrophobic descriptors, 3st component symmetry directional WHIM index weighted by atomic masses (G3m), 2st component symmetry directional WHIM index by atomic polarizabilities (G2p), and 3st component symmetry directional WHIM directional index/weighted by atomic polarizabilities (G3p) from class of WHIM (Weighted Holistic Invariant Molecular) descriptors had the influence upon antitumor activity of acridinones. The WHIM descriptors are molecular descriptors based on statistical indices calculated on the projections of the atoms along principal axes. They are built in such a way as to capture relevant molecular 3D information regarding molecular size shape, symmetry, and atom distribution with respect to invariant reference frames (Todeschini et al., 2000). In general, the obtained data indicate that hydrophobic and total molecular symmetry properties are important for antitumor activity of acridinones. These observations are in partial agreement with the data obtained by Mazerska (Mazerska et al., 1996), for which antitumor activity of imidazoacridinones is dependent on lipophilicity. However, impact of lipophilicity on the biological activity of these compounds was observed only in the case of derivatives with 8-hydroxyl group, which undergo metabolic activation (Mazerska et al., 1999, 2003). Moreover, non-hydroxyl or 9-hydroxyl derivatives also exhibited lipophilic properties, but its effect was not crucial when metabolic activation did not occur. Relocation of hydroxyl group from position 8 to 9 drastically decreases antitumor activity (C-1311 8-hydroxyl, C-1419 9-hydroxyl). In addition, hydrophobic properties of acridinones can play important role in transport and accumulation of these compounds in cells in view of fastening of metabolic activation (Składanowski et al., 1996). On the other hand, diaminoalkyl side chain has also crucial influence on antitumor activity of acridinones. For compounds without 8-hydroxyl group, the increase in number of carbon atoms between nitrogen atoms from two to three or five (C-1415, C-1176, and C-1233 two; C-1212 and C-1296 three; and C-1266 five) generally decreases antitumor activity of imidazo- and triazoloacridinones. In case of derivatives bearing 8-hydroxyl group, the increase in number of carbon atoms (C-1311, C-1263 two, C-1371 three, and C-1492 five) rather do not augment antitumor activity for imidazoacridinones, while good increase in antitumor activity is observed in case of triazoloacridinones (C-1303, C-1410 two, and C-1305 three carbon atoms). These observations indicate that length and conformation of diaminoalkyl side chain attached to flat aromatic acridinone ring have some influence on proper conformation of acridinone derivatives molecule and is in accordance with earlier modeling studies (Mazerski and Muchniewicz, 2000) The observations also proved our suggestion that total molecular symmetry properties in context of interaction with DNA as intercalation and DNA groove binding are crucial for antitumor activity of acridinones.
Table 3.
Values of molecular descriptors used in QSAR analysis
| Compound | Molecular descriptors | |||||||
|---|---|---|---|---|---|---|---|---|
| GATS7e | μi | H-047 | Mp | G3m | logP | G2p | G3p | |
| C-1310 | 1.07 | 3.70 | 13 | 0.66 | 0.16 | −1.98 | 0.15 | 0.15 |
| C-1311 | 0.92 | 3.06 | 16 | 0.66 | 0.15 | −2.19 | 0.15 | 0.15 |
| C-1330 | 1.19 | 3.16 | 16 | 0.66 | 0.15 | −2.15 | 0.15 | 0.15 |
| C-1415 | 0.90 | 2.32 | 14 | 0.67 | 0.15 | −1.16 | 0.15 | 0.15 |
| C-1419 | 0.89 | 2.01 | 13 | 0.66 | 0.15 | −2.19 | 0.15 | 0.16 |
| C-1558 | 2.13 | 2.28 | 13 | 0.65 | 0.15 | 0.15 | 0.15 | 0.15 |
| C-1176 | 0.94 | 2.50 | 16 | 0.68 | 0.16 | −1.12 | 0.16 | 0.16 |
| C-1263 | 0.90 | 3.34 | 15 | 0.67 | 0.16 | −2.87 | 0.16 | 0.16 |
| C-1212 | 1.01 | 2.61 | 16 | 0.67 | 0.16 | −1.79 | 0.16 | 0.16 |
| C-1371 | 0.94 | 2.11 | 15 | 0.67 | 0.15 | −2.82 | 0.15 | 0.15 |
| C-1554 | 0.83 | 2.66 | 13 | 0.66 | 0.15 | −1.01 | 0.15 | 0.15 |
| C-1266 | 0.86 | 2.60 | 13 | 0.66 | 0.15 | −0.95 | 0.15 | 0.16 |
| C-1492 | 0.86 | 3.10 | 15 | 0.66 | 0.15 | −1.97 | 0.15 | 0.15 |
| C-1233 | 0.99 | 2.99 | 16 | 0.68 | 0.17 | −1.12 | 0.17 | 0.16 |
| C-1303 | 0.87 | 2.48 | 15 | 0.67 | 0.16 | −2.14 | 0.16 | 0.16 |
| C-1533 | 0.91 | 1.11 | 15 | 0.67 | 0.16 | −1.78 | 0.17 | 0.16 |
| C-1567 | 2.15 | 3.53 | 15 | 0.66 | 0.15 | 0.2 | 0.15 | 0.15 |
| C-1410 | 0.86 | 2.39 | 11 | 0.67 | 0.16 | −2.16 | 0.16 | 0.16 |
| C-1296 | 0.94 | 3.08 | 19 | 0.67 | 0.16 | −1.06 | 0.17 | 0.16 |
| C-1305 | 0.81 | 2.44 | 18 | 0.67 | 0.17 | −2.09 | 0.16 | 0.16 |
On the other hand, statistically significant parameters—values of molecular descriptors are presented in the Table 3—such as dipole moment (μi) from class of electronic descriptors, mean atomic polarizability scaled on carbon atom (Mp) from class of constitutional descriptors, Geary autocorrelation-lag 7 weighted by atomic Sanderson electronegativities (GATS7e) from class of 2D autocorrelations descriptors, and H attached to C1(sp3)/CO(sp2) (H-047) from class of atom-centered fragments descriptors had the influence upon physicochemical (noncovalent) DNA-duplexes stabilization of acridinone derivatives. The obtained data indicate that electronic and topological properties are important for noncovalent DNA-duplexes stabilization of these compounds. It is known that drug–DNA binding induces changes in DNA structure and topology and is closely connected with conformation of drug molecule and its electronic and topological properties. The presence of a hydroxyl group in position 8 of acridinone ring slightly increases the affinity for DNA compared to unsubstituted or alkyl-substituted derivatives, possibly because of additional hydrogen bonds with the DNA phosphate backbone. As it was mentioned earlier (Mazerski and Muchniewicz, 2000), the charged diaminoalkyl side chain of acridinone compounds can interact with DNA in the minor groove, in addition to intercalation. In addition, some other data (Koba and Konopa, 2007) indicated that intercalation is not involved in stabilization of secondary structure of DNA. However, for the biologically non-active compounds, C-1558 and C-1567, bearing a t-butyl group in position 8, the ΔT m values were 2.4 and 6.8°C, respectively, indicating that stabilization of the DNA duplex by these compounds especially by C-1567 probably derived from electrostatic interactions of the side chain with DNA and not from its intercalation to DNA. This means that intercalation to DNA is necessary for the biological activity of acridinones via positioning the drug molecules within DNA before the covalent reaction and formation of interstrand DNA crosslink (Koba and Konopa, 2007). This also indicated that topological and electronic properties of acridinone derivatives are important for their physicochemical interactions with DNA. Moreover, the molecular modeling studies (Mazerski and Muchniewicz, 2000) evidenced that when acridinone C-1311 is intercalated between GC, the highly reactive position 8 on acridinone core is in close proximity to nucleophilic N7 position on guanine. It is plausible to postulate that drug molecule first intercalates into DNA and then, after in situ activation, binds covalently to the neighboring base. These observations are compatible with recent findings demonstrating that electrochemically activated C-1311 forms covalent adducts with deoxyguanine (Mazerska et al., 2003). On the other hand, the structure of acridinones suggests that there are at least two possible sites for enzymatic oxidation/activation, which potentially could be involved in the covalent binding to DNA. One is the diaminoalkyl side chain at position 5 which is necessary for covalent binding of mitoxantrone to DNA (Składanowski and Konopa, 2000). The other one is the potential quinone–imine group formed by hydroxyl group in position 8 (8-OH) and heterocyclic nitrogen atom in acridinone nucleus (Mazerska et al., 2003). Recently proposed mechanism of oxidation involves highly unstable carbocations generated in these two positions (Mazerska et al., 2003). It is suggested that C-1311 carbocations react rapidly with nucleophiles present in the environment, including DNA bases forming covalent adducts. These observations indicate that topological and electronic properties of acridinone derivatives are also important for their covalent interactions with DNA.
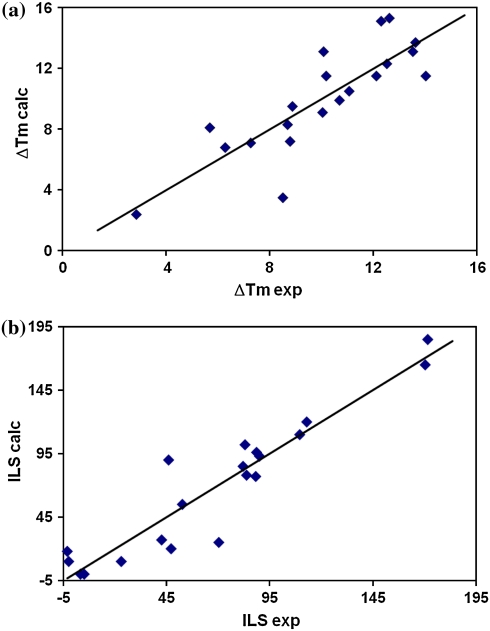
Moreover, the calculated values of ILS and ΔT m obtained for other compounds (Table 4) and the plots of the experimental data versus the calculated data (Fig. 1a–b) for DNA-duplexes stabilization of acridinones expressed as ΔT m (the increase in DNA melting temperature at drug to DNA base pairs 0.25 M ratio) and antitumor activity of acridinones expressed as ILS (survival time of treated to control mice with P388 leukemia at optimal dose) proved good correlation and predictive potency of proposed QSAR models. In addition, the RMSECV as value, which quantifies the predictive power of the proposed QSAR model, are calculated by the leave-one-out and the leave-ten-out methods and presented in the Table 5. The obtained values of RMSECV test (22.79 and 22.27 for quantitative structure–antitumor activity relationships as well as 2.39 and 2.41 for quantitative structure–ability to DNA-duplexes stabilization relationships) performed for all the four statistically significant independent variables proved the predictive power of the derived QSAR models.
Table 4.
Values of experimental and calculated data for DNA-duplexes stabilization and antitumor activity of acridinones
| Compound | ΔT m exp.a | ΔT m calc. | Δb | ILS exp.c | ILS calc. | Δ |
|---|---|---|---|---|---|---|
| C-1310 | 15.3 | 12.6 | 2.7 | 185 | 172 | 13 |
| C-1311 | 13.7 | 13.6 | 0.1 | 93 | 90 | 3 |
| C-1330 | 11.5 | 12.1 | 0.6 | 96 | 89 | 7 |
| C-1415 | 7.2 | 8.8 | 1.6 | 55 | 53 | 2 |
| C-1419 | 8.3 | 8.7 | 0.4 | 27 | 43 | 16 |
| C-1558 | 2.4 | 2.8 | 0.4 | 0 | 5 | 5 |
| C-1176 | 9.5 | 8.9 | 0.6 | 90 | 46 | 44 |
| C-1263 | 12.3 | 12.5 | 0.2 | 110 | 110 | 0 |
| C-1212 | 11.5 | 10.2 | 1.3 | 25 | 70 | 45 |
| C-1371 | 3.5 | 8.5 | 5.0 | 120 | 113 | 7 |
| C-1554 | 10.5 | 11.1 | 0.6 | 20 | 47 | 27 |
| C-1266 | 9.9 | 10.7 | 0.8 | 10 | −2 | 8 |
| C-1492 | 13.1 | 13.5 | 0.4 | 85 | 82 | 3 |
| C-1233 | 9.1 | 10.0 | 0.9 | 77 | 88 | 11 |
| C-1303 | 13.1 | 10.1 | 3.0 | 102 | 83 | 19 |
| C-1533 | 8.1 | 5.7 | 2.4 | 10 | 23 | 13 |
| C-1567 | 6.8 | 6.3 | 0.5 | 0 | 3 | 3 |
| C-1410 | 7.1 | 7.3 | 0.2 | 78 | 84 | 6 |
| C-1296 | 11.5 | 14.0 | 2.5 | 18 | −3 | 15 |
| C-1305 | 15.1 | 12.3 | 2.8 | 165 | 170 | 5 |
| Mean value of Δ | 1.4 | 13 | ||||
aThe increase in DNA melting temperature (expressed in centigrade degrees) at drug to DNA base pairs 0.25 M ratio
bDifference between experimental and calculated values
cThe percentage of increase in survival time of treated to control mice with P388 leukemia at optimal dose
Fig. 1.
Correlation between the experimental data and the calculated data from the derived multiple regression QSAR equation for a DNA-duplexes stabilization of acridinones expressed as ΔT m (the increase in DNA melting temperature at drug to DNA base pairs 0.25 M ratio) and b antitumor activity of acridinones expressed as ILS (survival time of treated to control mice with P388 leukemia at optimal dose)
Table 5.
Values of the cross-validated root-mean-square error RMSECV test
| QSAR model for dependent variable | RMSECV test | |||||||
|---|---|---|---|---|---|---|---|---|
| Leave-one-out method | Leave-ten-out method | |||||||
| 1a | 2 | 3 | 4 | 1 | 2 | 3 | 4 | |
| ΔT m | 3.36 | 2.53 | 2.56 | 2.39 | 3.44 | 2.63 | 2.64 | 2.41 |
| ILS | 53.39 | 42.10 | 28.48 | 22.79 | 54.23 | 42.35 | 28.74 | 22.27 |
a1–4 represents RMSECV test performed only for one, combined two and three, and for all the four significance descriptors in QSAR models, respectively. In the case of QSAR model, for ΔT m as dependent-variable values, 1–4 were obtained for only GATS7e, GATS7e combined with μi, GATS7e combined with μi and H-047, GATS7e combined with μi, H-047, and Mp descriptors. In the case of QSAR model for ILS as dependent-variable values, 1–4 were obtained for only G3m, G3m combined with logP, G3m combined with logP and G2p, and G3m combined with logP, G2p and G3p descriptors
Conclusions
Statistically significant equations describing structure–antitumor activity relationships and structure–ability to physicochemical (noncovalent) interaction with DNA relationships in acridinone derivatives group were derived. It has been found that hydrophobic and total molecular symmetry properties are important for antitumor activity of acridinone derivatives, and electronic and topological properties are important for physicochemical (noncovalent) DNA-duplexes stabilization of these compounds.
Open Access
This article is distributed under the terms of the Creative Commons Attribution Noncommercial License which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
References
- Alami N, Paterson J, Belanger S, Juste S, Grieshaber CK, Leyland-Jones B. Comparative cytotoxicity of C-1311 in colon cancer in vitro and in vivo using the hollow fiber assay. J Chemother. 2007;19:546–553. doi: 10.1179/joc.2007.19.5.546. [DOI] [PubMed] [Google Scholar]
- Augustin E, Plocka E, Konopa J. Induction of cell death (apoptosis) by antitumor triazoloacridinones in tumor cells. Drug Metab Rev. 2004;32(suppl. 1):33. [Google Scholar]
- Augustin E, Mos-Rompa A, Skwarska A, Witkowski JM, Konopa J. Induction of G2/M phase arrest and apoptosis of human leukemia cells by potent antitumor triazoloacridinone C-1305. Biochem Pharmacol. 2006;72:1668–1679. doi: 10.1016/j.bcp.2006.07.035. [DOI] [PubMed] [Google Scholar]
- Berger B, Marguardt H, Westendorf J. Pharmacological and toxicological aspects of new imidazoacridinone antitumor agents. Cancer Res. 1996;56:2094–2104. [PubMed] [Google Scholar]
- Bram EE, Ifergan I, Grimberg M, Lemke K, Składanowski A, Assaraf YG. C421 allele-specific ABCG2 gene amplification confers resistance to the antitumor triazoloacridone C-1305 in human lung cancer cells. Biochem Pharmacol. 2007;74:41–53. doi: 10.1016/j.bcp.2007.03.028. [DOI] [PubMed] [Google Scholar]
- Burger AM, Double JA, Konopa J, Bibby MC. Preclinical evaluation of novel imidazoacridinone derivatives with potent activity against experimental colorectal cancer. Br J Cancer. 1996;74:1369–1374. doi: 10.1038/bjc.1996.551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burger AM, Jenkins TC, Double JA, Bibby MC. Cellular uptake, cytotoxicity and DNA-binding studies of the novel imidazoacridinone antineoplastic agent C1311. Br J Cancer. 1999;81:367–375. doi: 10.1038/sj.bjc.6690702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calabrese CR, Bibby MC, Double JA, Loadman PM. Pharmacokinetics and tissue distribution of the imidazoacridinone C1311 in tumour-bearing mice. Cancer Chemother Pharmacol. 1998;42:379–385. doi: 10.1007/s002800050833. [DOI] [PubMed] [Google Scholar]
- Calabrese CR, Loadman PM, Lim LS, Bibby MC, Double JA, Brown JE, Lamb JH. In vivo metabolism of the antitumor imidazoacridinone C1311 in the mouse and in vitro comparison with humans. Drug Metab Dispos. 1999;27:240–245. [PubMed] [Google Scholar]
- Cholody WM, Martelli S, Konopa J. 8-substituted 5-[(aminoalkyl)amino]-6H-v-triazolo[4,5,1-de]acridin-6-ones as potential antineoplastic agents. J Med Chem. 1990;33:2852–2856. doi: 10.1021/jm00172a028. [DOI] [PubMed] [Google Scholar]
- Cholody WM, Martelli S, Konopa J. Chromophore-modified antineoplastic imidazoacridinones. Synthesis and activity against murine leukemias. J Med Chem. 1992;35:378–382. doi: 10.1021/jm00080a026. [DOI] [PubMed] [Google Scholar]
- Cholody WM, Horowska B, Paradziej-Łukowicz J, Martelli S, Konopa J. Structure-activity relationship for antineoplastic imidazoacridinones: synthesis and antileukemic activity against murine leukemias. J Med Chem. 1996;39:1028–1032. doi: 10.1021/jm950564r. [DOI] [PubMed] [Google Scholar]
- De Marco C, Zaffaroni N, Comijn E, Tesei A, Zoli W, Peters GJ. Comparative evaluation of C1311 cytotoxic activity and interference with cell cycle progression in a panel of human solid tumour and leukaemia cell lines. Int J Oncol. 2007;31:907–913. [PubMed] [Google Scholar]
- Den Brok MW, Nuijen B, Harms R, Buluran JN, Harvey MD, Grieshaber CK, Beijnen JH. Compatibility and stability of the novel anti-cancer agent C1311 in infusion devices and its in vitro biocompatibility. J Oncol Pharm Pract. 2005;11:13–19. doi: 10.1191/1078155205jp142oa. [DOI] [PubMed] [Google Scholar]
- Den Brok MW, Nuijen B, Hillebrand MJ, Grieshaber CK, Harvey MD, Beijnen JH. Development and validation of an LC-UV method for the quantification and purity determination of the novel anticancer agent C1311 and its pharmaceutical dosage form. J Pharm Biomed Anal. 2005;39:46–53. doi: 10.1016/j.jpba.2005.02.038. [DOI] [PubMed] [Google Scholar]
- Den Brok MW, Nuijen B, Kettenes-Van Den Bosch JJ, Van Steenbergen MJ, Buluran JN, Harvey MD, Grieshaber CK, Beijnen JH. Pharmaceutical development of a parenteral lyophilised dosage form for the novel anticancer agent C1311. PDA J Pharm Sci Technol. 2005;59:285–297. [PubMed] [Google Scholar]
- Dziegielewski J, Konopa J. Interstrand crosslinking of DNA induced in tumor cells by a new group of antitumor imidazoacridinones. Proc Am Assoc Cancer Res. 1996;37:410. [Google Scholar]
- Dziegielewski J, Slusarski B, Konitz A, Skladanowski A, Konopa J. Intercalation of imidazoacridinones to DNA and its relevance to cytotoxic and antitumor activity. Biochem Pharmacol. 2002;63:1653–1662. doi: 10.1016/S0006-2952(02)00916-4. [DOI] [PubMed] [Google Scholar]
- Hyzy M, Bozko P, Konopa J, Skladanowski A. Antitumour imidazoacridone C-1311 induces cell death by mitotic catastrophe in human colon carcinoma cells. Biochem Pharmacol. 2005;69:801–809. doi: 10.1016/j.bcp.2004.11.028. [DOI] [PubMed] [Google Scholar]
- Ivanciuc O. HyperChem release 4.5 for Windows. Inf Comput Sci. 1996;36:612–614. doi: 10.1021/ci950190a. [DOI] [Google Scholar]
- Kaliszan R, Turowski M, Buciński A, Hartwick RA. Quantitative structure-retention relationships in capillary electrophoresis of inorganic cations and β-adrenolytic and sulfonamided compomids. Quant Struct Act Relat. 1995;14:356–361. doi: 10.1002/qsar.19950140406. [DOI] [Google Scholar]
- Koba M, Konopa J. Interactions of antitumor triazoloacridinones with DNA. Acta Biochim Pol. 2007;54:297–306. [PubMed] [Google Scholar]
- Koba M, Koba K, Bączek T. Is DNA minor groove binding crucial for biological activity of triazoloacridinones with cytotoxic and antitumour properties? Lett Drug Des Discov. 2009;6:242–245. doi: 10.2174/157018009788452465. [DOI] [Google Scholar]
- Kusnierczyk H, Cholody WM, Paradziej-Łukowicz J, Radzikowski C, Konopa J. Experimental antitumor activity and toxicity of the selected triazolo- and imidazoacridinones. Arch Immunol Ther Exp. 1994;42:414–423. [PubMed] [Google Scholar]
- Lamb J, Wheatley DN. Cell killing by the novel imidazoacridinone antineoplastic agent, C-1311, is inhibited at high concentrations coincident with dose-differentiated cell cycle perturbation. Br J Cancer. 1996;74:1359–1368. doi: 10.1038/bjc.1996.550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemke K, Poindessous V, Składanowski A, Larsen AK. The antitumor triazoloacridone C-1305 is a topoisomerase II poison with unusual properties. Mol Pharmacol. 2004;66:1035–1042. doi: 10.1124/mol.104.000703. [DOI] [PubMed] [Google Scholar]
- Lemke K, Wojciechowski M, Laine W, Bailly C, Colson P, Baginski M, Larsen AK, Skladanowski A. Induction of unique structural changes in guanine-rich DNA regions by the triazoloacridone C-1305, a topoisomerase II inhibitor with antitumor activities. Nucleic Acids Res. 2005;33:6034–6047. doi: 10.1093/nar/gki904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazerska Z, Augustin E, Dziegielewski J, Chołody MW, Konopa J. QSAR of acridines, III. Structure-activity relationship for antitumour imidazoacridinones and intercorrelations between in vivo and in vitro tests. Anticancer Drug Des. 1996;11:73–88. [PubMed] [Google Scholar]
- Mazerska Z, Augustin E, Składanowski A, Bibby MC, Double JA, Konopa J. C-1311. Drugs Future. 1998;23:702–706. doi: 10.1358/dof.1998.023.07.463751. [DOI] [Google Scholar]
- Mazerska Z, Gorlewska K, Kraciuk A, Konopa J. The relevance of enzymatic oxidation by horseradish peroxidase to antitumour potency of imidazoacridinone derivatives. Chem Biol Interact. 1999;115:1–22. doi: 10.1016/S0009-2797(98)00042-8. [DOI] [PubMed] [Google Scholar]
- Mazerska Z, Sowiński P, Konopa J. Molecular mechanism of the enzymatic oxidation investigated for imidazoacridinone antitumor drug, C-1311. Biochem Pharmacol. 2003;66:1727–1736. doi: 10.1016/S0006-2952(03)00477-5. [DOI] [PubMed] [Google Scholar]
- Mazerski J, Muchniewicz K. The intercalation of imidazoacridinones into DNA induces conformational changes in their side chain. Acta Biochim Pol. 2000;47:65–78. [PubMed] [Google Scholar]
- Put R, Daszykowski M, Bączek T, Vander Heyden Y. Retention prediction of peptides based on uninformative variable elimination by partial least squares. J Proteome Res. 2006;5:1618–1625. doi: 10.1021/pr0600430. [DOI] [PubMed] [Google Scholar]
- Składanowski A, Konopa J. Mitoxantrone and ametantrone induce interstrand cross-links in DNA of tumour cells. Br J Cancer. 2000;82:1300–1304. doi: 10.1054/bjoc.1999.1095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Składanowski A, Plisov SY, Konopa J, Larsen AK. Inhibition of DNA topoisomerase II by imidazoacridinones, new antineoplastic agents with strong activity against solid tumor. Mol Pharmacol. 1996;49:772–780. [PubMed] [Google Scholar]
- Składanowski A, Larsen AK, Konopa J, Lemke K. Inhibition of DNA topoisomerase II by antitumor triazoloacridinones in vitro and in tumor cells. Proc Am Assoc Cancer Res. 1999;40:681. [Google Scholar]
- Skwarska A, Augustin E, Konopa J. Sequential induction of mitotic catastrophe followed by apoptosis in human leukemia MOLT4 cells by imidazoacridinone C-1311. Apoptosis. 2007;12:2245–2257. doi: 10.1007/s10495-007-0144-y. [DOI] [PubMed] [Google Scholar]
- Todeschini R, Consonni V, Mannhold R, Kubinyi H, Timmerman H. Handbook of molecular descriptors. Weinheim: Wiley-VCH; 2000. [Google Scholar]
- Wesierska-Gadek J, Schloffer D, Gueorguieva M, Uhl M, Skladanowski A. Increased susceptibility of poly(ADP-ribose) polymerase-1 knockout cells to antitumor triazoloacridone C-1305 is associated with permanent G2 cell cycle arrest. Cancer Res. 2004;64:4487–4497. doi: 10.1158/0008-5472.CAN-03-3410. [DOI] [PubMed] [Google Scholar]
- Zaffaroni N, De Marco C, Villa R, Riboldi S, Daidone MG, Double JA. Cell growth inhibition, G2M cell cycle arrest and apoptosis induced by the imidazoacridinone C1311 in human tumour cell lines. Eur J Cancer. 2001;37:1953–1962. doi: 10.1016/S0959-8049(01)00227-1. [DOI] [PubMed] [Google Scholar]