Abstract
Flavonoid pigments are known to accumulate in red grains and coleoptiles of wheat and are synthesized through the flavonoid biosynthetic pathway. Flavanone 3-hydroxylase (F3H) is a key enzyme at a diverging point of the flavonoid pathway leading to production of different pigments: phlobaphene, proanthocyanidin, and anthocyanin. We isolated three F3H genes from wheat and examined a relationship between their expression and tissue pigmentation. Three F3Hs are located on the telomeric region of the long arm of chromosomes 2A, 2B, and 2D, respectively, designated as F3H-A1, F3H-B1, and F3H-D1. The telomeric regions of the long arms of the chromosomes of homoeologous group 2 of wheat showed a syntenic relationship to the telomeric region of the long arm of rice chromosome 4, on which rice F3H gene was also located. All three genes were highly activated in the red grains and coleoptiles and appeared to be controlled by flavonoid regulators in each tissue.
1. Introduction
Flavonoid pigments are well known to play an important role in pigmentation of tissues such as flowers, fruits, and grains. These pigments not only render the tissues as more conspicuous but also add physiological function to tissues, such as protection against UV damage [1] and increased level of grain dormancy [2].
In wheat, red pigmentation was observed in many tissues including grain coats, coleoptiles, anthers, culms, and pericarps. Several genes affecting anthocyanin pigmentation (i.e., R-1 (R in former notation) for red grain, Rc for red coleoptile, Pan for purple anthers, Ra for red auricles, Pc for purple culms, and Pp for purple pericarp) have been reported [3]. Red-grained wheat has been reported to contain red flavonoid pigments, phlobaphene or proanthocyanidin (condensed tannin), in grain coat tissues [4]. In contrast, pigments of red coleoptiles were anthocyanin [5]. Phlobaphene, proanthocyanidin, and anthocyanin are synthesized through the common flavonoid biosynthetic pathway [6] (Figure 1). Phlobaphenes are compounds produced by polymerization of flavan-4-ols, which are synthesized by three enzymes: chalcone synthase (CHS), chalcone isomerase (CHI), and dihydroflavonol 4-reductase (DFR) in the early steps of the flavonoid pathway. On the other hand, proanthocyanidin and anthocyanin are produced via 3,4-deoxy flavonoids, which are synthesized by four enzymes: CHS, CHI, F3H, and DFR. A step of F3H is a diverging point in the flavonoid pathway leading to the production of either phlobaphene or proanthocyanidin.
Figure 1.

Flavonoid biosynthetic pathway. The F3H gene is indicated in bold letters. ANS, FS, and FLS, which belong to 2OG-Fe(II) oxygenase superfamily, are shown in italic. Enzyme names are abbreviated as follows: ANS: anthocyanidin synthase, CHI: chalcone isomerase, CHS: chalcone synthase, DFR: dihydroflavonol 4-reductase, F3H: flavanone 3-hydroxylase, FS: and flavone synthase.
Activation of flavonoid biosynthetic genes is required for pigmentation of plant tissue. Transcription factors involved in expression of flavonoid genes have been studied extensively and identified in several plant species, including Arabidopsis and maize. The transcription factors, which activate flavonoid genes, are mainly classified into two gene families: one with an MYB domain and the other with a basic helix-loop-helix (bHLH) domain [7]. In maize, C1 (MYB-type) and R (bHLH-type) factors work together leading to the production of anthocyanin. The P (MYB-type) factor of maize alone is responsible for the synthesis of phlobaphene [7]. The TT2 (MYB-type) factor of Arabidopsis is required for proanthocyanidin production in the seed coat [8]. Recently, our group showed that R-1 gene which regulates grain color in wheat was considered to be an MYB-type transcription factor [9].
Transcription factor binding elements have also been studied in promoters of flavonoid biosynthetic genes. Hartmann et al. [10] identified light regulatory units (LRUs) in promoters of CHS, CHI, F3H, and flavonol synthase (FLS). The LRU consists of two elements: an MYB-recognition element (MRE) and an ACGT-containing element (ACE). Himi and Noda [11] also found a unit of MRE and ACE that was repeated in promoters of wheat DFRs.
This paper describes three full sequences of F3H genes (F3H-A1, F3H-B1, and F3H-D1), along with their promoters, isolated from hexaploid wheat. These genes were located on the telomeric regions of the long arm of the chromosomes of homoeologous group 2. We also studied a relationship between F3H expression and tissue pigmentation of lines with red grain and red coleoptile (R/Rc), red grain and white coleoptile (R/rc), white grain and red coleoptile (r/Rc), and white grain and white coleoptile (r/rc). Expressions of F3H-A1, F3H-B,1 and F3H-D1 were associated with pigmentation and appeared to be enzymes that are required for pigment synthesis.
2. Materials and Methods
2.1. Plant Materials
Triticum aestivum cvs. Norin 61, Norin 17, Novosibirskaya 67 (NS67), ANK-1C, Chinese Spring (CS), three deletion and three ditelosomic lines of CS were grown under a semitransparent plastic roof in a field (Table 1). The CS spikes were tagged at anthesis and harvested at 5-days postanthesis (DPA). Grains at 5 DPA and the mature stage (water content 15%) were collected from primary and secondary florets of the central spikelets of spikes.
Table 1.
Wheat lines used in the experiments.
| Lines | Grain color | Coleoptile color |
|---|---|---|
| Norin 61 | Red | White |
| Norin 17 | White | White |
| Chinese Spring (CS) | Red | White |
| Deletion lines of CS | ||
| 2AL-2 | Red | White |
| 2BL-6 | Red | White |
| 2DL-6 | Red | White |
| Ditelosomic lines of CS | ||
| ditelo 2AS | Red | White |
| ditelo 2BS | Red | White |
| ditelo 2DS | Red | White |
| Novosibirskaya 67 (NS67) | White | Red |
| ANK-1C | Red | Red |
2.2. DNA and RNA Extraction
Seedlings were grown at 20°C under darkness for a week and used for DNA preparation. The DNA and RNA were isolated using methods of Himi and Noda [11]. Total RNA was extracted from grains and 3-day-old coleoptiles grown at 20°C under 12 h of UV light (about 100 moL m−2 s−1, UV lamp) by the sodium dodecyl sulfate-phenol method [11]. Poly(A)+ RNA was isolated from 10 mg of the total RNA using an mRNA isolation kit according to the supplier's instructions (Roche Diagnostic Systems Inc., Tokyo, Japan).
2.3. Primers Designed for Isolation of Wheat F3H Genes
Primers used in this study were listed in Table 2. Two primers, F3H3LP and F3H2RP, were designed based on the cDNA sequences of barley F3H (Accession no. X58138 [12]) and a wheat EST clone, whe24e20 (Accession no. BJ237068) in wheat EST database (http://www.shigen.nig.ac.jp/wheat/komugi/ests/tissueBrowse.jsp), which is similar in nucleotide sequence to barley F3H. Partial sequences of wheat F3H were amplified using the above primers. New primers were designed in the amplified F3H sequences for further amplifications of F3H by 3′ RACE, inverse PCR, and RT-PCR.
Table 2.
Names and sequences of the primers used in the experiments.
| Primer | Sequence (5′-3′) | Sequence source | Accession No. |
|---|---|---|---|
| Primers for genomic DNA | |||
| F3H3LP | GCGACACAAGTGGACGAT | whe24e20 | BJ237068 |
| F3H2RP | GAACGTCGCGATCGACAG | barley F3H (Meldgaard, 1992) | X58138 |
| Primers for 3′ region | |||
| F3HLP | CCTACTTCTCGTACCCGGTG | barley F3H (Meldgaard, 1992) | X58138 |
| F3H2LP | ATTCGTCGTCAACCTCGG | barley F3H (Meldgaard, 1992) | X58138 |
| Oligo (dT) with 3′ adapter | GGCCACGCGTCGACTAGTACTTTTTTTTTTTTTTTTT | ||
| 3′ adapter | GGCCACGCGTCGACTAGTAC | ||
| F3H-3UTRRP | TCTGTCAGACACATGCACACA | 3′ region obtained by 3′ RACE | |
| F3Hint1LP | ACTGTCTTGTAGCCGCTTCC | 1st intron of F3H-A1, B1 | |
| Primers for RT-PCR | |||
| F3H5LP | CAAGAAGCAGGCCAAGGAC | 3rd exon of F3H-A1, B1, D1 | |
| F3HARP | CCAAACTCACGATAACTCCTTATTTAC | 3′ regions of F3H-A1 | |
| F3HBRP | GGAGAATAATCAATCCCACCA | 3′ regions of F3H-B1 | |
| F3HDRP | CTGCTACACACGTACGGATACC | 3′ regions of F3H-D1 | |
| Primers for inverse PCR and | |||
| chromosomal location analysis | |||
| F3H1stintAspLP | TGCTAGAATGGCGGTGGGT | 1st intron of F3H-A1 | |
| F3H1stintBspLP | GATGATGGTGGGGAACGGT | 1st intron of F3H-B1 | |
| F3Hint2LP | GCCATGCCACGTAAAATGAT | 1st intron of F3H-D1 | |
| F3HABDRP | CTTCACCGGGTACGAGAAGT | 2nd exon of F3H-A1, B1, D1 | |
| F3HBLP | GCAGGTATACACGCATTCATTT | 1st exon and intron of F3H-B1 | |
| F3H3RP | GTGGCTGGAGACGATGAAG | whe24e20 | BJ237068 |
| F3H4LP | CGATACAGCGAGCGACTCAT | 2nd exon of F3H-A1, B1, D1 | |
| F3H4RP | AGGAACGTCTCGTTGCTCAC | 1st exon of F3H-A1, B1, D1 | |
| F3H5RP | TTGTGGTTTTCTGGACGTTG | 5′ regions of F3H-A1, B1, D1 | |
| F3HABDLP | GACAAGCTCCGGTACGACAT | 1st exon of F3H-A1, B1, D1 | |
| Primers for Actin | |||
| TaActinLP | GAGGGATACACGCTTCCTCA | wheat actin | AB181991 |
| TaActinRP | GAAAGTGCTAAGAGAGGCCAAA | wheat actin | AB181991 |
PCR conditions were as follows: 5 min denaturation at 94°C followed by 30 cycles of 1 min at 94°C, 1 min at 58°C, and 1 min at 72°C, except the RT-PCR and PCR for chromosomal location analysis of F3H, in which annealing temperatures used were mentioned later. PCR products were cloned into the pGEM-T vector (Promega Corp., Madison, USA). DNA sequences were determined using the ABI 3100 sequencer (PerkinElmer Inc.) and were analyzed using two software programs: GENETYX (Version 7.0; Software Development, Tokyo, Japan) and CLUSTALW (Bioinformatic Center, Institute for Chemical Research, Kyoto University, available from the web site at http://www.genome.jp/tools/clustalw/).
2.4. 3′ RACE and Inverse PCR
The 3′ regions of F3H were amplified in mRNA of grains at 5 DPA of Norin 61 using the 3′ RACE method with F3HLP and F3H2LP primers and an oligo (dT) primer with an adaptor sequence (Table 2, Figure 2).
Figure 2.

Alignment of the genomic nucleotide sequences of wheat F3H-A1, F3H-B1, and F3H-D1. Uppercase letters in gray boxes indicate exons: 5′ and 3′ untranslated region and introns are in lowercase letters. Putative translation initiation codon (ATG) is shown in italic and A of ATG is labeled as +1, and stop codons (TGA and TAG) are shown as underlined. Primer sequences are written in bold with the primer name and arrow (right arrow: sense primer; left arrow: antisense primer).
For inverse PCR, genomic DNA (1.5 μg) of CS was digested with 15 U of one of the following enzymes, BspT104I (for F3H-A1), BspT104I (for F3H-B1), or Sac I (for F3H-D1) (Figure 2). The DNA was ligated using a ligation high solution (Toyobo Co. Ltd., Japan); the DNA was subsequently used as a template for PCR. The 5′ upstream regions of F3H were amplified using inverse PCR method in 20 μL of reaction solution with 30 ng of ligated DNA and 0.5 μM of the primers for inverse PCR listed in Tables 2 and 3. Inverse PCR was carried out first with the first primers and then with the nested primers (2nd and 3rd primers). The transcription factor binding site in the amplified 5′ region was sought using the MOTIF program (Bioinformatic Center, Institute for Chemical Research, Kyoto University, available from the web site at http://motif.genome.ad.jp/).
Table 3.
Primers and annealing temperature for each PCR.
| F3H-A1 | F3H-B1 | F3H-D1 | ||
|---|---|---|---|---|
| Location analysis | ||||
| Sense | F3H1stintAspLP | F3H1stintBspLP | F3Hint2LP | |
| Antisense | F3HABDRP | F3HABDRP | F3HABDRP | |
| Temp.∗1 | 62.0 | 62.0 | 62.0 | |
| Expression analysis | ||||
| Sense | F3H5LP | F3H5LP | F3H5LP | |
| Antisense | F3HARP | F3HBRP | F3HDRP | |
| Temp.∗1 | 55.0 | 60.0 | 62.0 | |
| Inverse PCR | ||||
| Sense | F3HBLP | F3HABDLP | F3H4LP | 1st PCR |
| Antisense | F3H3RP | F3H4RP | F3HABDRP | |
| Temp.∗1 | 55.0 | 55.0 | 55.0 | |
| Sense | F3H1stintAspLP | F3HBLP | F3H2LP | 2nd PCR |
| Antisense | F3H4RP | F3H5RP | F3H3RP | |
| Temp.∗1 | 58.0 | 58.0 | 58.0 | |
| Sense | F3H1stintAspLP | F3H1stintBspLP | F2H2LP | 3rd PCR |
| Antisense | F3H5RP | F3H5RP | F3H4RP | |
| Temp.∗1 | 60.0 | 60.0 | 60.0 |
∗1Annealing temperature at PCR reaction.
2.5. RT-PCR (Reverse Transcription-PCR)
The first-strand cDNA synthesis and subsequent quantitative RT-PCR assay were carried out according to the method of Himi and Noda [11]. Concentration of cDNA was standardized after evaluating the amount of actin mRNA in samples by PCR with the actin primers (Table 2). The F3H-A1 was amplified with F3H5LP and F3HARP primers at annealing temperature of 55°C, F3H-B1 with F3H5LP and F3HBRP primers at annealing temperature of 60°C, and F3H-D1 with F3H5LP and F3HDRP primers at annealing temperature of 62°C (Table 3).
2.6. Southern Blot Analysis
Genomic DNA of Norin 61 and CS was digested with one of the following restriction enzymes: Bgl II, EcoR I, Xba I, and Xho I. Then it was separated on 0.7% (w/v) agarose gel and transferred onto a nylon membrane (Hybond-N+; Amersham Pharmacia Biotech Co. Ltd., Japan). The membranes were prehybridized in a solution of 50% formamide, 5 × SSC (0.75 M NaCl, 75 mM trisodium citrate dihydrate; pH 7.5), 0.1% (w/v) N-lauroylsarcosine, 0.02% SDS, and 2% blocking reagent (Roche Diagnostic Systems Inc.) at 42°C and hybridized for 16 h with a solution containing DIG-labeled probe. The probe was labeled using a PCR DIG Labeling Mix (Roche Diagnostic Systems Inc.) with a pair of primers: F3HLP and F3H2RP (Table 2). The membranes were washed at 65°C with a solution of 0.5 × SSC and 0.1% SDS.
2.7. Chromosomal Location
We examined chromosomal locations of F3H-A1, F3H-B1, and F3H-D1 by amplifying these genes in three ditelosomic lines and three deletion lines of CS, which, respectively, lacked the chromosome arm and a part of chromosome (Table 1). Specific primers for F3H-A1 were F3H1stintAspLP and F3HABDRP. Those for F3H-B1 were F3H1stintBspLP and F3HABDRP; those for F3H-D1 were F3Hint2LP and F3H2RP (Table 3). Annealing temperature of 62°C was applied in the PCR to increase specificity of these primers to each F3H.
3. Results
3.1. Copy Number of the F3H Gene
Four fragments of wheat F3H genes were detected by Southern blot method in genomic DNA of CS digested with Bgl II (Figure 3). Identical numbers of fragments were also observed in Norin 61 DNA digested with Bgl II, Xba I, or Xho I (data not shown). Hexaploid wheat appears to have 4 copies of F3H gene.
Figure 3.
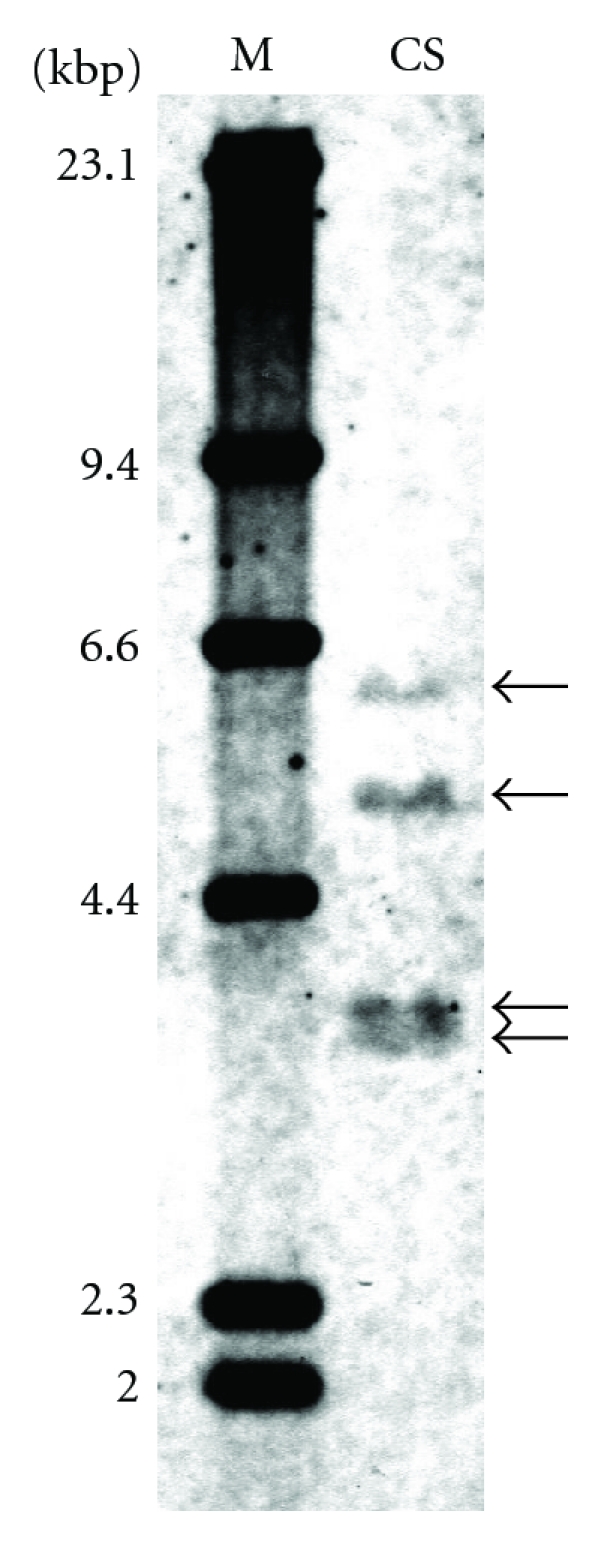
Southern blot of F3H gene in genomic DNA of CS digested with an enzyme Bgl II. Wheat F3H fragment (580-bp) used as a probe was amplified with a pair of F3HLP and F3H2RP primers.
3.2. Identification of F3H-A1, F3H-B1, and F3H-D1
An EST clone of wheat, whe24e20 (Accession no. BJ23706), which is similar in nucleotide sequence to barley F3H cDNA (Accession no. X58138 [12]), was found in the integrated wheat science database, KOMUGI (http://www.shigen.nig.ac.jp/wheat/komugi/top/top.jsp), by blast search. The F3H3LP and F3H2RP primers were designed, respectively, in the 5′ UTR near the start codon (ATG) and in the middle region of the F3H cDNA sequences (Table 2, Figures 2 and 4). Two PCR products of about 1.6 kbp and 1.2 kbp were obtained in CS genomic DNA using a pair of F3H3LP and F3H2RP primers (see Supplementary Figure 1 in Supplementary Material available online at doi: 10.1155/2011/369460). These products were similar to barley F3H and included two putative intron regions. Similarity in the exon region between the 1.6 kbp fragment and barley F3H was 94.3%; that between the 1.2 kbp fragment and barley F3H was 93.8%.
Figure 4.

The 5′ region sequences of F3H-A1, F3H -B1, and F3H -D1. Sequences in box indicate the binding sites of transcription factors; the box of broken lines is a putative TATA-box. Exons are indicated in uppercase letters with gray box; the 5′ untranslated region and promoter are in lowercase letters. The putative translation initiation codon (ATG) is shown in italic typeface. Primer sequences are written in bold with primer names and arrows (right arrow: sense primer; left arrow: antisense primer). Elements for transcription factor binding are as follows: bZIP911: bZIP transcription factor of Antirrhinum majus; G-box: bZIP-type transcription factor; P: MYB-type transcription factor, P of maize; RAV1: transcription factor with AP2 and B3 domains.
The 1.2 kbp fragment was not amplified in the ditelo 2DS line in preliminary experiments using 40 ditelosomic lines. This fragment appears to be located on the long arm of chromosome 2D. The results also suggested that the 1.6 kbp fragment was also on the long arms of chromosomes 2A and 2B. We amplified and cloned two 1.6 kbp fragments in ditelo 2AS and 2BS lines, which, respectively, lack the long arms of chromosomes 2A and 2B. The 1.6 kbp fragments amplified in ditelo 2AS and 2BS mutually differed in their nucleotide sequences. These sequences on chromosomes 2AL, 2BL, and 2DL were, respectively, designated tentatively as F3H-A1, F3H-B1, and F3H-D1.
3.3. 3′ Regions of F3H-A1, F3H-B1, and F3H-D1
The 3′ regions of F3H amplified by the 3′ RACE included about 200 bp of 3′ UTR. The F3H-3UTRRP primer was designed in the 3′ UTR region (Table 2, Figure 2). The 3′ regions with the second intron were amplified using a pair of primers, F3H2LP and F3H-3UTRRP, in genomic DNA of CS. One amplified fragment was similar in the second intron to F3H-A1. The F3Hint1LP and F3Hint2LP primers were newly designed in intron 1 of F3H-B1 and F3H-D1 (Table 2, Figure 2). The 3′ regions of F3H-B1 and F3H-D1 were amplified with F3Hint1LP and F3H-3UTR primers for F3H-B1 and F3Hint2LP and F3H-3UTRRP primers for F3H-D1 (Table 2, Figure 2). The F3HARP primer specific to F3H-A1, the F3HBRP primer specific to F3H-B1, and the F3HDRP primer specific to F3H-D1 were designed in the 3′ UTRs (Table 2, Figure 2). These primers were used to amplify cDNA sequences of F3Hs and to elucidate the expression of each gene.
3.4. Nucleotide Sequences after the Start Codon of F3H-A1, F3H-B1, and F3H-D1
The full sequences of F3H-A1, F3H-B1, and F3H-D1 between the start codon and 3′ UTR region were amplified with F3HARP, F3HBRP, F3HDRP, and F3H3LP primers in genomic DNA and cDNAs to examine whether the partial sequences and 3′ regions isolated are in cis position (Table 2, Figures 2 and 4). All three F3Hs had 3 exons and 2 introns; these introns were inserted in the same position as the introns of rice and Arabidopsis F3Hs (Figure 5(a)). The ORF sequences of wheat F3H-A1, F3H-B1, and F3H-D1 were similar to each other at more than 96% (F3H-A1 versus F3H-B1: 96.7%, F3H-A1 versus F3H-D1: 96.9%, and F3H-B1 versus F3H-D1: 97.4%), whereas differences exist in sequence and length between the introns, as observed in Figure 2. An F3H enzyme has been reported to have a unique motif of pfam03171 that is maintained commonly in a superfamily of 2-oxoglutarate (2OG) and Fe (II)-dependent oxygenase. Deduced amino acid sequences of wheat F3Hs also have this pfam03171 motif (Figure 5(a)). Phylogenic relationships among F3Hs of wheat and other plant species were calculated using the UPGMA method of GENETYX software ver. 7.0. Wheat is grouped into the monocotyledon species, including barley, rice, and maize (Figure 5(b)).
Figure 5.
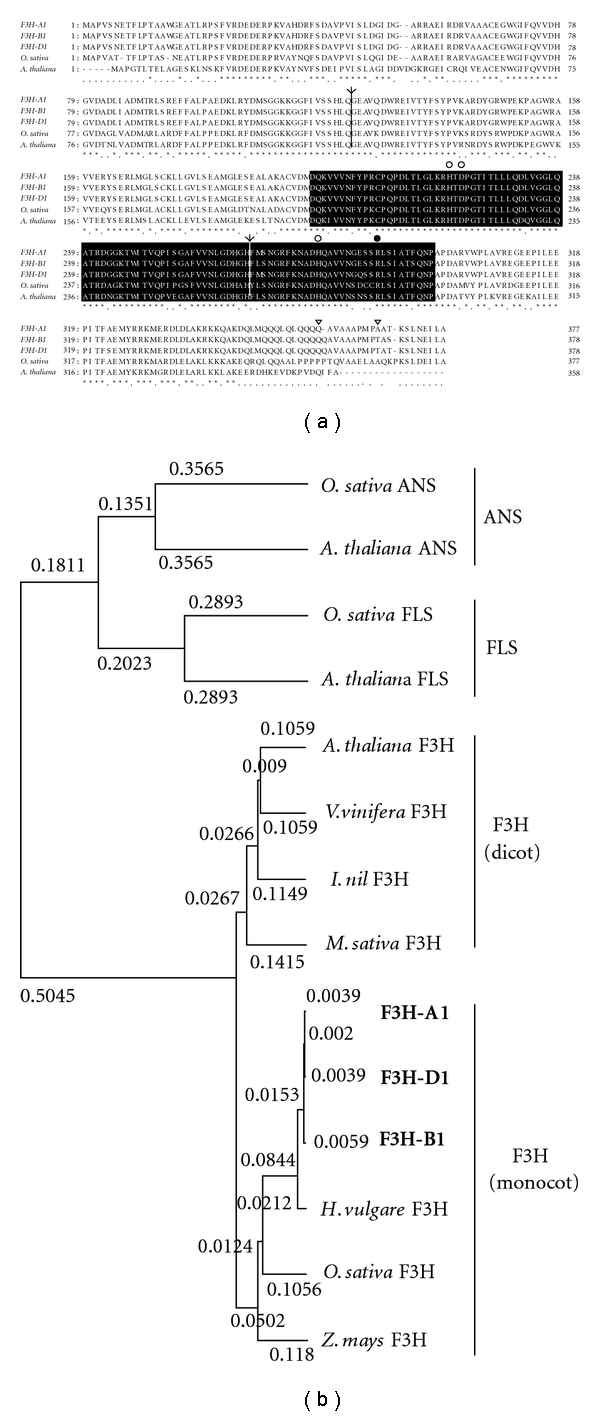
(a) Deduced amino acid sequences of wheat, rice, and Arabidopsis F3H. Lines with an arrowhead indicate the intron insertion sites. The region in the black box indicates a characteristic motif, pfam03171, of 2OG-Fe(II) oxygenase superfamily. White circles indicate conserved histidine (H) and aspartic acid (D) residues for ferrous-iron coordination, and a black circle indicates arginine (R) residue for a binding site of 2-oxoglutarate. Triangles show a site of glutamine (Q) residue lacked and an amino acid substitution from threonine (T) to alanine (A) in F3H-A1. (b) UPGMA tree depicted using GENETYX software ver. 7.0, using the following genes belonging to 2OG-Fe(II) oxygenase superfamily: ANSs of Arabidopsis thaliana (Q96323) and Oryza sativa (CAA69252), FLSs of Arabidopsis thaliana (Q96330) and Oryza sativa (XP_467968), F3Hs of Arabidopsis thaliana (Q9S818), Hordeum vulgare (CAA41146), Ipomea nil (BAA21897), Medicago sativa (S71772), Oryza sativa F3H (XP_474226), Vitis vinifera (P41090), and Zea mays (AAA91227), and F3H-A1 (AB223024), F3H-B1 (AB223025), and F3H-D1 (AB223026) of Triticum aestivum.
3.5. Nucleotide Sequences of 5′ Region
The 5′ upstream regions of F3H-A1, F3H-B1, and F3H-D1 were amplified by the inverse PCR. The respective sequences of 734 bp, 731 bp, and 504 bp from the start codon (ATG) of F3H-A1, F3H-B1, and F3H-D1 were isolated and identified as the 5′ regions of respective F3H based on the intron sequences of the PCR products (Figure 4). Three P elements to which an MYB-type transcription factor, P, was able to bind were found in these 5′ upstream sequences. A G-box core sequence and an element for RAV1 transcription factor were also found in all three promoters. Only F3H-A1 had a unique element for a leucine zipper-type transcription factor, bZIP911, in the promoter (Figure 4).
3.6. Chromosomal Locations of F3H-A1, F3H-B1, and F3H-D1
Chromosomal locations of F3H-A1, F3H-B1, and F3H-D1 were examined in three deletion lines of CS with primers that were specific to each F3H (Tables 2 and 3). No amplification of F3H-A1, F3H-B1, and F3H-D1 was observed in deletion lines 2AL-1 (FL = 0.85), 2BL-6 (FL = 0.89), and 2DL-6 (FL = 0.94), which lack only the small telomeric region of the long arm of chromosomes 2A, 2B, and 2D (Figure 6(a)). These results suggest that F3H-A1, F3H-B1, and F3H-D1 were located, respectively, on the telomeric regions of the long arms of chromosomes 2A, 2B, and 2D.
Figure 6.
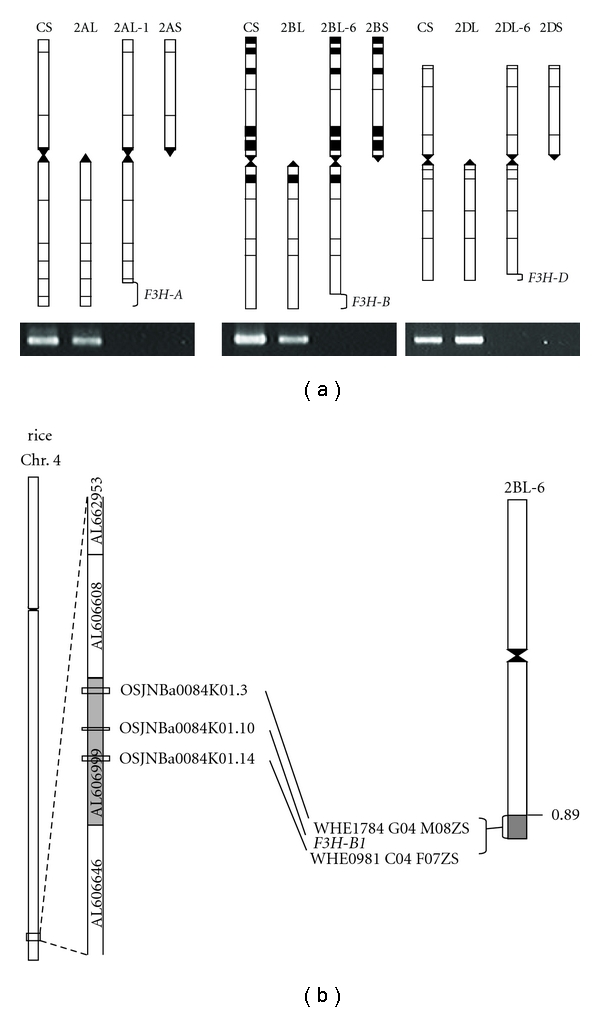
Amplification of F3H with the primers specific to F3H-A1, F3H-B1, and F3H-D1 in CS, ditelosomic and deletion lines of chromosomes 2A, 2B, and 2D: (a) amplification of F3H-A1 in CS, ditelo 2AL, ditelo 2AS, and a partial deletion line of 2AL-1 (FL = 0.85) (left), F3H-B1 in CS, ditelo 2BL, ditelo 2BS, and a partial deletion line of 2BL-6 (FL = 0.89) (center), and F3H-D1 in CS, ditelo 2DL, ditelo 2DS, and a partial deletion line of 2DL-6 (FL = 0.94) (right). (b) Locations of two wheat EST clones (WHE1784 G04 M08ZS and WHE0981 C04 F07ZS) and F3H-B1 on the 0.89–1.00 region of wheat chromosome 2BL, and putative rice F3H (OSJNBa0084K01.10) and two rice genes (OSJNBA0084K01.3 and OSJNBa0084K01.14) corresponding to the two wheat ESTs on rice chromosome 4.
3.7. Expressions of F3H-A1, F3H-B1, and F3H-D1
Respective expressions of F3H-A1, F3H-B1, and F3H-D1 in grains harvested at 5 DPA and 3-day-old coleoptiles were investigated with ANK-1C (red grain and red coleoptile; R/Rc), CS (red grain and white coleoptile; R/rc), NS67 (white grain and red coleoptile; r/Rc), and Norin 17 (white grain and white coleoptile; r/rc). F3H5LP and F3HARP primers for F3H-A1, F3H5LP and F3HBRP primers for F3H-B1, and F3H5LP and F3HDRP primers for F3H-D1 were used for RT-PCR (Table 2). All F3H-A1, F3H-B1, and F3H-D1 were highly expressed in red grains of ANK-1C and CS and red coleoptile of ANK-1C and NS67 (Figure 7). On the other hand, no expression of F3H-A1, F3H-B1, and F3H-D1 was detected in white tissues, although F3H-A1 of grains and coleoptiles of Norin 17 was slightly expressed.
Figure 7.

Expression of F3H-A1, F3H-B1, and F3H-D1 in grains and coleoptiles. Respective expressions of F3H-A1, F3H-B1, and F3H-D1 in grains harvested at 5 DPA and 3-day-old coleoptiles were investigated with ANK-1C (red grain and red coleoptile; R/Rc), CS (red grain and white coleoptile; R/rc), NS67 (white grain and red coleoptile; r/Rc), and Norin 17 (white grain and white coleoptile; r/rc). Actin was used as an internal control.
4. Discussion
4.1. Sequences of Wheat F3H Genes
The deduced amino acid sequences of three wheat F3Hs were similar to those of F3Hs of the other monocotyledon species (Figure 5(b)). Three F3Hs of wheat had a characteristic motif (pfam03171) found in the enzymes of 2OG-Fe (II) oxygenase superfamily (Figure 5(a)). Furthermore, 2-oxoglutarate- (2OG-) dependent and Fe (II)-dependent dioxygenases are known to catalyze oxidation of organic substrates using a dioxygen molecule [13]. Flavonol synthase (FLS), anthocyanidin synthase (ANS), and flavone synthase I (FS I) of the flavonoid pathway are also known to have the pfam03171 motif. Two histidines and one aspartic acid of the motif have been identified as sites for putative iron binding; an arginine residue has been identified as a 2-oxoglutarate binding site [14] (Figure 5(a)). These amino acid residues were conserved in wheat F3Hs (Figure 5(a)). Three wheat F3H enzymes appear to function as oxygenases. Recently, two sequences of F3H genes on B genome in wheat were isolated and reported as one of these sequences was not detected in red coleoptiles [15]. It is consistent with the result of southern blot which showed four fragments (Figure 3). It appears that the fourth gene F3H-B2, a nonhomologous duplication of F3H-B1, remains to be characterized.
4.2. Chromosomal Location of F3H Genes of Wheat
The results suggest that the F3H genes of wheat are located on the telomeric regions of the long arms of chromosomes 2A, 2B, and 2D (Figure 6(a)). Reportedly, there is a syntenic relationship between the chromosomes of wheat homoeologous group 2 and rice chromosomes 4 and 7 [16, 17]. Rice F3H (OSJNBa0084K01.10) in BAC clone OSJNBa0084K01 (Accession no. AL606999) was located on the telomeric region of rice chromosome 4 (Figure 6(b)). The BAC clone (OSJNBa0084K01) also included two putative genes, OSJNBa0084K01.3 and OSJNBa0084K01.14, that, respectively, showed high degrees of similarity (more than 80%) to wheat EST clones: WHE1784 G04 M08ZS (Accession no. BF202800) and WHE0981 C04 F07ZS (Accession no. BE500307). These two wheat EST clones were also located on the telomeric region of the long arm of the chromosome 2B (2BL6 0.89–1.00) in the physical bin map of wheat EST clones (http://wheat.pw.usda.gov/wEST/binmaps/) (Figure 6(b)). These results confirmed a high syntenic relationship between the telomeric region of the long arm of the chromosome of wheat homoeologous group 2 and the telomeric region of rice chromosome 4.
4.3. Expression of F3H and Pigmentation of Wheat Grain and Coleoptile
F3H-A1, F3H-B1, and F3H-D1 were all activated in red grain and coleoptile. On the other hand, only F3H-A1 of Norin 17 was slightly expressed in white grain and coleoptile. These results suggest that at least F3H-B1 and F3H-D1 are involved in pigmentation of the grain and coleoptile. Winkel-Shirley [6] showed that F3H took part in the biosynthesis of proanthocyanidin (condensed tannin) and anthocyanins, but not in that of phlobaphene. Pigment synthesized in wheat grain appeared to be proanthocyanidin.
The F3H-A1 of Norin 17 was expressed in the nonpigmented tissues. Comparison of the deduced amino acid sequence of F3H-A1 with that of F3H-B1 and F3H-D1, F3H-A1, showed an amino acid substitution from threonine to alanine and a lack of glutamine residue in the C-terminal region (Figure 5(a)). If these residues play an important role in function, F3H-A1 protein might not be involved in flavonoid biosynthesis. However, this possibility is not probable because these changes in the amino acid sequence occurred far from the characteristic motif, pfam03171, which is a key domain for enzyme function. Himi et al. [5] suggested that DFR was not expressed in white grains and coleoptiles and played a critical role in pigment production. Without DFR expression, F3H expression might not be decisive for pigmentation.
4.4. 5′ Region of F3Hs
Three P (MYB-type transcription factor) binding motifs were found in the 5′ region of F3H-A1, F3H-B1, and F3H-D1 (Figure 4). Himi and Noda [11] showed that DFRs of wheat also had three P motifs in their promoters. The F3H and DFR of wheat appeared to be controlled by a P-like transcription factor. Recently, we isolated an MYB-type gene of wheat, Tamyb10, which might control grain color [9, 18]. It is possible that Tamyb10 protein interacts with the P-binding motifs in promoters of F3H and DFR.
An RAV1 binding site was also found in the promoters of F3H-A1, F3H-B1, and F3H-D1. Rice F3H (OSJNBa0084K01.10) also had an RAV1 binding site in its promoter. In addition, RAV1 (RAV: for related to ABI3/VP1) was identified as a DNA binding protein possessing an N-terminal AP2/ERF- (or EREBP-) type DNA binding domain and a C-terminal B3 domain [19]; RAV1 is cold-responsive [20]. In Arabidopsis and petunia, cold stress (4°C) is known to activate PAL and CHS expression and to induce the accumulation of anthocyanin in leaves, stems, and flowers [21, 22]. It is likely that an RAV1-like transcription factor also activates wheat F3Hs because wheat coleoptiles showed more reddish color after chilling.
F3H-A1 was expressed in white grain and coleoptile. A recognition site of bZIP-type transcription factor (bZIP911) was found only in the promoter of F3H-A1. In Antirrhinum, bZIP911 reportedly modifies gene expression of histone H4 and chlorophyll a/b (CAB) binding protein [23]. Although no reports have described the bZIP911-like gene in wheat, a bZIP protein such as bZIP911 might activate F3H-A1 in white grain and coleoptile. Differences in promoter sequences of the F3Hs increase variation of F3H expression and appear to have occurred in evolution during differentiation of three wheat genomes (A, B, and D).
Acknowledgments
We thank Dr. Takashi R. Endo, Kyoto University, Japan, the National BioResource Project (NBRP), WHEAT, Japan, and Dr. John Flintham, John Innes Centre, Norwich, UK, for providing the grains of deletion lines and Novosibirskaya 67. We also extend our appreciation for grants from the Ohara Foundation for Agricultural Science, Ofu-Kai for the Promotion of Education and Culture at Japan Women's University, the Elizabeth Arnold Fuji Foundation, and the Ministry of Agriculture, Forestry and Fisheries of Japan.
References
- 1.Holton TA, Cornish EC. Genetics and biochemistry of anthocyanin biosynthesis. The Plant Cell. 1995;7(7):1071–1083. doi: 10.1105/tpc.7.7.1071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Himi E, Mares DJ, Yanagisawa A, Noda K. Effect of grain colour gene (R) on grain dormancy and sensitivity of the embryo to abscisic acid (ABA) in wheat. Journal of Experimental Botany. 2002;53(374):1569–1574. doi: 10.1093/jxb/erf005. [DOI] [PubMed] [Google Scholar]
- 3.McIntosh RA, Yamazaki Y, Dubcovsky J, et al. Catalogue of Gene Symbols for Wheat. 2010. [Google Scholar]
- 4.Miyamoto T, Everson EH. Biochemical and physiological studies of wheat seed pigmentation. Agronomy Journal. 1958;50:733–734. [Google Scholar]
- 5.Himi E, Nisar A, Noda K. Colour genes (R and Rc) for grain and coleoptile upregulate flavonoid biosynthesis genes in wheat. Genome. 2005;48(4):747–754. doi: 10.1139/g05-026. [DOI] [PubMed] [Google Scholar]
- 6.Winkel-Shirley B. Flavonoid biosynthesis. A colorful model for genetics, biochemistry, cell biology, and biotechnology. Plant Physiology. 2001;126(2):485–493. doi: 10.1104/pp.126.2.485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mol J, Grofewold E, Koes R. How genes paint flowers and seeds. Trends in Plant Science. 1998;3(6):212–217. [Google Scholar]
- 8.Nesi N, Jond C, Debeaujon I, Caboche M, Lepiniec L. The Arabidopsis TT2 gene encodes an R2R3 MYB domain protein that acts as a key determinant for proanthocyanidin accumulation in developing seed. The Plant Cell. 2001;13(9):2099–2114. doi: 10.1105/TPC.010098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Himi E, Maekawa M, Miura H, Noda K. Development of PCR markers for Tamyb10 related to R-1, red grain color gene in wheat. doi: 10.1007/s00122-011-1555-2. Theoretical and Applied Genetics. In press. [DOI] [PubMed] [Google Scholar]
- 10.Hartmann U, Sagasser M, Mehrtens F, Stracke R, Weisshaar B. Differential combinatorial interactions of cis-acting elements recognized by R2R3-MYB, BZIP, and BHLH factors control light-responsive and tissue-specific activation of phenylpropanoid biosynthesis genes. Plant Molecular Biology. 2005;57(2):155–171. doi: 10.1007/s11103-004-6910-0. [DOI] [PubMed] [Google Scholar]
- 11.Himi E, Noda K. Isolation and location of three homoeologous dihydroflavonol-4-reductase (DFR) genes of wheat and their tissue-dependent expression. Journal of Experimental Botany. 2004;55(396):365–375. doi: 10.1093/jxb/erh046. [DOI] [PubMed] [Google Scholar]
- 12.Meldgaard M. Expression of chalcone synthase, dihydroflavonol reductase, and flavanone-3-hydroxylase in mutants of barley deficient in anthocyanin and proanthocyanidin biosynthesis. Theoretical and Applied Genetics. 1992;83(6-7):695–706. doi: 10.1007/BF00226687. [DOI] [PubMed] [Google Scholar]
- 13.Aravind L, Koonin EV. The DNA-repair protein AlkB, EGL-9, and leprecan define new families of 2-oxoglutarate- and iron-dependent dioxygenases. Genome biology. 2001;2(3) doi: 10.1186/gb-2001-2-3-research0007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lukacin R, Britsch L. Identification of strictly conserved histidine and arginine residues as part of the active site in Petunia hybrida flavanone 3β-hydroxylase. European Journal of Biochemistry. 1997;249(3):748–757. doi: 10.1111/j.1432-1033.1997.t01-2-00748.x. [DOI] [PubMed] [Google Scholar]
- 15.Khlestkina EK, Roder MS, Salina EA. Relationship between homoeologous regulatory and structural genes in allopolyploid genome—a case study in bread wheat. BMC Plant Biology. 2008;8, article 88 doi: 10.1186/1471-2229-8-88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kurata N, Moore G, Nagamura Y, et al. Conservation of genome structure between rice and wheat. Bio/Technology. 1994;12(3):276–278. [Google Scholar]
- 17.Sorells ME, La Rota M, Bermudez-Kandianis CE, et al. Comparative DNA sequence analysis of wheat and rice genomes. Genome Research. 2003;13(8):1818–1827. doi: 10.1101/gr.1113003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Himi E, Noda K. Red grain colour gene (R) of wheat is a Myb-type transcription factor. Euphytica. 2005;143(3):239–242. [Google Scholar]
- 19.Kagaya Y, Ohmiya K, Hattori T. RAV1, a novel DNA-binding protein, binds to bipartite recognition sequence through two distinct DNA-binding domains uniquely found in higher plants. Nucleic Acids Research. 1999;27(2):470–478. doi: 10.1093/nar/27.2.470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fowler S, Thomashow MF. Arabidopsis transcriptome profiling indicates that multiple regulatory pathways are activated during cold acclimation in addition to the CBF cold response pathway. The Plant Cell. 2002;14(8):1675–1690. doi: 10.1105/tpc.003483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Leyva A, Jarillo JA, Salinas J, Martinez-Zapater JM. Low temperature induces the accumulation of Phenylalanine ammonia-lyase and Chalcone synthase mRNAs of Arabidopsis thaliana in a light-dependent manner. Plant Physiology. 1995;108(1):39–46. doi: 10.1104/pp.108.1.39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shvarts M, Borochov A, Weiss D. Low temperature enhances petunia flower pigmentation and induced chalcone synthase gene expression. Physiologia Plantarum. 1997;99(1):67–72. [Google Scholar]
- 23.Martinez-Garcia JF, Moyano E, Alcocer MJ, Martin C. Two bZIP proteins from Antirrhinum flowers preferentially bind a hybrid C-box/G-box motif and help to define a new sub-family of bZIP transcription factors. Plant Journal. 1998;13(4):489–505. doi: 10.1046/j.1365-313x.1998.00050.x. [DOI] [PubMed] [Google Scholar]


