Abstract
Cancer genomes frequently undergo genomic instability resulting in accumulation of chromosomal rearrangement. To date, one of the main challenges has been to confidently and accurately identify these rearrangements using short-read massively parallel sequencing. We were able to improve cancer rearrangement detection by combining two distinct massively parallel sequencing strategies: fosmid-sized (36 Kilobases on average) and standard 5 Kilobase mate pair libraries. We applied this strategy to map rearrangements in two breast cancer cell lines, MCF7 and HCC1954. We detect and validate a total of 91 somatic rearrangements in MCF7 and 25 in HCC1954, including genomic alterations corresponding to previously reported transcript aberrations in these two cell lines. Each of the genomes contains two types of breakpoints – clustered and dispersed. In both cell lines, the dispersed breakpoints show enrichment for low copy repeats, while the clustered breakpoints associate with high-copy number amplifications. Comparing the two genomes, we observe highly similar structural mutational spectra affecting different sets of genes, pointing to similar histories of genomic instability against the background of very different gene network perturbations.
Keywords: fosmid ditag, massively parallel sequencing, gene fusion, copy number variation, genomic instability
Introduction
End sequence profiling of clonal libraries have been used extensively to discover structural variation in both normal and cancer genomes [1–5]. Recently, the adoption of massively parallel sequencing has supplemented structural variation detection by identifying rearrangements at fine-scale resolution for both normal and cancer genomes [6–14]. These massively parallel sequencing studies have significantly added to the catalog of genomic rearrangements, but the limited insert sizes between paired ends have provided less power than larger insert clones to map across duplications and repeat-rich regions in the genome, thereby missing a large fraction of variation [2, 15]. Massively parallel mate pair sequencing is also hindered by high false positive rearrangement discovery rates, requiring additional breakpoint validation. Commonly employed techniques for breakpoint validation include: optical mapping based on restriction enzyme maps or incorporated fluorochrome-labelled nucleotide [2, 16], hybridization of fluorescent probes that span rearrangements [5, 17], and polymerase chain reaction amplification across aberrant fusions followed by Sanger-based sequencing of the breakpoint amplicons [1, 7, 11, 12, 14]. Although these validation techniques offer proof of genomic rearrangement, none are currently amenable to high-throughput workflows.
We supplement the limited insert size of standard massively parallel mate pair sequencing by incorporating fosmid-sized insert libraries, thereby providing additional validation of detected rearrangements. Our fosmid-sized mate pair libraries, called fosmid diTags, leverage the affordable costs and high-throughput capacity of massively parallel sequencing while providing clone-sized inserts able to span long repetitive sequence elements. Fosmid diTags are well-suited for rearrangement detection either in stand-alone or complementary fashion with other mate pair libraries; fosmid diTags are also advantageous for de novo genome assembly where larger insert size facilitates greater continuity [18]. Fosmid diTags are an extension of paired end tag methods [14, 19–21], where short paired tags from the ends of DNA fragments are enzymatically extracted and covalently linked as ditag constructs for high-throughput sequencing, see Supplemental Figures 1 and 2 for fosmid diTag workflow details.
Materials and Methods
Sequencing library preparation
Paired end sequencing methods exploit the fact that structural abnormalities consist of two chromosomal segments that are in a relative position, orientation, or at a relative distance that is not consistent with the reference genome assembly. Construction of paired end sequencing libraries that adequately cover the genome of interest allows for comprehensive identification of structural abnormalities.
1.55 million MCF7 (ATCC HTB-22) and 1.50 million HCC1954 (ATCC CRL-2338) fosmids were cloned using the novel pFosDT1.2 vector (derived from the Epicentre pCC1FOS™ plasmid). The pFosDT1.2 vector contains two EcoP15I restriction sites that flank the site of insertion. EcoP15I, a type III restriction endonuclease, cuts 25/27 bp downstream of its recognition site and requires two separated and inversely oriented recognition sites in supercoiled DNA. Addition of sinefungin in the EcoP15I digest reaction facilitates cleavage at all recognition sites independent of DNA topology [22]. Starting with 10 μg of purified pooled fosmid DNA from each breast cancer cell line, two independent long-range clonal insert, fosmid diTag massively parallel sequencing libraries were produced. For each fosmid library, 26 bp end-tags from the insert termini are isolated and concatenated as illustrated in Supplemental Figure 2.
Illumina mate pair whole-genome shotgun libraries, of insert sizes ranging from 4 to 6 Kb, were additionally constructed using 10 μg of genomic DNA from each of the MCF7 (ATCC HTB-22) and HCC1954 (ATCC CRL-2338) cell lines. Mate pair libraries were prepared according to the manufacturer’s instructions (Illumina PE-112-1002). Two separate MCF7 mate pair libraries with 4 Kb and 6 Kb inserts were constructed, and a single HCC1954 mate pair library with 5 Kb inserts was constructed.
The fosmid diTag and Illumina mate pair libraries were sequenced on an Illumina Genome Analyzer II massively parallel sequencing system following the manufacturer’s instructions. Raw sequence data for the fosmid diTag and standard Illumina mate pair libraries is available online at www.genboree.org/breastCellLineReads/.
Mapping to reference genome
Novocraft-V2.05.02 was used to align quality-filtered paired end reads to the reference human genome (March 2006 assembly, NCBI Build 36.1, UCSC Build HG18). Novoalign parameters used for mapping are described in Supplementary Materials and mapping coordinates are available for viewing and download through the Genboree open-hosting genome browser at www.genboree.org.
Structural rearrangement calling
Fosmid diTags and Illumina mate pair sequences that align discordantly were used to call putative structural rearrangements. The combined fosmid diTag and standard Illumina mate pair structural rearrangements that validated as cancer-specific somatic mutations are available in the Supplemental Table.
Determining structural variants from Illumina mate pair and fosmid diTag sequences is complicated by two factors: the contamination of inward facing reads and the formation of chimeric clones, respectively. Inward facing reads are paired end sequences from a contiguous piece of DNA sized equal to the final sequencing library length, approximately 400 bp. Formation of chimeric clones during the fosmid diTag procedure introduce false information about the distance and orientation between two reads, complicating structural variant calling.
False positive breakpoints called by inward facing reads were removed prior to reporting structural variants by filtering the discordant read clusters. Inward facing read clusters were filtered based on size and their inward facing read orientation. Because inward facing read clusters are limited by the final sequencing library length, they are easily identified and maybe removed from further analysis, see Supplementary Materials for filter parameters. However, inward facing read clusters that span true positive rearrangements are unable to be filtered, thereby introducing persistent confounding contamination. To overcome fosmid diTag chimera noise, discordant reads supporting the same structural variant were clustered. Clusters are formed if there are at least two uniquely mapping paired end signatures with corroborating genomic positions, sizes and read orientations, such a strategy is called standard clustering and is commonly employed [6, 8, 10, 23–25].
Breakpoint spanning PCR primer design pipeline
PCR primers were designed for amplification across aberrant fusions using the human reference genome (March 2006 assembly, NCBI Build 36.1, UCSC Build HG18). The Primer3 primer design algorithm was employed to obtain a set of nested primers using two categories or parameters, stringent and relaxed. Primer pairs in each category are scored and the highest-scoring primer pair is selected for PCR assay validation. Priority is given to the stringent category using the repeat-masked human reference genome. In cases of PCR amplification failure, additional lower-scoring primer pairs were utilized. More details, including Primer3 parameters, can be found in the Supplementary Materials; the automated primer design pipeline code is available for download at http://github.com/oliverhampton/Breakpoint-Primer-Design.
Copy number variation calling
Uniquely mapping reads were used as input for the readDepth R package [26] which calls copy number alterations by evaluating depth of sequence coverage. The package’s default parameters were used including an overdispersion value of 3 and a false-discovery rate of 0.01. The readDepth package also provided a breakpoint refinement tool that allowed us to adjusted copy number segment ends to matched breakpoint positions.
Breakpoint clustering algorithm
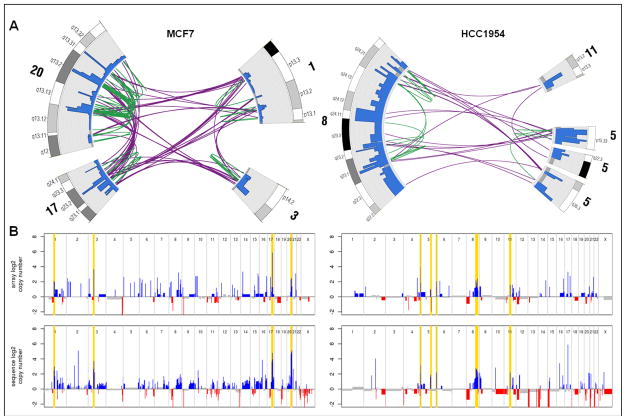
Combined fosmid-sized and 5 Kb breakpoints that map within 2 Mb in MCF7 and 5 Mb in HCC1954 were clustered. Chromosome segment annotations were retained if five or more breakpoints in MCF7 or two or more breakpoints in HCC1954 were contained within the cluster. For each set of MCF7 and HCC1954 breakpoint clusters, the cluster containing the highest number of breakpoints served as a seed for a connected graph (or clique) where the chromosome segments are nodes and spanning breakpoints are edges. In this manner, cliques of four breakpoint clusters in MCF7 on chromosomes 1, 3, 17, and 20, and five breakpoint clusters in HCC1954 on chromosomes 5, 8, and 11 were constructed as shown in Figure 2.
Figure 2.
(A) Arc visualizations of the largest MCF7 and HCC1954 breakpoint cliques and their association with copy number amplification. Chromosome cytobands are shaded and labeled. The colored rearrangements depict breast tumor intrachromosomal (green) and interchromosomal (purple) mutations. Copy number counts are plotted with gains in blue, losses in red, and normal diploid in grey (count scales are from zero to MCF7: max = 60; HCC1954: max = 15). (B) MCF7 and HCC1954 log2 copy number plots of Affymetrix 100k SNP chip arrays [34] (top) and Illumina mate pair mapped sequence counts (bottom); gains are plotted in blue, losses in red, and normal diploid in grey. Highlighted regions correspond to the largest breakpoint cliques from A.
Identification of low copy repeat regions
Each of the fosmid diTag and Illumina mate pair breakpoints from MCF7 and HCC1954 breast cancer cell lines were examined for the presence of low copy repeats (LCRs). Intra- and inter-chromosomal homologous LCRs were detected by applying a novel algorithmic method to the human reference genome sequence (March 2006 assembly, NCBI Build 36.1, UCSC Build HG18). The method achieved higher sensitivity than previously applied methods [27] by using k-mer frequency sequence information to detect, parse and cluster LCRs, without removing high copy number repetitive elements (repeat-masking). The LCRs detected by this method covered 6% of the whole genome in length, of which 19% were gene-containing regions. A detailed description of the algorithm is available in the Supplementary Materials.
PCR amplification of genomic DNA from cell lines
Breast cancer cell line breakpoint confirmation employed PCR amplification of MCF7 (ATCC HTB-22) and a pool of negative control genomic DNA isolated from human female (Novagen 70605-3) and from two different cell lines, MCF10A (ATCC CRL-10317) and HCC1599-BL (ATCC CRL-2332); and PCR amplification of genomic DNA from HCC1954 (ATCC CRL-2338) and negative control HCC1954-BL (ATCC CRL-2339) cell lines. Genomic cell line DNA was isolated with the DNAeasy kit (Qiagen). PCR bands were visualized on a 2% agarose gel.
Results
Combining fosmid diTag and 5 Kb mate pair sequencing libraries increases specificity to detect chromosomal rearrangements
The Illumina standard mate pair libraries, with an average 5 Kb insert size, generated 2.9 and 1.9 Gigabases of sequence data for MCF7 and HCC1954, respectively. Upon mapping to the reference genome, the relatively short distance between the paired ends is compatible for PCR primer design across aberrant fusions, and the density of mapped reads allow measurement of segment copy number. The fosmid diTag libraries generated 93.3 and 56.9 Megabases of sequence data for MCF7 and HCC1954, respectively. Because of the larger insert size, mapping of fosmid diTags provided near identical percent-coverage of the reference genome (81% ± 7%) as observed from the Illumina mate pair libraries.
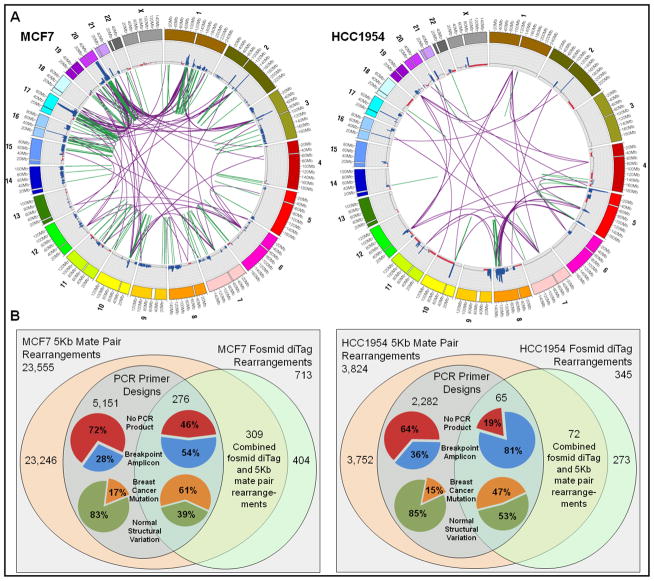
Rearrangements are reported where at least two independent pairs of ends show discrepancy by their predicted size and/or orientation. Discordant mate pairs and diTags are reported when the distance between the mapped ends is in excess of two standard deviations from the insert mean. Discordant ends are clustered based on mapping position and orientation discrepancies, thereby refining the position of the detected breakpoint. From the Illumina 5 Kb mate pair libraries, we identify 23,555 putative rearrangements in MCF7 and 3,824 in HCC1954. Breakpoint spanning PCR primers designs were able to be created for 23% of these rearrangements in MCF7 and 61% in HCC1954. From the fosmid diTag libraries, we identify 713 putative rearrangements in MCF7 and 345 in HCC1954 – because of the much longer fosmid diTag insert size and relatively low fold-coverage, standard and long-range PCR primer design was incompatible.
The high percentage of failed PCR primer designs from the MCF7 mate pair data is due to increased prevalence of repetitive sequence elements surrounding aberrant fusions. Closer inspection of the PCR primer design failure sites reveals overlap with repeat-masked sections of the human genome and disproportionate calling of small indels (2–4 Kb) at a rate ten times more than expected. We speculate that MCF7 has unique defects in its DNA repair pathways, which explains the imbalance of mutations between the two cell lines. We have previously shown RAD51C to be mutated in MCF7 [28]; such a mutation could affect the Holliday junction (HJ) [29] resolution machinery causing misrecognition of HJs, cruciforms, and other homology-driven secondary structures leading to double-strand breaks and accumulation of such indels [30].
Breakpoint spanning primers from the Illumina mate pairs were applied to their respective breast cancer cell line genomes and normal controls. In most cases, the PCR assay failed to produce an amplification product, indicating a high rate of false positive rearrangement detection. In the cases where a breakpoint amplicon was produced, the majority identified normal structural polymorphisms – only a small percentage identified breast cancer-specific somatic mutation, see Figure 1B Venn diagrams. Interestingly, combining fosmid diTag and Illumina mate pair data, and selecting rearrangements detected by both methods show 3-fold enrichment for cancer-specific somatic mutation and 2-fold reduction in false positives when compared to the Illumina mate pair libraries alone. Combining fosmid-sized and 5 Kb mate pairs provides cross validation to rearrangement detection; moreover, the incorporation of longer fosmid-sized inserts increases specificity to detect breast cancer-specific somatic mutation and decreases the reporting of false positives when compared to the shorter 5 Kb inserts alone. Combining fosmid diTag and 5 Kb mate pair libraries, we identify 309 chromosomal rearrangements in MCF7 and 72 in HCC1954, and design breakpoint spanning PCR primers for approximately 90% of them, see Figure 1A for the positions of the combined rearrangements. While it is desirable to increase the specificity of chromosomal rearrangement detection, it must be noted that a corresponding loss of sensitivity is associated with this improvement.
Figure 1.
(A) Circular visualizations of the MCF7 and HCC1954 genomes obtained using Circos [52] software. Chromosomes are individually colored with centromeres in white. Copy number variation is plotted with gains in blue and losses in red. The colored rearrangements depict breast cancer-specific somatic mutations from the combined fosmid-sized and 5Kb mate pair libraries. Green lines denote intrachromosomal and purple lines denote interchromosomal rearrangements. (B) Venn diagrams comparing the numbers of fosmid-sized and 5Kb mate pair rearrangements, PCR primer designs, PCR assays that produced breakpoint amplicon versus no amplification product, and rearrangements that are validated as breast cancer-specific mutation versus normal structural variation in the MCF7 and HCC1954 genomes.
Corresponding genomic DNA fusions exist for upwards of half of the gene fusions and truncations previously detected by transcript mapping
Chimeric gene transcripts have been previous identified in MCF7 [31, 32] and HCC1954 [33] using transcript mapping. Transcript mapping is analogous to targeted paired end sequencing; however, instead of investigating aberrant genomic fusions, chimeric mRNA transcripts are queried. Transcript mapping delivers a gene-centric view of rearrangements that encompass post-transcriptional modifications, but can’t detect genomic rearrangements outside of gene coding regions. We therefore sought to comprehensively identify rearrangement events at the genomic DNA level that may have caused chimeric or truncated mRNA transcripts.
In MCF7, we identified ten out of nineteen and nine out of thirty genomic rearrangements that are correlated with corresponding chimeric mRNA transcripts reported by Maher, CA et. al. [9, 31] and Inaki, K et. al. [32], respectively. These genomic lesions involve oncogenes (TMEM49), tumor suppressors (SULF2, PTPRG), constituents of DNA double-strand break repair (RAD51C.2, BRIP1), and other genes related to cell cycle, growth, and survival (RPS6KB1, ELOVL7, ABCA5), see Table I for functional details.
Table I.
Validated chimeric protein fusions and gene truncations in MCF7 and HCC1954 breast cancer cell lines.
| Cell line | Genomic changes | Chromosome locations | Genes affected | Effect on coding | Somatic changes reported in cancers | Validation method |
|---|---|---|---|---|---|---|
| MCF7 | Inter-chromosomal translocation | t(17;20)(q23.2;q13.13) | BCAS3 and BCAS4 | chimeric protein | Fusion is recurrently present in MCF7 breast and HCT116 colon cancer cell lines. | cDNA, genomic and published [9, 20, 28, 32, 53] |
| MCF7 | Inter-chromosomal translocation | t(17;3)(q22;p14.1) | RAD51C.2 and ATXN7 | chimeric protein | Decreased expression of RAD51C found in majority of breast cancer cell lines. | FISH, cDNA, genomic and published [28, 32] |
| MCF7 | Intra-chromosomal inversion | t(20;20)(q13.13;q13.13) | SULF2 and ARFGEF2 | chimeric protein | SULF2 is a known tumor suppressor. SULF2 siRNA silencing is tumorigenic in vivo. | cDNA, genomic and published [9, 28, 32] |
| MCF7 | Inter-chromosomal translocation | t(3;20)(p14.1;q13.13) | SULF2 and PRICKLE2 | chimeric protein | see above SULF2 function and phenotype | Genomic and published [9, 28, 32] |
| MCF7 | Intra-chromosomal indel | t(19;19)(p13.11;p13.11) | MYO9B and FCHO1 | chimeric protein | MYO9B mutations associate with different inflammatory or autoimmune diseases. | Genomic and published [9, 32] |
| MCF7 | Intra-chromosomal indel | t(17;17)(q22;q23.1) | BC017255 and TMEM49 | chimeric protein | High-level amplification at TMEM49 induces high expression of miR-21, which targets PTEN and results in an aggressive breast cancer phenotype. [54] | Genomic and published [9] |
| MCF7 | Intra-chromosomal inversion | t(17;17)(q23.1;q23.1) | RPS6KB1 and TMEM49 | chimeric protein | See above TMEM49 function and phenotype. RPS6KB1 is amplified and overexpressed in 10–30% of primary breast cancers and cell lines. RPS6KB1 is regulated by the mTOR pathway which regulates cell cycle, growth and survival. [55] | Genomic and published [9, 32] |
| MCF7 | Intra-chromosomal inversion | t(5;5)(q12.1;q12.1) | DEPDC1B and ELOVL7 | chimeric protein | ELOVL7 is over expressed in bladder, breast, colorectal, esophageal, gastric, and prostate cancers. High-fat diet promotes growth of in vivo tumors of ELOVL7-expressed prostate cancer. [56] | cDNA, genomic and published [9, 28, 32] |
| MCF7 | Inter-chromosomal translocation | t(3;17)(p14.2;q22) | TEX14 and PTPRG | chimeric protein | PTPRG is a known tumor suppressor in kidney, lung and breast cancers. PTPRG has been shown to inhibit MCF7 anchorage-independent growth and reduce estrogenic response cell proliferation. [57] | Genomic and published [9, 32] |
| MCF7 | Inter-chromosomal translocation | t(17;20)(q24.3;q13.32) | ABCA5 and PPP4R1L | chimeric protein | Induction of ABCA5 correlates with differentiation state of human colon tumor. | Genomic and published [9] |
| MCF7 | Intra-chromosomal inversion | t(X;X)(p22.2;p22.2) | CXorf15 and SYAP1 | chimeric protein | no known cancer phenotype | Genomic and published [9, 32] |
| MCF7 | Inter-chromosomal translocation | t(3;17)(p14.1;q23.2) | BRIP1 | truncation | BRIP1 truncations confer a two-fold increased risk of developing breast cancer. Truncation mutants block double stranded break repair. | Genomic and published [9] |
| HCC1954 | Inter-chromosomal translocation | t(5;8)(q23.1;q13.13) | EIF3E | truncation | Truncation is tumorigenic in vivo. Decreased expression found in one third of all human breast carcinomas. | cDNA, genomic and published [12, 33] |
| HCC1954 | Inter-chromosomal translocation | t(5;8)(q35.3;q24.21) | NSD1 | truncation | Fusion protein in acute myeloid leukemia. | FISH, cDNA, genomic and published [33] |
| HCC1954 | Inter-chromosomal translocation | t(5;8)(p15.33;q24.21) | CLPTM1L and PVT1 | truncation | Amplification of PVT1 linked to pathophysiology of ovarian and breast cancers. | Genomic and published [33] |
| HCC1954 | Inter-chromosomal translocation | t(5;8)(p15.35.2;q24.21) | UIMC1 or RAP80 | truncation | Recurrent RAP80 missense mutations identified in breast cancer patients. [45, 46] | Genomic |
In HCC1954, we identified three out of seven genomic rearrangements resulting in chimeric or truncated gene transcripts reported by Zhao et. al. [33]. These three gene truncations (EIF3E, NSD1, PVT1) are implicated in differing aspects of breast and ovarian cancers, and acute myeloid leukemia pathophysiologies, see Table I for functional details. In addition, we discovered a novel genomic rearrangement of UIMC1 (RAP80), a DNA double-stranded break repair accessory protein and suspected tumor suppressor, resulting in the loss of its last 5 exons (exons 11–15), which would eliminate its DNA recognition and binding abilities. The fused DNA (8q24.21) downstream of the UIMC1 breakpoint does not contain any exons or introns, and it remains unclear whether the truncated mRNA would be stable as there is no transcription stop site or polyA tail.
High-level amplifications of distinct driver oncogenes in MCF7 and HCC1954 are detected from mapped read density
The luminal-type MCF7 and ERBB2-overexpressing HCC1954 breast cancer cell lines are both highly amplified and display complex structural mutability phenotypes; exhibiting distinct profiles of genome structural rearrangement and copy number variation. We integrated read density and breakpoint information from mapped fosmid-sized and 5 Kb mate pair libraries to accurately identify copy number variation (CNV) using the readDepth R package [26], see Figures 1A and 2 for visualized copy number counts.
For comparison, we obtained data for both breast cancer cell lines run on Affymetrix 100K SNP chips segmented with the GLAD algorithm [34]. Even with approximately 1-fold sequence coverage, our results provide higher resolution than the Affymetrix arrays and allow for CNV calls to be made in many regions where no array probes exist. A look at gross features shows good concordance between the two platforms, including detection of previously described high-level amplifications on MCF7 [28]; and on HCC1954 [12], see highlighted regions in Figure 2B. Notably, our sequence-based approach provides higher dynamic range and reveals multiple regions in both cell lines that have been copied 50 to 100 times. Due to saturation effects and lower resolution, these regions are called with far lower copy number on the Affymetrix arrays. There are a small number of aberrations, including regions on chromosomes 2 and 9 in the MCF7 genome, which we believe to be biological differences between different passages and/or sublines of MCF7. Most other discordant events are likely attributable to increased coverage, resolution, and dynamic range from the sequence-based assays.
In MCF7, a 20 Kb segment on cytoband 20q13.31 shows the highest level of amplification with a copy number count of 70. This region encompasses the BMP7 gene, a member of the transforming growth factor-beta superfamily, and corresponds to the fusion of the BMP7 promoter upstream of ZNF217 oncogene, which is overexpressed in breast cancer [35]. ZNF217 can attenuate apoptotic signals resulting from telomere dysfunction and may promote neoplastic transformation during later stages of malignancy [36].
In HCC1954, a 51 Kb segment on cytoband 17q12 showed the highest level of amplification with a copy number count of 117. This region encompasses HER2/neu (also known as ERBB-2) which is known to be overexpressed in this cell line. HER2 overexpression in breast cancer is associated with an aggressive tumor phenotype, increased disease recurrence, and overall worse prognosis. HER2 overexpression serves not only as a prognostic marker, but also as a drug target for the monoclonal antibody trastuzumab. Also of interest is a 59 Kb segment on cytoband 11q13.2 showing 9-fold amplification and encompassing the gene CCND1. The CCND1 gene, a key cell-cycle regulator, is often overexpressed in breast cancer patients, and correlates with shorter relapse-free survival times [37].
MCF7 and HCC1954 exhibit defects in the homologous double-strand break repair pathway
In the MCF7 and HCC1954 breast cancer cell lines, we identified rearrangements in genes that code for members of protein complexes involved in DNA double-stranded break repair (DSBR), raising the possibility that distinct defects in DSBR genes may have contributed to different patterns of genomic instability. For example, in MCF7 we identified the gene-gene fusion of RAD51C exons 1–7 to the neuronal-specific gene ATXN7 exons 6–13 resulting in an expressed chimeric transcript. RAD51C is a paralog of RAD51 a gene central to DNA DSBR. RAD51C is an essential component of a complex reported to be involved in resolving Holliday junctions (HJs) [29] formed during DSBR [38] and, as such, is integral to the maintenance of genomic stability. The translocation we have identified eliminates the domain of RAD51C that binds other family members such as RAD51D and Xrcc3 [39], possibly disrupting formation of the complex responsible for resolving HJs.
Also in MCF7, we identified truncation of the BRIP1 gene, BRCA1-interacting protein-1. BRIP1 was originally identified as a helicase-like protein that interacts directly with BRCA1 and contributes to its DNA repair function. BRIP1 binds to the BCRT repeat in BRCA1. The C-terminus of BRIP1 is critical for its interaction with BRCA1, and a truncation mutant has been shown to block DSBR [40–42]. Clinically, germline truncation mutations of BRIP1 have been identified in familial breast cancer without mutations of BRCA1/2, and BRIP1 truncations confer a two-fold increased risk of developing breast cancer. We identified a translocation that results in the loss of the last three exons (exons 18–20), however the fused DNA (3p14) downstream of BRIP1 does not contain any exons or introns. The truncation at exon 17 of BRIP1 would eliminate the C-terminal third of BRIP1 and eliminate binding to BRCA1. However, it is unclear at present whether the truncated mRNA would be stable as there is no transcription stop site or polyA tail.
In HCC1954 we discovered a novel gene truncation of UIMC1 (also referred to as the BRCA1-A complex subunit RAP80). RAP80 has been extensively studied because of its roles in localizing BRCA1 to DNA double strand break sites, regulating BRCA1-dependent DNA damage checkpoint function, and as a potential tumor suppressor [43, 44]. Whereas many RAP80 missense SNP mutations have been identified in non-BRCA1/2 multi-ethnic breast cancer cases [45, 46], no truncating mutation of the RAP80 gene in breast cancer has been previously published. Interestingly, Dr. Xiaochun Yu has identified a truncating SNP mutation on RAP80 cDNA (G1107A) in the ovarian adenocarcinoma cell line TOV21G that results in a premature stop codon at Trp369. This truncation product disrupts the RAP80 interaction with BRCA1 and fails to localize to nuclear foci following DNA damage [47]. The UIMC1 truncation we identified cleaves the native transcript after exon 10 and results in loss of the C-terminus exons 11–15, similarly eliminating DNA recognition and binding capability.
While our fosmid diTags and Illumina mate pairs do not detect the previously published HCC1954 gene truncation of MRE11A [33], we do confirm the existence of the t(4;11)(q32;q21) genomic lesion involving MRE11A in another study using 2 Kb Life Technologies SOLiD mate pairs (unpublished). MRE11A is involved in homologous recombination, telomere length maintenance and DNA DSBR; and this truncation eliminates its DNA binding domain in the HCC1954 breast cancer cell line.
Coinciding occurrences of rearrangement clustering and amplification point to similar histories of genomic instability in MCF7 and HCC1954
As evidenced from Figure 1A, the breakpoints in MCF7 and HCC1954 are not evenly distributed across the genome. A number of clusters of closely spaced breakpoints are evident. To formally delineate clustered breakpoints from the remainder, breakpoints within 2 Mb in MCF7 and 5 Mb in HCC1954 were clustered. In each cell line, the cluster containing the highest number of breakpoints was selected to seed a connected graph where chromosome segments are nodes and spanning breakpoints are edges. In MCF7, four clusters emerge at cytobands 1p13.1-p21.1, 3p14.1-p14.2, 17q22-q24.3 and 20q12-q13.33. In HCC1954, five clusters emerge at cytobands 5p15.3, 5q22.3-q23.2, 5q35.2-q35.3, 8q22.2-q24.22, and 11q13.2-q12.3. Moreover, the four MCF7 and five HCC1954 clustered breakpoint locations exactly coincide with high-level amplifications in their respective genomes, indicating possible positive selection and functional significance, see Figure 2A and 2B.
The amplification patterns found in MCF7 and HCC1954 are consistent with the complex firestorm pattern described by Hicks et al. that associate with breast cancer prognostic markers and correspond with poor patient outcomes [48]. Interestingly, the most often detected recurrent locations of firestorm amplification identified by Hicks et al. within the 243 breast tumors studied, reside on chromosomal arms 11q and 17q. These loci contain the genes CCND1 on 11q and ERBB2 on 17q, noted previously to be highly amplified in HCC1954, and may drive selection for these amplifying mutations.
In both cell lines, the remaining non-clustered or dispersed breakpoints are highly associated with low copy repeats (LCRs). The dispersed breakpoints in MCF7 show a 9.8-fold enrichment for LCRs, while the trend is reiterated in HCC1954 with a 9.1-fold enrichment for LCRs. LCR enrichment at dispersed breakpoints is a characteristic previously described in MCF7 [28], and is recurrently identified in HCC1954. This finding is in contrast to the clustered breakpoints, that do not exhibit enrichment for LCRs.
Discussion
It is known that chromosomal rearrangements are highly associated with repetitive sequences in genomic disorders and cancer. Up to a quarter of entries in the Gross Rearrangement Breakpoint Database show presence of repetitive elements [49]. The repetitive elements range in size and may be as large as 6 Kb in the case of Long Interspersed Nuclear Elements (LINEs) and may cluster, creating long stretches of non-unique sequence. Breakpoints that overlap repetitive sequence elements may not be detected by 5 Kb (or shorter-range) mate pair libraries. Even if the breakpoint is detected, the non-unique sequence surrounding the rearrangement may make validation by PCR challenging. Having large clonal-sized inserts, fosmid diTags overcome this problem by spanning repetitive sequences and correctly identifying aberrant fusions. For example, in our previous study of MCF7 cells, we identified the expressed DEPDC1B-ELOVL2 chimeric mRNA transcript which is formed by a 5q12.1 intra-chromosomal inversion [28]. This breakpoint is detected using fosmid diTags, but not 5 Kilobase-sized mate pairs due the presence of LINEs, SINEs, and microsatellites surrounding the site of rearrangement.
In many cases, optimal PCR primer design is hindered by the presence of repetitive sequence surrounding the join. This is common when rearrangements are facilitated by homologous recombination [50, 51]. Short repetitive elements or longer segmental duplications (also referred to as low copy repeats) at sites of rearrangement severely limit the number of unique priming positions. Fosmid-sized inserts are able to span such repetitive regions, thus providing means of validating breakpoints even in cases of PCR assay failure. For example, there are two previously published gene truncations identified by our fosmid diTag and 5 Kb mate pair libraries that fail breakpoint spanning PCR assay confirmation. First is the t(5;8)(q35.3;q24.21) translocation in HCC1954 involving the truncation of NSD1, a fusion protein also found in myeloid leukemia [33]. Second is the t(3;15)(p14.1;q23.2) translocation in MCF7 involving the truncation of BRIP1, a BRCA1-interacting protein that contributes to DNA repair [28]. Although these two gene truncations are cross validated by fosmid and 5 Kb sized inserts, PCR assay across the breakpoint results in amplification failure. In these cases, breakpoint spanning primer design was severely hindered due to the presence of interspersed nuclear elements and long terminal repeats across the aberrant joins.
We show that fosmid-sized inserts are adept at spanning repetitive sequences known to exist at sites of gross rearrangement and low copy repeats associated with homologous recombination. Combining fosmid diTag and 5Kb Illumina mate pair libraries we were able to detect and validate aberrant fusions involving repetitive genomic sequence where detection by shorter end sequence profiles alone or validation by breakpoint spanning PCR assays failed. In addition, we observe that those rearrangements detected by both insert size ranges exhibit 3-fold enrichment for cancer-specific somatic mutation and 2-fold reduction in false positives when compared to the 5 Kb mate pairs alone.
For those breast cancer-specific somatic mutations involving genes, we queried transcriptome fusion and truncation literature to corroborate our finding and assess the extent to which our combined fosmid diTag and 5 Kb mate pair libraries rediscovered known chimeric transcripts reported in MCF7 and HCC1954. We identified genomic alterations corresponding to upwards of approximately half of the published MCF7 and HCC1954 chrimeric mRNA transcripts, but it is difficult to assess the lower bound of our sensitivity since it is unclear if the undetected transcript mutations are due to trans-splicing or similar post-transcriptional modifications.
We integrated read density and breakpoint information from mapped fosmid diTags and 5 Kb mate pairs to accurately identify distinct copy number variation in MCF7 and HCC1954. We discovered distinct driver oncogenes associated with high-copy number amplifications in MCF7 and HCC1954. The distinct structural mutability profiles between MCF7 and HCC1954 correlate to their phenotypic differences. Amplified chromosomal segments, breakpoint clusters, and affected genes are located at different positions across the MCF7 and HCC1954 genomes; and correspond to overexpression of different oncogenes, silencing of diverse tumor suppressors, and distinct defects in DNA repair machinery responsible for homology-driven repair of double-stranded DNA breaks. It is intriguing that in conjunction with mutations in the same DNA repair pathway we also find similar patterns of structural mutability in the two cell lines. Both have clustered and dispersed breakpoints; both exhibit clustered breakpoints in regions of high copy number amplification and dispersed breakpoints that are enriched for the presence of low copy repeats.
Supplementary Material
Acknowledgments
This project was funded by the NIH-NHGRI grant 1 R01 HG02583 and NIH-NCI grants R33 CA114151 and R21 CA128496 to AM.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Bignell GR, Santarius T, Pole JC, Butler AP, Perry J, Pleasance E, Greenman C, Menzies A, Taylor S, Edkins S, Campbell P, Quail M, Plumb B, Matthews L, McLay K, Edwards PA, Rogers J, Wooster R, Futreal PA, Stratton MR. Architectures of somatic genomic rearrangement in human cancer amplicons at sequence-level resolution. Genome Res. 2007;17:1296–303. doi: 10.1101/gr.6522707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kidd JM, Cooper GM, Donahue WF, Hayden HS, Sampas N, Graves T, Hansen N, Teague B, Alkan C, Antonacci F, Haugen E, Zerr T, Yamada NA, Tsang P, Newman TL, Tuzun E, Cheng Z, Ebling HM, Tusneem N, David R, Gillett W, Phelps KA, Weaver M, Saranga D, Brand A, Tao W, Gustafson E, McKernan K, Chen L, Malig M, Smith JD, Korn JM, McCarroll SA, Altshuler DA, Peiffer DA, Dorschner M, Stamatoyannopoulos J, Schwartz D, Nickerson DA, Mullikin JC, Wilson RK, Bruhn L, Olson MV, Kaul R, Smith DR, Eichler EE. Mapping and sequencing of structural variation from eight human genomes. Nature. 2008;453:56–64. doi: 10.1038/nature06862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tuzun E, Sharp AJ, Bailey JA, Kaul R, Morrison VA, Pertz LM, Haugen E, Hayden H, Albertson D, Pinkel D, Olson MV, Eichler EE. Fine-scale structural variation of the human genome. Nat Genet. 2005;37:727–32. doi: 10.1038/ng1562. [DOI] [PubMed] [Google Scholar]
- 4.Volik S, Raphael BJ, Huang G, Stratton MR, Bignel G, Murnane J, Brebner JH, Bajsarowicz K, Paris PL, Tao Q, Kowbel D, Lapuk A, Shagin DA, Shagina IA, Gray JW, Cheng JF, de Jong PJ, Pevzner P, Collins C. Decoding the fine-scale structure of a breast cancer genome and transcriptome. Genome Res. 2006 doi: 10.1101/gr.4247306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Volik S, Zhao S, Chin K, Brebner JH, Herndon DR, Tao Q, Kowbel D, Huang G, Lapuk A, Kuo WL, Magrane G, De Jong P, Gray JW, Collins C. End-sequence profiling: sequence-based analysis of aberrant genomes. Proc Natl Acad Sci U S A. 2003;100:7696–701. doi: 10.1073/pnas.1232418100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bentley DR, Balasubramanian S, Swerdlow HP, Smith GP, Milton J, Brown CG, Hall KP, Evers DJ, Barnes CL, Bignell HR, Boutell JM, Bryant J, Carter RJ, Keira Cheetham R, Cox AJ, Ellis DJ, Flatbush MR, Gormley NA, Humphray SJ, Irving LJ, Karbelashvili MS, Kirk SM, Li H, Liu X, Maisinger KS, Murray LJ, Obradovic B, Ost T, Parkinson ML, Pratt MR, Rasolonjatovo IM, Reed MT, Rigatti R, Rodighiero C, Ross MT, Sabot A, Sankar SV, Scally A, Schroth GP, Smith ME, Smith VP, Spiridou A, Torrance PE, Tzonev SS, Vermaas EH, Walter K, Wu X, Zhang L, Alam MD, Anastasi C, Aniebo IC, Bailey DM, Bancarz IR, Banerjee S, Barbour SG, Baybayan PA, Benoit VA, Benson KF, Bevis C, Black PJ, Boodhun A, Brennan JS, Bridgham JA, Brown RC, Brown AA, Buermann DH, Bundu AA, Burrows JC, Carter NP, Castillo N, Chiara ECM, Chang S, Neil Cooley R, Crake NR, Dada OO, Diakoumakos KD, Dominguez-Fernandez B, Earnshaw DJ, Egbujor UC, Elmore DW, Etchin SS, Ewan MR, Fedurco M, Fraser LJ, Fuentes Fajardo KV, Scott Furey W, George D, Gietzen KJ, Goddard CP, Golda GS, Granieri PA, Green DE, Gustafson DL, Hansen NF, Harnish K, Haudenschild CD, Heyer NI, Hims MM, Ho JT, Horgan AM, Hoschler K, Hurwitz S, Ivanov DV, Johnson MQ, James T, Huw Jones TA, Kang GD, Kerelska TH, Kersey AD, Khrebtukova I, Kindwall AP, Kingsbury Z, Kokko-Gonzales PI, Kumar A, Laurent MA, Lawley CT, Lee SE, Lee X, Liao AK, Loch JA, Lok M, Luo S, Mammen RM, Martin JW, McCauley PG, McNitt P, Mehta P, Moon KW, Mullens JW, Newington T, Ning Z, Ling Ng B, Novo SM, O’Neill MJ, Osborne MA, Osnowski A, Ostadan O, Paraschos LL, Pickering L, Pike AC, Chris Pinkard D, Pliskin DP, Podhasky J, Quijano VJ, Raczy C, Rae VH, Rawlings SR, Chiva Rodriguez A, Roe PM, Rogers J, Rogert Bacigalupo MC, Romanov N, Romieu A, Roth RK, Rourke NJ, Ruediger ST, Rusman E, Sanches-Kuiper RM, Schenker MR, Seoane JM, Shaw RJ, Shiver MK, Short SW, Sizto NL, Sluis JP, Smith MA, Ernest Sohna Sohna J, Spence EJ, Stevens K, Sutton N, Szajkowski L, Tregidgo CL, Turcatti G, Vandevondele S, Verhovsky Y, Virk SM, Wakelin S, Walcott GC, Wang J, Worsley GJ, Yan J, Yau L, Zuerlein M, Mullikin JC, Hurles ME, McCooke NJ, West JS, Oaks FL, Lundberg PL, Klenerman D, Durbin R, Smith AJ. Accurate whole human genome sequencing using reversible terminator chemistry. Nature. 2008;456:53–9. doi: 10.1038/nature07517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Campbell PJ, Stephens PJ, Pleasance ED, O’Meara S, Li H, Santarius T, Stebbings LA, Leroy C, Edkins S, Hardy C, Teague JW, Menzies A, Goodhead I, Turner DJ, Clee CM, Quail MA, Cox A, Brown C, Durbin R, Hurles ME, Edwards PA, Bignell GR, Stratton MR, Futreal PA. Identification of somatically acquired rearrangements in cancer using genome-wide massively parallel paired-end sequencing. Nat Genet. 2008;40:722–9. doi: 10.1038/ng.128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Korbel JO, Urban AE, Affourtit JP, Godwin B, Grubert F, Simons JF, Kim PM, Palejev D, Carriero NJ, Du L, Taillon BE, Chen Z, Tanzer A, Saunders AC, Chi J, Yang F, Carter NP, Hurles ME, Weissman SM, Harkins TT, Gerstein MB, Egholm M, Snyder M. Paired-end mapping reveals extensive structural variation in the human genome. Science. 2007;318:420–6. doi: 10.1126/science.1149504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Maher CA, Palanisamy N, Brenner JC, Cao X, Kalyana-Sundaram S, Luo S, Khrebtukova I, Barrette TR, Grasso C, Yu J, Lonigro RJ, Schroth G, Kumar-Sinha C, Chinnaiyan AM. Chimeric transcript discovery by paired-end transcriptome sequencing. Proc Natl Acad Sci U S A. 2009;106:12353–8. doi: 10.1073/pnas.0904720106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.McKernan KJ, Peckham HE, Costa GL, McLaughlin SF, Fu Y, Tsung EF, Clouser CR, Duncan C, Ichikawa JK, Lee CC, Zhang Z, Ranade SS, Dimalanta ET, Hyland FC, Sokolsky TD, Zhang L, Sheridan A, Fu H, Hendrickson CL, Li B, Kotler L, Stuart JR, Malek JA, Manning JM, Antipova AA, Perez DS, Moore MP, Hayashibara KC, Lyons MR, Beaudoin RE, Coleman BE, Laptewicz MW, Sannicandro AE, Rhodes MD, Gottimukkala RK, Yang S, Bafna V, Bashir A, MacBride A, Alkan C, Kidd JM, Eichler EE, Reese MG, De La Vega FM, Blanchard AP. Sequence and structural variation in a human genome uncovered by short-read, massively parallel ligation sequencing using two-base encoding. Genome Res. 2009;19:1527–41. doi: 10.1101/gr.091868.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pleasance ED, Cheetham RK, Stephens PJ, McBride DJ, Humphray SJ, Greenman CD, Varela I, Lin ML, Ordonez GR, Bignell GR, Ye K, Alipaz J, Bauer MJ, Beare D, Butler A, Carter RJ, Chen L, Cox AJ, Edkins S, Kokko-Gonzales PI, Gormley NA, Grocock RJ, Haudenschild CD, Hims MM, James T, Jia M, Kingsbury Z, Leroy C, Marshall J, Menzies A, Mudie LJ, Ning Z, Royce T, Schulz-Trieglaff OB, Spiridou A, Stebbings LA, Szajkowski L, Teague J, Williamson D, Chin L, Ross MT, Campbell PJ, Bentley DR, Futreal PA, Stratton MR. A comprehensive catalogue of somatic mutations from a human cancer genome. Nature. 2010;463:191–6. doi: 10.1038/nature08658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Stephens PJ, McBride DJ, Lin ML, Varela I, Pleasance ED, Simpson JT, Stebbings LA, Leroy C, Edkins S, Mudie LJ, Greenman CD, Jia M, Latimer C, Teague JW, Lau KW, Burton J, Quail MA, Swerdlow H, Churcher C, Natrajan R, Sieuwerts AM, Martens JW, Silver DP, Langerod A, Russnes HE, Foekens JA, Reis-Filho JS, van’t Veer L, Richardson AL, Borresen-Dale AL, Campbell PJ, Futreal PA, Stratton MR. Complex landscapes of somatic rearrangement in human breast cancer genomes. Nature. 2009;462:1005–10. doi: 10.1038/nature08645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Berger MF, Lawrence MS, Demichelis F, Drier Y, Cibulskis K, Sivachenko AY, Sboner A, Esgueva R, Pflueger D, Sougnez C, Onofrio R, Carter SL, Park K, Habegger L, Ambrogio L, Fennell T, Parkin M, Saksena G, Voet D, Ramos AH, Pugh TJ, Wilkinson J, Fisher S, Winckler W, Mahan S, Ardlie K, Baldwin J, Simons JW, Kitabayashi N, MacDonald TY, Kantoff PW, Chin L, Gabriel SB, Gerstein MB, Golub TR, Meyerson M, Tewari A, Lander ES, Getz G, Rubin MA, Garraway LA. The genomic complexity of primary human prostate cancer. Nature. 2011;470:214–20. doi: 10.1038/nature09744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hillmer AM, Yao F, Inaki K, Lee WH, Ariyaratne PN, Teo AS, Woo XY, Zhang Z, Zhao H, Ukil L, Chen JP, Zhu F, So JB, Salto-Tellez M, Poh WT, Zawack KF, Nagarajan N, Gao S, Li G, Kumar V, Lim HP, Sia YY, Chan CS, Leong ST, Neo SC, Choi PS, Thoreau H, Tan PB, Shahab A, Ruan X, Bergh J, Hall P, Cacheux-Rataboul V, Wei CL, Yeoh KG, Sung WK, Bourque G, Liu ET, Ruan Y. Comprehensive long-span paired-end-tag mapping reveals characteristic patterns of structural variations in epithelial cancer genomes. Genome Res. 2011 doi: 10.1101/gr.113555.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Alkan C, Sajjadian S, Eichler EE. Limitations of next-generation genome sequence assembly. Nat Methods. 2011;8:61–5. doi: 10.1038/nmeth.1527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Teague B, Waterman MS, Goldstein S, Potamousis K, Zhou S, Reslewic S, Sarkar D, Valouev A, Churas C, Kidd JM, Kohn S, Runnheim R, Lamers C, Forrest D, Newton MA, Eichler EE, Kent-First M, Surti U, Livny M, Schwartz DC. High-resolution human genome structure by single-molecule analysis. Proc Natl Acad Sci U S A. 2010;107:10848–53. doi: 10.1073/pnas.0914638107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Das SK, Austin MD, Akana MC, Deshpande P, Cao H, Xiao M. Single molecule linear analysis of DNA in nano-channel labeled with sequence specific fluorescent probes. Nucleic Acids Res. 38:e177. doi: 10.1093/nar/gkq673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gnerre S, Maccallum I, Przybylski D, Ribeiro FJ, Burton JN, Walker BJ, Sharpe T, Hall G, Shea TP, Sykes S, Berlin AM, Aird D, Costello M, Daza R, Williams L, Nicol R, Gnirke A, Nusbaum C, Lander ES, Jaffe DB. High-quality draft assemblies of mammalian genomes from massively parallel sequence data. Proc Natl Acad Sci U S A. 2011;108:1513–8. doi: 10.1073/pnas.1017351108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fullwood MJ, Wei CL, Liu ET, Ruan Y. Next-generation DNA sequencing of paired-end tags (PET) for transcriptome and genome analyses. Genome Res. 2009;19:521–32. doi: 10.1101/gr.074906.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ruan Y, Ooi HS, Choo SW, Chiu KP, Zhao XD, Srinivasan KG, Yao F, Choo CY, Liu J, Ariyaratne P, Bin WG, Kuznetsov VA, Shahab A, Sung WK, Bourque G, Palanisamy N, Wei CL. Fusion transcripts and transcribed retrotransposed loci discovered through comprehensive transcriptome analysis using Paired-End diTags (PETs) Genome Res. 2007;17:828–38. doi: 10.1101/gr.6018607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chen J, Kim YC, Jung YC, Xuan Z, Dworkin G, Zhang Y, Zhang MQ, Wang SM. Scanning the human genome at kilobase resolution. Genome Res. 2008;18:751–62. doi: 10.1101/gr.068304.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Raghavendra NK, Rao DN. Exogenous AdoMet and its analogue sinefungin differentially influence DNA cleavage by R. EcoP15I--usefulness in SAGE. Biochem Biophys Res Commun. 2005;334:803–11. doi: 10.1016/j.bbrc.2005.06.171. [DOI] [PubMed] [Google Scholar]
- 23.Korbel JO, Abyzov A, Mu XJ, Carriero N, Cayting P, Zhang Z, Snyder M, Gerstein MB. PEMer: a computational framework with simulation-based error models for inferring genomic structural variants from massive paired-end sequencing data. Genome Biol. 2009;10:R23. doi: 10.1186/gb-2009-10-2-r23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chen K, Wallis JW, McLellan MD, Larson DE, Kalicki JM, Pohl CS, McGrath SD, Wendl MC, Zhang Q, Locke DP, Shi X, Fulton RS, Ley TJ, Wilson RK, Ding L, Mardis ER. BreakDancer: an algorithm for high-resolution mapping of genomic structural variation. Nat Methods. 2009;6:677–81. doi: 10.1038/nmeth.1363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ye K, Schulz MH, Long Q, Apweiler R, Ning Z. Pindel: a pattern growth approach to detect break points of large deletions and medium sized insertions from paired-end short reads. Bioinformatics. 2009;25:2865–71. doi: 10.1093/bioinformatics/btp394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Miller CA, Hampton O, Coarfa C, Milosavljevic A. ReadDepth: A Parallel R Package for Detecting Copy Number Alterations from Short Sequencing Reads. PLoS One. 2011;6:e16327. doi: 10.1371/journal.pone.0016327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bailey JA, Yavor AM, Massa HF, Trask BJ, Eichler EE. Segmental duplications: organization and impact within the current human genome project assembly. Genome Res. 2001;11:1005–17. doi: 10.1101/gr.187101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hampton OA, Den Hollander P, Miller CA, Delgado DA, Li J, Coarfa C, Harris RA, Richards S, Scherer SE, Muzny DM, Gibbs RA, Lee AV, Milosavljevic A. A sequence-level map of chromosomal breakpoints in the MCF-7 breast cancer cell line yields insights into the evolution of a cancer genome. Genome Res. 2009;19:167–77. doi: 10.1101/gr.080259.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mootha VK, Lepage P, Miller K, Bunkenborg J, Reich M, Hjerrild M, Delmonte T, Villeneuve A, Sladek R, Xu F, Mitchell GA, Morin C, Mann M, Hudson TJ, Robinson B, Rioux JD, Lander ES. Identification of a gene causing human cytochrome c oxidase deficiency by integrative genomics. Proc Natl Acad Sci U S A. 2003;100:605–10. doi: 10.1073/pnas.242716699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Inoue K, Lupski JR. Molecular mechanisms for genomic disorders. Annu Rev Genomics Hum Genet. 2002;3:199–242. doi: 10.1146/annurev.genom.3.032802.120023. [DOI] [PubMed] [Google Scholar]
- 31.Maher CA, Kumar-Sinha C, Cao X, Kalyana-Sundaram S, Han B, Jing X, Sam L, Barrette T, Palanisamy N, Chinnaiyan AM. Transcriptome sequencing to detect gene fusions in cancer. Nature. 2009;458:97–101. doi: 10.1038/nature07638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Inaki K, Hillmer AM, Ukil L, Yao F, Woo XY, Vardy LA, Zawack KF, Lee CW, Ariyaratne PN, Chan YS, Desai KV, Bergh J, Hall P, Putti TC, Ong WL, Shahab A, Cacheux-Rataboul V, Karuturi RK, Sung WK, Ruan X, Bourque G, Ruan Y, Liu ET. Transcriptional consequences of genomic structural aberrations in breast cancer. Genome Res. 2011 doi: 10.1101/gr.113225.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhao Q, Caballero OL, Levy S, Stevenson BJ, Iseli C, de Souza SJ, Galante PA, Busam D, Leversha MA, Chadalavada K, Rogers YH, Venter JC, Simpson AJ, Strausberg RL. Transcriptome-guided characterization of genomic rearrangements in a breast cancer cell line. Proc Natl Acad Sci U S A. 2009;106:1886–91. doi: 10.1073/pnas.0812945106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hu X, Stern HM, Ge L, O’Brien C, Haydu L, Honchell CD, Haverty PM, Peters BA, Wu TD, Amler LC, Chant J, Stokoe D, Lackner MR, Cavet G. Genetic alterations and oncogenic pathways associated with breast cancer subtypes. Mol Cancer Res. 2009;7:511–22. doi: 10.1158/1541-7786.MCR-08-0107. [DOI] [PubMed] [Google Scholar]
- 35.Collins C, Volik S, Kowbel D, Ginzinger D, Ylstra B, Cloutier T, Hawkins T, Predki P, Martin C, Wernick M, Kuo WL, Alberts A, Gray JW. Comprehensive genome sequence analysis of a breast cancer amplicon. Genome Res. 2001;11:1034–42. doi: 10.1101/gr.174301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Huang G, Krig S, Kowbel D, Xu H, Hyun B, Volik S, Feuerstein B, Mills GB, Stokoe D, Yaswen P, Collins C. ZNF217 suppresses cell death associated with chemotherapy and telomere dysfunction. Hum Mol Genet. 2005;14:3219–25. doi: 10.1093/hmg/ddi352. [DOI] [PubMed] [Google Scholar]
- 37.Bieche I, Olivi M, Nogues C, Vidaud M, Lidereau R. Prognostic value of CCND1 gene status in sporadic breast tumours, as determined by real-time quantitative PCR assays. Br J Cancer. 2002;86:580–6. doi: 10.1038/sj.bjc.6600109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Liu Y, Tarsounas M, O’Regan P, West SC. Role of RAD51C and XRCC3 in genetic recombination and DNA repair. J Biol Chem. 2007;282:1973–9. doi: 10.1074/jbc.M609066200. [DOI] [PubMed] [Google Scholar]
- 39.Miller KA, Sawicka D, Barsky D, Albala JS. Domain mapping of the Rad51 paralog protein complexes. Nucleic Acids Res. 2004;32:169–78. doi: 10.1093/nar/gkg925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lewis AG, Flanagan J, Marsh A, Pupo GM, Mann G, Spurdle AB, Lindeman GJ, Visvader JE, Brown MA, Chenevix-Trench G. Mutation analysis of FANCD2, BRIP1/BACH1, LMO4 and SFN in familial breast cancer. Breast Cancer Res. 2005;7:R1005–16. doi: 10.1186/bcr1336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yu X, Chini CC, He M, Mer G, Chen J. The BRCT domain is a phospho-protein binding domain. Science. 2003;302:639–42. doi: 10.1126/science.1088753. [DOI] [PubMed] [Google Scholar]
- 42.Cantor SB, Bell DW, Ganesan S, Kass EM, Drapkin R, Grossman S, Wahrer DC, Sgroi DC, Lane WS, Haber DA, Livingston DM. BACH1, a novel helicase-like protein, interacts directly with BRCA1 and contributes to its DNA repair function. Cell. 2001;105:149–60. doi: 10.1016/s0092-8674(01)00304-x. [DOI] [PubMed] [Google Scholar]
- 43.Kim H, Chen J, Yu X. Ubiquitin-binding protein RAP80 mediates BRCA1-dependent DNA damage response. Science. 2007;316:1202–5. doi: 10.1126/science.1139621. [DOI] [PubMed] [Google Scholar]
- 44.Liu Z, Wu J, Yu X. CCDC98 targets BRCA1 to DNA damage sites. Nat Struct Mol Biol. 2007;14:716–20. doi: 10.1038/nsmb1279. [DOI] [PubMed] [Google Scholar]
- 45.Akbari MR, Ghadirian P, Robidoux A, Foumani M, Sun Y, Royer R, Zandvakili I, Lynch H, Narod SA. Germline RAP80 mutations and susceptibility to breast cancer. Breast Cancer Res Treat. 2009;113:377–81. doi: 10.1007/s10549-008-9938-z. [DOI] [PubMed] [Google Scholar]
- 46.Novak DJ, Sabbaghian N, Maillet P, Chappuis PO, Foulkes WD, Tischkowitz M. Analysis of the genes coding for the BRCA1-interacting proteins, RAP80 and Abraxas (CCDC98), in high-risk, non-BRCA1/2, multiethnic breast cancer cases. Breast Cancer Res Treat. 2009;117:453–9. doi: 10.1007/s10549-008-0134-y. [DOI] [PubMed] [Google Scholar]
- 47.Yu X. Characterize RAP80, a Potential Tumor Suppressor Gene. University of Michigan: U.S. Army Medical Research and Materiel Command; 2009. p. 24. [Google Scholar]
- 48.Hicks J, Krasnitz A, Lakshmi B, Navin NE, Riggs M, Leibu E, Esposito D, Alexander J, Troge J, Grubor V, Yoon S, Wigler M, Ye K, Borresen-Dale AL, Naume B, Schlicting E, Norton L, Hagerstrom T, Skoog L, Auer G, Maner S, Lundin P, Zetterberg A. Novel patterns of genome rearrangement and their association with survival in breast cancer. Genome Res. 2006;16:1465–79. doi: 10.1101/gr.5460106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Abeysinghe SS, Chuzhanova N, Krawczak M, Ball EV, Cooper DN. Translocation and gross deletion breakpoints in human inherited disease and cancer I: Nucleotide composition and recombination-associated motifs. Hum Mutat. 2003;22:229–44. doi: 10.1002/humu.10254. [DOI] [PubMed] [Google Scholar]
- 50.Bashir A, Liu YT, Raphael BJ, Carson D, Bafna V. Optimization of primer design for the detection of variable genomic lesions in cancer. Bioinformatics. 2007;23:2807–15. doi: 10.1093/bioinformatics/btm390. [DOI] [PubMed] [Google Scholar]
- 51.Bashir A, Lu Q, Carson D, Raphael BJ, Liu YT, Bafna V. Optimizing PCR assays for DNA-based cancer diagnostics. J Comput Biol. 17:369–81. doi: 10.1089/cmb.2009.0203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Krzywinski M, Schein J, Birol I, Connors J, Gascoyne R, Horsman D, Jones SJ, Marra MA. Circos: an information aesthetic for comparative genomics. Genome Res. 2009;19:1639–45. doi: 10.1101/gr.092759.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Barlund M, Monni O, Weaver JD, Kauraniemi P, Sauter G, Heiskanen M, Kallioniemi OP, Kallioniemi A. Cloning of BCAS3 (17q23) and BCAS4 (20q13) genes that undergo amplification, overexpression, and fusion in breast cancer. Genes Chromosomes Cancer. 2002;35:311–7. doi: 10.1002/gcc.10121. [DOI] [PubMed] [Google Scholar]
- 54.Huang GL, Zhang XH, Guo GL, Huang KT, Yang KY, Shen X, You J, Hu XQ. Clinical significance of miR-21 expression in breast cancer: SYBR-Green I-based real-time RT-PCR study of invasive ductal carcinoma. Oncol Rep. 2009;21:673–9. [PubMed] [Google Scholar]
- 55.Heinonen H, Nieminen A, Saarela M, Kallioniemi A, Klefstrom J, Hautaniemi S, Monni O. Deciphering downstream gene targets of PI3K/mTOR/p70S6K pathway in breast cancer. BMC Genomics. 2008;9:348. doi: 10.1186/1471-2164-9-348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Tamura K, Makino A, Hullin-Matsuda F, Kobayashi T, Furihata M, Chung S, Ashida S, Miki T, Fujioka T, Shuin T, Nakamura Y, Nakagawa H. Novel lipogenic enzyme ELOVL7 is involved in prostate cancer growth through saturated long-chain fatty acid metabolism. Cancer Res. 2009;69:8133–40. doi: 10.1158/0008-5472.CAN-09-0775. [DOI] [PubMed] [Google Scholar]
- 57.Liu L, Gong G, Liu Y, Natarajan S, Larkin DM, Everts-van der Wind A, Rebeiz M, Beever JE. Multi-species comparative mapping in silico using the COMPASS strategy. Bioinformatics. 2004;20:148–54. doi: 10.1093/bioinformatics/bth018. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.