Abstract
Degradation of nuclear DNA is a hallmark of programmed cell death. Epidermal keratinocytes die in the course of cornification to function as the dead building blocks of the cornified layer of the epidermis, nails and hair. Here we investigated the mechanism and physiological function of DNA degradation during cornification in vivo. Targeted deletion of the keratinocyte-specific endonuclease DNase1L2 in the mouse resulted in the aberrant retention of DNA in hair and nails as well as in epithelia of the tongue and the esophagus. In contrast to our previous studies in human keratinocytes, ablation of DNase1L2 did not compromise the cornified layer of the epidermis. Quantitative PCRs showed that the amount of nuclear DNA was dramatically increased in both hair and nails and that mitochondrial DNA was increased in the nails of DNase1L2-deficient mice. The presence of nuclear DNA disturbed the normal arrangement of structural proteins in hair corneocytes and caused a significant decrease in the resistance of hair to mechanical stress. These data identify DNase1L2 as an essential and specific regulator of programmed cell death in skin appendages and demonstrate that the breakdown of nuclear DNA is crucial for establishing the full mechanical stability of hair.
Introduction
Hair, nails, and the cornified layer of the epidermis, i.e. the stratum corneum, consist of cornified keratinocytes, also known as corneocytes. The process of cornification involves the cross-linking of structural proteins and leads to the high mechanical stability of corneocytes which, along with the formation of stable desmosomal links between cells, is essential for the resistance of epidermal structures to insults from the environment (Holbrook, 1994; Candi et al, 2005; Paus and Cotsarelis, 1999; Popescu and Höcker, 2009). A characteristic step of cornification is the removal of the nucleus and the degradation of nuclear DNA (McCall and Cohen, 1991; Lippens et al, 2005; Kroemer et al, 2009). The mechanism of breakdown of the nucleus is unclear. Cornification does not involve the apoptotic members of the caspase family (Lippens et al, 2005, Lippens et al, 2008) and genetic deletion of caspase-activated DNase (CAD) does not result in a skin phenotype (Kawane et al, 2003).
By screening the expression pattern of human DNases, we have recently identified the endonuclease, DNase1-like2 (DNase1L2), as keratinocyte-specific DNase (Fischer et al, 2007). DNase1L2 is expressed in terminally differentiated keratinocytes of the human interfollicular epidermis, hair follicles and the nail matrix (Fischer et al, 2007; Jäger et al, 2007). siRNA-mediated knockdown of DNase1L2 in a human in vitro skin model suppressed the degradation of nuclear DNA and caused retention of nuclei in the stratum corneum, which is known as parakeratosis (Fischer et al, 2007). Moreover, parakeratotic lesions of skin diseases such as psoriasis were found to be associated with lack of DNase1L2 expression (Fischer et al, 2007).
Here we investigated cornification-associated DNA degradation in vivo by comparing DNase1L2-deficient and normal control mice. We demonstrate an essential role of DNase1L2 in the breakdown of the nucleus in skin appendages and distinct epithelia whereas cornification of the interfollicular epidermis was DNase1L2-independent in the mouse. In addition, our results reveal a contribution of DNA degradation to the establishment of the mechanical properties of hair and, therefore, suggest a novel physiological function of cell death-associated DNA degradation.
Results
DNase1L2 is expressed in terminally differentiated keratinocytes of the mouse
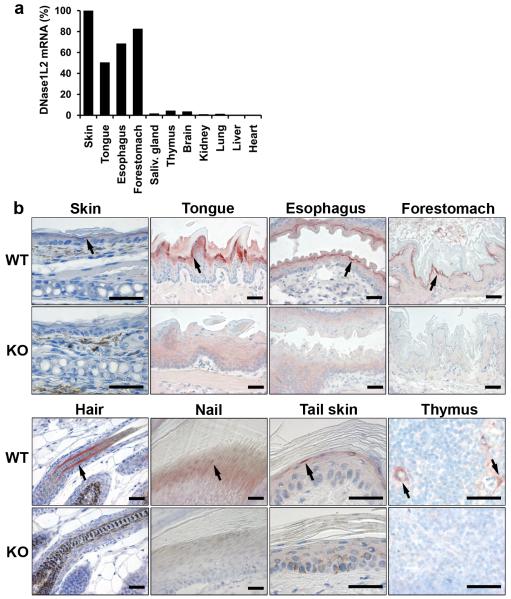
To determine the role of DNase1L2 in vivo, we generated DNase1L2 knockout mice. Exons 2 to 6 of Dnase1l2 were replaced by a neomycin resistance cassette thus abolishing the expression of DNase1L2 constitutively (Supplementary Figure S1). DNase1L2 knockout mice were fertile and macroscopically indistinguishable from wild-type mice up to an age of 2 years. Samples from these mice served as negative controls to determine the tissue expression pattern of DNase1L2 in wild-type mice. Quantitative RT-PCR showed that DNase1L2 mRNA was strongly expressed in keratinizing tissues of the mouse, such as the skin and the esophagus, but only at low levels in tissues that do not contain keratinocytes (Figure 1a). By immunohistochemical analysis, DNase1L2 was detected in hair follicles, the nail unit, the epithelia of the tongue and of the esophagus, the forestomach, the epidermoid cells of the Hassalĺs corpuscules of the thymus, and occasionally in the granular layer of adult interfollicular epidermis (Figure 1b, upper panels). Sebaceous glands which consist of keratinocytes of the sebocyte differentiation lineage (Fuchs, 2007) were immuno-negative for DNase1L2. In the hair follicle, DNase1L2 was expressed in the differentiating keratinocytes forming the hair cuticle and cortex, and, more weakly, in the medulla, i.e. the central part of the hair shaft (Supplementary Figure S2).
Figure 1. Expression of DNase1L2 is associated with cornification.
(a) Quantification of DNase1L2 mRNA expression in mouse organs using real-time PCR. Note that the skin sample included both interfollicular epidermis and hair follicles. The results of one of two experiments with similar results are shown. (b) Immunohistochemical detection of DNase1L2 in the mouse. Tissues of DNase1L2-deficient mice were used as negative controls. In some panels, arrows point to sites of DNase1L2 expression. Scale bars: 40 μm.
Ablation of DNase1L2 is associated with retention of nuclear DNA in skin appendages
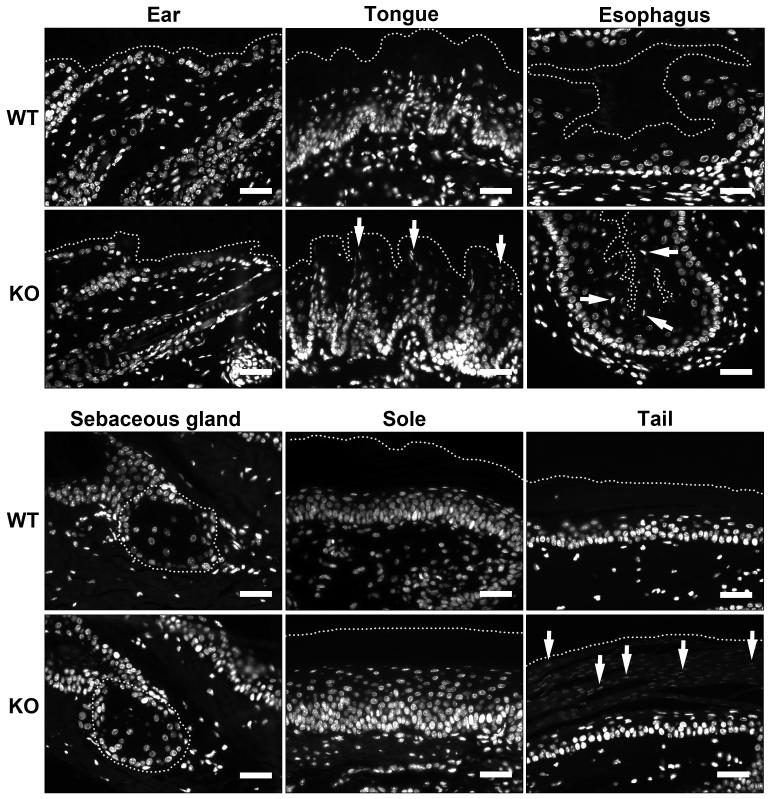
To compare the degree of DNA breakdown in cornified keratinocytes of DNase1L2-deficient and wild-type mice, thin sections of tissues and permeabilized hair were labeled with a DNA-specific fluorescent dye. The stratum corneum of the interfollicular epidermis and of the soles of both wild-type and DNase1L2-deficient mice lacked DNA, suggesting that DNA degradation at these sites did not depend on DNase1L2 (Figure 2a and 2b). Sebaceous glands also showed normal DNA breakdown in the absence of DNase1L2 (Figure 2c). By contrast, aberrant retention of nuclear DNA was detected in the papillae of the tongue, the epithelium of the esophagus and the tail scales of DNase1L2-deficient mice (Figure 2d-f). Moreover, hair and nails contained massively increased amounts of DNA (Figure 3). Virtually every cellular remnant in the medulla of DNase1L2-deficient hair, irrespective of body site and stage of the hair cycle, retained nuclear DNA (Figure 3d, e). DNA was consistently detected, although at weaker and more variable labeling intensity, in the cortex and cuticle of DNase1L2-deficient hair (Figure 3e).
Figure 2. Deletion of DNase1L2 leads to retention of DNA in distinct pathways of terminal differentiation of keratinocytes.
Thin sections of ear, sole, sebaceous glands, tongue, esophagus and the tail of DNase1L2 knockout (KO) and wild-type (WT) mice were labeled with the DNA-binding dye Hoechst 33258 (white). Arrows point to nuclear remnants within the corneocytes. Dotted lines mark the outer border of the cornified layer in all tissues except the sebaceous glands where the border of the gland is indicated. Scale bars: 40 μm.
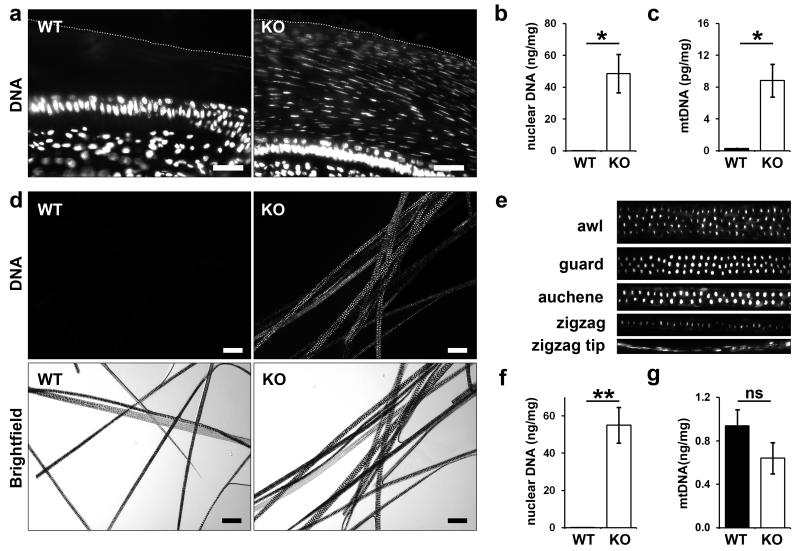
Figure 3. DNA is aberrantly retained in the hair and nails of DNase1L2-deficient mice.
(a) Hoechst 33258 labeling (white) of nails from DNase1L2 knockout (KO) and wild-type (WT) mice. The outer border of the nail is indicated by a dotted line. (b, c) Nuclear and mitochondrial DNA extracted from nails was quantified by quantitative real-time PCR. Data represent means ± s.e.m. (n = 3; asterisk, P < 0.05). (d) Hair from the back of DNase1L2 wild-type and DNase1L2-deficient mice was permeabilized and labeled with Hoechst 33258 (white). Fluorescent and bright field images of the same areas are shown. (e) Hoechst 33258 labeling of various hair types of DNase1L2-null mice. Note that the hair types differ in the number of cell rows in the medulla. (f) Quantification of nuclear DNA from hair by real-time PCR. Data represent means ± s.e.m. (n = 5; double asterisk, P < 0.01). (g) Quantification of mitochondrial DNA from hair by real-time PCR. Data represent means ± s.e.m. (n = 5; n.s., not significant). DNA quantifications were repeated in an independent experiment with similar results. KO, DNase1L2 knockout; WT, wild-tpye. Scale bars: (a) 40 μm; (d) 100 μm.
The amount of DNA in hair and nails of mutant and control mice was quantified by real-time PCR using primers specific for nuclear and mitochondrial DNA. Hair from the back of DNase1L2-deficient mice contained 400-fold more nuclear DNA than hair from wild-type mice (Figure 3f) whereas the content of mitochondrial DNA was not significantly different in DNase1L2-deficient and normal hair (Figure 3g). Nails of knockout mice contained 2000-fold more nuclear DNA and 30-fold more mitochondrial DNA than nail from control mice (Figure 3b, c). Taken together, these data demonstrate that the breakdown of nuclear DNA in both hair and nails requires DNase1L2. Furthermore, DNase1L2 is also essential for the degradation of mitochondrial DNA in nails but not in hair.
Failure to degrade DNA in hair of DNase1L2-deficient mice disturbs the structural maturation of the hair shaft
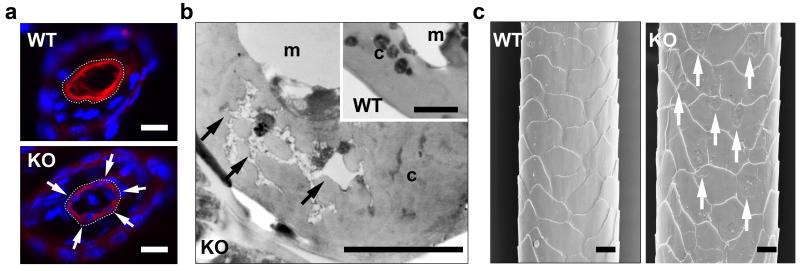
To test how the retention of DNA affected the maturation of the hair shaft, we analyzed the protein composition and the structure of hair. Histones H2B and H3, which directly bind nuclear DNA, were absent from wild-type hair but were retained in hair of mutant mice as revealed by Western blot analysis of hair lysates (Supplementary Figure S3a). By contrast, the most prominent protein species in hair, i.e. keratins and keratin-associated proteins, were not altered in their abundance in DNase1L2-deficient mice (Supplementary Fig. S3b). However, co-labeling of keratin 31, a key structural protein of the hair cortex (Moll et al, 2008), and DNA showed that DNA-positive nuclear remnants prevented keratin 31 from filling the cytosol of corneocytes of DNase1L2-deficient hair (Figure 4a). Ultrastructural analysis of hair cross sections showed that nuclear remnants with an atypical low electron density interdigitated the scaffold of structural proteins in the corneocytes of DNase1L2-deficient hairs (Figure 4b). When hairs were investigated by scanning electron microscopy under vacuum, the corneocytes of the hair cuticle of normal mice appeared smooth whereas the surface of the hair cuticle of mutant mice contained indentions corresponding in size to the nucleus (Figure 4c). These data indicated that the retention of nuclear DNA disturbed the tight packing of keratin filaments in hair corneocytes and altered the structure of hair corneocytes.
Figure 4. DNA disturbs the structural maturation of hair corneocytes.
(a) Cross sections of hair follicles were double-stained for DNA using Hoechst 33258 (blue) and keratin 31 (K31, red), a marker specific for the hair cortex, using a specific antiserum. Arrows point to cavities in the keratin 31 scaffold of DNase1L2 knockout mice that are filled with DNA. The outer border of the hair cortex is indicated by a dotted line. (b) Transmission electron microscopy of hair cross sections. Arrows point to nuclear remnants that interdigitate structural proteins in the hair cortex. (c) Scanning electron microscopy of hairs. Arrows point to indentions on the surface. c, cortex, m, medulla. Scale bars: (a) 10 μm; (b) 0.2 μm and 1 μm (inset); (c) 20 μm.
DNase1L2-deficient hair is less resistant to mechanical stress than normal hair
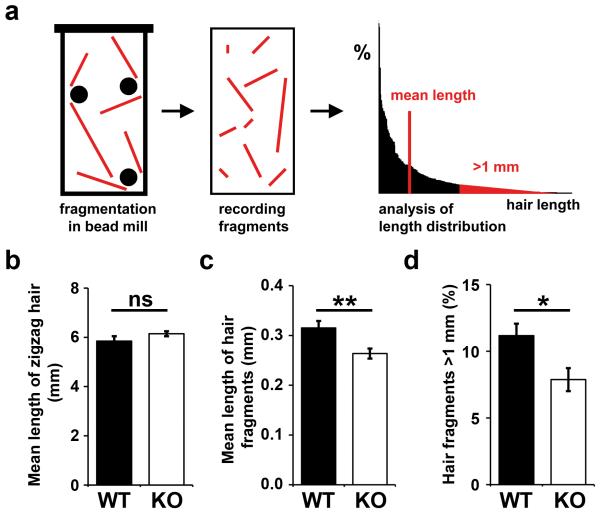
We hypothesized that the aberrant maturation of hair corneocytes might compromise a key physical property of the hair shaft, i.e. its mechanical resilience (Popescu and Höcker, 2009). Therefore, we devised an assay for the quantification of hair breakdown under mechanical stress (Figure 5a). Hair was shaved from the back of each mouse and ground with ceramic beads to yield hair fragments with an approximately exponential length distribution. In two independent experiments, hair from at least 5 wild-type and 5 DNase1L2-deficient mice was analyzed with regard to the degree of fragmentation under stress. Indeed, hair from mutant mice, which appeared normal on the skin of mice being housed under standard conditions and did not differ in length from wild-type hair (Figure 5b), was broken into fragments with a significantly smaller mean length (Figure 5c). Specifically, the portion of fragments maintaining a length of more than 1 mm was decreased as compared to wild-type mice (Figure 5d). This implies that DNase1L2-deficient hair was more fragile than normal hair under mechanical stress.
Figure 5. Retention of DNA increases the fragility of hair from DNase1L2-deficient mice.
(a) Schematic depiction of the mechanical stress test. Shaved hairs from wild-type and DNase1L2-deficient mice were exposed to mechanical stress in a bead mill containing ceramic beads. The resulting fragments were spread on microscope slides, photographed and measured using an automatic software algorithm. 3 slides were analyzed per bead mill treatment. (b) Untreated zigzag hairs of wild-type and mutant mice were photographed and their length was determined manually (n = 5; ns, not significant). (c, d) After exposure to mechanical stress the mean length of hair fragments (c) and the percentage of hair fragments longer than 1 mm was determined (d). Data represent means ± s.e.m. (n = 7 (WT) and 8 (KO); double asterisk, P < 0.01; asterisk, P < 0.05). Similar results were obtained in a second independent bead mill experiment using hair samples from n = 5 mice per genotype.
Discussion
This study provides key insights into the molecular regulation of a previously uncharacterized part of cornification, i.e. the breakdown of the nucleus. We show that DNase1L2 degrades nuclear DNA in terminally differentiated keratinocytes when they are converted to dead, but functional corneocytes of nail, hair, the cornified layer on the surface of the tongue and the esophagus and in the scales on the tail of the mouse. Similar to the cell-specific breakdown of nuclear DNA by DNase2-like acid DNase (DLAD) in the eye lens (Nishimoto et al, 2003), nuclear DNA of differentiated keratinocytes in the nail matrix and the hair shaft is degraded by a DNase not involved in classical apoptosis. This finding provides the molecular underpinning for the classification of cornification as a distinct mode of programmed cell death (Kroemer et al, 2009).
However, our results also show that the cornification of keratinocytes of murine interfollicular epidermis proceeds in a DNase1L2-independent manner. Notably, DNase1L2 is expressed at much lower levels in the interfollicular epidermis of the mouse than in human epidermis and in human in vitro skin models, in which the degradation of nuclear DNA can be suppressed by siRNA-mediated knockdown of DNase1L2 (Fischer et al, 2007). Hence, the relative contribution of DNase1L2 to DNA degradation in the epidermis is likely to differ between these species. At least one DNase different from DNase1L2 must be involved in stratum corneum formation in the murine interfollicular epidermis.
The data on the expression of DNase1L2 in murine (this study) and human tissues (Fischer et al, 2007) suggest that DNase1L2 is subject to tight transcriptional regulation. In addition, DNase1L2 appears to be regulated at the levels of intracellular localization and activity. DNase1L2 lacks a nuclear targeting signal (Shiokawa and Tanuma, 2001) and preferentially localizes to the endoplasmic reticulum upon overexpression in cell culture (our unpublished data). Nevertheless, DNase1L2 gains access to nuclear DNA during keratinocyte differentiation in vivo as evidenced by the essential role of DNase1L2 in the degradation of nuclear DNA in skin appendages of mice. Moreover, DNase1L2 degrades mitochondrial DNA of the nail but not of the hair. Interestingly, differences in the amount of residual mitochondrial DNA in hair and nails of wildtype mice also indicate that mitochondria are degraded by different mechanisms in these skin appendages. It is plausible to assume that degradation of DNA by DNase1L2 depends on the disruption of intracellular compartments during cornification (Karasek 1988). The sequences of intracellular remodeling events of cornification in the interfollicular epidermis, hair follicles and nails remain to be investigated in future studies.
Our data demonstrate that the failure to degrade DNA in the absence of DNase1L2 disturbs the processing of histones and the filling of the corneocyte cytoplasm with keratins during terminal differentiation of hair keratinocytes. The failure to modify or degrade histones H2B and H3 in the presence of DNA demonstrates that the processing of these histones, and possibly other nuclear components, occurs downstream of DNA degradation in normal hair keratinocytes. Aberrantly retained DNA, probably in conjunction with DNA-associated proteins, caused the suboptimal arrangement of keratins in DNase1L2-deficient corneocytes, as detected by immunolabeling of hair cross sections, and an abnormal intracellular organization of hair corneocytes, that was evident in ultrastructural analysis. These data indicate that the breakdown of nuclear DNA is not an epiphenomenon of cornification but has a central role in this process.
The in vitro stress experiments of this study suggest a function of DNA breakdown in establishing the mechanical properties of corneocytes. Although hair of DNase1L2 knockout mice appeared normal under standard housing conditions, the enhanced mechanical stress in the bead mill assay revealed significant differences between hair containing DNA and normal hair. The fragility of DNase1L2-deficient hair is very likely to be caused by the abovementioned disturbance in the structural organisation of the hair keratins. Previous studies have shown that defects in hair keratins are associated with hair fragility (Arin, 2009; Winter et al, 1997; Shimomura et al, 2010). In contrast to those cases of hair fragility, the mechanical properties of DNase1L2-deficient hair cannot be attributed to the absence of a structural component of hair but rather to the aberrant presence of a component of living cells. Thus, this study defines a novel role of DNA degradation in establishing the mechanical resilience of cell-derived structures.
Materials and Methods
Targeted deletion of the Dnase1l2 gene in the mouse
For construction of the targeting vector, the Dnase1l2 gene and chromosomal regions comprising approximately 4 and 5 kbp on the 5′-side (left arm) and the 3′-side (right arm) of the gene were amplified by long-range PCR from genomic DNA of mouse W4 ES cells (strain 129). The left arm was amplified with the forward primer AGATACGGGCCCTTAGAGCACTGGGCTGGAGCATGG and the reverse primer AGAAAACCTCGAGACAGAAATGTCCCAGAGTAA. The restriction sites of ApaI and the XhoI are underlined. The right arm was amplified with the forward primer AATCTAATCGATCTGTGGAAGTGACTTTCAAGACTCACTGAGAG and the reverse primer TATTCAGCGGCCGCCCGACACCATCTTCTGGCTTCTGC. The restriction sites of ClaI and the NotI are underlined. Both PCR products were digested with the above-mentioned restriction enzymes and cloned into the vector pBluescript II KS+. Exons 2-6 of the Dnase1l2 gene were replaced by a neomycin resistance cassette, and the vector was transfected into W4 ES cells of the mouse strain 129SvEv. ES clones were karyotyped and screened by Southern-blotting of genomic DNA digested with BstEII and SalI using a radioactively labeled probe annealing to a region indicated in Supplementary Figure S1. The chromosomal regions flanking the Neo resistance cassette in the selected clones were amplified by long-range PCR and sequenced to confirm that the coding sequences of the flanking genes, E4f1 and Dci were free of mutations. 2 ES clones were injected into blastocysts of the mouse strain C57BL6 to generate two independent mouse lines of mixed background. Chimeric, heterozygous and ultimately homozygous DNase1L2-targeted mice were obtained by breeding according to standard procedures. In addition mice carrying the Dnase1l2 deletion in the pure background of the strain 129 were generated by crossing chimeric mice and mice of the strain 129SvEv. For PCR-screening of DNA from DNase1L2+/+, DNase1L2+/−, and DNase1L2−/− mice, the Expand Long Template PCR System (Roche Applied Science, Mannheim, Germany) was used (forward primer: GTTTGGTGTTACTCTGGGACA, reverse primer: GAGTCTTGAAAGTCACTTCCAC).
Quantification of mRNA by real-time PCR
RNA from mouse tissues was isolated using TRIzol reagent (Invitrogen, Carlsbad, CA) and reverse-transcribed using the Gene Amp RNA PCR kit (Applied Biosystems, Foster City, CA). Quantitative real-time PCR was performed with the LightCycler technology (Roche Applied Science, Mannheim, Germany) as described previously (Gruber et al, 2010) using a primer pair specific for mouse DNase1L2 (forward primer: GACGCGCTCTACGATGTGTA, reverse primer: ATCCGGTCATAGGCACAGTC). Relative expression of the target gene was normalized to the expression of the housekeeping gene aminolevulinate synthase 1 (ALAS1).
Histological, immunohistochemical and immunofluorescence analysis
Tissues were fixed in phosphate-buffered 4.5% formaldehyde for 24 h and embedded in paraffin. Sections of 4 μm were stained with haematoxylin-eosin or processed for immunohistochemical and immunofluorescence analysis according to a protocol published previously (Fischer et al, 2007). Polyclonal antisera against DNase1L2 were produced by immunizing rabbits with purified recombinant DNase1L2 preparations produced in the yeast Pichia pastoris and affinity-purified against the antigen (Fischer et al, 2007). The antisera were used at a dilution of 1:400 for staining. Guinea pig antisera against keratin 28 and keratin 31 (1:200, kindly provided by Lutz Langbein, German Cancer Research Center, Heidelberg, Germany) were used to specifically label the inner root sheath and the cortex of hair, respectively.
Fluorescent dye labeling of DNA of hair in situ
To label DNA in situ, 2 mg of shaved mouse hair was washed three times with 0.04% SDS in 70% ethanol, dried over night at room temperature and subsequently permeabilized with 1% ammonia solution (pH 10.5) for 5 hours at room temperature. After washing three times with water, hair was incubated with DNA-specific Hoechst 33258 (1:5000, Invitrogen, Carlsbad, CA) over night at room temperature. Hair was washed again with water and mounted onto glass slides using Fluorprep (Biomerieux, France).
Quantification of nuclear and mitochondrial DNA by real-time PCR
Hair was shaved from the back of mice. Nails were clipped carefully to avoid contamination with tissue. Hair and nail samples were washed 3 times in 500 μl PBS with 0.1 % Tween 20 under vigorous shaking at 750 rpm for 10 minutes. Five mg of each sample were lysed at 56°C in a buffer containing 50 mM Tris-HCl, pH 8.0, 100 mM NaCl, 2 % SDS, 0.27 mg/ml protease K, 40 mM DDT and 2.5 mM EDTA. DNA was purified using the Qiagen DNeasy Blood and Tissue Kit (Qiagen, Hilden, Germany) according to the manufacture’s instructions.
To quantify nuclear and mitochondrial DNA, real-time PCR was performed using the LightCycler® technology and the LightCycler® 480 SYBR Green I Master Kit (Roche Applied Science) according to the manufacturer’s protocol. Mouse mitochondrial DNA was amplified with the primers M-mito-for, 5′-CCTCCGAATGATTATAACCTAGACTT-3′, and M-mito-rev, 5′-AGGGTAACTTGGTCCGTTGA -3′ which anneal to nucleotides 2277-2302 and 2352-2371, respectively, of the mouse mitochondrial genome (GenBank accession number: NC_005089). Mouse genomic DNA was amplified with the primers M-L1-for, 5′-TGGTAGAGGACATCAAGAAGGAC-3′, and M-L1-rev, 5′-TTCCTGTTTTTCTTTAAGGACTTGTAA-3′ which anneal to nucleotides 1147-1169 and 1215-1241, respectively, of the mouse L1 retrotransposon, Tf subfamily L1Md-Tf30 (GenBank accession number: AF081114). In all assays DNA was amplified with the following program:denaturation for 10 minutes, 50 cycles of 5 s at 95°C, 15 s at 65°C, and 15 s at 72°C, and melting point analysis in steps of 0.1°C.
Western blot analysis and silver staining
Hair was homogenized using a bead mill (Precellys®24, PEQLAB Biotechnology, Germany) containing 10 ceramic beads of a diameter of 1.4 mm for 2 × 50 seconds at 6500 rpm. The resulting powder was lysed in 50 mM Tris pH 7.4, 4% SDS, 5% 2-mercaptoethanol (Merck Schuchardt OHG, Hohenbrunn, Germany) at 95°C for 10 minutes. Insoluble debris was removed by centrifugation. For extraction of proteins from adult mouse skin, tissue was homogenized under liquid nitrogen in lysis buffer containing phosphate-buffered saline, 1% NP-40 (Sigma-Aldrich, St. Louis, MO) and complete protease inhibitor cocktail (Roche Applied Science). Insoluble debris was removed by centrifugation. SDS-PAGE and Western blot analysis was performed as described previously (Rendl et al, 2002). Rabbit polyclonal anti-DNase1L2 (1:1000) (Fischer et al, 2007), rabbit anti-histone H2B (kindly provided by Marc Monestier, Temple University, Philadelphia, PA) and rabbit anti-histone H3 (1:1000, Cell Signaling Technology, Danvers, MA) and were used as first step antibodies for the specific detection of antigens. Silver staining was performed according to Blum et al, 1987.
Ultrastructural analysis
For transmission electron microscopy, tissue samples were fixed in half-strength Karnovsky’s fixative for 3 hours, post-fixed in osmium tetroxide and processed as previously described (Tobin et al, 1991). Serial semi-thin sections were stained with toluidine blue, while ultrathin sections were stained with uranyl acetate and Reynold’s lead citrate and investigated in a transmission electron microscope (Jeol 1200EX, Tokyo, Japan) (Tobin et al, 1991). For scanning electron microscopy, hairs were mounted on stubs and coated with gold in a Polaron 5800 sputter coater (Biorad, Hercules, CA). The samples were examined using a Philips XL 20 (Eindhoven, Nederland) scanning electron microscope.
Mechanical stress test for hair and determination of hair length
Hair of the lower back of DNase1L2 null and wild-type mice, both in the pure background of the strain 129, was shaved. 10 mg hair per mouse was exposed to mechanical stress in a bead mill containing 10 ceramic beads of a diameter of 1.4 mm for 20 seconds at 5000 rpm (Precellys®24). After the treatment the hair fragments were dispersed in 100 μl 70% ethanol and aliquots were spread onto 3 microscope slides per sample. After drying for 10 - 15 minutes the slides containing hair fragments were mounted with Eukitt mounting medium (O. Kindler GmbH, Freiburg, Germany) and analyzed with a Leica MZ 16 stereomicroscope (Leica Microsystems, Wetzlar, Germany). Two images per sample were acquired with a Leica IC 3D CCD camera (Leica Microsystems, Wetzlar, Germany) using exactly the same camera settings for all samples (magnification of 7.11x, exposure time: 62.1 ms, saturation 1.5, gamma 0.7, histogramm 40-225). Using the Adobe Photoshop software (Adobe, San Jose, CA), the images were converted to greyscale, adjusted to contrast +40 and brightness -30 and sharpened by high pass filtering (radius 10 pixels). The fragment lengths were determined by the automatic algorithm of the Meta Morph software version 4.5 (Universal Imaging Corporation, West Chester, PA). According to the standard settings, all fragments larger than 0.02 mm were recorded. 2 independent experiments were performed with at least 5 and 7 mice per group, respectively.
For the determination of the length of untreated hair, the different hair types were separated under a stereo microscope. Since zigzag hairs can be identified unambiguously (Schlake, 2007), they were selected for length analysis. Shaved hairs from the lower back of 5 wild-type and 5 DNase1L2-deficient mice were investigated. The hairs were spread on microscope slides, embedded in Eukitt mounting media and photographed. The lengths of at least 15 hairs per mouse were measured manually.
Statistical analysis
Statistical analysis was performed using non-directional two-tailed Student’s t-test and P values < 0.05 were considered as significant. Data are shown as arithmetic means and the error bars represent the standard error of the mean.
Supplementary Material
Acknowledgments
The vector for the targeted deletion of the DNase1L2 gene was cloned and transfected into ES cells by the Center of Functional Genomics, University of Albany, NY. We thank Uta Möhle-Steinlein for culturing ES cells, Veronika Mlitz, Michael Mildner, Ramona Gmeiner, and Claudia Ballaun for helpful discussions and technical support, Lutz Langbein (German Cancer Research Center, Heidelberg, Germany) for providing anti-keratin 28 and 31 antisera, and Marc Monestier, Temple University, Philadelphia, PA for providing the anti-histone H2B antibody. The authors are grateful to Harald Höger for expert maintenance of mice and Crisan Popescu (DWI Aachen, Germany) for helpful discussions. We thank Peter Bauer and Gerald Hlavin (Institute of Medical Statistics, Medical University of Vienna) for support in statistical analyses. This work was supported by grants P20043 and P21312 of the Austrian Science Fund (FWF) to L.E.
Abbrevations
- DNase1L2
DNase1-like 2
- CAD
caspase-activated DNase
- DLAD
DNase2-like acid DNase
- K28
keratin 28
- K31
keratin 31
Footnotes
Conflict of interest
The authors declare no conflict of interest.
References
- Arin MJ. The molecular basis of human keratin disorders. Hum Genet. 2009;125:355–73. doi: 10.1007/s00439-009-0646-5. [DOI] [PubMed] [Google Scholar]
- Blum H, Beier H, Gross HJ. Improved silver staining of plant proteins, RNA and DNA in polyacrylamide gels. Electrophoresis. 1987;8:93–99. [Google Scholar]
- Candi E, Schmidt R, Melino G. The cornified envelope: a model of cell death in the skin. Nat Rev Mol Cell Biol. 2005;6:328–40. doi: 10.1038/nrm1619. [DOI] [PubMed] [Google Scholar]
- Fischer H, Eckhart L, Mildner M, et al. DNase1L2 degrades nuclear DNA during corneocyte formation. J Invest Dermatol. 2007;127:24–30. doi: 10.1038/sj.jid.5700503. [DOI] [PubMed] [Google Scholar]
- Fuchs E. Scratching the surface of skin development. Nature. 2007;445:834–42. doi: 10.1038/nature05659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gruber F, Mayer H, Lengauer B, et al. NF-E2-related factor 2 regulates the stress response to UVA-1-oxidized phospholipids in skin cells. FASEB J. 2010;24:39–48. doi: 10.1096/fj.09-133520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holbrook KA. Ultrastructure of the epidermis. In: Leigh IM, Lane EB, Watt F, editors. The keratinocyte handbook. Cambridge University Press; Cambridge: 1994. pp. 3–39. [Google Scholar]
- Jäger K, Fischer H, Tschachler E, et al. Terminal differentiation of nail matrix keratinocytes involves up-regulation of DNase1L2 but is independent of caspase-14 expression. Differentiation. 2007;75:939–46. doi: 10.1111/j.1432-0436.2007.00183.x. [DOI] [PubMed] [Google Scholar]
- Kawane K, Fukuyama H, Yoshida H, et al. Impaired thymic development in mouse embryos deficient in apoptotic DNA degradation. Nat Immunol. 2003;4:138–44. doi: 10.1038/ni881. [DOI] [PubMed] [Google Scholar]
- Karásek J. Nuclear morphology of transitional keratinocytes in normal human epidermis. J Invest Dermatol. 1988;91:243–6. doi: 10.1111/1523-1747.ep12470372. [DOI] [PubMed] [Google Scholar]
- Kroemer G, Galluzzi L, Vandenabeele P, et al. Classification of cell death: recommendations of the Nomenclature Committee on Cell Death 2009. Cell Death Differ. 2009;16:3–11. doi: 10.1038/cdd.2008.150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lippens S, Denecker G, Ovaere P, et al. Death penalty for keratinocytes: apoptosis versus cornification. Cell Death Differ. 2005;12:1497–508. doi: 10.1038/sj.cdd.4401722. [DOI] [PubMed] [Google Scholar]
- Lippens S, Hoste E, Vandenabeele P, et al. Cell death in the skin. Apoptosis. 2009;14:549–69. doi: 10.1007/s10495-009-0324-z. [DOI] [PubMed] [Google Scholar]
- McCall CA, Cohen JJ. Programmed cell death in terminally differentiating keratinocytes: role of endogenous endonuclease. J Invest Dermatol. 1991;97:111–14. doi: 10.1111/1523-1747.ep12478519. [DOI] [PubMed] [Google Scholar]
- Moll R, Divo M, Langbein L. The human keratins: biology and pathology. Histochem Cell Biol. 2008;129:705–33. doi: 10.1007/s00418-008-0435-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishimoto S, Kawane K, Watanabe-Fukunaga R, et al. Nuclear cataract caused by a lack of DNA degradation in the mouse eye lens. Nature. 2003;424:1071–4. doi: 10.1038/nature01895. [DOI] [PubMed] [Google Scholar]
- Paus R, Cotsarelis G. The biology of hair follicles. N Engl J Med. 1999;341:491–497. doi: 10.1056/NEJM199908123410706. [DOI] [PubMed] [Google Scholar]
- Popescu C, Höcker H. Cytomechanics of hair: basics of the mechanical stability. Int Rev Cell Mol Biol. 2009;277:137–56. doi: 10.1016/S1937-6448(09)77004-2. [DOI] [PubMed] [Google Scholar]
- Rendl M, Ban J, Mrass P, et al. Caspase-14 expression by epidermal keratinocytes is regulated by retinoids in a differentiation-associated manner. J Invest Dermatol. 2002;119:1150–5. doi: 10.1046/j.1523-1747.2002.19532.x. [DOI] [PubMed] [Google Scholar]
- Schlake T. Determination of hair structure and shape. Semin Cell Dev Biol. 2007;18:267–73. doi: 10.1016/j.semcdb.2007.01.005. [DOI] [PubMed] [Google Scholar]
- Shimomura Y, Wajid M, Kurban M, et al. Mutations in the keratin 85 (KRT85/hHb5) gene underlie pure hair and nail ectodermal dysplasia. J Invest Dermatol. 2010;130:892. doi: 10.1038/jid.2009.341. [DOI] [PubMed] [Google Scholar]
- Shiokawa D, Tanuma S. Characterization of human DNase I family endonucleases and activation of DNase gamma during apoptosis. Biochemistry. 2001;40:143–52. doi: 10.1021/bi001041a. [DOI] [PubMed] [Google Scholar]
- Tobin DJ, Fenton DA, Kendall MD. Cell degeneration in alopecia areata. An ultrastructural study. Am J Dermatopathol. 1991;13:248–56. doi: 10.1097/00000372-199106000-00006. [DOI] [PubMed] [Google Scholar]
- Winter H, Rogers MA, Langbein L, et al. Mutations in the hair cortex keratin hHb6 cause the inherited hair disease monilethrix. Nat Genet. 1997;16:372–4. doi: 10.1038/ng0897-372. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.