Abstract
Bacterial pathogens in coastal sediments may pose a health risk to users of beaches. Although recent work shows that beach sands harbor both indicator bacteria and potential pathogens, it is not known how deep within beach sands the organisms may persist nor if they may be exposed during natural physical processes. In this study, sand cores of approximately 1 m depth were collected at three sites across the beach face in Kitty Hawk, North Carolina before, during and after large waves from an offshore hurricane. The presence of DNA from the fecal indicator bacterium Enterococci was detected in subsamples at different depths within the cores by PCR amplification. Erosion and accretion of beach sand at the three sites also was determined for each sampling day. The results indicate that ocean beach sands with persisting enterococci signals could be exposed and redistributed when wind, waves, and currents cause beach erosion or accretion.
Keywords: indicator microbes, erosion, accretion, enterococci, pollution
1. Introduction
Beach water quality is monitored to reduce the risk of recreational water-borne illness. The abundances of the bacteria Escherichia coli (freshwater) or enterococci (marine) are used to assess whether water conditions are suitable for swimming. The presence of these indicator bacteria at densities higher than the EPA recommended levels has been linked to swimmer illnesses in epidemiological studies (Haile et al. 1999, Wade et al. 2003, Sinigalliano et al. 2010). Indicator bacteria do not cause disease, but are used to monitor beach water quality because they are associated with fecal material, and their occurrence in the environment was thought to be transient, indicating relatively recent contamination with raw or treated sewage or animal wastes. However, indicator bacteria and potential pathogens also exist in beach sands of both freshwater and marine environments (Ghinsberg et al. 1995, Obiri-Danso and Jones 2000, Desmarais et al. 2002, Whitman and Nevers 2003, Beversdorf et al. 2007, Yamahara et al. 2007, Hartz et al. 2008), where cell numbers can be substantially higher than in nearby waters (Burton et al. 1987, Doyle et al. 1992, Irvine and Pettibone 1993, Oshiro and Fujioka 1995, Whitman and Nevers 2003, Yamahara et al. 2007). The origins of these organisms in sand often are unclear, but likely contain contributions from both exogenous (from sewage or runoff) and indigenous sources (Ferguson et al. 2005). The presence, persistence, and regrowth of indicator bacteria in sediments suggest that beach sands may serve as a reservoir for these microbes, and that they may have the ability to impact water quality monitoring (Byappanahalli et al. 2003, Boehm and Weisberg 2005, Beversdorf et al. 2007, Yamahara et al. 2007, Hartz et al. 2008, Halliday and Gast 2011).
Studies regarding the movement of indicator bacteria between the sand and ocean have focused primarily on the effects of tides and rain (Boehm and Weisberg 2005, Beversdorf et al. 2007, Yamahara et al. 2007, Zehms et al. 2008). These studies have observed increases in the water column levels of indicator organisms under high tide and rain events. Furthermore, a reduction in fecal indicator bacteria in sand approximately equal to the increase observed in the water column was reported (Yamaraha et al. 2007). Even pedestrian traffic may influence the distribution of bacterial pollution at recreational beaches by translocation of bacterial-sized particles deposited on the sand surface (Bonilla et al. 2007). Several studies have examined the correlation between increased fecal indicator bacteria in sand and water within the swash zone (Zehms et al. 2008), and the potential for stream bank and bed resuspension to contribute to indicator presence in stream and lake water (Byappanahalli et al. 2003, Jamieson et al. 2005). The results support the hypothesis that sand and sediment disruption could contribute to the release of fecal indicator bacteria into water, but it is not known how changes in beach topology affect indicator distribution within the sand.
Fecal indicator bacteria and pathogens from sewage spills or runoff introduced into the beach and adjacent shallow waters may pass into the subsurface sands. While many bacteria either will die or be grazed by protistan consumers, some may persist either as free particles or as particles incorporated into biofilms on grains of sand or small pebbles (Jeng et al. 2005), and may even regrow in this environment (Desmarais et al. 2002, Hartz et al. 2008, Yamahara et al. 2009). Waves can cause significant changes in the beach, through both accretion and erosion, and sediment that had been in contact with sewage could be buried and re-exposed. If pathogens also were present, human health hazards could be created by direct contact with contaminated sand and possibly by exposure to pathogens carried by spray from breaking waves in the surf. Eroded sediments from the beach can be transported long distances along the shoreline by surfzone currents (Smith and Largier 1995, Ciavola et al. 1998, Grant et al. 2005, Feddersen 2007), potentially distributing organisms as well.
To examine how physical forces may impact the distribution of fecal indicator bacteria, and potentially pathogens, on beaches, the effects of sand erosion and accretion on the distribution of enterococci in ocean beaches were investigated. Here, it is shown that beach sands that may contain persistent populations of enterococci were exposed and reworked when large waves from an offshore hurricane caused erosion and accretion, potentially resulting in release of the organisms to the water column.
2. Methods
2.1. Location and sand core sampling
Sand cores were collected on a beach near Kitty Hawk (Byrd St. public access, Figure 1), on the Outer Banks of North Carolina, between 11 and 15 September, 2006 as Hurricane Florence moved northward between Bermuda and the US coast (11 September, 2006). This site was selected because it is a well-attended public-access beach with numerous houses close to the shore (Figure 1A), and thus the presence of fecal indicator bacteria was likely. There also were nearby environmental observations, and relatively accurate local wave forecasts that allowed determination of when beach change was likely to occur.
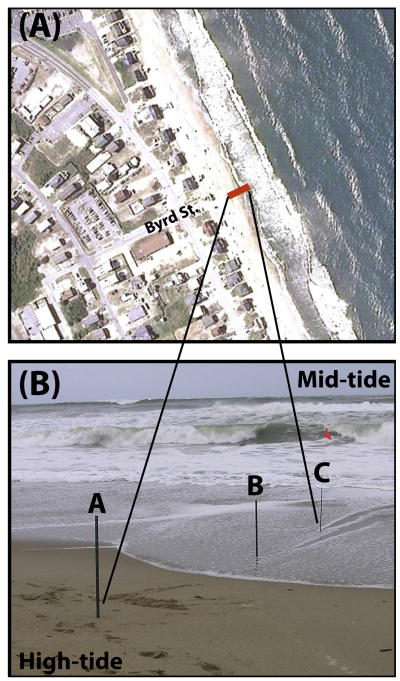
Figure 1.
Sampling site. A) Google Earth image of the Byrd St. public access and the surrounding neighborhood and beach near Kitty Hawk, NC. Samples were taken along the red line, latitude = 36.0983° N, longitude = 75.7093° W. B) Photograph of beach sites where cores were taken on September 11, 13, and 15, 2006.
Winds and waves were measured at the Army Corps of Engineers Field Research Facility located about 10 km north of the study site (http://www.frf.usace.army.mil/). Wind speeds (Figure 2A) and directions collected about 19 m above the water surface with an RM Young marine anemometer at the end of a pier (7-m water depth) were vector averaged and rotated so winds from true north have a direction of 0° (shore normal is 70°). Significant wave heights (defined as 4 times the standard deviation of the sea-surface elevation fluctuations, Figure 2B) and directions were estimated from bottom-pressure fluctuations observed with an array of 15 bottom-mounted Setra gages just offshore of the pier in 8-m water depth using linear theory and a maximum likelihood estimator. Beach erosion and accretion were determined from walking surveys conducted on each of the sampling days by the Army Corps of Engineers using a differential Global Positioning System. The system has an accuracy of 3 cm in both the horizontal and vertical dimensions (personal communication, Mike Forte, US Army Corps of Engineers, Duck, NC).
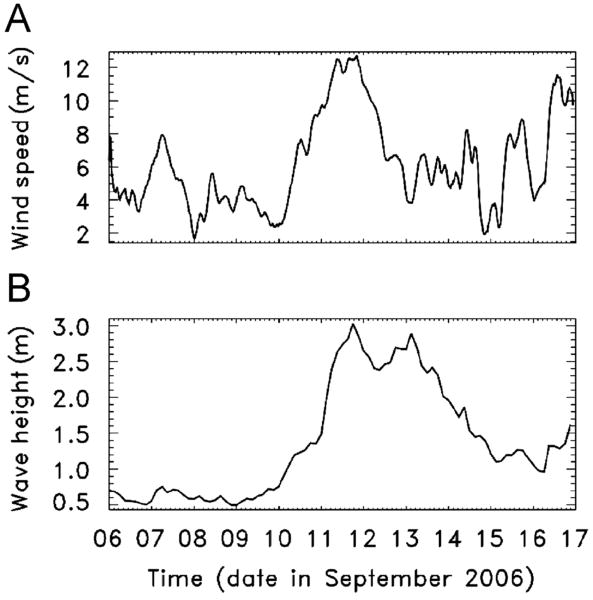
Figure 2.
(A) Wind speed and (B) significant wave height versus time from before, during, and after the sampling period. Waves were near shore normal throughout the study period, whereas winds were from north of shore normal until September 13, when wind directions became near shore normal then south of shore normal until September 15th (not shown).
Approximately 100-cm long sediment cores were collected in duplicate at 3 locations along a cross-shore transect between the high- and mid-tide regions (Figure 1B). Cores were collected using new, clean acrylic tubes that were pounded into the beach surface by hand. After recovery from the beach, core ends were trimmed to be even with the sand surface and capped using new, clean plastic caps that were taped in place. Cores were collected on September 11, 13, and 15, and kept at 4°C until sub-sampled on September 21, 2006 at the laboratory in Woods Hole, MA.
There were no on-site analysis facilities, and samples were preserved as best possible. The 7-day delay prior to processing would be inappropriate for culturing organisms, assessing community structure and diversity, or conducting quantitative PCR assays because there is the potential for loss of nucleic acids. For determining the presence or absence of a particular bacterial genus, the loss of target might result in a sample with an initially low signal to become negative in assays. However, DNA signals can persist for fairly long times in the environment. For example, PCR signals persisted for 28 days when seawater was spiked with enterococci, although the recovery of viable cells fell below the mEI agar plate detection level after 5 days (Walters et al. 2009). Seawater spiked with free DNA from enterococci yielded a PCR signal for 18 days (Walters et al. 2009), suggesting that whole cells in the dead or viable but not cultivable (VBNC) state may help preserve nucleic acids in the environment. Thus, it is expected that the results here are a conservative, but legitimate, assessment of the distribution of enterococci in the beach environment sampled.
Samples were extracted from the cores by drilling a 4-cm diameter hole through one side of the plastic core tube and inserting a 15 ml sterile tube through the hole to recover a small sand plug. Plugs from the same position in the duplicate cores were combined for extraction and amplification. All of the cores were sampled within 3-5 cm of the sand surface, and most also were sampled deeper in the core (between 14 and 70 cm below the surface). To ensure that subsequent samples from the same site were extracted at approximately the same vertical location (within 5 cm) as those in the first core (September 11), the depth of sampling was adjusted to account for erosion and accretion. For example, at site B there was 9 cm of erosion between September 11 and 13, and thus cores on September 11 were sampled at the surface, at 14 cm below the surface (corresponding to the surface of the core taken on September 13), and at 57 cm below the surface (corresponding to the September 13 sample 44 cm below the surface). An exception to sampling at approximately the same vertical location was made for the deeper samples at site A, which were taken from the sulfidic layer that was present between 22 and 25 cm below the surface, even as the site accreted. Subsamples of 0.25 grams were recovered from the combined duplicate samples and were extracted using the MoBio PowerSoil™ DNA Kit following the manufacturer's instructions.
2.2. Detection of indicator organisms
A total of 23 samples were analyzed for DNA using PCR detection methodology. PCR was used to amplify the Enterococcus genus 23S ribosomal RNA gene using the ECST784F and ENC854R primers (Ludwig et al. 1994), and to amplify the E. faecalis 16S ribosomal RNA gene using the 72F and 210R primers (Sedgley et al. 2005). E. faecalis genomic DNA was purchased from ATCC® (700802) and used as a positive control for both primer sets. No-template DNA negative controls were included in every set of PCR reactions to monitor for contamination.
The 50 μl PCR reaction mixture was 1X Taq polymerase buffer, 100 ng each primer, 2.5 mM MgCl2, 0.2 mM deoxynucleotides, 1.25 U GoTaq (Promega), 1 μl of template DNA, and water. PCR conditions for ECST784F/ENC854R were 1 cycle of 95°C for 2 minutes, followed by 40 cycles of 94°C for 10 seconds, 52°C for 30 seconds, 72°C for 10 seconds, followed by 4°C hold. The conditions were the same for primers 72F/210R, except that the annealing temperature was 62°C. An ABI 2700 Thermal Cycler was used.
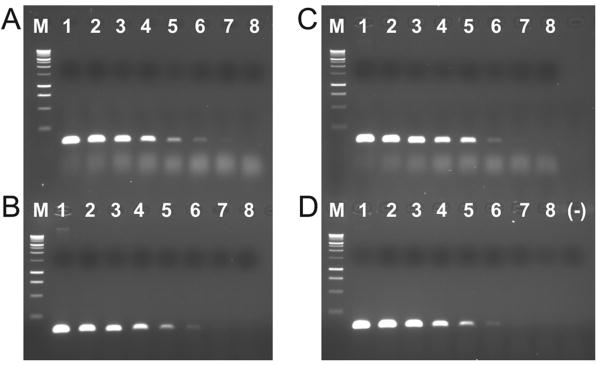
The detection limit of the PCR method for these samples was 1 picogram of genomic DNA, roughly equivalent to between 28 - 280 cells (Figure 3), determined using a 1:10 dilution series of E. faecalis genomic DNA (starting amount 100 ng). Extracted sand core DNA that was negative for the presence of enterococci using the genus primers was added to each reaction in the dilution series. Amplification inhibition was observed for reactions spiked with 1 μl of undiluted sand core DNA (not shown, detection limit 10 ng), but dilution of the extract 1:10, 1:100, or 1:1000 mitigated this effect (Figure 3). To reduce amplification inhibition, all DNA extracts were diluted 1:10 before amplification to test for the presence or absence of products for Enterococcus genus and E. faecalis.
Figure 3.
Test for detection limit of the PCR method. A) E. faecalis genomic DNA dilution alone. B) E. faecalis genomic DNA dilution plus 1:10 sand extract dilution. C) E. faecalis genomic DNA dilution plus 1:100 sand extract dilution. D) E. faecalis genomic DNA dilution plus 1:1000 sand extract dilution. M = Promega 1Kb ladder, 1 = 100 ng genomic DNA, 2 = 10 ng genomic DNA, 3 = 1 ng genomic DNA, 4 = 100 pg genomic DNA, 5 = 10 pg genomic DNA, 6 = 1 pg genomic DNA, 7 = 100 fg genomic DNA, 8 = 10 fg genomic DNA, (-) = no DNA negative control.
3. Results and Discussion
3.1. Beach dynamics and indicator presence
The offshore hurricane produced relatively light winds at the shoreline (Figure 2A) that likely did not impact the beach topology as much as the large waves (Figure 2B). On September 12, the waves and storm surge at high tide exceeded the top of the dunes at the public access parking lot area near the sampling site. At the high-tide site (A in Figure 1B and Figure 4) beach sediments accreted a total of 18 cm throughout the sampling period, whereas at sites B and C there was 8-9 cm of erosion during the storm, followed by 11-24 cm of accretion as the storm abated (Figure 4).
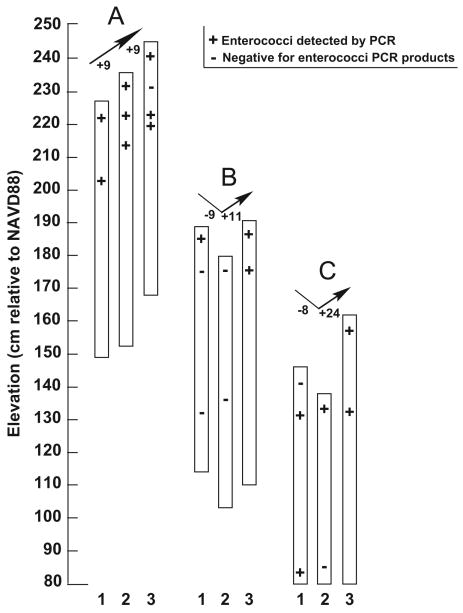
Figure 4.
Total enterococci PCR results from sand cores relative to overall beach change. Cores are labeled 1 (September 11), 2 (September 13), and 3 (September 15) at each site A, B, and C. ‘+’ sign indicates core samples where enterococci were detected by PCR. ‘-’ sign indicates core samples that were negative for enterococci PCR products. Elevations are cm relative to the North American Vertical Datum of 1988 (NAVD88, approximately 10 cm above Mean Sea Level). The arrows above each set of cores indicate erosion (downwards) and accretion (upwards), with the corresponding amounts of sand level change (cm) listed next to the arrows.
None of the samples were positive by PCR for E. faecalis, but many produced a positive signal using the Enterococcus genus primers (Figure 4). Surface samples at the high tide site (A, Figure 4) always had a positive signal for enterococci, as were samples collected deeper in the core. These results suggest the accreted sands could have contained enterococci.
The center site (B, Figure 4) yielded enterococcal signals from the surface sands on the first day of sampling and in accreted surface sands at the end of the storm. This site was the only one that gave negative results for enterococcal products in subsurface samples at the start of the sampling. These negatives persisted through the second day of sampling as the site eroded, but they became positive as sand accreted at the end of the storm, suggesting that deposited sands played a role in the redistribution of enterococcal signals.
At the mid-tide site (C, Figure 4), the surface samples on September 11 were negative for enterococcal product, but subsurface samples were positive. Erosion at this site may have allowed the exposure of these subsurface sands, which became the enterococci positive surface sand samples on September 13. Accreted surface sand and subsurface sands both gave positive signals for enterococci on the final day of sampling.
Site C (mid-tide) was the only site that showed potential reworking of the sands as far as 60 cm below the surface. On the first day of sampling, the 60-cm-deep samples at site C had layers of fine sand, and gave positive PCR results for enterococci, but in the subsequent samples at that depth much coarser sands that were negative for enterococci products replaced the fine-sand layers. The change in enterococci presence at this sample location could be related to the grain size, as discussed below. However, the change in grain type also suggests sands were moved to and from this location, and thus that enterococcal signals in the initial sample may have been transported to other sites. The erosion and subsequent cross-shore transport of enterococci-containing sediments from the middle (B) and mid-tide (C) sites may have contributed to the enterococcal signals in accreted sands at all three sites.
The presence of enterococcal signals from 25 - 70 cm deep in the cores suggests there is potential for these indicator organisms to persist or regrow at significant depth in the beach environment. Natural populations of enterococci grow in the marine environment (Hartz et al. 2008, Yamahara et al. 2009), and can confound indicator studies. Thus, the signals present at depth, and even at the surface, in the samples reported here might be owing to indigenous strains of enterococci rather than the persistence of ones introduced by beach contamination. However, fecal enterococci can persist in beach sands longer than in the water column (Hartz et al. 2008), so it is possible that subsurface sediments could harbor fecal bacteria and pathogens.
The presence and abundance of fecal indicator bacteria can be influenced by the sand grain size within the beach, with more indicator bacteria present in fine sand with a uniform distribution (Skalbeck et al. 2010). Although grain sizes were not measured, visual inspections of the cores showed similar medium to coarse-grained sands (0.25 – 1.00 mm diameter) at all sample locations except the deep samples at site A and the initial (September 11) deep sample at site C. Thus, while the presence of an enterococcal signal at the deepest location at site C might be related to the fine grain size present prior to the storm, medium-coarse grained sands from other core samples also often were positive for enterococci. Therefore, it seems likely that the differences in enterococci detection were not only owing to differences in grain size.
The PCR method detects the presence of nucleic acids, but not necessarily viable cells. Therefore, the results indicate only that enterococcal genetic material was present in the samples. Positive results could have been caused by free DNA in the sample, as well as by viable, dead, or viable but not cultivable (VBNC) enterococcal cells. However, DNA signals from added cells have been shown to persist longer in environmental samples (28 days) than signals from added free DNA (18 days) (Walters et al. 2009), suggesting that recovery of a DNA signal in the samples here likely indicates the presence of whole cells. Fecal enterococci have been found to persist in beach sands longer than in the water column (Hartz et al. 2008), so their presence in these samples would not be unprecedented. The extent to which human pathogens, such as Giardia or Staphylococcus, are able to persist within beach sands is unknown, although some of these pathogens have been detected (Abdelzaher et al. 2010, Plano et al. 2011).
4. Conclusions
The results from this pilot project demonstrate that there is the potential for redistribution of enterococci as beach sand erodes or accretes. Unlike traditional culture approaches, the PCR-based method is useful because it can detect viable but not cultivable (VBNC) cells in addition to live cells. Future work with both culture-dependent and culture-independent methods is necessary to determine the extent that beach reworking contributes to the live enterococcal signal in water and throughout the beach environment.
Acknowledgments
We thank the staff of the US Army Corps of Engineers Field Research Facility at Duck, North Carolina for assistance with beach surveys and for the use of their facilities. Support was provided by the Woods Hole Center for Oceans and Human Health Pilot Project Program, the National Science Foundation (OCE-0430724 & OCE-0622844), the National Institute of Environmental Health Sciences (P50ES012742), the Woods Hole Oceanographic Institution Interdisciplinary Study Award and a National Security Science and Engineering Faculty Fellowship.
Literature cited
- Abdelzaher AM, Wright ME, Ortega C, Solo-Gabriele HM, Miller G, Elmir S, Newman X, Shih P, Bonilla JA, Bonilla TD, Palmer CJ, Scott T, Lukasik J, Harwood VJ, McQuaig S, Sinigalliano CD, Gidley M, Plano LRW, Zhu XF, Wang JD, Flemming LE. Presence of pathogens and indicator microbes at a non-point source subtropical recreational marine beach. Applied and Environmental Microbiology. 2010;76:724–732. doi: 10.1128/AEM.02127-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beversdorf L, Borstein-Forst S, McLellan S. The potential for beach sand to serve as a reservoir for Escherichia coli and the physical influences on cell die-off. Journal of Applied Microbiology. 2007;102:1372–1381. doi: 10.1111/j.1365-2672.2006.03177.x. [DOI] [PubMed] [Google Scholar]
- Boehm AB, Weisberg SB. Tidal forcing of enterococci at marine recreational beaches at fortnightly and semidiurnal frequencies. Environmental Science and Technology. 2005;39:5575–5583. doi: 10.1021/es048175m. [DOI] [PubMed] [Google Scholar]
- Bonilla T, Nowosielski K, Cuvelier M, Hartz A, Green M, Esiobu N, McCorquodale D, Fleisher J, Rogerson A. Prevalence and distribution of fecal indicator organisms in South Florida beach sand and preliminary assessment of health effects associated with beach sand exposure. Marine Pollution Bulletin. 2007;54:1472–1482. doi: 10.1016/j.marpolbul.2007.04.016. [DOI] [PubMed] [Google Scholar]
- Burton G, Gunnison D, Lanza G. Survival of pathogenic bacteria in various freshwater sediments. Applied and Environmental Microbiology. 1987;53:633–638. doi: 10.1128/aem.53.4.633-638.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byappanahalli MN, Fowler M, Shively D, Whitman RL. Ubiquity and persistence of Escherichia coli in a midwestern coastal stream. Applied and Environmental Microbiology. 2003;69:4549–4555. doi: 10.1128/AEM.69.8.4549-4555.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciavola P, Dias N, Ferreira O, Taborda R, Dias JM. Fluorescent sands for measurements of longshore transport rates: a case study from Praia de Faro in southern Portugal. Geo-Marine Letters. 1998;18:49–57. [Google Scholar]
- Desmarais TR, Solo-Gabriele HM, Palmer CJ. Influence of soil on fecal indicator organisms in a tidally influenced subtropical environment. Applied and Environmental Microbiology. 2002;68:1165–1172. doi: 10.1128/AEM.68.3.1165-1172.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doyle J, Tunnicliff B, Kramer R, Kuehl R, Brickler S. Instability of fecal coliform populations in waters and bottom sediments at recreational beaches in Arizona. Water Research. 1992;26:979–988. [Google Scholar]
- Feddersen F. Breaking wave induced cross-shore tracer dispersion in the surf zone: model results and scalings. Journal of Geophysical Research 2007:C09012. [Google Scholar]
- Ferguson DM, Moore DF, Getrich MA, Zhowandai MH. Enumeration and speciation of enterococci found in marine and intertidal sediments and coastal water in southern California. Journal of Applied Microbiology. 2005;99:598–608. doi: 10.1111/j.1365-2672.2005.02660.x. [DOI] [PubMed] [Google Scholar]
- Ghinsberg R, Drasinover V, Sheinber Y, Nitzan Y. Seasonal distribution of Aeromonas hydrophila and Vibrio species in Mediterranean coastal water and beaches: a possible health hazard. Biomedical Letters. 1995;51:151–159. [Google Scholar]
- Grant SB, Kim JH, Jones BH, Jenkins SA, Wasyl J, Cudaback C. Surf zone entrainent, along-shore transport, and human health implications of pollution from tidal outlets. Journal of Geophysical Research. 2005;110:C10025. [Google Scholar]
- Haile RW, Witte JS, Gold M, Cressey R, McGee C, Millikan RC, Glasser A, Harawa N, Ervin C, Harmon P, Harper J, Dermand J, Alamillo J, Barrett K, Nides M, Wang G. The health effect of swimming in ocean water contaminated by storm drain runoff. Epidemiology. 1999;10:355–363. [PubMed] [Google Scholar]
- Halliday E, Gast RJ. Bacteria in beach sands: an emerging challenge in protecting coastal water quality and bather health. Environmental Science and Technology. 2011;45:370–379. doi: 10.1021/es102747s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartz A, Cuvelier M, Nowosielski K, Bonilla T, Green M, Esiobu N, McCorquodale D, Rogerson A. Survival potential of Escherichia coli and enterococci in subtropical beach sand: Implications for water quality managers. Journal of Environmental Quality. 2008;37:898–905. doi: 10.2134/jeq2007.0312. [DOI] [PubMed] [Google Scholar]
- Irvine K, Pettibone G. Dynamics of indicator bacteria populations in sediment and river water near combined sewer outfall. Environmental Technology. 1993;14:531–542. [Google Scholar]
- Jamieson RC, Joy DM, Kostaschuk R, Gordon RJ. Resuspension of sediment-associated Escherchia coli in a natural stream. Journal of Environmental Quality. 2005;34:581–589. doi: 10.2134/jeq2005.0581. [DOI] [PubMed] [Google Scholar]
- Jeng HWC, England AJ, Bradford HB. Indicator organisms associated with stormwater suspended particles and estuarine sediment. Journal of Environmental Science and Health Part A - Toxic/Hazardous Substances & Environmental Engineering. 2005;40:779–791. doi: 10.1081/ese-200048264. [DOI] [PubMed] [Google Scholar]
- Ludwig W, Dorn S, Springer N, Kirchhof G, Schleifer KH. PCR-based preparation of 23S rRNA-targeted group-specific polynucleotide probes. Applied and Environmental Microbiology. 1994;60:3236–3244. doi: 10.1128/aem.60.9.3236-3244.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Obiri-Danso K, Jones K. Intertidal sediment as reservoirs of hippurate negative campylobacters, salmonellae, and fecal indicators in three EU recognized bathing waters in north west England. Water Research. 2000;34:519–527. [Google Scholar]
- Oshiro R, Fujioka RS. Sand, soil and pigeon droppings - sources of indicator bacteria in the water of Hanauma Bay, Oahu, Hawaii. Water Science and Technology. 1995;31:251–254. [Google Scholar]
- Plano LRW, Garza AC, Shibata T, Elmir SM, Kish J, Sinigalliano CD, Gidley ML, Miller G, Withum K, Fleming LE, Solo-Gabriele HM. Shedding of Staphylococcus aureus and methicillin-resistant Staphylococcus aureus from adult and pediatric bathers in marine waters. BMC Microbiology. 2011;11:5. doi: 10.1186/1471-2180-11-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sedgley CM, Nagel AC, Shelburne CE, Clewell DB, Applebe O, Molander A. Quantitative real-time PCR detection of oral Enterococcus faecalis in humans. Archives of Oral Biology. 2005;50:575–583. doi: 10.1016/j.archoralbio.2004.10.017. [DOI] [PubMed] [Google Scholar]
- Sinigalliano CD, Fleisher J, Gidley ML, Solo-Gabriele HM, Shibata T, Plano LRW, Elmir SM, Wanless D, Bartowiak J, Boiteau R, Withum K, Abdelzaher AM, He GQ, Ortega C, Zhu XF, Wright ME, Kish J, Hollenbeck J, Scott T, Backer LC, Flemming LE. Traditional and molecular analyses for fecal indicator bacteri in non-point source subtropical recreational marine waters. Water Research. 2010;44:3763–3772. doi: 10.1016/j.watres.2010.04.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skalbeck JD, Kinzelman JL, Mayer GC. Fecal indicator organism density in beach sands: impact of sediment grain size, uniformity, and hydrologic factors on surface water loading. Journal of Great Lakes Research. 2010;36:707–714. [Google Scholar]
- Smith J, Largier J. Observations of nearshore circulation: Rip currents. Journal of Geophysical Research. 1995;100:10,967–910,975. [Google Scholar]
- Wade TJ, Pai N, Eisenberg JNS, Colford JM., Jr Do US Environmental Protection Agency water quality guidelines for recrational waters prevent gastrointestinal illness? A systematic review and meta-analysis. Environmental Health Perspectives. 2003;111:1102–1109. doi: 10.1289/ehp.6241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walters SP, Yamahara KM, Boehm AB. Persistence of nucleic acid markers of health-relevant organisms in seawater microcosms: Implications for their use in assessking risk in recreational waters. Water Research. 2009;43:4929–4939. doi: 10.1016/j.watres.2009.05.047. [DOI] [PubMed] [Google Scholar]
- Whitman R, Nevers M. Foreshore sand as a source of E. coli in nearshore water of a Lake Michigan beach. Applied and Environmental Microbiology. 2003;69:5555–5562. doi: 10.1128/AEM.69.9.5555-5562.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamahara KM, Layton BA, Santoro AE, Boehm AB. Beach sands along the California coast are diffuse sources of fecal bacteria to coastal waters. Environmental Science and Technology. 2007;41:5415–4521. doi: 10.1021/es062822n. [DOI] [PubMed] [Google Scholar]
- Yamahara KM, Walters SP, Boehm AB. Growth of enterococci in unaltered, unseeded beach sands subjected to tidal wetting. Applied and Environmental Microbiology. 2009;75:1517–1524. doi: 10.1128/AEM.02278-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zehms TT, McDermott CM, Kleinheinz GT. Microbial concentrations in sand and their effect on beach water in Door County, Wisconsin. Journal of Great Lakes Research. 2008;34:524–534. [Google Scholar]