Abstract
Messenger RNA localization involves the assembly of ribonucleoprotein particles (RNPs) and their subsequent transport along the cytoskeleton to their final destination. Here, we provide new evidence that microtubule-associated protein 2 (MAP2), calcium/calmodulin-dependent protein kinase II (CaMKIIα) and β-actin RNAs localize to dendrites in distinct RNPs, which contain—unexpectedly—very few RNA molecules. The number of MAP2 molecules per particle is affected by synaptic activity and Staufen 2, indicating that RNP composition is tightly controlled. Our data suggest that the independent localization of individual RNAs in low copy numbers could contribute to tighter temporal and spatial control of expression in neurons and synapse-specific plasticity.
Keywords: RNA localization, β-actin , CaMKIIα , MAP2 mRNA, Staufen 2
Introduction
Several RNAs localize and get locally translated in growth cones of developing neurites and mature dendrites (Holt & Bullock, 2009; Martin & Ephrussi, 2009) and have key roles in the development and plasticity of the nervous system, as well as in learning and memory.
The mechanism underlying the transport of RNAs to dendrites is unclear. RNAs associate with RNA-binding proteins (RBPs) to form ribonucleoprotein particles (RNPs), which are transported by molecular motors (Kiebler & Bassell, 2006). Several classes of neuronal RNPs have been purified and characterized by mass spectrometry and microarray analysis (Kanai et al, 2004; Anderson & Kedersha, 2006; Kiebler & Bassell, 2006; Martin & Ephrussi, 2009). On the basis of the large number of proteins and RNAs identified in various types of RNP, a hypothesis has been put forward that neurons might transport their RNAs in large and complex neuronal RNA granules containing various RNAs and RBPs, possibly including ribosomes (Krichevsky & Kosik, 2001; Kanai et al, 2004). This view has been reinforced by overexpression and microinjection experiments in mammalian cells including neurons, in which RNAs or RBPs form bright particles. In addition, homologous targeting elements have been identified, arguing for conserved mechanisms (Shan et al, 2003; Gao et al, 2008). For most localization pathways, however, we still lack essential information on the heterogeneity and complexity of RNPs (Holt & Bullock, 2009). Even in systems where mRNAs are found in an oligomeric state, this has been considered an early, intermediate step in the formation of large localization complexes (Chekulaeva et al, 2006; Besse et al, 2009; Martin & Ephrussi, 2009). In neurons, the assembly of dendritic mRNAs into ‘large’ RNPs and their subsequent co-transport to dendrites would imply that synapse-specific modifications associated with plasticity can only arise from on-site regulation of translation and not by selective delivery of each mRNA to individual synapses. We have recently investigated the localization of microinjected dendritically localized transcripts (Tübing et al, 2010). In that study, we observed that microtubule-associated protein 2 (MAP2) and calcium/calmodulin-dependent protein kinase II (CaMKIIα) RNAs appear to segregate into distinct neuronal RNA granules. Therefore, we decided to use novel assays to characterize in detail the RNA composition of neuronal RNPs. We now provide evidence that dendritic RNAs localize independently of each other in low copy numbers and are differentially affected by Staufen 2 (Stau2), as well as by synaptic activity, arguing for selective RNA transport to sites of demand.
Results And Discussion
Neuronal mRNAs localize in distinct dendritic RNPs
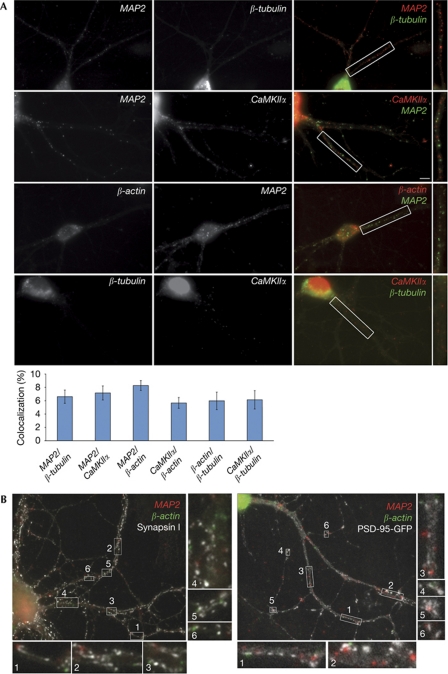
To investigate whether different dendritic RNAs are found in the same or distinct particles, we performed double in situ hybridization (ISH). Simultaneous detection of RNAs with differentially labelled probes, followed by sequential antibody detection and signal amplification allowed us to co-visualize two mRNAs at particle resolution with high specificity and no cross-reactivity. As proof-of-principle, we performed double ISH against β-tubulin mRNA, which is highly expressed and enriched in the soma but found at very low abundance in dendrites, and MAP2 mRNA, which is expressed at lower levels but localizes in dendrites. The different localization patterns are preserved in the double ISH illustrating the specificity of the signal and lack of cross-reactivity (Fig 1A). Building on our initial observation that MAP2 and CaMKIIα RNAs colocalized at low levels in dendrites of hippocampal neurons (Tübing et al, 2010), we here substantiate the initial hypothesis by testing additional pairs of candidate dendritic mRNAs and identify similarly infrequent colocalization events ranging from 5.7% (±0.8) to 8.3% (±0.7) (Fig 1A). To address whether this observation reflected cotransport of the respective mRNAs to dendrites or rather their stochastic colocalization, we analysed how often localizing and non-localizing RNAs are found in the same RNPs at proximal dendritic sites. As shown in Fig 1A, 6.6% (±1.0), 6.1% (±1.4) and 6.0% (±1.3) of proximal β-tubulin mRNA colocalize with MAP2, CaMKIIα and β-actin, respectively. This is not significantly different from the colocalization we observed between any two dendritic RNAs (Fig 1A). Our data indicate that RNAs rarely localize in the same dendritic RNPs, which might allow their differential regulation and selective delivery to individual synapses and consequently contribute to synapse-specific modifications and plasticity at the RNA level. Indeed, we found that MAP2 and β-actin mRNAs localize at distinct postsynaptic sites (Fig 1B). Preliminary results showed localization of CaMKIIα at synapses (data not shown). However, its relation to MAP2 or β-actin was not investigated. The age of the neurons or synaptic activity did not show any significant effect on colocalization between MAP2, CaMKIIα and β-actin (supplementary Fig S1B online), nor did it substantially interfere with their accumulation to dendrites.
Figure 1.
Neuronal mRNAs localize in distinct dendritic ribonucleoprotein particles. (A) Several pairs of candidate mRNAs (MAP2/β-tubulin; n=21 dendrites), MAP2/CaMKIIα (n=45), MAP2/β-actin (n=118), CaMKIIα/β-actin (n=65), β-actin/β-tubulin (n=17) and CaMKIIα/β-tubulin (n=11) were detected in 14–16 days in vitro rat hippocampal neurons by double in situ hybridization. Quantification of colocalization in dendrites is shown below. Error bars represent the s.e.m. of colocalization rates in individual dendrites. There was no significant difference between the groups (analysis of variance). Scale bars, 10 μm. (B) Hippocampal neurons were stained for MAP2/β-actin and synapsin I (left) or MAP2/β-actin and GFP, on transfection with PSD-95-GFP (right). Boxes in A and B represent high-magnification insets. GFP, green fluorescent protein; PSD-95, postsynaptic density 95.
As a positive control for colocalization, we performed double ISH using two probes targeting the same transcript (Fig 2). In this case, we could detect up to 55% of colocalization. The lack of 100% colocalization does not result from low hybridization efficiency, as addition of the second probe (in this case labelled identically) in single ISH resulted in the expected increase in particle intensity (supplementary Fig S2C online; supplementary Methods online), indicating that most RNA molecules are bound by both probes. We rather attribute this to lower efficiency of the second detection. In double ISH experiments, we detected fewer particles of a given mRNA, whenever it was detected second (supplementary Fig S1C,D online). This suggests that the rates reported in double ISH experiments are an underestimate of colocalization. However, they are still much lower than those observed in the positive control, indicating that dendritic mRNAs are indeed mostly found in distinct RNPs. To confirm our observations by another approach, we tagged CaMKIIα mRNA with yellow fluorescent protein (YFP)—using the MS2 reporter system (Bertrand et al, 1998) consisting of CaMKIIα 3′-untranslated region fused to 24 × MS2 binding sites and MS2 coat protein (MCP)–YFP—and detected MAP2 by ISH (supplementary Fig S1E online). Consistent with our previous results, we found that only 9.1% (±1.5) of all YFP puncta contained MAP2 mRNA in contrast to 85.8% (±5.7) of all YFP puncta positive for CaMKIIα.
Figure 2.
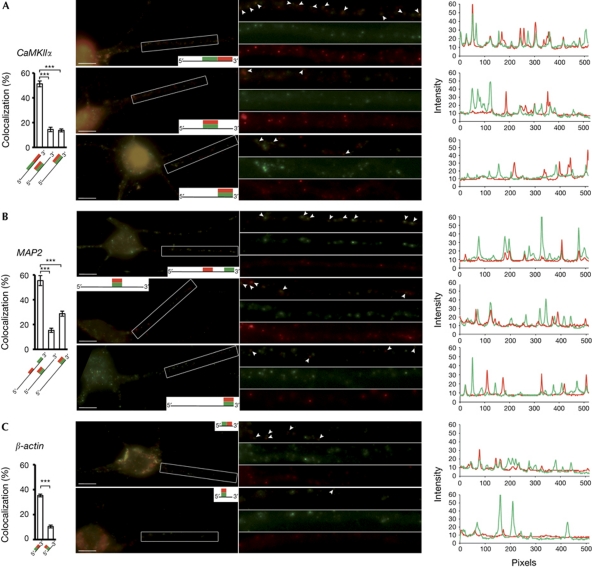
MAP2, CaMKIIα and β-actin ribonucleoprotein particles contain few RNA molecules. Detection of CaMKIIα (A; n=26, 18 and 21 dendrites from top to bottom), MAP2 (B; n=22, 17 or 38 dendrites) and β-actin (C; n=56 or 99 dendrites) in hippocampal neurons with differentially labelled probes targeting non-overlapping sequences (upper panels of A–C) or the same sequence (middle panels of A,B and lower panels of A–C) of each transcript. Scale bars, 10 μm. Colocalization of the signals significantly increased when non-overlapping sequences were detected. Error bars represent the s.e.m. of colocalization rates. ***P<0.001. Linescans show intensity of each signal along a single dendrite; x values are number of pixels.
Dendritic RNPs contain from one to a few RNA molecules
To investigate whether many molecules of a given RNA localize together in single dendritic particles, we performed double ISH with probes that target the same sequence within one RNA and would therefore compete for binding. We predicted the following outcome: if the RNPs contained one RNA molecule, then only one of the probes would bind per particle and we would not observe any colocalization. If, conversely, the particles contained many RNA molecules, then both probes would bind to RNAs within the same particle, resulting in up to 100% colocalization. For this experiment, we used sets of probes targeting the same regions of CaMKIIα, MAP2 or β-actin, respectively, and detected colocalization rates of 10.5% (±1.2) for β-actin (Fig 2C), 14.4% (±1.8) or 13.7% (±1.2) for CaMKIIα (Fig 2A) and 15.3% (±1.7) or 28.8% (±2.1) for MAP2 (Fig 2B). When we used combinations of probes targeting non-overlapping sequences in the respective mRNAs, the observed colocalization rates—although not 100% because of limitations imposed by the second detection (see above and supplementary Fig S1D online)—were significantly higher: 51.2 (±2.4)% for CaMKIIα, 55.5 (±4.0)% for MAP2 and 35.3 (±1.1)% for β-actin (Fig 2). We obtained similar results with septin 7 (data not shown). Our findings indicate that these mRNAs are not present in ‘large’ complexes containing many molecules of each RNA. Normalization of the values obtained with two probes targeting the same sequence to the colocalization ratio of the respective positive control (see above) suggests that the average RNA content per particle ranges from one to two molecules (see supplementary Methods online).
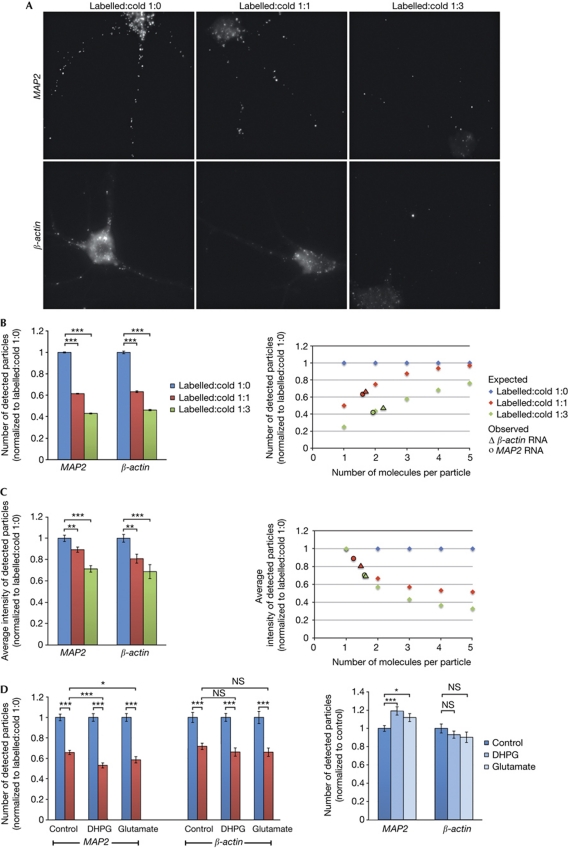
To substantiate our results, we used a competition assay. Here, instead of two differentially labelled probes, we used labelled and non-labelled (termed ‘cold’) probes in the same hybridization mix. These target the same sequence and should compete for binding. If the particles contained one mRNA molecule, then either labelled or cold probe would bind to each particle and the number of observed particles would decrease to 50% in the case of equal amounts of cold and labelled probes or even further when using an excess of cold probe. The average intensity of the detected particles, however, should not be affected by the addition of the cold probe. If RNPs contained many copies of an RNA, then all particles would still be detected on the addition of the cold probe, but their signal intensity would be decreased to 50% for a 1:1 ratio of cold and labelled probe.
When we detected MAP2 and β-actin (Fig 3A), the number of detected particles was significantly reduced even with the addition of equal amounts of cold probe, confirming that these particles indeed contain a low number of RNA molecules (Fig 3B). Control competition experiments, in which probes with different labels and cold probes are shown to compete with each other with the same efficiency (supplementary Fig S2A online), confirm that labelled and cold probes bind to the target with the same efficiency. On the basis of the equal binding probabilities for each probe and the high efficiency of our method (supplementary Fig S2A,C online; supplementary Methods online), we calculated the expected numbers of detected particles on the assumption that they contain one, two, three, four or five RNA molecules. The observed values all fall around two molecules, indicating that dendritic particles contain very few molecules of a given mRNA (Fig 3B). Quantitative measurements of particle intensities (based on the linearity of detection, see supplementary Fig S2D online; supplementary Methods online) confirmed this conclusion (Fig 3C). As a negative control for competition, we used cold probe targeting a non-overlapping sequence on the same or another mRNA. In these cases, even the addition of a fivefold excess of cold probe did not interfere with binding of the labelled probe (supplementary Fig S2B online). Analysis of the distribution of single particle intensities suggests that the majority of MAP2 particles contain one molecule, with some particles containing two, three or more molecules, with decreasing frequency (supplementary Fig S2E online).
Figure 3.
Dendritic ribonucleoprotein particles contain on average two molecules of RNA. MAP2 and β-actin RNAs were detected in the presence (A, middle and right panels) or absence (A, left) of cold probe. (B, left) The number and (C, left) average intensity of detected dendritic particles in the presence (red and green) or absence (blue) of cold probe are shown. The error bars represent the corresponding s.e.m. (normalized to labelled:cold 1:0) for individual dendrites (B) or individual particles (C). The respective expected values (diamond) for particles containing 1–5 RNA molecules, as well as the observed values for MAP2 (circle) and β-actin (triangle), are shown on the right. In B, the numbers of dendrites analysed were 66, 65 and 31 for MAP2, and 29, 39 and 36 for β-actin. To determine average particle intensities in C, 1,381, 1,686 and 731 MAP2 particles and 1,863, 950 and 352 β-actin particles were measured. (D) MAP2 and β-actin RNAs were detected in the absence (blue) or presence (red) of equal amounts of cold probe in control cells (n=98 and 93 for MAP2, n=70 and 73 for β-actin) and neurons treated with DHPG (n=71 and 84 for MAP2, n=61 and 60 for β-actin) or glutamate (n=59 and 54 for MAP2, n=56 and 72 for β-actin). Numbers of detected particles are normalized to the respective labelled:cold 1:0 condition (left) or the control condition (right). *P<0.05, **P<0.01 and ***P<0.001. NS, not significant.
Our data argue that dendritic particles are not only more heterogeneous and less complex than previously thought, containing as little as one type of mRNA, but also much ‘smaller’ at least with respect to their RNA content. This is in line with previous studies determining the exact number of β-actin and other mRNAs in mammalian cells and yeast (Femino et al, 1998; Raj et al, 2006; Zenklusen et al, 2008). RNA localization in many small, rather than fewer larger particles might allow the independent transport and regulation of each molecule and their flexible localization to distinct synapses on demand. Similar experiments will allow us in the future to determine how many RNA molecules are found in a given compartment, for example, in a dendrite or even a dendritic spine, which will advance our quantitative understanding of gene expression in neurons (Larson et al, 2009). To address whether non-localizing RNAs also assemble into ‘small’ particles, we detected septin 7 (found mostly in proximal dendrites) and β-tubulin (highly expressed, at very low abundance in dendrites) in the presence of cold probe (supplementary Fig S2F online and data not shown). The observed effects on the number of particles suggest that non-localizing mRNAs are not found in ‘large’ granules either, at least in dendrites.
MAP2 RNP content is affected by neuronal activity
To test whether the RNA composition of MAP2 and β-actin RNPs is affected by neuronal activity, we performed competition experiments for MAP2 and β-actin on hippocampal neurons stimulated with glutamate or (S)-3,5-dihydroxyphenylglycine (DHPG), which are known to regulate RNA localization and local translation. Although for β-actin the competition effect does not change in either condition, DHPG treatment (and to a lesser extent stimulation with glutamate) resulted in a greater decrease in relative MAP2 particle numbers detectable in the presence of cold probe (0.53 compared with 0.66 in the control condition; Fig 3D, left). This brings the relative number of particles close to 0.5, the maximum competition effect corresponding to one molecule per particle (compared with almost two molecules per particle in the control condition). These results correlate with an observed increase in dendritic MAP2, but not β-actin particle numbers in DHPG- and glutamate-treated neurons (Fig 3D, right), suggesting disassembly of more complex particles upon synaptic activity.
Neuronal mRNAs are differentially affected by Staufen 2
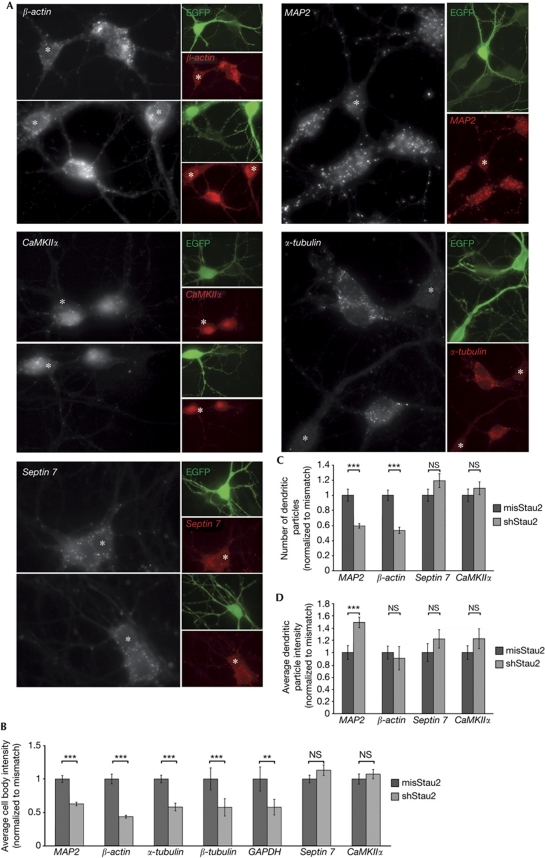
Our data are consistent with the idea that dendritic RNAs localize in distinct RNPs, potentially involving different factors, such as RBPs. We therefore investigated whether Stau2, an RBP involved in dendritic spine morphogenesis, synapse formation and β-actin localization in dendrites (Goetze et al, 2006; Lebeau et al, 2011) also affects the localization of other neuronal mRNAs. In Stau2-deficient neurons, the number of dendritic MAP2 RNPs was decreased (Fig 4A,C). Interestingly, this does not seem to result from reduced transport to dendrites, as the cell body levels of MAP2 and β-actin were also substantially lower in Stau2 downregulated compared with wild-type neurons, which were either non-transfected (Fig 4A) or expressing a Stau2 mismatch short hairpin RNA (Fig 4B). In contrast to β-actin and MAP2, the cell body levels of CaMKIIα or septin 7 and their localization to dendrites were not impaired by Stau2 depletion (Fig 4A–C). This is in agreement with recent evidence that MAP2 colocalizes with Stau2 in hippocampal neurons (Lebeau et al, 2011), and that β-actin, but not CaMKIIα or septin 7, is highly enriched in Stau2 particles from rat brain (Maher-Laporte & DesGroseillers, 2010). We extended our analysis to non-localizing RNAs and found that the levels of α-tubulin, β-tubulin and glyceraldehyde 3-phosphate dehydrogenase (GAPDH) were also significantly decreased, which could be explained by an effect of Stau2 on the stability of a broad range of target RNAs. Indeed, inhibition of transcription with actinomycin D resulted in a further decrease of MAP2 and β-actin levels in Stau2-deficient neurons (supplementary Fig S3B online), indicating that Stau2 affects the stability of these mRNAs.
Figure 4.
Neuronal mRNAs are differentially affected by Staufen 2. (A) Neuronal mRNAs were detected in wild-type (untransfected) or Stau2-deficient hippocampal neurons (transfected with shStau2 and coexpressing EGFP, marked with asterisks). Quantification of average cell body signal intensity of MAP2, β-actin, α-tubulin, β-tubulin, GAPDH, septin 7 and CaMKIIα RNAs (B) of neurons transfected with shStau2 or misStau2. For each respective condition, 31, 25, 26, 16, 16, 20, 9, 12, 18, 14, 16, 25, 12 or 18 cells were analysed (in the order they appear on the graph). (C) Numbers of dendritic particles containing MAP2, β-actin, septin 7 and CaMKIIα and (D) average fluorescence intensity of these dendritic particles in shStau2-transfected neurons compared with mismatch control. In panel C, 34, 106, 33, 42, 28, 26, 29 or 25 dendrites, and in panel D, 217, 436, 62, 115, 124, 211, 92 or 139 particles were analysed for each respective condition (in the order they appear on the graphs). Error bars are the s.e.m. of cell body intensities of individual cells (B), detected particles per dendrite (C) or intensities of individual particles (D). **P<0.01 and ***P<0.001. EGFP, enhanced green fluorescent protein; GAPDH, glyceraldehyde 3-phosphate dehydrogenase; misStau2, mismatch shRNA against Stau2; NS, not significant; shStau2, short hairpin Staufen 2.
Interestingly, Stau2 affects the composition of MAP2 but not of β-actin, septin 7 or CaMKIIα RNPs. We compared the average particle intensity in wild-type and Stau2-deficient hippocampal neurons (Fig 4D). Although there was no significant difference for β-actin, septin 7 or CaMKIIα, the intensity of MAP2 particles increased significantly in Stau2-deficient neurons (1.57±0.08, normalized to mismatch short hairpin RNA). This indicates an increase in the RNA content of dendritic MAP2 RNPs in the absence of Stau2. To corroborate this conclusion, we detected MAP2 or β-actin in Stau2-deficient or wild-type neurons in the presence of equal amounts of cold probe and compared the competition effect with the untransfected control. For β-actin, we did not detect any obvious changes (supplementary Fig S3C online, right). Although there are no differences for mismatch transfected cells, MAP2 particle numbers were less affected by the addition of cold probe in Stau2-deficient neurons (supplementary Fig S3C online, left), arguing for a higher number of RNA molecules per particle. Taken together, the low number of RNA molecules in MAP2 RNPs seems to be neither stochastic nor merely a consequence of mRNA abundance, but instead subject to regulation. We suggest that there are mechanisms in the cell that tightly control RNP composition, preventing the formation of large complexes. In contrast to the increase in the average number of MAP2 RNA molecules per particle, colocalization of MAP2 with either CaMKIIα or β-actin RNA was not affected by Stau2 depletion (supplementary Fig S3D online).
Conclusions
In summary, we provide evidence that MAP2, CaMKIIα and β-actin localize in distinct RNPs. This suggests the possibility of their independent regulation and selective localization to sites of demand. Previous work (Gao et al, 2008) showed the coassembly of CaMKIIα, neurogranin and Arc in heterogeneous nuclear ribonucleoprotein (hnRNP) A2-containing RNPs and their multiplex dendritic targeting. Our findings that MAP2, CaMKIIα and β-actin are differentially regulated by Stau2 suggests the existence of diverse pathways including (at least partly) distinct machineries for dendritic targeting. Moreover, we show that MAP2, CaMKIIα and β-actin RNPs contain a low number of RNA molecules. Interestingly, the amount of RNA in MAP2 RNPs is affected by synaptic activity. Our data support a model in which RNAs localize independently to synapses, where their expression can be individually regulated on-site. Such a mechanism would allow tight temporal and spatial control of expression in dendrites and synapse-specific plasticity. It would be interesting to investigate whether in other systems, for example, Drosophila embryos or oocytes, mRNAs that localize to the same intracellular compartment are transported in large, complex granules or independently in small localization complexes.
Methods
In situ hybridization and competition assays. Single and double detection of RNAs by ISH was performed as described (Tübing et al, 2010). To quantify the degree of colocalization, puncta in dendrites (from 10 μm from the cell body up to 75 μm) were counted manually. Colocalization rates were calculated as the ratio of overlapping signal to the number of particles of the less abundant colour in the investigated section of the dendrite. Linescan analysis was performed using Metamorph.
For competition, unlabelled in vitro transcribed probes targeting the same sequence as the labelled probe were added to the hybridization mix at equal amounts (1:1) to labelled probe or in threefold excess (1:3). Particles (10–75 μm from the cell body) were counted and used for fluorescence intensity measurements.
Statistical analysis. Typically, one representative dendrite per neuron was used for measuring ISH signal and assessing colocalization. Thus, dendrite numbers are usually numbers of cells analysed. Asterisks denote statistical significance as determined by using two-tailed Student's t-test for the complete data set. All data were obtained from at least three independent experiments.
Supplementary information is available at EMBO reports online (http://www.emboreports.org).
Supplementary Material
Acknowledgments
We thank S. Thomas, J. Heraud, M. Tolino, P. Pfeiffer, J. Riefler and K. Wieczorek for excellent technical assistance in the preparation of neuronal cultures; B. Ritschka and P. Pfeiffer for assistance in experiments; D. Bredt, K. Czaplinski, J. Deshler, W. Sieghart, R. Singer, O. Steward and F. Tübing for reagents; G. Ammer, C. Cowan, J. Heraud and S. Hüttelmaier for insightful comments; and S. Butter for assistance in figure preparation. This work was supported by the Austrian Science Fund (FWF; SFB F4314-B09, P20583-B12, I127-B12), the European Science Foundation Program RNAQuality and a Human Frontier Science Program network (all to M.A.K.), and a Hochschuljubiläumsfonds (to G.V.).
Footnotes
The authors declare that they have no conflict of interest.
References
- Anderson P, Kedersha N (2006) RNA granules. J Cell Biol 172: 803–808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertrand E, Chartrand P, Schaefer M, Shenoy SM, Singer RH, Long RM (1998) Localization of ASH1 mRNA particles in living yeast. Mol Cell 2: 437–445 [DOI] [PubMed] [Google Scholar]
- Besse F, Lopez de Quinto S, Marchand V, Trucco A, Ephrussi A (2009) Drosophila PTB promotes formation of high-order RNP particles and represses oskar translation. Genes Dev 23: 195–207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chekulaeva M, Hentze MW, Ephrussi A (2006) Bruno acts as a dual repressor of oskar translation, promoting mRNA oligomerization and formation of silencing particles. Cell 124: 521–533 [DOI] [PubMed] [Google Scholar]
- Femino AM, Fay FS, Fogarty K, Singer RH (1998) Visualization of single RNA transcripts in situ. Science 280: 585–590 [DOI] [PubMed] [Google Scholar]
- Gao Y, Tatavarty V, Korza G, Levin MK, Carson JH (2008) Multiplexed dendritic targeting of alpha calcium calmodulin-dependent protein kinase II, neurogranin, and activity-regulated cytoskeleton-associated protein RNAs by the A2 pathway. Mol Biol Cell 19: 2311–2327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goetze B, Tuebing F, Xie Y, Dorostkar MM, Thomas S, Pehl U, Boehm S, Macchi P, Kiebler MA (2006) The brain-specific double-stranded RNA-binding protein Staufen2 is required for dendritic spine morphogenesis. J Cell Biol 172: 221–231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holt CE, Bullock SL (2009) Subcellular mRNA localization in animal cells and why it matters. Science 326: 1212–1216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanai Y, Dohmae N, Hirokawa N (2004) Kinesin transports RNA: isolation and characterization of an RNA-transporting granule. Neuron 43: 513–525 [DOI] [PubMed] [Google Scholar]
- Kiebler MA, Bassell GJ (2006) Neuronal RNA granules: movers and makers. Neuron 51: 685–690 [DOI] [PubMed] [Google Scholar]
- Krichevsky AM, Kosik KS (2001) Neuronal RNA granules: a link between RNA localization and stimulation-dependent translation. Neuron 32: 683–696 [DOI] [PubMed] [Google Scholar]
- Larson DR, Singer RH, Zenklusen D (2009) A single molecule view of gene expression. Trends Cell Biol 19: 630–637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lebeau G, Miller LC, Tartas M, McAdam R, Laplante I, Badeaux F, Desgroseillers L, Sossin WS, Lacaille JC (2011) Staufen 2 regulates mGluR long-term depression and Map1b mRNA distribution in hippocampal neurons. Learn Mem 18: 314–326 [DOI] [PubMed] [Google Scholar]
- Maher-Laporte M, DesGroseillers L (2010) Genome wide identification of Staufen2-bound mRNAs in embryonic rat brains. BMB Rep 43: 344–348 [DOI] [PubMed] [Google Scholar]
- Martin KC, Ephrussi A (2009) mRNA localization: gene expression in the spatial dimension. Cell 136: 719–730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raj A, Peskin CS, Tranchina D, Vargas DY, Tyagi S (2006) Stochastic mRNA synthesis in mammalian cells. PLoS Biol 4: e309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shan J, Munro TP, Barbarese E, Carson JH, Smith R (2003) A molecular mechanism for mRNA trafficking in neuronal dendrites. J Neurosci 23: 8859–8866 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tübing F, Vendra G, Mikl M, Macchi P, Thomas S, Kiebler M (2010) Dendritically localized transcripts are sorted into distinct RNPs that display fast directional motility along dendrites of hippocampal neurons. J Neurosci 30: 4160–4170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zenklusen D, Larson DR, Singer RH (2008) Single-RNA counting reveals alternative modes of gene expression in yeast. Nat Struct Mol Biol 15: 1263–1271 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.