Abstract
The nicotinamide adenine dinucleotide (NAD+)-dependent deacetylase SIRT1 is a major metabolic regulator activated by energy stresses such as fasting or calorie restriction. SIRT1 activation during fasting not only relies on the increase in the NAD+/NADH ratio caused by energy deprivation but also involves an upregulation of SIRT1 mRNA and protein levels in various metabolic tissues. We demonstrate that SIRT1 expression is controlled systemically by the activation of the cyclic AMP response-element-binding protein upon low nutrient availability. Conversely, in the absence of energetic stress, the carbohydrate response-element-binding protein represses the expression of SIRT1. Altogether, these results demonstrate that SIRT1 expression is tightly controlled at the transcriptional level by nutrient availability and further underscore that SIRT1 is a crucial metabolic checkpoint connecting the energetic status with transcriptional programmes.
Keywords: SIRT1, CREB, ChREBP, glucagon, nutrient availability
Introduction
SIRT1 is a nicotinamide adenine dinucleotide (NAD+)-dependent deacetylase that regulates important metabolic processes such as lipid oxidation, mitochondrial activity, cholesterol homeostasis and gluconeogenesis (Schwer & Verdin, 2008; Yu & Auwerx, 2010), through the deacetylation of key metabolic enzymes, and the coordination of gene-expression programmes, through the deacetylation of transcriptional regulators (Feige & Auwerx, 2008). SIRT1 activity is stimulated by energy stresses, such as fasting or calorie restriction, when NAD+ levels are high (Cohen et al, 2004; Rodgers et al, 2005; Canto et al, 2010). In addition to the role that NAD+ might have on intrinsic SIRT1 activity (Houtkooper et al, 2010), SIRT1 function can also be increased simply by upregulating its expression levels (Rodgers et al, 2005). Although some mechanisms have been characterized in cellular models such as the activation of SIRT1 expression through a derepression mediated by the hypermethylated in cancer (HIC1)–C-terminal binding protein (CtBP) complex (Zhang et al, 2007), the events underlying the regulation of SIRT1 expression in response to metabolic status in vivo remain largely unknown.
In response to low nutrient availability, glucagon promotes glucose production through protein-kinase-A-mediated activation of the cyclic AMP (cAMP) response-element-binding protein (CREB; Montminy et al, 2004). Conversely, when energy levels rise on food consumption, the carbohydrate response-element-binding protein (ChREBP) orchestrates the transcriptional response to nutrient availability that shifts metabolism towards energy utilization and storage (Uyeda & Repa, 2006).
In this study, we explored the mechanisms governing SIRT1 expression in response to metabolic status and demonstrated that SIRT1 levels are transcriptionally regulated by nutrient availability through opposite actions mediated by CREB and ChREBP. These data establish that SIRT1 expression is modulated according to energetic needs.
Results And Discussion
CREB induces SIRT1 expression in response to fasting
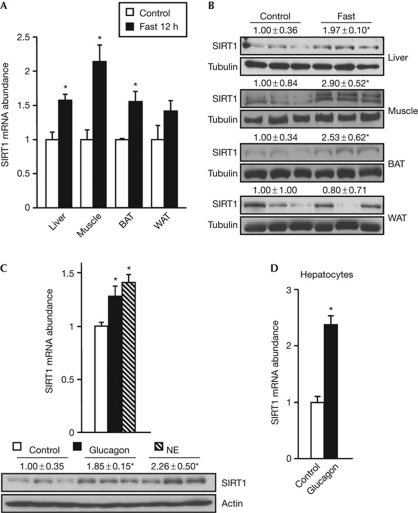
To investigate how physiological changes in nutrient availability affect SIRT1 expression, we measured SIRT1 expression levels in various tissues from either fed or fasted mice. SIRT1 mRNA abundance (Fig 1A) and protein levels (Fig 1B) were significantly increased in the liver, muscle and brown adipose tissue of fasted mice. Interestingly, the increase in the liver SIRT1 mRNA reached its maximum at 18 h and returned to basal levels after 24 h fasting (supplementary Fig S1A online). As metabolic adaptation to fasting is systemically regulated by the release of glucagon and norepinephrine in the blood, we analysed whether these hormones could recapitulate the action of fasting on SIRT1 expression. Norepinephrine administration significantly increased SIRT1 mRNA abundance in the liver, muscle, and brown and white adipose tissues, whereas the action of glucagon on SIRT1 expression was restricted to tissues such as the liver and brown adipose tissue (Fig 1C; supplementary Fig S1B online), where glucagon signalling is active (Christophe, 1996). In line with the increase in mRNA levels, glucagon and norepinephrine increased SIRT1 protein content in the liver (Fig 1C). Importantly, the regulation of SIRT1 expression by glucagon was also evident in primary hepatocytes (Fig 1D). All these results indicate that SIRT1 expression is stimulated at the transcriptional level as a response to humoral factors released during fasting, suggesting an alternative mechanism to those previously described, such as the miR-34a-mediated inhibition of SIRT1 expression (Yamakuchi et al, 2008) and the Jun amino-terminal kinase-2-mediated regulation of SIRT1 protein stability (Ford et al, 2008).
Figure 1.
SIRT1 expression is induced by low nutrient availability and in response to humoral factors released during fasting. (A) SIRT1 mRNA levels in the liver, gastrocnemius muscle, BAT and epididymal WAT of mice fasted for 12 h. The control group had free access to food (n=4). (B) SIRT1 protein levels in the liver, gastrocnemius, BAT and epididymal WAT of control or 24 h-fasted mice. (C) SIRT1 mRNA and protein levels in the liver of mice 1 h after the intraperitoneal administration of PBS (control), glucagon (50 μg/kg) or NE (1 mg/kg; n=5). (D) Quantitative PCR analysis of SIRT1 mRNA levels in primary mouse hepatocytes stimulated for 2 h with 100 nM glucagon (n=4). Protein levels were measured by western blot using tubulin or actin as loading control. Values are presented as the average±s.e.m. and asterisk indicates a statistical difference compared with control (P-value <0.05). BAT, brown adipose tissue; NE, norepinephrine; WAT, white adipose tissue.
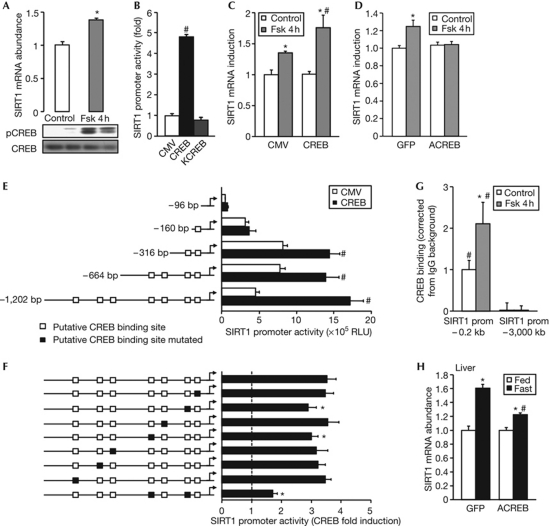
Next, we investigated the molecular mechanisms by which fasting-related hormones promote SIRT1 expression. In the liver, glucagon and norepinephrine promote gluconeogenesis by activating CREB (supplementary Fig S2 online; Montminy et al, 2004). Stimulation of cultured hepatic cells with forskolin, which specifically increases cAMP and activates CREB, was sufficient to increase SIRT1 mRNA abundance (Fig 2A), suggesting that the actions of glucagon and norepinephrine might converge on SIRT1 expression at the level of the cAMP–CREB axis. By analysing the SIRT1 promoter, we identified seven putative half CREB-binding sites in the proximal promoter region (supplementary Fig S3 online), indicating that CREB could possibly bind to the SIRT1 promoter to regulate its transcription. To test this hypothesis, a 1.2-kb fragment of the proximal human SIRT1 promoter was co-transfected with wild-type CREB or the inactive KCREB variant in HepG2 cells. SIRT1 promoter activity was robustly induced by CREB, but not by KCREB (Fig 2B). In addition, the induction of endogenous SIRT1 mRNA expression by forskolin was further enhanced by overexpression of CREB (Fig 2C), but totally blunted by the dominant-negative ACREB form (Fig 2D). Serial deletions of the SIRT1 promoter revealed that the stimulatory effect of CREB on SIRT1 expression required at least the first 316 bp of the proximal SIRT1 promoter region (Fig 2E). Individual single-point mutation of the potential CREB-binding sites located at −175 and −330 bp modestly reduced the CREB-mediated induction of SIRT1 promoter activity (Fig 2F). However, a double mutant of these two sites significantly decreased the activity of SIRT1 promoter (Fig 2F), confirming the relevance of these binding sites for CREB-mediated induction of the SIRT1 promoter and indicating that CREB binds to several CREB-binding sites on the SIRT1 promoter. To confirm further that CREB directly targets the SIRT1 promoter, we performed chromatin immunoprecipitation (ChIP) experiments. In the absence of stimulation, CREB binding could be detected in the proximal region of the SIRT1 promoter, but not in the distal region devoid of potential CREB-binding sites (Fig 2G). Moreover, forskolin treatment robustly stimulated CREB binding to the proximal SIRT1 promoter (Fig 2G). Finally, the induction of hepatic SIRT1 expression in response to fasting was impaired in mice infected intravenously with an adenovirus expressing ACREB (Fig 2H), providing in vivo evidence that fasting regulates SIRT1 expression through CREB activation. Our findings differ from previous reports of an increase in SIRT1 protein levels, but not in SIRT1 mRNA (Rodgers et al, 2005; Kanfi et al, 2008). This difference is probably because of the use of different cell lines and the time of harvesting; the latter being particularly important during an in vivo situation, as we have observed that the induction of SIRT1 mRNA peaks between 12 and 18 h fasting but returns to normal levels at 24 h (supplementary Fig S1A online). Increased SIRT1 levels in liver can participate in the activation of gluconeogenic genes through the deacetylation of several transcriptional regulators, such as peroxisome proliferator-activated receptor gamma coactivator 1α (PGC1α; Rodgers et al, 2005) and the FOXO family of transcription factors (Brunet et al, 2004). Consequently, the induction of SIRT1 expression levels by CREB might constitute a primordial step of a physiological amplification loop in the hepatic fasting response. Interestingly, prolonged stimulation of SIRT1 expression might itself tone down the gluconeogenic programme through the deacetylation and inhibition of CREB-regulated transcription coactivator 2 and thereby favour energy-sparing processes such as ketogenesis (Liu et al, 2008).
Figure 2.
SIRT1 expression is transcriptionally activated by CREB. (A) SIRT1 mRNA in HepG2 cells stimulated for 4 h with 10 μM Fsk (n=4). pCREB and CREB protein levels were analysed by western blot analysis. (B) HepG2 cells were transfected with a SIRT1 promoter luciferase reporter, a β-galactosidase normalization plasmid and 10 ng of pCMV–CREB or pCMV–KCREB. Luciferase activity was measured 48 h after transfection and values were normalized to β-galactosidase levels (n=4). Values are presented as fold increase over the empty vector (CMV; n=4). SIRT1 mRNA levels in (C) HepG2 cells transfected with pCMV–CREB or an empty vector (CMV; n=4) and (D) HepG2 infected with adenoviruses encoding GFP or the dominant-negative ACREB (n=6), after 4 h stimulation with 10 μM Fsk. Data are relative to control group. HepG2 cells were transfected with either (E) 10 ng of pCMV–CREB and serial deletions of the SIRT1 promoter luciferase reporter or (F) the SIRT1 promoter luciferase reporter containing site-directed mutagenesis of the indicated putative CREB-binding site. Luciferase activity was measured 48 h after transfection. (G) HepG2 cells were treated for 4 h with Fsk, and ChIP was performed using an IgG control or a CREB-specific antibody. Recruitment of CREB to the SIRT1 promoter was measured by qPCR using primers against the proximal −0.2 kb and the distal −3 kb promoter, and corrected for background measured in IgG immunoprecipitates (n=4). (H) SIRT1 mRNA abundance in liver from fed or 18 h-fasted mice infected with adenovirus expressing GFP or ACREB. Values are presented as the average±s.e.m., asterisk indicates statistical difference compared with control (A,C,D), WT SIRT1 promoter luciferase reporter (F) or fed group (G) and hash symbol indicates statistical difference compared with empty vector (B,E), distal promoter (G) or GFP (H) at P<0.05. ChIP, chromatin immunoprecipitation; CMV, cytomegalovirus; CREB, cyclic AMP response-element-binding protein; Fsk, forskolin; GFP, green fluorescent protein; IgG, immunoglobulin G; pCMV, CMV promoter; pCREB, CREB phosphorylation; prom, promoter; qPCR, quantitative PCR; RLU, relative luciferase units; WT, wild type.
ChREBP represses SIRT1 transcription during feeding
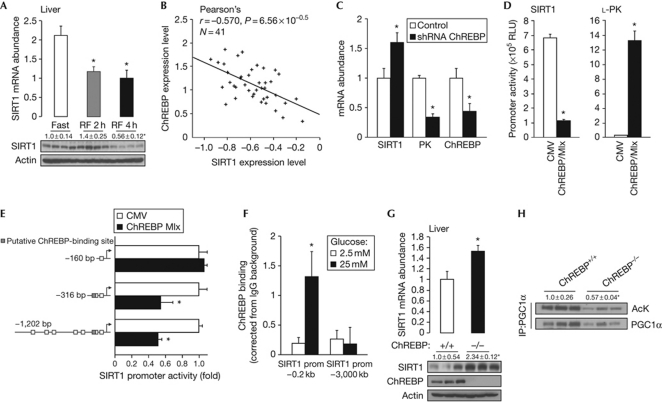
Oppositely to the activation of SIRT1 expression by fasting, SIRT1 mRNA and protein levels rapidly decayed after re-feeding, down to basal levels (Fig 3A; supplementary Fig S4A online), suggesting the existence of a mechanism that actively represses SIRT1 transcription when nutrients become available. Given the role of ChREBP in the hepatic transcriptional response to nutrient availability (Uyeda & Repa, 2006), we speculated that ChREBP could be a negative regulator of SIRT1 expression. Reinforcing this possibility, we identified a conserved binding site for ChREBP in the human SIRT1 promoter (supplementary Fig S3 online). Furthermore, ChREBP and SIRT1 mRNA expression were inversely correlated in the livers of 41 recombinant inbred mouse strains of the BxD genetic reference population (Fig 3B; http://www.genenetwork.org/). The inverse correlation between ChREBP and SIRT1 mRNA levels was further corroborated by the use of a short hairpin RNA against ChREBP in HepG2 cells, which significantly increased SIRT1 mRNA expression while decreasing that of L-pyruvate kinase, a well-known target induced by ChREBP (Fig 3C). Further consolidating the hypothesis that ChREBP inhibits SIRT1 transcription was the fact that ChREBP robustly repressed SIRT1 promoter activity when co-transfected with its functional partner Max-like protein (Mlx) in HepG2 cells, whereas it significantly increased L-pyruvate kinase promoter activity (Fig 3D). Moreover, SIRT1 promoter activity was stimulated when ChREBP was co-transfected with a dominant-negative form of Mlx that blocks the binding of ChREBP to DNA (supplementary Fig S4B online). Interestingly, ChREBP-meditated repression of the SIRT1 promoter was lost in a −160-bp fragment of the SIRT1 promoter devoid of the putative ChREB-binding site (Fig 3E). To evaluate whether ChREBP repressed SIRT1 expression by directly binding to its promoter, we performed ChIP analysis in HepG2 cells in which ChREBP was activated by incubation in high glucose. ChREBP binding was significantly enhanced in the proximal region of the SIRT1 promoter (Fig 3F). However, no ChREBP binding was observed in a more distal region of the SIRT1 promoter. In addition, SIRT1 mRNA and protein levels were higher in the livers of ChREBP−/− mice (Fig 3G), providing evidence that ChREBP functions as a repressor of SIRT1 expression in vivo. Finally, the deacetylase activity of SIRT1 was enhanced in the absence of ChREBP, as the acetylation levels of PGC1α, a target of SIRT1, were decreased in the livers of ChREBP−/− mice (Fig 3H). Altogether, these results demonstrate that ChREBP represses SIRT1 expression and activity when nutrients are available. Although the exact mechanism by which ChREBP represses genes is poorly defined at present, the repressive activity of ChREBP has already been observed in several situations, such as the regulation of COUP-TFII and ARNT/HIF-1β (Perilhou et al, 2008; Noordeen et al, 2009).
Figure 3.
ChREBP transcriptionally represses SIRT1 expression. (A) SIRT1 mRNA and protein levels in the liver of mice fasted for 24 h or refed for 2 or 4 h. (B) Correlation plots for the liver mRNA expression of SIRT1 and ChREBP in the mouse BxD genetic reference population. (C) SIRT1, L-PK and ChREBP mRNA levels in HepG2 cells transfected with a ChREBP shRNA (n=4). (D) HepG2 cells were transfected with a SIRT1 or L-PK promoter luciferase reporter, β-galactosidase, and 100 ng of an empty vector (CMV) or 50 ng of ChREBP and 50 ng of Mlx expression vectors. Luciferase activity was measured as described (n=4). (E) HepG2 cells were transfected with −1,202, −316 or −160 bp fragment of the SIRT1 promoter luciferase reporter and ChREBP and Mlx plasmid. Luciferase activity was measured 48 h after transfection and values were normalized to total protein (n=6). Values are presented as fold increase over the empty vector (CMV). (F) HepG2 cells were incubated 20 h with 2.5 or 25 mM glucose; further, ChIP was performed using an IgG control or a ChREBP-specific antibody. Recruitment of ChREBP to the SIRT1 promoter was measured as described (n=4). (G) SIRT1 mRNA and protein levels in liver lysates of fed ChREBP+/+ or ChREBP−/− mice. Actin was used as loading control. (H) PGC1α acetylation levels were examined in liver from fed ChREBP+/+ or ChREBP−/− mice after immunoprecipitation. The quantification represents the ratio of acetylated to total PGC1α. Values are the fold induction over control/empty vector and are presented as the average±s.e.m., and asterisk indicates a statistical difference compared with fast/control/empty vector/ChREBP+/+ or 2.5 mM glucose at P<0.05. Ack, acetylated lysine; ChIP, chromatin immunoprecipitation; ChREBP, carbohydrate response-element-binding protein; CMV, cytomegalovirus; IP-PGC1α, immunoprecipitated PGC1α; IgG, immunoglobulin G; PGC1α, peroxisome proliferator-activated receptor gamma coactivator 1α; PK, pyruvate kinase; prom, promoter; RF, refed; RLU, relative luciferase units; shRNA, short hairpin RNA.
Interdependent regulation of SIRT1 by CREB and ChREBP
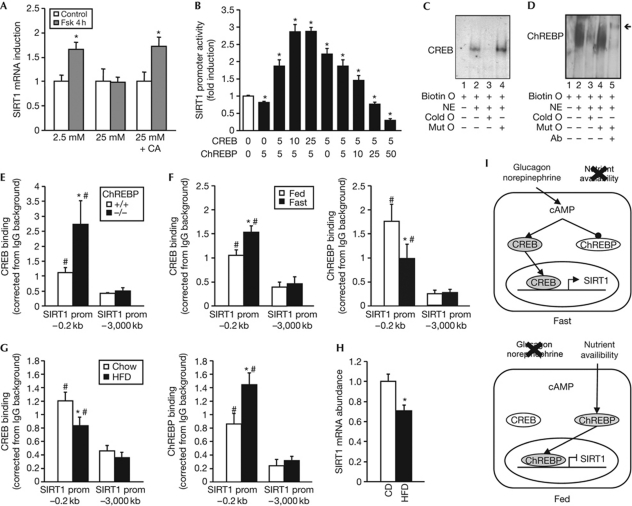
We next evaluated how the regulation of SIRT1 expression by CREB and ChREBP is inter-regulated according to the physiological status. Interestingly, the induction of SIRT1 expression by forskolin was blunted when HepG2 cells were incubated in high glucose to promote the nuclear translocation and activation of ChREBP (Fig 4A). When the nuclear translocation of ChREBP was blocked with cantharidic acid (supplementary Fig S5 online; Kawaguchi et al, 2001), the induction of SIRT1 by forskolin was restored despite the high-glucose medium (Fig 4A), suggesting that ChREBP present in the nucleus under high-glucose conditions can compete with CREB on the SIRT1 promoter. As both ChREBP and CREB bind to the proximal region of the SIRT1 promoter, we hypothesized that ChREBP could potentially compete with CREB and therefore interfere with the CREB-mediated activation of the SIRT1 promoter. Consistent with this hypothesis, transient transfection assays in HepG2 cells showed that the ChREBP-mediated repression of the SIRT1 promoter activity was completely overcome by increasing amounts of CREB (Fig 4B). Conversely, increasing amounts of ChREBP attenuated the CREB-mediated activation of the SIRT1 promoter (Fig 4B). To assess whether the competition between CREB and ChREBP results from binding to an overlapping binding site, we performed an electrophoretic mobility shift assay with a 25-bp oligonucleotide containing the region of the SIRT1 promoter with overlapping putative CREB- and ChREBP-binding sites (supplementary Fig S3 online). A slower-migrating complex was observed when the oligonucleotide was incubated in the presence of nuclear extracts from CHO cells transfected with CREB (Fig 4C). Addition of an excess of cold oligonucleotide competed out the labelled probe, whereas an oligonucleotide carrying a mutation of the core-binding sequence did not influence the shift of the oligonucleotide, demonstrating the specificity of this binding. Similar results were obtained using nuclear extracts from CHO cells expressing FLAG-tagged ChREBP and Mlx (Fig 4D). Additional proof of the presence of ChREBP in the retarded complex was provided by the presence of a supershifted band using a FLAG antibody. Together, these results demonstrate that CREB and ChREBP are able to bind to the same sequence of the SIRT1 promoter. Importantly, the competition between CREB and ChREBP also occurs in vivo as the binding of CREB to the SIRT1 promoter evaluated by ChIP was significantly enhanced in the liver of ChREBP−/− mice (Fig 4E). We then analysed how the binding of CREB and ChREBP to the SIRT1 promoter was regulated in vivo upon various metabolic challenges. Consistent with the activation of CREB and inhibition of ChREBP in response to fasting, CREB recruitment to the proximal SIRT1 promoter was significantly enhanced during fasting, whereas that of ChREBP was reduced (Fig 4F). Conversely, nutrient excess induced by feeding mice with a high-fat diet decreased CREB recruitment and increased the binding of ChREBP to the SIRT1 promoter (Fig 4G), resulting in a reduction of SIRT1 expression (Fig 4H). Altogether, these results demonstrate that an interchange between CREB and ChREBP on the SIRT1 promoter regulates SIRT1 expression under both conditions of nutrient shortage and excess (Fig 4I). The interdependent connection between ChREBP and CREB suggests that the regulation of SIRT1 expression by these two transcription factors take place in two layers. First, during situations of nutrient stress such as fasting, protein kinase A activity is high and leads to both ChREBP nuclear export (Denechaud et al, 2008) and CREB nuclear import by phosphorylation. Second, the subsequent nuclear increase of activated CREB helps to compete the residual ChREBP away from the SIRT1 promoter, leading to SIRT1 transcription. In the opposite scenario, when nutrients are available, ChREBP is imported to the nucleus and CREB shuttled out, which leads to ChREBP binding to the SIRT1 promoter and SIRT1 transcriptional repression. ChREBP-mediated repression of SIRT1 transcription could have a major physiological impact, as it represents a mechanism to blunt gluconeogenesis and other energy-producing processes when nutrients are abundant. In line with this, livers from ChREBP−/− mice show major alterations in energy substrate utilization (Burgess et al, 2008), which probably derive from the inflexibility of these livers to adapt to nutrient availability. Finally, a recent report demonstrated that mice carrying a liver-specific SIRT1 null mutation showed increased ChREBP expression and liver steatosis (Wang et al, 2010), suggesting a potential feedback regulation loop in which SIRT1 might inhibit ChREBP by decreasing its expression.
Figure 4.
The regulation of SIRT1 by CREB and ChREBP is interdependent and coordinated by energy availability. (A) HepG2 cells were stimulated for 4 h with 10 μM forskolin in 2.5 mM or 25 mM glucose. A third group was treated with CA 1 h before treatment of forskolin in 25 mM glucose. SIRT1 mRNA levels were then measured by qPCR. Data are relative to the control group. (B) HepG2 cells were transfected with the SIRT1 promoter luciferase reporter and with the indicated amount (ng) of pCMV–CREB and ChREBP/Mlx. Luciferase activity was measured as described (n=6). (C) EMSA was performed using nuclear extracts from CHO cells transfected with CREB and a 25-bp biotinylated oligonucleotide with the overlapping core sequence for CREB and ChREBP. (D) EMSA was performed as in C, using nuclear extracts from CHO cells transfected with Flag-tagged ChREBP and its partner Mlx. Arrow indicates the supershifted band. (E) ChIP was performed in the liver from fed ChREBP+/+ or ChREBP−/− mice using an IgG control or CREB-specific antibody. (F) ChIP was performed in the liver from fed or 18 h-fasted mice using an IgG control, CREB- or ChREBP-specific antibody. (G) ChIP was performed in the liver from mice fed chow or HFD using an IgG control or CREB- or ChREBP-specific antibody as described previously. (H) SIRT1 mRNA abundance in the liver of mice fed chow or HFD. Values are presented as the average±s.e.m., asterisk indicates a statistical difference compared with control/empty vector/ChREBP+/+/FED or chow, and hash symbol indicates statistical difference compared with distal promoter at P<0.05. (I) Scheme illustrating how nutrient availability is integrated by CREB and ChREBP to coordinately regulate SIRT1 expression. During fasting, glucagon and norepinephrine lead to an increase in cAMP and PKA activity, in turn leading to CREB activation and ChREBP inactivation. CREB then induces SIRT1 expression. Conversely, during the fed state, ChREBP binds to the SIRT1 promoter to downregulate its expression, potentially through competition with CREB binding (bottom panel). Ab, flag-specific antibody; biotin O, biotinylated oligonucleotide; CA, cantharidic acid; cAMP, cyclic AMP; ChIP, chromatin immunoprecipitation; ChREBP, carbohydrate response-element-binding protein; cold O, cold oligonucleotide excess; CREB, cAMP response-element-binding protein; EMSA, electrophoretic mobility shift assay; Fsk, forskolin; HFD, high-fat diet; IgG, immunoglobulin G; mut O, cold mutant oligonucleotide excess; NE, nuclear extract; pCMV, CMV promoter; PKA, protein kinase A; prom, promoter; qPCR, quantitative PCR.
Altogether, our results demonstrate that the SIRT1 promoter is responsive to nutrient availability through a CREB–ChREBP interchange in promoter occupancy. SIRT1 gene expression is therefore finely tuned to hormonal inputs and glucose availability. The fact that both CREB and ChREBP divergently regulate SIRT1 expression further underscores the crucial role of SIRT1 as a metabolic mediator linking environmental cues to metabolic adaptations.
Methods
Plasmids, adenovirus and reagents. Complete information about plasmids, adenovirus and reagents is provided in the supplementary information online.
Animal experiments. Animal experiments were conducted in accordance with Swiss, French and EU ethical laws and standard operating procedures were followed as described previously (Champy et al, 2004, 2008). Seven-week-old C57Bl6/J male mice were obtained from Charles River Laboratories and maintained in a 12 h light–dark cycle with unrestricted access to regular diet and water. Animals were fasted for 12, 18 or 24 h starting 3 h after the beginning of the dark cycle, with free access to water. Another group was re-fed with chow diet (2018S, Harlan Teklad) for 2 and 4 h after 12 h fasting. Mice were injected intraperitoneally with glucagon (50 μg/kg body weight), norepinephrine (1 mg/kg) or PBS. For virus injections, animals were anaesthetized with isoflurane and a total of 1 × 109 plaque-forming units per recombinant virus were administered by a systemic jugular vein injection. In each experiment, at least six animals received the same treatment. Liver, muscle, brown and white adipose tissues were immediately frozen in liquid nitrogen for subsequent RNA and protein isolation. ChREBP knockout animals were described previously (Iizuka et al, 2004). For animal experiments, we used 6–8 animals per group. In cases in which we used fewer, the experiment was repeated twice.
Cell culture and treatments. HepG2 cells were cultured in DMEM supplemented with 10% fetal bovine serum. For experiments using forskolin, cells were depleted of serum for 12 h and then stimulated with 10 μM forskolin for 4 h. Unless otherwise stated, the glucose concentration of DMEM was 5 mM. Primary mouse hepatocytes were prepared from 7-week-old C57Bl6/J male mice as described previously (Berry & Friend, 1969). Cells were maintained overnight in serum-free Williams’ E medium and then exposed to 100 nM glucagon for 2 h.
Other experimental procedures. Genenetwork data, Genomatix analysis, gene expression, protein analysis, transfection, ChIP, electrophoretic mobility shift assays and adenoviral infection of cells were performed as described in supplementary information online.
Statistical analyses. All data are reported as mean±s.e.m. Results were considered statistically significant when P-values were <0.05 using an unpaired two-tailed Student's t-test.
Supplementary information is available at EMBO reports online (http://www.emboreports.org).
Supplementary Material
Acknowledgments
We thank Charles Vinson for the gift of the ACREB plasmid, Howard Towle for the ChREBP and Mlx constructs, and Kosaku Uyeda for the ChREBP knockout mice and the members of the Auwerx laboratory for discussion and technical help. Work in the authors’ laboratory was supported by grants from the Ecole Polytechnique Fédérale de Lausanne (EPFL), Swiss National Science Foundation, EU (ERC-Ideas–Sirtuins-231138 for J.A.) and National Institutes of Health (NIH/NIDDK grant DK 59820 to J.A., DK43051 to B.B.K. and K08DK076726 to M.A.H.). L.G.N., J.N.F., C.C. and H.Y. were supported by fellowships from Consejo Ñacional de Ciencia y Tecnología (CONACYT), Federation of European Biochemical Societies (FEBS), EMBO and Fondation pour la Recherche Médicale (FRM).
Footnotes
The authors declare that they have no conflict of interest.
References
- Berry MN, Friend DS (1969) High-yield preparation of isolated rat liver parenchymal cells: a biochemical and fine structural study. J Cell Biol 43: 506–520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brunet A et al. (2004) Stress-dependent regulation of FOXO transcription factors by the SIRT1 deacetylase. Science 303: 2011–2015 [DOI] [PubMed] [Google Scholar]
- Burgess SC, Iizuka K, Jeoung NH, Harris RA, Kashiwaya Y, Veech RL, Kitazume T, Uyeda K (2008) Carbohydrate-response element-binding protein deletion alters substrate utilization producing an energy-deficient liver. J Biol Chem 283: 1670–1678 [DOI] [PubMed] [Google Scholar]
- Canto C, Jiang LQ, Deshmukh AS, Mataki C, Coste A, Lagouge M, Zierath JR, Auwerx J (2010) Interdependence of AMPK and SIRT1 for metabolic adaptation to fasting and exercise in skeletal muscle. Cell Metab 11: 213–219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Champy MF, Selloum M, Piard L, Zeitler V, Caradec C, Chambon P, Auwerx J (2004) Mouse functional genomics requires standardization of mouse handling and housing conditions. Mamm Genome 15: 768–783 [DOI] [PubMed] [Google Scholar]
- Champy MF, Selloum M, Zeitler V, Caradec C, Jung B, Rousseau S, Pouilly L, Sorg T, Auwerx J (2008) Genetic background determines metabolic phenotypes in the mouse. Mamm Genome 19: 318–331 [DOI] [PubMed] [Google Scholar]
- Christophe J (1996) Glucagon and its receptor in various tissues. Ann NY Acad Sci 805: 31–42; discussion 42–33 [DOI] [PubMed] [Google Scholar]
- Cohen HY, Miller C, Bitterman KJ, Wall NR, Hekking B, Kessler B, Howitz KT, Gorospe M, de Cabo R, Sinclair DA (2004) Calorie restriction promotes mammalian cell survival by inducing the SIRT1 deacetylase. Science 305: 390–392 [DOI] [PubMed] [Google Scholar]
- Denechaud PD, Bossard P, Lobaccaro JM, Millatt L, Staels B, Girard J, Postic C (2008) ChREBP, but not LXRs, is required for the induction of glucose-regulated genes in mouse liver. J Clin Invest 118: 956–964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feige JN, Auwerx J (2008) Transcriptional targets of sirtuins in the coordination of mammalian physiology. Curr Opin Cell Biol 20: 303–309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ford J, Ahmed S, Allison S, Jiang M, Milner J (2008) JNK2-dependent regulation of SIRT1 protein stability. Cell Cycle 7: 3091–3097 [DOI] [PubMed] [Google Scholar]
- Houtkooper RH, Canto C, Wanders RJ, Auwerx J (2010) The secret life of NAD+: an old metabolite controlling new metabolic signaling pathways. Endocr Rev 31: 194–223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iizuka K, Bruick RK, Liang G, Horton JD, Uyeda K (2004) Deficiency of carbohydrate response element-binding protein (ChREBP) reduces lipogenesis as well as glycolysis. Proc Natl Acad Sci USA 101: 7281–7286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanfi Y, Peshti V, Gozlan YM, Rathaus M, Gil R, Cohen HY (2008) Regulation of SIRT1 protein levels by nutrient availability. FEBS Lett 582: 2417–2423 [DOI] [PubMed] [Google Scholar]
- Kawaguchi T, Takenoshita M, Kabashima T, Uyeda K (2001) Glucose and cAMP regulate the L-type pyruvate kinase gene by phosphorylation/dephosphorylation of the carbohydrate response element binding protein. Proc Natl Acad Sci USA 98: 13710–13715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y et al. (2008) A fasting inducible switch modulates gluconeogenesis via activator/coactivator exchange. Nature 456: 269–273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montminy M, Koo SH, Zhang X (2004) The CREB family: key regulators of hepatic metabolism. Ann Endocrinol (Paris) 65: 73–75 [DOI] [PubMed] [Google Scholar]
- Noordeen NA, Khera TK, Sun G, Longbottom ER, Pullen TJ, da Silva Xavier G, Rutter GA, Leclerc I (2009) ChREBP is a negative regulator of ARNT/HIF-1β gene expression in pancreatic islet β-cells. Diabetes 59: 153–160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perilhou A, Tourrel-Cuzin C, Kharroubi I, Henique C, Fauveau V, Kitamura T, Magnan C, Postic C, Prip-Buus C, Vasseur-Cognet M (2008) The transcription factor COUP-TFII is negatively regulated by insulin and glucose via Foxo1- and ChREBP-controlled pathways. Mol Cell Biol 28: 6568–6579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodgers JT, Lerin C, Haas W, Gygi SP, Spiegelman BM, Puigserver P (2005) Nutrient control of glucose homeostasis through a complex of PGC-1alpha and SIRT1. Nature 434: 113–118 [DOI] [PubMed] [Google Scholar]
- Schwer B, Verdin E (2008) Conserved metabolic regulatory functions of sirtuins. Cell Metab 7: 104–112 [DOI] [PubMed] [Google Scholar]
- Uyeda K, Repa JJ (2006) Carbohydrate response element binding protein, ChREBP, a transcription factor coupling hepatic glucose utilization and lipid synthesis. Cell Metab 4: 107–110 [DOI] [PubMed] [Google Scholar]
- Wang RH, Li C, Deng CX (2010) Liver steatosis and increased ChREBP expression in mice carrying a liver specific SIRT1 null mutation under a normal feeding condition. Int J Biol Sci 6: 682–690 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamakuchi M, Ferlito M, Lowenstein CJ (2008) miR-34a repression of SIRT1 regulates apoptosis. Proc Natl Acad Sci USA 105: 13421–13426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu J, Auwerx J (2010) Protein deacetylation by SIRT1: an emerging key post-translational modification in metabolic regulation. Pharmacol Res 62: 35–41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Q, Wang SY, Fleuriel C, Leprince D, Rocheleau JV, Piston DW, Goodman RH (2007) Metabolic regulation of SIRT1 transcription via a HIC1:CtBP corepressor complex. Proc Natl Acad Sci USA 104: 829–833 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.