Abstract
Exposure of yeast to high osmolarity induces a transient activation of the Hog1 stress-activated protein kinase (SAPK), which is required for cell survival under these conditions. However, sustained activation of the SAPK results in a severe growth defect. We found that prolonged SAPK activation leads to cell death, which is not observed in nma111 cells, by causing accumulation of reactive oxygen species (ROS). Mutations of the SCFCDC4 ubiquitin ligase complex suppress cell death by preventing the degradation of Msn2 and Msn4 transcription factors. Accumulation of Msn2 and Msn4 leads to the induction of PNC1, which is an activator of the Sir2 histone acetylase. Sir2 is involved in protection against Hog1-induced cell death and can suppress Hog1-induced ROS accumulation. Therefore, cell death seems to be dictated by the balance of ROS induced by Hog1 and the protective effect of Sir2.
Keywords: cell death, Hog1, SCF, SAPK, Sir2
Introduction
Mitogen-activated protein kinase pathways convert extracellular stimuli into cellular responses. Mitogen-activated protein kinase activation must be appropriately regulated because the biological outcome is different depending on the intensity and duration of the activation (Marshall, 1995). For instance, transient activation of the p38 stress-activated protein kinase (SAPK) leads to cell proliferation, whereas sustained activation leads to apoptosis-like cell death (Williamson et al, 2004; Wagner & Nebreda, 2009). In yeast, transient activation of Hog1 SAPK is essential for cell adaptation and survival of osmostress and controls from cell-cycle progression to gene expression (Clotet et al, 2006; Chen & Thorner, 2007; Hohmann et al, 2007; de Nadal & Posas, 2010). In contrast to its role in cell survival, sustained activation of the SAPK results in a severe growth defect that is prevented by overexpression of protein tyrosine phosphatase 2 (Maeda et al, 1994; Wurgler-Murphy et al, 1997). However, little is known about the molecular basis of this severe growth defect.
Here, we demonstrate that sustained activation of Hog1 induces cell death by promoting high levels of reactive oxygen species (ROS). This is suppressed by mutations of the SCFCDC4 (SCF; Skp1/Cul1/F-box protein) ubiquitin–ligase complex. Accumulation of Msn2 and Msn4 transcription factors induces PNC1, an activator of the Sir2 histone deacetylase.
Results And Discussion
Sustained activation of Hog1 leads to cell death
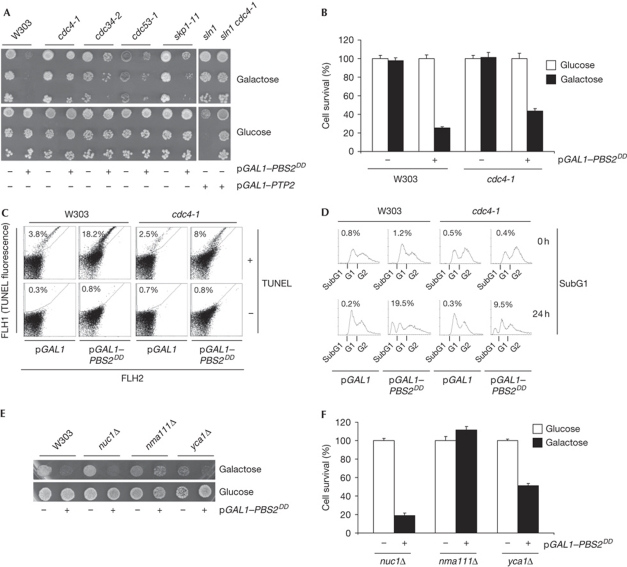
Inactivation of SLN1 or overexpression of PBS2DD impairs cell growth (Maeda et al, 1994; Wurgler-Murphy et al, 1997; Fig 1A). We tested whether this was associated with a decrease of cell survival, by assessing colony-forming units. Only 24% of wild-type cells survived 24 h of PBS2DD expression (Fig 1B). A decrease in cell viability was also observed in a cell permeability assay with propidium iodide (supplementary Fig S1A online).
Figure 1.
Sustained activation of Hog1 causes cell death that is partly suppressed by SCFCDC4 mutations. (A) Mutations on the SCFCDC4 complex prevent cell death induced by PBS2DD or SLN1 inactivation. Cells expressing the PBS2DD allele under the GAL1 promoter (pGAL1–PBS2DD) were spotted on glucose (control) or galactose. The sln1Δ and sln1Δ cdc4-1 strains carrying a plasmid expressing the protein tyrosine phosphatase 2 (PTP2) gene under the GAL1 promoter (pGAL1–PTP2). (B) Hog1-mediated cell death is improved in a cdc4-1 mutant strain. Cells as in A were grown in galactose for 24 h. Viability was monitored by counting the colony-forming units in glucose plates. Data represent the mean and standard deviation of three independent experiments. (C) DNA single-strand breaks caused by Hog1 activation are reduced in cdc4-1 mutant cells. Cells were processed with the TdT-mediated dUTP nick end labeling (TUNEL) assay (+) and the presence of single-strand DNA breaks was detected by FACS analysis. Data shown are representative of three independent experiments. (D) Induction of the appearance of SubG1 population of cells on Hog1 activation is reduced in cdc4-1 mutant cells. Cells were grown as in A and analysed for SubG1 population by FACS analysis. Data shown are representative of three independent experiments. (E) Cell death caused by sustained PBS2DD expression is mediated by Nma111. The indicated strains expressing the PBS2DD allele were grown in glucose (control) or galactose. (F) Cell viability on permanent Hog1 activation is fully restored in the absence of NMA111. Strains as in E were grown in glucose (control) or galactose for 24 h and colony-forming units were assessed in glucose plates. Data represent the mean and standard deviation of three independent experiments. FACS, fluorescence-activated cell sorting.
Activation of p38 has been associated with apoptosis (Dolado & Nebreda, 2008). Yeast can undergo cell death accompanied by features that are diagnostic of apoptosis or programmed cell death (PCD), and it is associated with characteristics of apoptosis (Galluzzi et al, 2009; Madeo et al, 2009). Activation of Hog1 by overexpression of Pbs2DD resulted in 18.2% of TdT-mediated dUTP nick end labelling-positive cells, whereas only 3.8% of cells expressing a control plasmid were positive (Fig 1C). Activation of the SAPK resulted in 19.5% of cells having a SubG1 DNA content, indicating DNA fragmentation (Fig 1D). Correspondingly, sustained activation of Hog1 induced an increase in the number of cells with metacaspase activation in the highly sensitive FAM–FLICA Apoptosis Detection Kit (Immunochemistry technologies) (supplementary Fig S1B online). Thus, several independent assays indicated that activation of the Hog1 SAPK induced cell death. Several genes have been involved as mediators of apoptosis-like cell death in yeast (Carmona-Gutierrez et al, 2010). Deletion of NUC1 did not prevent cell death on Hog1 activation. By contrast, deletion of YCA1 partly suppressed cell death and deletion of NMA111—the Omi/HtrA2 homologue (Walter et al, 2006)—completely abolished it (Fig 1E and F). Thus, Hog1 activation leads to PCD mediated by Nma111.
SCF mutations suppress Hog1-mediated cell death
Permissive mutations of the CDC4 gene—the E3 ligase of the SCFCDC4 complex—suppressed the growth defect associated with HOG hyperactivation (Fig 1A,B; supplementary Fig S1A online). SCFCDC4 is a complex containing Skp1, Cdc53 and Cdc34 proteins. Permissive mutations in any of these genes suppressed the growth defect caused by activation of Hog1 (Fig 1A), although to different extents. The cdc4-1 cells overexpressing PBS2DD showed a survival rate almost two times higher than wild-type cells and reduced apoptosis (Fig 1C and D; supplementary Fig S1B online). Thus, mutations of the SCFCDC4 partly suppress cell death caused by Hog1 activation.
cdc4-1 shows reduced levels of Hog1-induced ROS
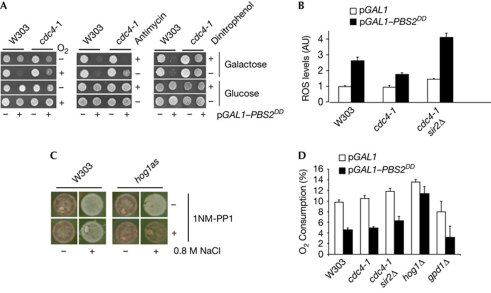
The induction of ROS is the most common cause of apoptosis-like cell death in yeast (Madeo et al, 1999). In the absence of O2, which prevents ROS formation, cell death caused by Hog1 was abolished (Fig 2A, left panel). Antimycin A inhibits respiration and provokes an increase of ROS formation. Interestingly, antimycin A abolished the anti-apoptotic effect of cdc4-1 on Hog1 activation (Fig 2A, middle panel). Incubation of cells with dinitrophenol—which inhibits adenosine triphosphate formation without affecting ROS levels—did not alter the ability of cdc4-1 cells to suppress cell death caused by Hog1 (Fig 2A, right panel). Therefore, cell death caused by Hog1 activation is not due to a deficit of adenosine triphosphate, but is probably due to an increase in ROS formation.
Figure 2.
Hog1-induced reactive oxygen species accumulation is reduced in a cdc4-1 mutant. (A) Reactive oxygen species (ROS) production causes cell death under sustained Hog1 activation. Indicated strains plated on glucose or galactose, and anaerobiosis (left panel) in the presence of antimycin at 2.5 μg/ml (middle panel) or dinitrophenol at 25 μg/ml (right panel). (B) Hog1-induced ROS accumulation is reduced in a cdc4-1 mutant. Strains were grown in galactose for 12 h. Cells were incubated with DCFH-DA for 30 min and ROS was measured. Data represent the mean and standard deviation of three independent experiments. (C) Hog1 inhibits mitochondrial respiration in response to osmostress. Wild-type and hog1as-mutant strains in YPD plates with or without 0.8 M NaCl were grown for 12 h at 25 °C, and tetrazolium chloride was added as an overlay in the presence (+) of the kinase inhibitor 1NM-PP1 (5 μM). (D) Activation of Hog1 causes a reduction in oxygen consumption independently of SIR2, cdc4-1 and GPD1. Oxygen consumption was measured as in B. Data represent the mean and standard deviation of three independent experiments. DCFH-DA, dichloro-dihydro-fluorescein diacetate.
We then quantitatively assessed ROS production. Overexpression of PBS2DD caused a 2.6-fold increase in ROS formation in wild-type cells, whereas it was lower than 1.8-fold in a cdc4-1 mutant (Fig 2B). An increase in ROS levels might be caused by inhibition of mitochondrial respiration. We assessed mitochondrial respiration in a plate assay in the presence of triphenyl tetrazolium (Kobayashi et al, 1974) and found that it was strongly inhibited in reponse to stress (white cells; Fig 2C). Correspondingly, growth on a non-fermentable carbon source was impaired in the presence of osmostress (supplementary Fig S5B online). To establish the role of Hog1 in the inhibition of mitochondrial respiration, we used an analogue-sensitive hog1as strain (Macia et al, 2009) and found that inhibition of hog1as prevented the inhibition of respiration (Fig 2C). Sustained activation of Hog1 by Pbs2DD resulted in a similar reduction of oxygen consumption in wild-type and cdc4-1 mutant, which was dependent on HOG1 (Fig 2D). It is noteworthy that deletion of GPD1 did not prevent the reduction of oxygen consumption in response to Hog1 activation, indicating that glycerol metabolism is not the source of the inhibition of respiration (Fig 2D). Therefore, activation of Hog1 decreases mitochondrial respiration and leads to an increase in ROS formation.
SCFCDC4 restricts Msn2-mediated gene expression
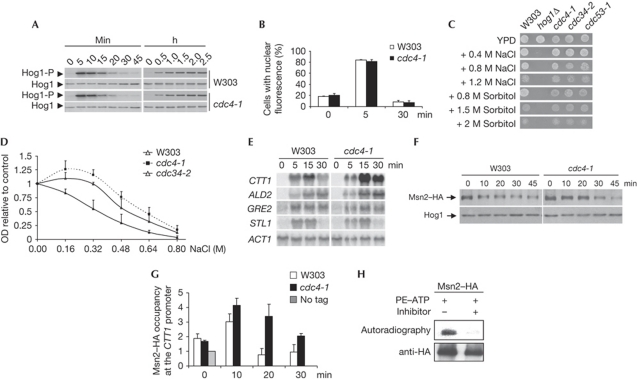
We then assessed HOG signalling. Phosphorylation of Hog1 and its nuclear accumulation were identical in a wild-type and cdc4-1 mutant in response to osmostress or PBS2DD induction (Fig 3A,B). It is noteworthy that mutations in SCFCDC4 (cdc4-1, cdc34-2 and cdc53-1) were slightly more resistant to osmotic stress than wild-type cells, despite normal Hog1 signalling (Fig 3C,D).
Figure 3.
cdc4-1 mutation increases Msn2- and Msn4-dependent gene expression. (A) Hog1 phosphorylation in response to osmostress or PBS2DD induction is not altered in a cdc4-1 mutant. Cells were subjected to osmostress (0.4 M NaCl; left panel). Cells were grown in raffinose for 4 h and then shifted to galactose. Hog1 phosphorylation was followed by using antibodies against phospho-p38 mitogen-activated protein kinase or Hog1. (B) Hog1 nuclear accumulation in response to osmostress is not affected in a cdc4-1 mutant. Cells carrying pRS416–GFP–Hog1 were subjected to osmostress (0.4 M NaCl). Data represent the mean and standard deviation of three independent experiments. (C) SCFCDC4 mutants are slightly osmoresistant in a plate assay. Wild-type and the indicated mutant strains were spotted on YPD plates in the presence of NaCl and sorbitol. (D) SCFCDC4 mutants are slightly osmoresistant in liquid assays. The indicated mutant strains were grown in the presence of NaCl. Data represent the mean and standard deviation of three independent experiments. (E) Mutations in the SCFCDC4 complex increase Msn2- and Msn4-dependent gene expression in response to osmostress. Cells were grown in YPD and treated with 0.4 M NaCl for the indicated times. Total RNA collected was assayed by northern blot analysis for transcript levels of CTT1, ALD3, GRE2, STL1 and ACT1 (as a loading control). (F) Degradation of Msn2 is delayed in a cdc4-1 mutant strain in response to osmostress. Wild-type and cdc4-1 mutant cells carrying a monocopy plasmid with haemagglutinin (HA)-tagged MSN2 under the ADH1 promoter were grown to mid-log phase and treated with 0.4 M NaCl. (G) Recruitment of Msn2 at osmoresponsive promoters is extended in a SCFCDC4 mutant on osmostress. Cells as in F were treated with 0.4 M NaCl. Chromatin immunoprecipitation of Msn2 was assessed by immunoprecipitation with HA antibody. Binding to CTT1 promoter was determined by real-time polymerase chain reaction. Data represent the mean and standard deviation of three independent experiments. (H) Hog1 phosphorylates Msn2 in vitro. Msn2-HA was phosphorylated in an in vitro kinase assay using Hog1as with or without the presence of the kinase inhibitor 1NM-PP1 (5 μM). In both cases, radioactively labelled PE-adenosine triphosphate (ATP) was used. GFP, green fluorescent protein; OD, optical density; PE, phycoerythrin.
The Hog1 SAPK is a key regulator of gene expression by controlling several transcription factors and chromatin-associated proteins (de Nadal & Posas, 2010). The SCFCDC4 complex is involved in control of the turnover of several transcription factors (Pal et al, 2007; Olson et al, 2008). In a cdc4-1 strain, expression of genes under the control of Msn2 and Msn4 transcription factors, but not Sko1 or Hot1 (that is, CTT1 and ALD3 compared with GRE2 and STL1), was stronger than that in wild-type cells (Fig 3E). Therefore, these data indicate that Msn2 and Msn4 might be the target for SCFCDC4. It is noteworthy that Msn2 is ubiquitinated in vitro by Cdc4 (Chi et al, 2001).
Degradation of Msn2 in response to osmostress and by overexpression of PBS2DD was slower in a cdc4-1 strain than in the wild type (Fig 3F; supplementary Fig S2 online). Correspondingly, Msn2 occupancy at the CTT1 promoter, analysed by chromatin immunoprecipitation (ChIP), was extended in a cdc4-1 strain in response to stress (Fig 3G). It is noteworthy that Msn2 was phosphorylated by Hog1 in an in vitro assay (Fig 3H). Thus, cdc4-1 cells have an increased amount of Msn2 at stress-responsive promoters, which leads to an increase in MSN2-dependent gene expression.
cdc4-1 suppression depends on PNC1 expression
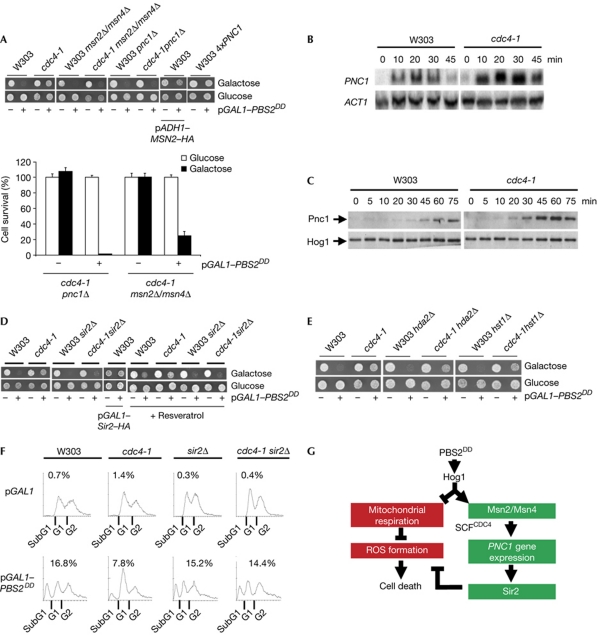
Cell death due to Hog1 was not suppressed in a cdc4-1 msn2 msn4 strain (Fig 4A). Correspondingly, overexpression of MSN2 under the ADH1 promoter prevents cell death Hog1 activation in wild-type cells (Fig 4A). Expression of PNC1 is induced in response to osmostress by Msn2 and Msn4 (Posas et al, 2000; Causton et al, 2001), and the role of Msn2 and Msn4 in cell longevity is mediated by PNC1, an activator of Sir2 (Bitterman et al, 2002; Anderson et al, 2003; Gallo et al, 2004; Medvedik et al, 2007). Either PBS2DD overexpression or osmostress induced PNC1 expression that was stronger and more extended in a cdc4-1 mutant (Fig 3B,C; supplementary Fig S3A,B online). Similarly to a cdc4-1 msn2 msn4 strain, overexpression of PBS2DD induced cell death in a cdc4-1 pnc1 strain (Fig 3A). Correspondingly, overexpression of PNC1 prevented cell death in response to Hog1 activation (Fig 3B). Thus, Pnc1 mediates the effect of Msn2 and Msn4 to prevent cell death in response to sustained Hog1 activation.
Figure 4.
MSN2/MSN4, PNC1 and SIR2 counteract Hog1-mediated cell death. (A) Deletion of MSN2/MSN4 and PNC1 eliminates the ability of cdc4-1 mutation to prevent cell death. Cells were spotted onto glucose or galactose plates. Viability was monitored by counting colony-forming units in glucose plates (lower panel). Data represent the mean and standard deviation of three independent experiments. (B) cdc4-1 mutant shows an increase in PNC1 gene expression in response to osmostress. Strains were treated with 0.4 M NaCl and total RNA was probed with PNC1 and ACT1. (C) Mutation in the SCFCDC4 complex increases Pnc1 protein levels in response to osmostress. Pnc1 and Hog1 were assessed in cells treated as in B. (D) SIR2 deletion eliminates the ability of cdc4-1 mutation to prevent cell death. Strains were grown in the presence of resveratrol in the plates (5 μM). Cells that expressed haemagglutinin (HA)-tagged SIR2 under the GAL1 promoter (pGAL1–SIR2–HA) and those that expressed PBS2DD were spotted on glucose (control) or galactose plates. (E) Hda2 and Hst1 deacetylases do not mediate the effect of cdc4-1. Strains were spotted onto glucose or galactose plates. (F) Sir2 prevents Hog1-induced apoptosis-like cell death. Strains were grown from raffinose to galactose for 24 h. DNA content was assessed by flow cytometry. Data shown are representative of three independent experiments. (G) Tentative model that depicts the effect of Hog1 and Sir2 in dictating cell-fate determination. Hog1 inhibits mitochondrial respiration, which results in an increase in reactive oxygen species (ROS) accumulation that leads to cell death. In parallel, Hog1 induces PNC1 expression through Msn2 and Msn4 transcription factors, which are regulated by SCFCDC4. Pnc1 activates Sir2, which mediates a decrease of ROS accumulation. SIR2 activation by the stress-activated protein kinase Hog1 relieves the Hog1-induced oxidative stress to prevent apoptotic cell death. ROS, reactive oxygen species.
Sir2 activation suppresses Hog1-mediated apoptosis
A cdc4-1 sir2 strain was unable to prevent cell death caused by Hog1 activation (Fig 4D). This effect was specific for Sir2, as deletion of HDA2 or SIR2 did not abolish the effect of cdc4-1 (Fig 4E). Correspondingly, cell death, as measured by the presence of SubG1 cells, was partly suppressed in a cdc4-1 strain, but not in a cdc4-1 sir2 strain (Fig 4F).
We then analysed whether resveratrol, a drug that was suspected to induce Sir2 activity, improved cell growth (Howitz et al, 2003). Resveratrol did not prevent cell death in a sir2 strain. Correspondingly, overexpression of Sir2 suppressed cell death on Hog1 activation (Fig 4D). The cdc4-1 sir2 strain showed an increase in ROS levels on Hog1 activation, which was similar to that of the wild-type strain and twofold higher than that of the cdc4-1 strain (Fig 2B). There was a slight increase of ROS on 2 h incubation in the presence of NaCl in the wild type, which was further increased in a sir2 strain (supplementary Fig S5 online). Therefore, Sir2 is required for protection from Hog1-induced cell death, by preventing Hog1-induced ROS accumulation.
A main role for Sir2 in cell survival is the suppression of ribosomal DNA recombination and the formation of toxic extra-chromosomal ribosomal DNA circles in the nucleus of mother cells (Sinclair & Guarente, 1997). SIR2 also affects lifespan by increasing silencing at telomeric regions (Dang et al, 2009). Interestingly, deletion of NET1, a component of the ribosomal DNA-localized Sir2 complex (Straight et al, 1999), abolished the effect of cdc4-1 on Hog1 activation (supplementary Fig S4B online). By contrast, deletion of either SIR4 or deletion of HM loci (Aparicio et al, 1991) did not affect cell viability (supplementary Fig S4B online). Therefore, although this is genetic evidence that will require further characterization, our data indicate that the function of Sir2 at ribosomal DNAs might dictate the level of cell death on Hog1 activation.
Activation of SAPK signalling is essential for cell adaptation to stress. However, sustained activation of the pathway unravels a more-complex hypothesis. When it is not restricted, SAPK activation causes an inhibition of mitochondrial respiration, which results in an increase of ROS formation that can only be counteracted by the Hog1-dependent activation of Sir2 and the lifespan extension pathway (Fig 4G). Although a decrease in mitochondrial respiration might be important for cell adaptation, an extended reduction of respiration leads to excessive ROS formation. To prevent cell damage, Hog1 induces PNC1 gene expression and concomitantly activates Sir2 to balance excessive ROS accumulation. Therefore, cell fate is dictated by the balance between ROS induced by Hog1 SAPK and the protective effects of Sir2.
Methods
Yeast strains and plasmids. A complete list is included in the supplementary information online. The strains used in this study showed similar growth rates when grown in YPD. Expression of the Pbs2DD protein was similar in all strains tested (supplementary Fig S4A online).
Northern blot analysis. Yeast cultures were grown to early log phase (optical density at 660=0.6–0.8). Cells were either subjected to stress (0.4 M NaCl, indicated times), shifted to galactose or untreated. Total RNA was probed by using radiolabelled polymerase chain reaction fragments containing labelled CTT1 (1.7 kb), ALD3 (1.5 kb), GRE2 (1.1 kb), STL1 (1.7 kb), PNC1 (0.8 kb) and ACT1 (1.4 kb). Signals were quantified by a Typhoon 8600 phosphorimager and the ImageQuant software.
Chromatin immunoprecipitation. ChIP was performed as described previously (Alepuz et al, 2003). Yeast cultures were grown to early log phase before osmostress (0.4 M NaCl). For crosslinking, yeast cells were treated with 1% formaldehyde for 20 min at 25°C. Primer mixes were adjusted for balanced signals.
PCD measurements. Apoptosis-like cell death was measured by using FLICA assay adapted to flow cytometry, a TdT-mediated dUTP nick end labelling assay adapted to flow cytometry or following the presence of a SubG1 population of cells by flow cytometry, as described in the supplementary information online.
Colony-forming unit assay. Yeast cultures were grown to early log phase in raffinose and then shifted to galactose or glucose for 24 h before plating. The number of colonies was determined after 3 days at 25 °C in replicas. Data are the result of three independent experiments with replicas.
ROS and mitochondrial respiration assays. ROS amounts in liquid growing cells were detected by using 2′,7′-dichloro-dihydro-fluorescein diacetate (H2DCF-DA) from Invitrogen (Carlsbad, CA). In plate, mitochondrial analysis was performed using a tetrazolium overlay assay. Measurement of oxygen consumption was recorded by direct measurement using oxygen measurer, as indicated in the supplementary information online.
Supplementary information is available at EMBO reports online (http://www.emboreports.org).
Supplementary Material
Acknowledgments
We thank L. Subirana, S. Ovejas and O. Fornas for technical support; G. Ammerer, J. Clotet, X. Escoté and G. Mas for their helpful advice and contribution; and E. de Nadal for constant advice and support. This work was supported by grants from the Ministerio de Ciéncia y Innovación (BIO2009-07762) and Consolider Ingenio 2010 programme (grant CSD2007-0015), UNICELLSYS from FP7, as well as supported by Fundación Marcelino Botín. F.P. is the recipient of the Institució Catalana de Recerca i Estudis Avançats Acadèmia (Generalitat de Catalunya). D.A.S. is supported by grants from the National Institute on Aging/National Institutes of Health, the Ellison Medical Foundation and the Glenn Foundation for Medical Research.
Footnotes
The authors declare that they have no conflict of interest.
References
- Alepuz PM, de Nadal E, Zapater M, Ammerer G, Posas F (2003) Osmostress-induced transcription by Hot1 depends on a Hog1-mediated recruitment of the RNA Pol II. EMBO J 22: 2433–2442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson RM, Bitterman KJ, Wood JG, Medvedik O, Sinclair DA (2003) Nicotinamide and PNC1 govern lifespan extension by calorie restriction in Saccharomyces cerevisiae. Nature 423: 181–185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aparicio OM, Billington BL, Gottschling DE (1991) Modifiers of position effect are shared between telomeric and silent mating-type loci in S. cerevisiae. Cell 66: 1279–1287 [DOI] [PubMed] [Google Scholar]
- Bitterman KJ, Anderson RM, Cohen HY, Latorre-Esteves M, Sinclair DA (2002) Inhibition of silencing and accelerated aging by nicotinamide, a putative negative regulator of yeast Sir2 and human SIRT1. J Biol Chem 277: 45099–45107 [DOI] [PubMed] [Google Scholar]
- Carmona-Gutierrez D, Eisenberg T, Büttner S, Meisinger C, Kroemer G, Madeo F (2010) Apoptosis in yeast: triggers, pathways, subroutines. Cell Death Diff 17: 763–773 [DOI] [PubMed] [Google Scholar]
- Causton HC, Ren B, Koh SS, Harbison CT, Kanin E, Jennings EG, Lee TI, True HL, Lander ES, Young RA (2001) Remodeling of yeast genome expression in response to environmental changes. Mol Biol Cell 12: 323–337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen RE, Thorner J (2007) Function and regulation in MAPK signaling pathways: lessons learned from the yeast Saccharomyces cerevisiae. Biochim Biophys Acta 1773: 1311–1340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chi Y, Huddleston MJ, Zhang X, Young RA, Annan RS, Carr SA, Deshaies RJ (2001) Negative regulation of Gcn4 and Msn2 transcription factors by Srb10 cyclin-dependent kinase. Genes Dev 15: 1078–1092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clotet J, Escote X, Adrover MA, Yaakov G, Gari E, Aldea M, de Nadal E, Posas F (2006) Phosphorylation of Hsl1 by Hog1 leads to a G2 arrest essential for cell survival at high osmolarity. EMBO J 25: 2338–2346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dang W, Steffen KK, Perry R, Dorsey JA, Johnson FB, Shilatifard A, Kaeberlein M, Kennedy BK, Berger SL (2009) Histone H4 lysine 16 acetylation regulates cellular lifespan. Nature 459: 802–807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Nadal E, Posas F (2010) Multilayered control of gene expression by stress-activated protein kinases. EMBO J 29: 4–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dolado I, Nebreda AR (2008) Regulation of tumorigenesis by p38 α MAP kinase. Top Cur Genet 20: 99–128 [Google Scholar]
- Gallo CM, Smith DL Jr, Smith JS (2004) Nicotinamide clearance by Pnc1 directly regulates Sir2-mediated silencing and longevity. Mol Cell Biol 24: 1301–1312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galluzzi L et al. (2009) Guidelines for the use and interpretation of assays for monitoring cell death in higher eukaryotes. Cell Death Differ 16: 1093–1107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hohmann S, Krantz M, Nordlander B (2007) Yeast osmoregulation. Methods Enzymol 428: 29–45 [DOI] [PubMed] [Google Scholar]
- Howitz KT et al. (2003) Small molecule activators of sirtuins extend Saccharomyces cerevisiae lifespan. Nature 425: 191–196 [DOI] [PubMed] [Google Scholar]
- Kobayashi GS, Cheung SS, Schlessinger D, Medoff G (1974) Effects of rifamycin derivatives, alone and in combination with amphotericin B, against histoplasma capsulatum. Antimicrob Agents Chemother 5: 16–18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macia J, Regot S, Peeters T, Conde N, Sole R, Posas F (2009) Dynamic signaling in the Hog1 MAPK pathway relies on high basal signal transduction. Sci Signal 2: ra13. [DOI] [PubMed] [Google Scholar]
- Madeo F, Frohlich E, Ligr M, Grey M, Sigrist SJ, Wolf DH, Frohlich KU (1999) Oxygen stress: a regulator of apoptosis in yeast. J Cell Biol 145: 757–767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madeo F, Carmona-Gutierrez D, Ring J, Buttner S, Eisenberg T, Kroemer G (2009) Caspase-dependent and caspase-independent cell death pathways in yeast. Biochem Biophys Res Commun 382: 227–231 [DOI] [PubMed] [Google Scholar]
- Maeda T, Wurgler-Murphy SM, Saito H (1994) A two-component system that regulates an osmosensing MAP kinase cascade in yeast. Nature 369: 242–245 [DOI] [PubMed] [Google Scholar]
- Marshall CJ (1995) Specificity of receptor tyrosine kinase signaling: transient versus sustained extracellular signal-regulated kinase activation. Cell 80: 179–185 [DOI] [PubMed] [Google Scholar]
- Medvedik O, Lamming DW, Kim KD, Sinclair DA (2007) MSN2 and MSN4 link calorie restriction and TOR to sirtuin-mediated lifespan extension in Saccharomyces cerevisiae. PLoS Biol 5: e261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olson BL, Hock MB, Ekholm-Reed S, Wohlschlegel JA, Dev KK, Kralli A, Reed SI (2008) SCFCdc4 acts antagonistically to the PGC-1α transcriptional coactivator by targeting it for ubiquitin-mediated proteolysis. Genes Dev 22: 252–264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pal B, Chan NC, Helfenbaum L, Tan K, Tansey WP, Gething MJ (2007) SCFCdc4-mediated degradation of the Hac1p transcription factor regulates the unfolded protein response in Saccharomyces cerevisiae. Mol Biol Cell 18: 426–440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Posas F, Chambers JR, Heyman JA, Hoeffler JP, de Nadal E, Ariño J (2000) The transcriptional response of yeast to saline stress. J Biol Chem 275: 17249–17255 [DOI] [PubMed] [Google Scholar]
- Sinclair DA, Guarente L (1997) Extrachromosomal rDNA circles—a cause of aging in yeast. Cell 91: 1033–1042 [DOI] [PubMed] [Google Scholar]
- Straight AF, Shou W, Dowd GJ, Turck CW, Deshaies RJ, Johnson AD, Moazed D (1999) Net1, a Sir2-associated nucleolar protein required for rDNA silencing and nucleolar integrity. Cell 97: 245–256 [DOI] [PubMed] [Google Scholar]
- Wagner EF, Nebreda AR (2009) Signal integration by JNK and p38 MAPK pathways in cancer development. Nat Rev Cancer 9: 537–549 [DOI] [PubMed] [Google Scholar]
- Walter D, Wissing S, Madeo F, Fahrenkrog B (2006) The inhibitor-of-apoptosis protein Bir1 protects against apoptosis in S. cerevisiae and is a substrate for the yeast homologue of Omi/HtrA2. J Cell Sci 119: 1843–1851 [DOI] [PubMed] [Google Scholar]
- Williamson AJ, Dibling BC, Boyne JR, Selby P, Burchill SA (2004) Basic fibroblast growth factor-induced cell death is effected through sustained activation of p38MAPK and up-regulation of the death receptor p75NTR. J Biol Chem 279: 47912–47928 [DOI] [PubMed] [Google Scholar]
- Wurgler-Murphy SM, Maeda T, Witten EA, Saito H (1997) Regulation of the Saccharomyces cerevisiae HOG1 mitogen-activated protein kinase by the PTP2 and PTP3 protein tyrosine phosphatases. Mol Cell Biol 17: 1289–1297 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.