EMBO Rep (2011) advance online publication. doi:; DOI: 10.1038/embor.2011.171
We were all taught that proteins have to fold correctly to be active and that the primary sequence of amino acids acts as the ‘blueprint’ for successful, productive folding. In recent years, we have also learnt how sensitive that blueprint is to change. For example, a single amino-acid change in the protein sequence, or a subtle change in temperature at which folding takes place, can lead to the formation and accumulation of non-native species, which have a tendency to self-associate and deposit in and around tissues, thereby triggering disease. To understand protein misfolding and its links with disease, we need information about the structures of all the key players and their relationships with each other. This has proven exceptionally challenging due to the transient nature of many of the species involved and the heterogeneity of the final misfolded product.
For one misfolding disorder, α1-antitrypsin deficiency—a devastating disease that affects approximately 1 in 2,500 individuals—we are a step closer to characterizing all of the main culprits. In this issue of EMBO reports, Huntington and colleagues present the X-ray crystal structure of an α1-antitrypsin trimer that sheds light on the structure of a potentially pathogenic form of α1-antitrypsin and provides new insights into the molecular mechanism of α1-antitrypsin deficiency (Yamasaki et al, 2011).
Attempts to understand the molecular basis for α1-antitrypsin deficiency began in 1963 when Laurell & Eriksson noticed its absence from the serum of a cohort of patients with obstructive lung disease (Laurell & Eriksson, 1963). Sharp and co-workers subsequently described periodic acid Schiff-positive inclusions of α1-antitrypsin within hepatocytes of α1-antitrypsin-deficient patients (Sharp et al, 1969). These two findings linked the major clinical outcomes of the disease. α1-Antitrypsin inhibits elastase in the lower respiratory tract and, therefore, a plasma deficiency leads to early onset emphysema due to uncontrolled elastase activity. The aggregation of α1-antitrypsin at its site of production—the hepatocyte—leads to liver damage and cirrhosis. In the early 1990s, elegant work by Lomas and colleagues revealed that the key molecular event leading to the deficiency was the misfolding and formation of α1-antitrypsin polymers within the endoplasmic reticulum of hepatocytes (Lomas et al, 1992).
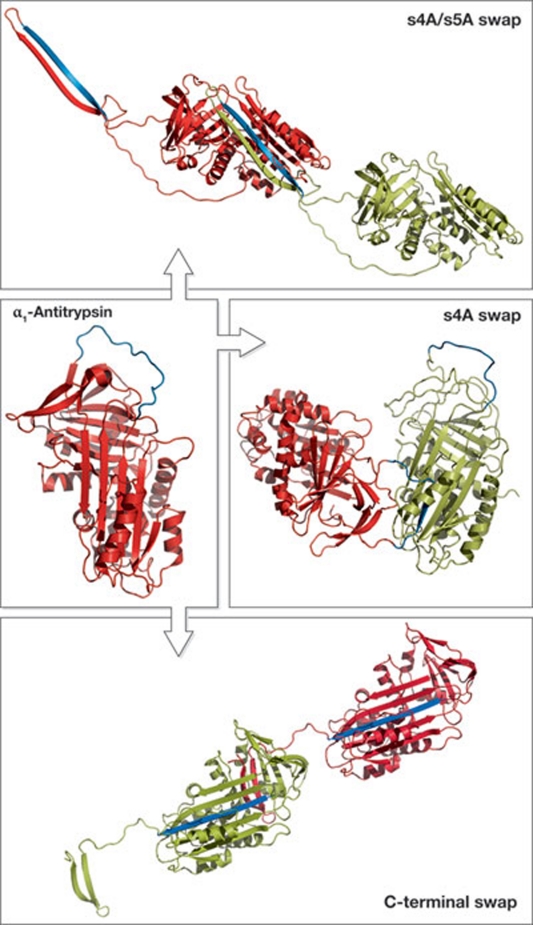
α1-Antitrypsin, like all members of the serpin superfamily, is a large single-domain protein consisting of 394 amino acids that fold into three β-sheets surrounded by nine α-helices (Fig 1; Elliott et al, 2000). Protruding from the core structure is the flexible reactive centre loop, containing the scissile bond that dictates the inhibitory specificity of a serpin. Similar to all serpins, α1-antitrypsin undergoes a marked conformational change to inhibit proteinases, which involves the insertion of the reactive centre loop into the middle of β-sheet A. This conformational change is possible because the native state of the serpin superfamily is metastable. However, the instability of the native state of α1-antitrypsin makes it extremely susceptible to misfolding and polymerization, which results in the formation of more thermodynamically stable conformations. The most common mutation that causes polymerization in α1-antitrypsin is the Z mutation (Glu342Lys), which is present in approximately 4% of northern Europeans. This mutation does not alter the stability of the native molecule but slows down its folding rate such that a polymerization-prone intermediate state persists longer, favouring polymerization (Knaupp et al, 2010). In addition, the native state of Z α1-antitrypsin is more easily polymerized than its wild-type counterpart (Lomas et al, 1992). Although we have known the structure of native wild-type α1-antitrypsin for more than 10 years (Elliott et al, 2000), and despite a wealth of biochemical and biophysical studies, we do not know the structure of the final polymeric form.
Figure 1.
Structures of native α1-antitrypsin and the known three types of polymer. The reactive centre loop region is highlighted in blue. Antitrypsin is deposited in the Protein Data Bank, ID no. 1QLP. Models of the s4A/s5A swap polymer and carboxy-terminal swap polymer were kindly provided by Professor J. Huntington; the model of the s4A swap polymer was kindly provided by Professor J. Whisstock.
Over the past 20 years, various linkages between α1-antitrypsin monomers have been reported and/or suggested, which involve different sheets and extents of interaction. Until recently, the most widely accepted model for α1-antitrypsin polymerization involved the reactive centre loop of one molecule entering the β-sheet A of another in the strand 4 position (s4A swap polymer; Fig 1; Sivasothy et al, 2000), which was supported by extensive biochemical and biophysical analyses. This model was recently challenged when Yamasaki and colleagues described a new polymeric linkage in which monomers were linked by the insertion of two strands (strands 4 and 5) into β-sheet A of another serpin (s4A/s5A swap polymer; Fig 1; Yamasaki et al, 2008). Notably, these polymers were formed in the presence of the denaturant guanidine hydrochloride and so their physiological relevance was unclear. Extensive experimental work by the Lomas group has shown that heat-induced polymerization of α1-antitrypsin produces polymers with a structure similar to those formed in hepatocellular inclusions (Ekeowa et al, 2010). The key to their studies was the identification and characterization of the antibody 2C1, which recognizes only polymerized α1-antitrypsin formed both in vitro and in vivo (Miranda et al, 2010).
“One of the most striking results of this work is the heterogeneity of polymer formation, which has important implications…”
In this current paper, Yamasaki and colleagues set out to determine the structure of these heat-induced polymers. The authors initially compare heat- and denaturant-induced polymers by using native PAGE. They find that the s4A/s5A swap polymers dominate when denaturant is used to induce polymerization, whereas the heat-induced polymers are made up of a mixture of s4A/s5A swap polymers and another polymer morphology that binds to the 2C1 antibody. By using this information, they construct a recombinant α1-antitrypsin molecule that cannot undergo the s4A/s5A swap reaction. By heating this α1-antitrypsin variant at 60 °C, they form and subsequently purify short polymers that bind to the 2C1 antibody, indicating that they contain the pathological structure. The resultant polymer is a trimer that cannot be extended. Structural analysis showed that the monomers are linked through a swap of secondary structure involving the carboxyl terminus of the protein, specifically the first strand from the β-sheet C and strands 4 and 5 from β-sheet B were swapped from one serpin to another (C-terminal swap polymer; Fig 1).
“…more than one polymer structure might be formed simultaneously during polymerization […] toxic and non-toxic polymers might coexist”
The C-terminal swap polymer was obtained by heating recombinant protein at 60 °C, raising questions about its biological relevance, which the authors tried to answer by using protein engineering. For the C-terminal swap polymer to form, the C-terminus must move. Thus, the authors engineered a disulphide bond between the third strand of β-sheet C and the C-terminus, effectively preventing the conformational change. Notably, when this disulphide-bonded protein was polymerized by using heat, only s4A/s5A swap polymers—which cannot bind to the 2C1 antibody—were formed. In further experiments, they used a protein engineering approach to examine what happens in two cell models—the yeast Pichia pastoris and monkey COS-7 cells—of Z α1-antitrypsin production. Both models produce Z α1-antitrypsin polymers that react with the 2C1 antibody. By using the introduced cysteine residues to probe for the polymeric linkage, they were able to show that the C-terminal swap mechanism dominated in both cell models.
The study by Yamasaki and colleagues provides another important piece of the puzzle in identifying, characterizing and linking all the players involved in α1-antitrypsin deficiency. One of the most striking results of this work is the heterogeneity of polymer formation, which has important implications for understanding this disease and the development of therapeutic strategies. α1-Antitrypsin is a flexible molecule capable of extreme conformational change and this inherent ‘desire’ for a more stable conformation makes it susceptible to polymerization. Numerous parallel pathways of polymer production probably occur, but the structural and/or environmental factors involved in deciding which pathway is followed remain unknown. In addition, does this ability to form different polymers affect which type of polymer is cleared from the cell? The kinetics of these polymerization reactions need to be elucidated and previous studies by several groups might need to be re-evaluated, as it is possible that heterogeneous populations of polymers were being formed, which would confuse the data analysis.
From a biomedical perspective, identifying the pathological species and determining how it damages the liver is critical. The C-terminal swap polymer is probably found in hepatocytes, but this does not mean that it is the toxic agent. The results of the Yamasaki study suggest that more than one polymer structure might be formed simultaneously during polymerization, which raises the possibility that toxic and non-toxic polymers might coexist. Therefore, another polymer form—such as the s4A/s5A swap polymer or one not yet identified—could be damaging to cells. Answers to these questions will come in time and hopefully will lead to successful therapeutic approaches to prevent α1-antitrypsin deficiency.
References
- Ekeowa UI et al. (2010) Proc Natl Acad Sci USA 107: 17146–17151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elliott PR et al. (2000) Protein Sci 9: 1274–1281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knaupp AS et al. (2010) J Mol Biol 396: 375–383 [DOI] [PubMed] [Google Scholar]
- Laurell C-B, Eriksson S (1963) Scand J Clin Lab Invest 15: 132–140 [DOI] [PubMed] [Google Scholar]
- Lomas DA et al. (1992) Nature 357: 605–607 [DOI] [PubMed] [Google Scholar]
- Miranda E et al. (2010) Hepatology 52: 1078–1088 [DOI] [PubMed] [Google Scholar]
- Sharp HL et al. (1969) J Lab Clin Med 73: 934–939 [PubMed] [Google Scholar]
- Sivasothy P et al. (2000) J Biol Chem 275: 33663–33668 [DOI] [PubMed] [Google Scholar]
- Yamasaki M et al. (2008) Nature 455: 1255–1258 [DOI] [PubMed] [Google Scholar]
- Yamasaki M et al. (2011) EMBO Rep [Epub 9 September 2011] doi:; DOI: 10.1038/embor.2011.171 [DOI] [Google Scholar]