Abstract
Jaagsiekte sheep retrovirus (JSRV) can induce rapid, multifocal lung cancer, but JSRV is a simple retrovirus having no known oncogenes. Here we show that the envelope (env) gene of JSRV has the unusual property that it can induce transformation in rat fibroblasts, and thus is likely to be responsible for oncogenesis in animals. Retrovirus entry into cells is mediated by Env interaction with particular cell-surface receptors, and we have used phenotypic screening of radiation hybrid cell lines to identify the candidate lung cancer tumor suppressor HYAL2/LUCA2 as the receptor for JSRV. HYAL2 was previously described as a lysosomal hyaluronidase, but we show that HYAL2 is actually a glycosylphosphatidylinositol (GPI)-anchored cell-surface protein. Furthermore, we could not detect hyaluronidase activity associated with or secreted by cells expressing HYAL2, whereas we could easily detect such activity from cells expressing the related serum hyaluronidase HYAL1. Although the function of HYAL2 is currently unknown, other GPI-anchored proteins are involved in signal transduction, and some mediate mitogenic responses, suggesting a potential role of HYAL2 in JSRV Env-mediated oncogenesis. Lung cancer induced by JSRV closely resembles human bronchiolo-alveolar carcinoma, a disease that is increasing in frequency and now accounts for ≈25% of all lung cancer. The finding that JSRV env is oncogenic and the identification of HYAL2 as the JSRV receptor provide tools for further investigation of the mechanism of JSRV oncogenesis and its relationship to human bronchiolo-alveolar carcinoma.
Jaagsiekte sheep retrovirus (JSRV) is the causative agent of a contagious lung cancer of sheep called ovine pulmonary carcinoma or sheep pulmonary adenomatosis (1). Tumors originate from type II secretory alveolar and nonciliated bronchiolar epithelial cells, and late stages of the disease are accompanied by the secretion of copious lung fluid containing the virus. Purified virus induces multifocal tumors in as little as 10 days (2), suggesting the role of a viral oncogene. However, JSRV is a simple retrovirus with typical gag, pol, and env genes and no known oncogenes. The viral structural (Gag) and enzymatic (Pol) proteins are unlikely to be responsible, because they interact primarily with viral components, but there is precedent for alteration of cellular functions by the envelope (Env) protein of retroviruses, which interacts with cellular components to mediate virus entry. For example, the deleted Env protein of the Friend spleen focus-forming virus causes erythroleukemia by binding to and activating the erythropoietin receptor (3). If the JSRV Env protein was indeed oncogenic, identification of the host cell receptor for JSRV would provide key insights into the oncogenic mechanism of this highly pathogenic retrovirus. Furthermore, the contagious nature of JSRV and its ability to survive exposure to proteases and surfactants present in lung fluid suggest that vectors based on JSRV might be useful for gene therapy targeted to the lung, provided that the pathogenic features of the virus can be controlled. Identification of the JSRV receptor and determination of the pattern of receptor expression in the lung will help identify cells that can be targeted by JSRV vectors.
To facilitate identification of the JSRV receptor, we made retrovirus packaging cells that produce a retrovirus vector with Gag and Pol proteins from Moloney murine leukemia virus, Env protein from JSRV, and a retroviral vector genome (LAPSN) that encodes human placental alkaline phosphatase (AP) as a marker for vector transduction (4). This vector was able to transduce sheep and human cells, with the exception of HeLa cells, but was unable to transduce cells from mice, rats, and hamsters because of the lack of functional receptors for JSRV Env.
The presence of a functional receptor in human cells and its absence in hamster cells allowed us to use phenotypic screening of human/hamster radiation hybrid cell lines (5) to localize the receptor in the human genome. We assayed the Stanford G3 panel of 83 human/hamster radiation hybrid clones for susceptibility to transduction by the JSRV vector. Each hybrid contains a full complement of hamster chromosomes and multiple radiation-fragmented pieces of human DNA with average size of ≈4 megabases and representing ≈18% of the human genome. These hybrids have been screened by PCR for the presence or absence of >15,000 unique human DNA markers to generate a high resolution map of the human genome. Assuming that vector transduction correlates with the presence of the receptor gene, we identified several markers that were closely linked to the JSRV receptor and thereby localized the receptor to the p21.3 band of human chromosome 3 (4).
Here, we show that the JSRV receptor maps to a region of 3p21.3 that had previously been sequenced in a search for lung cancer tumor suppressor genes localized to this region by analysis of chromosomal deletions in lung and breast cancers (6, 7), and show that the receptor is HYAL2. We have characterized the receptor and show that it is a glycosylphosphatidylinositol (GPI)-anchored cell-surface protein. Although HYAL2 is related to known hyaluronidases, HYAL2 exhibited no hyaluronidase activity, and its normal function is currently unknown. Finally, we show that the env gene of JSRV can transform cultured fibroblasts, setting the stage for further work to determine the role of the receptor in JSRV Env-mediated oncogenesis.
Materials and Methods
Plasmid Expression Vectors.
All cDNAs were expressed by using a cytomegalovirus (CMV) immediate early promoter and a bovine growth hormone gene polyadenylation signal. By convention, human protein names are capitalized whereas only the initial letter is capitalized for those of mouse. Most were expressed by using pCR3.1 (Invitrogen), with the exception of mouse Hyal1, which was expressed by using pcDNA3.1 (Invitrogen). Expression vectors for human HYAL4 and SPAM1 and for mouse Hyal1 and Hyal3 were kind gifts from Antonei Csoka and Robert Stern (University of California, San Francisco). Accession numbers for hyaluronidase family member cDNAs are as follows: HYAL1, U03056; HYAL2, U09577; HYAL3, AF036035; HYAL4, AF009010; human SPAM1, L13781; Hyal1, AF011567; and Hyal2, AF302843. Amino-terminal Flag-tagged proteins were made by cloning the cDNAs into pFLAG-CMV2 (Flag tag without an endoplasmic reticulum signal sequence) or pFLAG-CMV1 (preprotrypsin signal sequence upstream of the Flag tag; Sigma-Aldrich). After construction, the coding regions in the expression plasmids were checked for accuracy by complete sequencing.
Retrovirus Vectors.
The Moloney murine leukemia virus-based retroviral vectors LXSN (8) and LAPSN (9) have been described. LNCZ was constructed by inserting the bacterial β-galactosidase (β-gal) cDNA into the LNCX vector (8), and the vector and packaging cells producing the vector were gifts from Clarissa Dirks (Fred Hutchinson Cancer Research Center, Seattle, WA). Vectors expressing HYAL2 and HYAL1 were made by cloning the respective cDNAs into LXSN to make LHYAL2SN and LHYAL1SN. Helper virus-free vector stocks were made by using PJ14 JSRV-pseudotype (4), PT67 10A1-pseudotype (10), PG13 GALV-pseudotype (11), or FLYRD RD114-pseudotype (12) packaging cell lines as described (13). Pseudotype here refers to the Env protein present in the vector virions, and all of these packaging cell lines produce virions containing Gag-Pol proteins from Moloney murine leukemia virus.
Analysis of HYAL2 Expression in 293 Cells Transfected with Flag-Tagged HYAL2 Expression Plasmids.
Human embryonic kidney 293 cells were transfected with the HYAL2 cDNA constructs SS-Flag-M2HYAL2 or Flag-M2HYAL2 or the empty vector as a control by using SuperFect (Qiagen, Chatsworth, CA). The cells were collected 48 h later and were washed with PBS. Phosphatidylinositol-specific phospholipase C (PI-PLC; Sigma or Molecular Probes) was added at final concentrations of 0, 0.05, 0.2, and 1 unit/ml, and the cells were incubated at 4°C for 4 h. Cells were pelleted, and the supernatants were transferred into new tubes containing protease inhibitor mixture (Sigma) for Western blotting analysis. The pelleted cells were washed two times with PBS and divided into two fractions for immunostaining and Western blot analysis.
For Western blot analysis, the cells were lysed in 50 mM Hepes (pH 7.4), 150 mm NaCl, 10% glycerol, 1 mM EDTA, 1 mM sodium orthovanadate, 10 mM sodium pyrophosphate, 100 mM NaF, 1% Triton X-100, 10 μg/ml leupeptin, 10 units/ml aprotinin, and 1 mM PMSF. Insoluble material was removed by centrifugation. Flag-tagged HYAL2 was immunoprecipitated from the clarified cell lysates and supernatants from PI-PLC-treated cells by using anti-Flag goat antibodies (Santa Cruz Biotechnology) and protein-G-agarose conjugate (GIBCO/BRL). Precipitated proteins were separated by electrophoresis in 8–16% polyacrylamide gels containing SDS and were transferred to a nitrocellulose membrane; Flag-tagged HYAL2 was detected by using anti-Flag monoclonal antibodies (Upstate Biotechnology, Lake Placid, NY).
For immunofluorescence analysis, the transfected 293 cells were placed on coverslips precoated with polylysine (molecular weight 70,000–150,000, Sigma) and were fixed in 3.7% formaldehyde in PBS for 30 min at room temperature. For some experiments, the cells were permeabilized by treatment with 0.5% Triton X-100 in PBS for 5 min. Permeabilized and nonpermeabilized cells were treated with SuperBlock solution (Pierce) for 30 min at room temperature to block nonspecific binding. The coverslips were incubated with anti-Flag monoclonal antibodies (dilution 1:500, Upstate Biotechnology) for 1 h, washed with PBS, incubated with secondary goat anti-mouse antibodies labeled with Alexa488 (Molecular Probes, dilution 1:500), and were washed with PBS. Cells transfected with empty vector and cells stained only with secondary antibodies served as controls.
Electrophoretic Analysis of Hyaluronidase Activity.
Hyaluronic acid from human umbilical chord (Calbiochem) was incubated with bovine testes hyaluronidase (Calbiochem), conditioned medium from cultured cells, or with cell lysates prepared by exposing confluent cells in 6-well dishes (105 to 9 × 105 cells per well) to 500 μl of 0.5% Triton X-100 per well. After incubation at pH 3.8 (using formate buffer) or at pH 7.5 (using phosphate buffer) at 37°C, the pH of the samples was brought above 10 by addition of NaOH, and the samples were boiled for ≥10 min to inactivate the enzymes. Loading buffer (2 M sucrose, 7×) was added to the samples, and they were electrophoresed in standard 0.5% agarose Tris-acetate-EDTA gels at 1.67 V/cm (50 V for a 30-cm box) for 8–10 hr. After electrophoresis, gels were incubated in the dark with 0.005% Stains-All (Sigma) in 50% ethanol with moderate agitation overnight (14). Gels were destained by exposure to light and photographed.
Oncogenic Transformation Assay.
208F rat embryo fibroblasts (15) were seeded at 5 × 105 per 6-cm dish in DMEM plus 10% FBS on day 1. On day 2, the cells were transfected with 10 μg of plasmid DNA by using the CaPO4 coprecipitation method as described (16). On day three, the cells were trypsinized and divided 1:5 into 6-cm dishes. After the cells became confluent, they were fed with DMEM plus 5% FBS and 1 μM dexamethasone, and were fed with this medium every 3 to 4 days thereafter. Foci were quantitated and photographed ≈2 weeks after transfection.
Results
Identification of HYAL2 as the Receptor for JSRV.
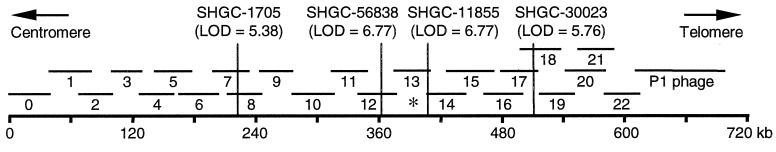
We previously used phenotypic screening of radiation hybrid cell lines to localize the receptor for JSRV to chromosome 3p21.3 (4). A search of nucleotide databases revealed that the four unique-sequence markers that mapped closest to the receptor were present in a 700-kb cosmid and phage contig (Fig. 1) that had been sequenced as part of a search for lung cancer tumor suppressor genes localized to this region by analysis of chromosomal deletions in lung and breast cancers (6, 7). Transfection of the cosmids from this region into the parental A23 hamster cells used to make the radiation hybrid clones revealed that only the Luca 13 cosmid conferred susceptibility to JSRV vector transduction (Table 1 and data not shown). Of the three genes in this cosmid, FUS1, HYAL1 and HYAL2, only an expression vector containing the HYAL2 cDNA promoted transduction of nonpermissive A23, NIH 3T3, and HeLa cells (Table 1), identifying HYAL2 as the gene that confers susceptibility to JSRV.
Figure 1.
JSRV receptor localization to the human chromosome 3p21.3 lung cancer cosmid and phage contig. The human genome is depicted as a bold line with overlapping cosmid and phage clones indicated above. Luca cosmid numbers are indicated, and the asterisk indicates the cosmid that contains the JSRV receptor. The positions of Stanford Human Genome Center (SHGC) markers are shown by vertical lines, and the degree of linkage to the JSRV receptor is indicated by LOD (logarithm of odds) scores that were calculated by the radiation hybrid web server at the SHGC.
Table 1.
Cell susceptibility to JSRV vector transduction after transfection of expression plasmids and cosmids
| Transfected DNA | JSRV vector transduction of cells, transducing units
per μg DNA
|
||
|---|---|---|---|
| NIH 3T3 | A23 | HeLa | |
| β-gal | 3 | 60 | nd |
| Luca 13 cosmid | 5,100 | >10,000 | 1,500 |
| Human FUS1 | <2 | 10 | nd |
| Human HYAL1 | 12 | 120 | 10 |
| Human HYAL2 | >10,000 | >10,000 | 7,400 |
| Human HYAL3 | 13 | 90 | 20 |
| Human HYAL4 | <2 | 70 | 9 |
| Human SPAM1 | <2 | 20 | 14 |
| Mouse Hyal1 | 6 | 20 | 10 |
| Mouse Hyal2 | 130 | 660 | 570 |
| Mouse Hyal3 | <2 | 140 | 10 |
| M2HYAL2 | 5 | 170 | 12 |
| SS-Flag-M2HYAL2 | >10,000 | >10,000 | 6,900 |
| Flag-M2HYAL2 | 14 | 120 | 12 |
| HYAL2 d440-453 | 7 | 40 | 8 |
| HYAL2 d440-473 | <4 | 50 | 12 |
Cells were seeded into 6-cm dishes on day 1, transfected (16) on day 2, trypsinized and replated on day 3, exposed to 1 ml LAPSN vector (NIH 3T3 and A23 cells) or 1 ml LNCZ vector (HeLa cells) produced by JSRV packaging cells on day 4, and stained for AP (NIH 3T3 and A23 cells) or β-gal (HeLa cells) on day 6. A vector encoding AP (LAPSN) was used for the rodent cells, whereas a lower-titer vector encoding β-gal (LNCZ) was used for the HeLa cells because of their high endogenous heat-stable AP levels. Results are means from two to seven experiments.
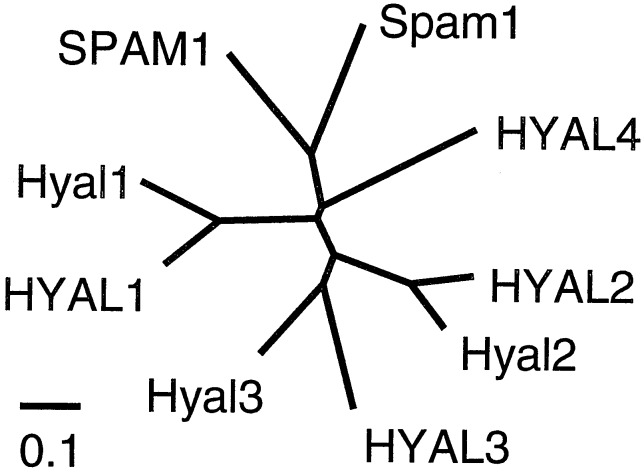
HYAL2 is a member of a protein family that includes the hyaluronidase present on the sperm surface (SPAM1, also called PH-20) that enables sperm to penetrate the hyaluronic acid-rich cumulus cells surrounding the oocyte, and a hyaluronidase found in lysosomes and serum, HYAL1 (17; Fig. 2). However, none of the human hyaluronidase family members besides HYAL2 mediated JSRV transduction (Table 1). HYAL2 is widely expressed in most tissues, with the notable exception of brain (7, 17, 18), and closely related orthologs of the HYAL2 gene are found in rodents. Thus, the resistance of rodent cells to JSRV vector transduction suggests that these cells express a nonfunctional receptor. Indeed, mouse Hyal2 showed very low receptor activity compared with human HYAL2 (Table 1). As with the human orthologs, the mouse hyaluronidase family members Hyal1 and Hyal3 did not promote JSRV vector transduction (Table 1).
Figure 2.
Dendrogram (clustal w) of relationships between hyaluronidase family members. Scale bar indicates 10% amino acid sequence difference.
HYAL2 Is a GPI-Anchored Cell Surface Protein.
Analysis of the 473-aa sequence of HYAL2 indicated an amino-terminal 20 residue endoplasmic reticulum signal sequence, no transmembrane domains, and a hydrophobic carboxyl terminus and upstream signal, suggesting that HYAL2 is anchored to the membrane by a GPI anchor (D. Buloz and J. Kronegg, http://members.nbci.com/jkronegg/unige/GPI-anchor/index.html; refs. 19 and 20; Fig. 3), as is the sperm hyaluronidase SPAM1 (21). We made several alterations to HYAL2 to test these predictions (Table 1). The putative 20-aa signal sequence is followed by a methionine in a good context for translation (Fig. 3), and expression of a truncated HYAL2 protein that began at this second methionine (M2HYAL2) did not confer JSRV vector susceptibility to nonpermissive cells. However, expression of an altered HYAL2 protein in which the 20-aa signal was replaced with a preprotrypsin signal sequence followed by a Flag epitope for facile protein detection (SS-Flag-M2HYAL2) did confer full JSRV vector susceptibility. The preprotrypsin signal sequence was required for receptor activity because replacement of the native signal sequence with the Flag epitope alone (Flag-M2HYAL2) abrogated receptor activity. Together, these results indicate that the amino-terminal amino acids of HYAL2 function as an endoplasmic reticulum signal sequence to direct HYAL2 to the cell surface where it can function as a JSRV receptor.
Figure 3.
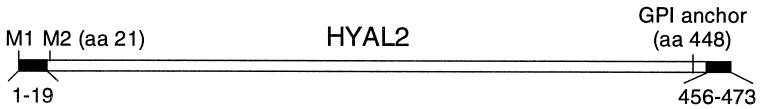
HYAL2 posttranslational processing signals.
To test for GPI linkage of HYAL2 to the cell membrane, we transfected human 293 cells with the SS-Flag-M2HYAL2 or Flag-M2HYAL2 expression vectors and examined the effects of treatment of the cells with bacterial PI-PLC, which can cleave GPI linkages. SS-Flag-M2HYAL2 was expressed on the surface of transfected cells (Fig. 4A), and treatment of the cells with PI-PLC resulted in removal of surface protein by immunofluorescence analysis (not shown) and total cell protein Western blot analysis (Fig. 4B). Flag-M2HYAL2 was found inside the cell and not on the cell surface (Fig. 4A), and treatment of cells with PI-PLC had no effect on the level (Fig. 4B) or localization (not shown) of Flag-M2HYAL2. These results show that SS-Flag-M2HYAL2 and, by inference, HYAL2 are linked to the cell surface by a GPI anchor. Lastly, deletion of the putative GPI linkage site (HYAL2 d440–453) or this site and the hydrophobic tail (HYAL2 d440–473) from HYAL2 (Fig. 3) abrogated its function as a receptor (Table 1), indicating the importance of this region.
Figure 4.
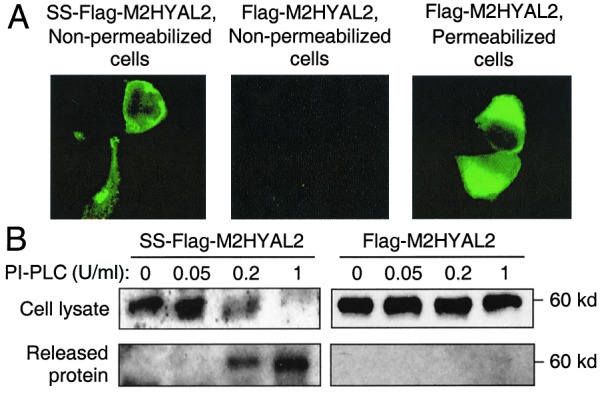
HYAL2 localizes to the cell surface and can be removed by PI-PLC treatment. Human embryonic kidney 293 cells were transfected with plasmids that express the Flag-tagged HYAL2 protein with (SS-Flag-M2HYAL2) or without (Flag-M2HYAL2) an endoplasmic reticulum signal sequence and were analyzed 2 days later. (A) Immunofluorescence analysis of Flag-tagged protein. (B) Western analysis of Flag-tagged protein in cell lysates and in the incubation medium after incubation of cells with PI-PLC at the indicated concentrations (units/ml).
Removal of HYAL2 by PI-PLC Treatment Blocks JSRV Vector Transduction.
To provide evidence for GPI linkage of the native HYAL2 protein and to confirm the role of this protein as a receptor for JSRV, we assayed JSRV vector transduction of cells before and after treatment with PI-PLC (Table 2). We tested HT-1080 human cells that are susceptible to JSRV vector transduction and HeLa cells that were rendered susceptible by transduction with a retroviral vector that expresses HYAL2. Treatment of both cell types with PI-PLC greatly reduced JSRV vector transduction rates, whereas transduction by an otherwise identical vector having a gibbon ape leukemia virus Env protein that recognizes the non-GPI-anchored integral membrane protein PIT1 (GLVR1) as a receptor (22) was not appreciably affected. These results indicate that both the native JSRV receptor and the introduced HYAL2 protein are attached to the cell surface by a GPI anchor. Our findings conflict with a previous report (18) that localized HYAL2 to lysosomes, but this study was done by tracking fluorescence of a fusion protein in which green fluorescent protein (GFP) was added to the carboxyl end of HYAL2, an arrangement that would either disrupt normal GPI processing or result in cleavage of the GFP tag from HYAL2 such that GFP fluorescence would not necessarily indicate the location of HYAL2.
Table 2.
Abrogation of HYAL2 receptor function by treatment of cells with PI-PLC
| Target cells | Vector pseudotype | Vector titer, FFU/ml
|
Ratio | |
|---|---|---|---|---|
| No PI-PLC treatment | With PI-PLC treatment | |||
| HT-1080 | JSRV | 1.0 × 104 | 35 | 290 |
| GALV | 1.2 × 106 | 9.6 × 105 | 1 | |
| HeLa/LHYAL2SN | JSRV | 2.2 × 103 | 80 | 28 |
| GALV | 320 | 280 | 1 | |
Cells were seeded at 1.5 × 104 per well of 24-well plates. One day later, the medium was removed, and 200 μl of medium with or without 0.04 (HT-1080) or 0.1 (HeLa/LHYAL2SN) unit of PI-PLC was added. One hour later, retroviral vectors and Polybrene (4 μg/ml final concentration) were added in a volume of 60 μl. A vector encoding AP (LAPSN) was used for the HT-1080 cells, whereas a vector encoding β-gal (LNCZ) was used for the HeLa cells. Vectors were produced using JSRV packaging cells (4) or gibbon ape leukemia virus-based packaging cells (11). Virus was removed 2 hr later by washing with 300 μl of medium, followed by addition of 250 μl of medium containing half the amount of PI-PLC used initially. Two days after virus addition, the cells were stained for AP (HT-1080 cells) or β-gal (HeLa cells). Results are means of two experiments with duplicate determinations in each experiment. FFU, focus-forming unit.
HYAL2 Has Low if Any Hyaluronidase Activity.
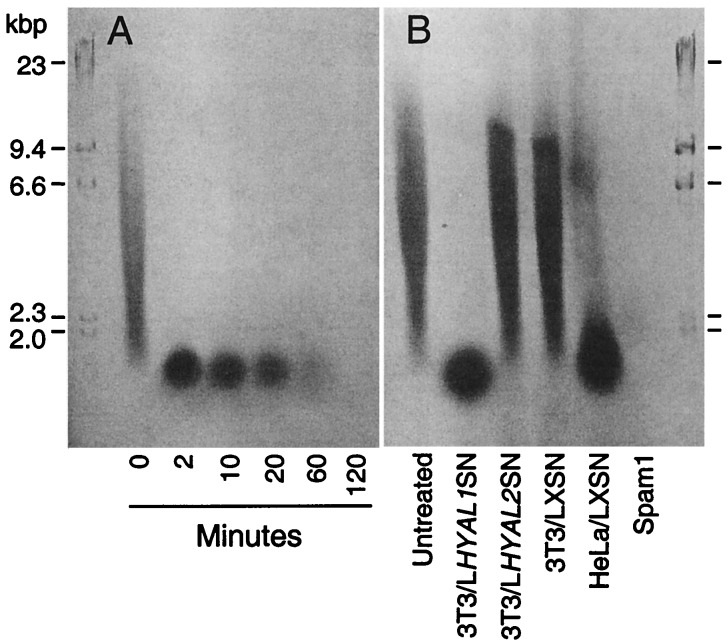
HYAL2 has been reported to have an atypical hyaluronidase activity that cleaves high molecular weight hyaluronic acid to a 20-kDa species that might have distinct biological functions in stimulating angiogenesis and in gene regulation (18), both relevant to a potential role for HYAL2 in cancer. To explore this finding further, we examined hyaluronic acid metabolism by electrophoresis of digestion products through agarose (14, 18). In control experiments, we found that commercially available bovine testes hyaluronidase (Spam1) digested human umbilical cord hyaluronic acid to undetectable products in two phases, the first resulting in production of a relatively stable intermediate that migrated slightly faster than a 2.0-kb DNA marker and that has been estimated to have a size of about 20 kDa (14, 18), followed by a slower second phase that resulted in the complete disappearance of this intermediate (Fig. 5A). The first phase proceeded through heterogeneous intermediates and was ≈25-fold more rapid than the second phase (data not shown). Spam1 was active both at pH 3.8 and at pH 7.5, as previously reported.
Figure 5.
Electrophoretic analysis of hyaluronidase activity in cell lysates. Fifty-microgram samples of human umbilical cord hyaluronic acid were incubated with 500 ng of hyaluronidase (Spam1) from bovine testes at pH 3.8 for the indicated times (A) or with cell lysates (prepared in 0.5% Triton X-100, ≈2 × 104 cells per reaction) at pH 3.8 for 12 h (B), and the digestion products were analyzed by agarose gel electrophoresis, followed by detection with the Stains-All. HindIII-digested λ-phage DNA size markers and their sizes are shown.
We next tested for hyaluronidase activity in cell lysates and conditioned medium from NIH 3T3 and HeLa cells that had been transduced with retroviral vectors that express HYAL1 (LHYAL1SN), HYAL2 (LHYAL2SN), or no insert (LXSN), made by inserting the cDNAs into the LXSN vector (8). Only the NIH 3T3 and HeLa cells containing the LHYAL2SN vector became susceptible to JSRV vector transduction, as expected (not shown). Mammalian hyaluronidases fall into two classes, those that show peaks of activity at acidic and at neutral pH, including Spam1, and those that are active only at acidic pH with a peak at pH 3.8, including HYAL1 and HYAL2; thus, we focused on detection of activity at pH 3.8. Lysates of HeLa cells transduced with the control LXSN vector (Fig. 5B) or no vector (not shown) exhibited high background levels of a hyaluronidase active at pH 3.8 and were not studied further. Lysates of NIH 3T3 cells expressing the control vector LXSN or the vector containing HYAL2 showed no hyaluronidase activity, whereas those from cells expressing the vector containing HYAL1 showed hyaluronidase activity at pH 3.8 (Fig. 5B). The same pattern was observed by assay of conditioned medium from this set of cells (not shown). Longer incubations at pH 3.8 showed that HYAL1 was capable of further degrading hyaluronic acid to undetectable forms, but, as with Spam1, this process was much slower than the conversion to the 20-kDa intermediate. Longer incubations at pH 3.8 with cell lysates from cells expressing HYAL2 still did not reveal any hyaluronidase activity. Furthermore, no activity was detected in cell lysates or conditioned medium from any of the cells at pH 7.5. We estimate, based on these analyses, that HYAL2 hyaluronidase activity, if any, is at least 50-times lower than that of HYAL1. In addition, the 20-kDa hyaluronic acid species appears to be a relatively stable intermediate of hyaluronic acid metabolism and not the atypical product of a particular hyaluronidase.
NCI-H524 Small Cell Lung Cancer Cells Are Resistant to JSRV Vector Transduction but Can Be Rendered Susceptible by HYAL2 Expression.
Most small cell lung cancers and many non-small cell lung (including bronchiolo-alveolar carcinoma), breast, and cervical cancers show loss of heterozygosity or deletion of sequences in the p21.3 region of chromosome 3 where HYAL2 is located (6, 7, 23, 24). The small cell lung cancer cell line NCI-H524 shows a very small homozygous deletion of ≈30 kb that removes most of HYAL2 and all of HYAL1 (7). We tested the prediction that these cells should be resistant to JSRV vector transduction. Indeed, H524 cells were resistant to JSRV vector transduction, but were susceptible to transduction by otherwise identical vectors having different Env proteins that recognize other receptors (Table 3). Transduction of these cells with a retroviral vector that expressed HYAL2, but not an empty vector or a vector that expressed HYAL1, rendered these cells fully susceptible to JSRV vector transduction (Table 4). These results support the hypothesis that HYAL2 is the sole receptor for JSRV, and that the HYAL2 gene is deleted in H524 cells. The JSRV vector transduction assay provides a means to screen tumors for lack of HYAL2 expression and possible deletion of the gene. In contrast to the majority of human cell lines that we have found to be susceptible to JSRV vector transduction, HeLa cells are resistant, indicating reduced expression of HYAL2 in this cervical carcinoma cell line and perhaps suggesting a role for HYAL2 in this cancer.
Table 3.
H524 lung cancer cells are resistant to JSRV vector transduction
| Target cells | Titer
(AP+ FFU/ml) for LAPSN pseudotype
|
||
|---|---|---|---|
| PJ14 | PG13 | FLYRD | |
| H524 | <25 | 4 × 104 | 4 × 104 |
| HT-1080 | 6 × 103 | 2 × 105 | 2 × 105 |
H524 cells were seeded at 5 × 105 to 106 per well in non-tissue culture-treated six-well plates coated with fibronectin fragment CH-296 (Takara Biomedicals, Shiga, Japan) in RPMI 1640 medium with 10% FBS, and HT-1080 cells were seeded at 5 × 104 per well in tissue culture-treated six-well plates in DMEM with 10% FBS. The next day, the media were replaced with media containing 4 μg/ml Polybrene and various amounts of the indicated pseudotypes of the LAPSN vector. Two days after vector addition, the cells were fixed and stained for AP expression. Results are means of two to four experiments. FFU, focus-forming unit.
Table 4.
H524 cells can be rendered susceptible to JSRV vector transduction by expression of HYAL2
| Target cells | LAPSN(PJ4) vector titer (AP+ FFU/ml) |
|---|---|
| H524/LXSN | <10 |
| H524/LHYAL1SN | <10 |
| H524/LHYAL2SN | 6 × 103 |
H524 cells were seeded at 5 × 105 to 106 per well in non-tissue culture-treated six-well plates coated with fibronectin fragment CH-296 (Takara Biomedicals, Shiga, Japan) in RPMI 1640 medium with 10% FBS on day 1. The next day, the cells were exposed to the LXSN, LHYAL1SN, or LHYAL2SN to generate H524 cells expressing no additional protein, HYAL1, or HYAL2, respectively. On day three, the cells were treated with trypsin and divided into two fibronectin fragment-coated wells of six-well plates. On day 4, the cells were exposed to LAPSN(PJ4), and, on day 6, the cells were fixed and stained for AP. The results are means of two experiments, each performed in duplicate. FFU, focus-forming unit.
The JSRV Env Gene Can Transform Rodent Fibroblasts.
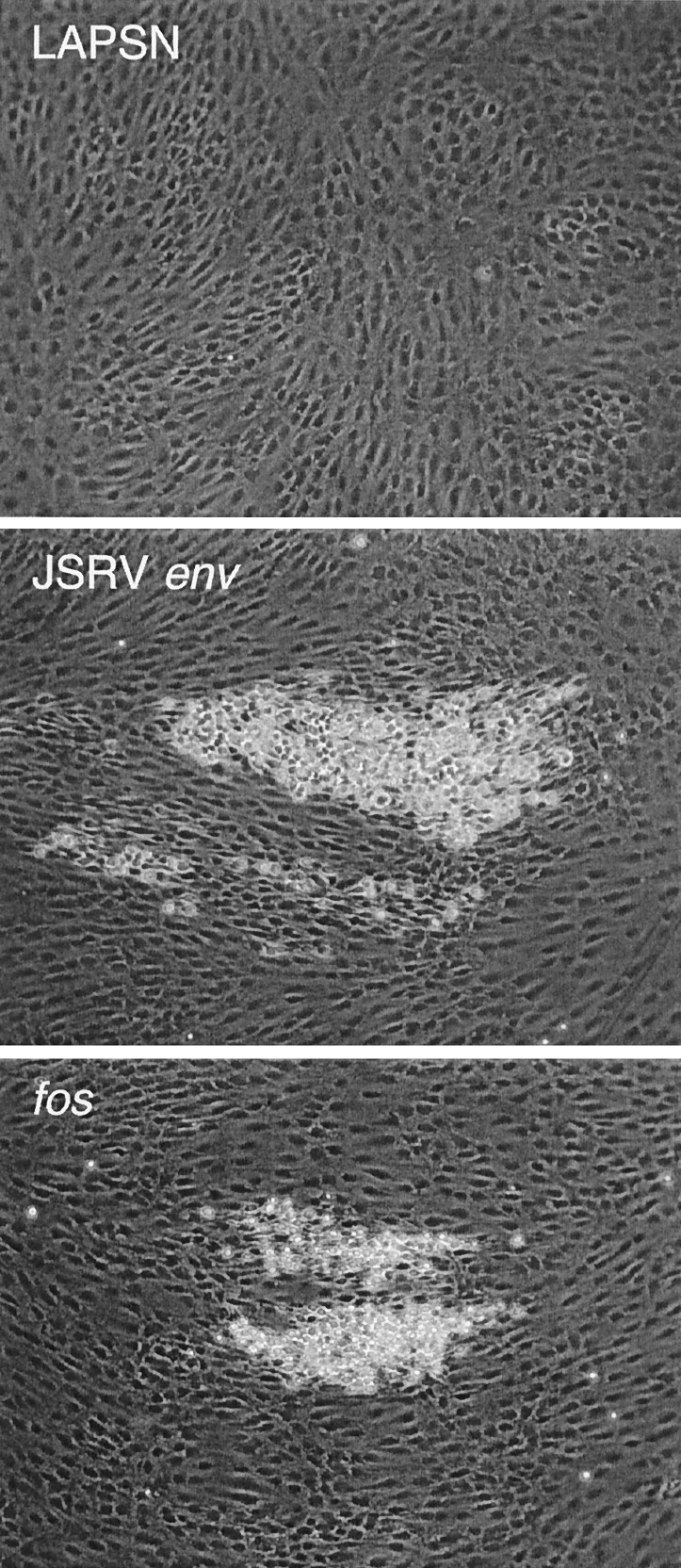
We tested whether the JSRV Env protein could transform 208F rat embryo fibroblasts (15), a cell line with a very flat morphology that provides a good assay for oncogenic transformation (25). We used the pSX2.Jenv plasmid that we used to generate the PJ14 JSRV-pseudotype packaging cells (4) for the test. Indeed, the pSX2.Jenv plasmid induced transformed foci in 208F cells with morphology and sizes similar to foci induced by a hybrid viral fos expression plasmid (pFBJ/R) that has previously been shown to have very strong transforming activity (25; Fig. 6). The pSX2.Jenv plasmid induced foci at a 6-fold lower rate than the plasmid carrying the fos oncogene (25 versus 150 foci per μg plasmid DNA, respectively). No foci were detected in negative control experiments by using the pLAPSN plasmid.
Figure 6.
Focus formation in 208F rat cells after transfection with JSRV env (pSX2.Jenv), fos (pFBJ/R), or a plasmid containing alkaline phosphatase and neo genes (pLAPSN).
Discussion
Our studies provide strong genetic evidence that HYAL2 is the receptor that mediates entry of JSRV. Thus, several uninfectable cell types from different species can be rendered fully susceptible to JSRV vector transduction by expression of HYAL2 alone. The fact that overexpression of mouse Hyal2 promotes only very low levels of JSRV vector transduction in mouse cells while human HYAL2 promotes very efficient transduction argues for a specific interaction of the JSRV Env with human HYAL2, rather than JSRV entry being dependent on a common functional property shared by the mouse and human proteins. Removal of HYAL2 by using PI-PLC renders cells resistant to infection, further indicating a direct interaction between the JSRV Env and HYAL2. To our knowledge, there have been no previous reports of a naturally occurring retrovirus receptor attached to the cell surface by a GPI anchor, although one alternatively spliced form of the quail receptor (Tva) for subgroup A avian sarcoma and leukosis viruses is predicted to have this linkage as well (26). GPI-anchored proteins are typically found on the apical surface of epithelial cells (20, 27), an appropriate location for the receptor for a virus such as JSRV that replicates in the airway. In addition, an apical location for HYAL2 would be advantageous for gene therapy directed to the lung using JSRV vectors.
Our results demonstrate that positional cloning of genes by phenotypic analysis of radiation hybrid cells is feasible. Use of this technique with the G3 radiation hybrid library allows localization of a gene within ≈250 kb, or about 8 genes. In the case of the JSRV receptor, phenotypic screening localized the gene to the middle of a 650-kb cosmid contig that had been completely sequenced, providing a relatively high likelihood that the receptor was within the contig. Our results show that the receptor lies directly between the two unique-sequence markers that mapped closest to the receptor, a space of ≈45 kb. We had previously used this technique to localize the receptors for three other retroviruses that can infect human cells but not hamster cells: the RD114 cat endogenous retrovirus, xenotropic murine leukemia virus, and feline leukemia virus type B (5). Unfortunately, in these instances, neither sequence information nor cosmid contigs were available in the regions of receptor localization. Ultimately we and others cloned these receptors by using a retroviral cDNA library approach (5). In the future, the completion of the human genome sequence and the availability of clones spanning the genome will make gene identification by phenotypic screening of radiation hybrids a reliable and quick method for gene identification.
We initially hypothesized that the transforming activity of JSRV would be mediated through the cell-surface receptor for the virus, and, because rodent cells do not express a functional receptor, we did not immediately test for transformation by env using standard rodent cell-based transformation assays. Recently, Hung Fan and coworkers observed that the whole JSRV virus could transform NIH 3T3 cells, provided the 5′ retroviral promoter was replaced with a CMV immediate early promoter (personal communication), prompting us to test for JSRV env transforming activity. We found that the JSRV env protein could indeed transform rat cells (Fig. 6) and NIH 3T3 mouse cells (data not shown), neither of which express a functional receptor for JSRV (4). It is still possible that Hyal2 interacts with and mediates oncogenesis by the JSRV envelope even though it is unable to promote efficient virus entry. Overexpression of mouse Hyal2 renders cells somewhat susceptible to JSRV vector transduction (Table 1), showing that the JSRV Env can interact to some extent with mouse Hyal2. Alternatively, JSRV Env may cause transformation by interacting with proteins other than Hyal2.
Analysis of deletions and loss of heterozygosity indicate the presence of tumor suppressor(s) in the 3p21.3 region, but no clear example has been yet identified (7). Our finding that HYAL2 is the receptor for the highly oncogenic retrovirus JSRV raises the possibility that HYAL2 may be one of the long-sought tumor suppressor(s). Other GPI-anchored cell surface proteins participate in cell signaling and mitogenic activation (19, 20), providing a possible mechanism for HYAL2 involvement in oncogenesis. Study of JSRV oncogenesis in animals has been restricted to sheep and has not been possible in rodents because of the lack of functional virus receptors. The next step will be to generate transgenic and knock-in mice expressing human HYAL2, and these mice should provide convenient models for further study of cancer induction by JSRV and its relevance to common human epithelial cancers, including the phenotypically similar lung cancer, bronchiolo-alveolar carcinoma.
Acknowledgments
We thank T. Csoka and R. Stern for helpful discussions and for providing the HYAL4, SPAM1, Hyal1, and Hyal3 expression plasmids; C. Dirks for providing the LNCZ vector; N. Van Hoeven, J. Strickler, and S. Moe for assistance; and M. Emerman, P. Neiman, J. Overbaugh, and B. Zbar for comments on the manuscript. This work was supported by Grants DK47754 and HL54881 and Contract CO56000 from the National Institutes of Health.
Abbreviations
- JSRV
jaagsiekte sheep retrovirus
- GPI
glycosylphosphatidylinositol
- AP
alkaline phosphatase
- Env
envelope
- PI-PLC
phosphatidylinositol-specific phospholipase C
- CMV
cytomegalovirus
- β-gal
β-galactosidase
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
See commentary on page 4285.
References
- 1.Palmarini M, Sharp J M, de las Heras M, Fan H. J Virol. 1999;73:6964–6972. doi: 10.1128/jvi.73.8.6964-6972.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sharp J M, Angus K W, Gray E W, Scott F M. Arch Virol. 1983;78:89–95. doi: 10.1007/BF01310861. [DOI] [PubMed] [Google Scholar]
- 3.Li J P, D'Andrea A D, Lodish H F, Baltimore D. Nature (London) 1990;343:762–764. doi: 10.1038/343762a0. [DOI] [PubMed] [Google Scholar]
- 4.Rai S K, DeMartini J C, Miller A D. J Virol. 2000;74:4698–4704. doi: 10.1128/jvi.74.10.4698-4704.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rasko J E J, Battini J-L, Kruglyak L, Cox D R, Miller A D. Proc Natl Acad Sci USA. 2000;97:7388–7392. doi: 10.1073/pnas.130200097. . (First Published June 13, 2000; 10.1073/pnas.130200097) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wei M H, Latif F, Bader S, Kashuba V, Chen J Y, Duh F-M, Sekido Y, Lee C C, Geil L, Kuzmin I, et al. Cancer Res. 1996;56:1487–1492. [PubMed] [Google Scholar]
- 7.Lerman M I, Minna J D for The International Lung Cancer Chromosome 3p21.3 Tumor Suppressor Gene Consortium. Cancer Res. 2000;60:6116–6133. [PubMed] [Google Scholar]
- 8.Miller A D, Rosman G J. Biotechniques. 1989;7:980–990. [PMC free article] [PubMed] [Google Scholar]
- 9.Miller D G, Edwards R H, Miller A D. Proc Natl Acad Sci USA. 1994;91:78–82. doi: 10.1073/pnas.91.1.78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Miller A D, Chen F. J Virol. 1996;70:5564–5571. doi: 10.1128/jvi.70.8.5564-5571.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Miller A D, Garcia J V, von Suhr N, Lynch C M, Wilson C, Eiden M V. J Virol. 1991;65:2220–2224. doi: 10.1128/jvi.65.5.2220-2224.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cosset F-L, Takeuchi Y, Battini J L, Weiss R A, Collins M K. J Virol. 1995;69:7430–7436. doi: 10.1128/jvi.69.12.7430-7436.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Miller A D, Miller D G, Garcia J V, Lynch C M. Methods Enzymol. 1993;217:581–599. doi: 10.1016/0076-6879(93)17090-r. [DOI] [PubMed] [Google Scholar]
- 14.Lee H G, Cowman M K. Anal Biochem. 1994;219:278–287. doi: 10.1006/abio.1994.1267. [DOI] [PubMed] [Google Scholar]
- 15.Quade K. Virology. 1979;98:461–465. doi: 10.1016/0042-6822(79)90569-5. [DOI] [PubMed] [Google Scholar]
- 16.Chen C, Okayama H. Mol Cell Biol. 1987;7:2745–2752. doi: 10.1128/mcb.7.8.2745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Csoka A B, Scherer S W, Stern R. Genomics. 1999;60:356–361. doi: 10.1006/geno.1999.5876. [DOI] [PubMed] [Google Scholar]
- 18.Lepperdinger G, Strobl B, Kreil G. J Biol Chem. 1998;273:22466–22470. doi: 10.1074/jbc.273.35.22466. [DOI] [PubMed] [Google Scholar]
- 19.Low M G. FASEB J. 1989;3:1600–1608. doi: 10.1096/fasebj.3.5.2522071. [DOI] [PubMed] [Google Scholar]
- 20.Lublin D M. Curr Top Microbiol Immunol. 1992;178:141–162. doi: 10.1007/978-3-642-77014-2_9. [DOI] [PubMed] [Google Scholar]
- 21.Phelps B M, Primakoff P, Koppel D E, Low M G, Myles D G. Science. 1988;240:1780–1782. doi: 10.1126/science.3381102. [DOI] [PubMed] [Google Scholar]
- 22.O'Hara B, Johann S V, Klinger H P, Blair D G, Rubinson H, Dunn K J, Sass P, Vitek S M, Robins T. Cell Growth Differ. 1990;1:119–127. [PubMed] [Google Scholar]
- 23.Sekido Y, Ahmadian M, Wistuba I I, Latif F, Bader S, Wei M H, Duh F M, Gazdar A F, Lerman M I, Minna J D. Oncogene. 1998;16:3151–3157. doi: 10.1038/sj.onc.1201858. [DOI] [PubMed] [Google Scholar]
- 24.Wistuba I I, Montellano F D, Milchgrub S, Virmani A K, Behrens C, Chen H, Ahmadian M, Nowak J A, Muller C, Minna J D, et al. Cancer Res. 1997;57:3154–3158. [PubMed] [Google Scholar]
- 25.Miller A D, Verma I M, Curran T. J Virol. 1985;55:521–526. doi: 10.1128/jvi.55.3.521-526.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bates P, Young J A T, Varmus H E. Cell. 1993;74:1043–1051. doi: 10.1016/0092-8674(93)90726-7. [DOI] [PubMed] [Google Scholar]
- 27.Benting J H, Rietveld A G, Simons K. J Cell Biol. 1999;146:313–320. doi: 10.1083/jcb.146.2.313. [DOI] [PMC free article] [PubMed] [Google Scholar]