Abstract
Regulatory T-cells (Treg) play an essential role in the negative regulation of immune answers by developing an attenuated cytokine response that allows suppressing proliferation and effector function of T-cells (CD4+ Th). The transcription factor FoxP3 is responsible for the regulation of many genes involved in the Treg gene signature. Its ablation leads to severe immune deficiencies in human and mice. Recent developments in sequencing technologies have revolutionized the possibilities to gain insights into transcription factor binding by ChiP-seq and into transcriptome analysis by mRNA-seq. We combine FoxP3 ChiP-seq and mRNA-seq in order to understand the transcriptional differences between primary human CD4+ T helper and regulatory T-cells, as well as to study the role of FoxP3 in generating those differences. We show, that mRNA-seq allows analyzing the transcriptomal landscape of T-cells including the expression of specific splice variants at much greater depth than previous approaches, whereas 50% of transcriptional regulation events have not been described before by using diverse array technologies. We discovered splicing patterns like the expression of a kinase-dead isoform of IRAK1 upon T-cell activation. The immunoproteasome is up-regulated in both Treg and CD4+ Th cells upon activation, whereas the ‘standard’ proteasome is up-regulated in Tregs only upon activation.
INTRODUCTION
The recent years have shown that regulatory T-cells (Treg) play an essential role in the negative regulation of immune answers and the prevention of autoimmunity (1) by maintaining tolerance to self and controlling autoimmune deviation. Treg cells have an attenuated cytokine response and can suppress proliferation and effector function of other T-cells. One important regulator to develop the Treg specific gene expression signature is the forkhead box transcription factor FoxP3, which is highly expressed in Treg cells, although it is also present at lower levels in effector T-cell populations. Deficiency in FoxP3 has been shown to underlie the lympho-proliferation and multi-organ autoimmunity of mutant mice and is linked with immunodysregulation polyendocrinopathy and the X-linked syndrome (IPEX) in humans (2). Although being a key regulator in Treg development, it becomes also increasingly evident that FoxP3 alone only accounts for part of the Treg signature and, for example, the suppression of IL2 and activation of IL2RA in T-cells are also found in FoxP3-deficient Scurfy mice (3).
Treg cells differ significantly in their gene expression profile from CD4+ T helper cells, both, in resting and activated states. A large number of gene expression profiling studies (4–7) allowed for the identification of a signature of genes which are typically up- or down-regulated in Tregs. Those genes are involved in a variety of biologic processes and functions including cell surface and membrane proteins (TLR4, IL2RA, IL2RB or CTLA4), kinases (Map3K8), phosphatases (DUSP4) or transcription factors (FoxP3, IKZF2 and IKZF4).
The role of FoxP3 in regulating the expression of Treg specific genes has further been elucidated by three studies which used chromatin immunoprecipitation (ChiP) in conjunction with microarrays to identify chromosomal locations of FoxP3 binding in mouse (8,9) and in human (10). They described a set of ~1400 mouse genes and 5579 human genes which are bound by FoxP3. In combination with gene expression profiling, they identified genes which are subsequently de-regulated in their expression levels. Those studies showed that FoxP3 binding can explain activation or repression of a subset of genes involved in the Treg signature but also suggested that FoxP3 alone is not responsible for developing Treg cells.
In addition, major advances in sequencing technologies also known as next or second generation sequencing (NGS) (11) allow for applications which go far beyond what has been possible only a few years ago. Those applications include genome re-sequencing projects to identify genetic variation between parents and their children (12) or the analysis of folding principles of the human genome (13). To gain a deeper understanding of properties of a specific transcription factor, the combination of co-immunoprecipitation and subsequent DNA sequencing, also known as ChiP-seq (14,15), enables the unbiased identification of genomic regions bound by a transcription factor. NGS has also enabled us to perform gene expression profiling (mRNA-seq) at an unknown depth, sensitivity and resolution including the identification of splice variants (16), expression profiles of single cells (17) or organisms without prior genome sequence information (18).
In this study, we use Illumina's Genome Analyzer platform to perform ChiP-seq of FoxP3-bound genomic regions and mRNA-seq for transcriptome profiling in samples of resting and activated primary CD4+ Th and Treg cells from human donors. Using this data set, we are able to show the influence of FoxP3 on gene expression patterns in human and also in comparison to published data sets in the mouse. Furthermore, our analysis enables very detailed insights into the transcriptomal landscape of resting and activated Treg and Teff cells including differential expression of genes, splice variants and ncRNAs.
MATERIALS AND METHODS
Preparation of T-cell populations
Leukapheresis products were obtained from adult healthy volunteers with approval by the ethical committee (Landesaerztekammer Baden-Wuertemberg). Human naturally occurring CD4+CD25+ regulatory T-cells (Tregs) and untouched human CD4+ T helper cells (CD4+ Th) were isolated as described previously (19). Polyclonal T-cell activation was performed using soluble 1 µg/ml anti CD3 (clone OKT3) antibody (ebioscience) and 2 µg/ml anti CD28 (clone 28.2) antibody (ebioscience).
FoxP3 ChiP
Genpathway's FactorPath method was carried out as described by Labhart et al. (20). In brief, human T-cell populations, activated for 16 h, were fixed with 1% formaldehyde for 15 min and quenched with 0.125 M glycine. Chromatin was isolated by adding lysis buffer, followed by disruption with a Dounce homogenizer (cells). Lysates were sonicated (Misonix) to shear the DNA to an average length of 300–500 bp. Genomic DNA (Input) was purified from an aliquot of chromatin and quantified on a Nanodrop spectrophotometer. Extrapolation to the original chromatin volume allowed quantitation of the total chromatin yield.
ChiP assays of activated Th cells and nTregs were carried out in duplicate. An aliquot of chromatin (50 ug) was pre-cleared with protein G agarose beads (Invitrogen). FoxP3-bound genomic DNA regions were isolated using a goat polyclonal antibody against FoxP3 (Abcam ab2481). After incubation at 4°C overnight, protein G agarose beads were used to isolate the immune complexes. Complexes were washed, eluted from the beads with SDS buffer and subjected to RNase and proteinase K treatment. Crosslinks were reversed by incubation overnight at 65°C and ChiP DNA was purified by phenol-chloroform extraction and ethanol precipitation. To assay for the enrichment of positive control region in the ChiP DNA, quantitative PCR (qPCR) reactions were carried out in triplicate with primers specific for these regions using SYBR Green Supermix (Bio-Rad). The resulting signals were normalized for primer efficiency by carrying out qPCR for each primer pair using Input DNA (data not shown).
ChiP Sequencing (ChiP-seq)
Remaining ChiP DNA (90% of entire sample) was amplified using the Illumina ChiP-seq DNA Sample Prep Kit. In brief, DNA ends were polished and 5′-phosphorylated using T4 DNA polymerase, Klenow polymerase and T4 polynucleotide kinase. After addition of 3′-A to the ends using Klenow fragments (3′–5′ exo−), Illumina genomic adapters were ligated and the sample was size-fractionated (~175–225 bp) on a 2% agarose gel. After a final PCR amplification step (18 cycles, Phusion polymerase), the resulting DNA libraries were quantified and tested by qPCR at the same specific genomic regions as the original ChiP DNA to assess quality of the amplification reactions. DNA libraries were sent to Illumina Sequencing Services for sequencing on a Genome Analyzer II resulting in ~14 million quality-filtered sequences of length 35 per sample.
mRNA Sequencing
Total RNA from CD4+ Th and Treg cells was send to Fasteris SA, Switzerland for sample preparation and Illumina GA sequencing. Poly-A was purified with oligo-dT magnetic beads (Dynabeads) and full length cDNA was prepared using Superscript III reverse transcriptase and RNAseH. The cDNA was fragmented by nebulization and then treated as described above for the ChiP-seq samples. After PCR amplification the library was quality controlled by cloning 1 µl into a TOPO plasmid and eight clones per library were Sanger sequenced. Libraries were quantified using Q-bit picogreen assay (Invitrogen) and BioAnalyzer (Agilent). Sequencing was performed on the Genome Analyzer II for 2 × 36 cycles (paired-ends) using sequencing kits version 2.0, 100 tiles per lane and data analysis pipeline GAPipeline-1.0rc4.
Bioinformatics analysis workflow
Short read data from mRNA-seq and ChiP-seq experiments were mapped to the human genome (Ensembl 54) using the Genomatix Mapping Station (21) allowing up to 4 mismatches and no gaps. Furthermore, mRNA-seq reads were mapped to a database of artificial splice junctions consisting of all pairs of exons in the human Ensembl 54 release. Mapping to splice junctions was performed using Bowtie (22) allowing for a maximum of two mismatches on either side of the junction and requiring at least 10 bases to match to one part of the junction.
Expression levels for genes were obtained by summarizing all reads mapping to a gene in a sample and computing the RPKM value for a gene as proposed by Mortazavi et al. (23). RPKM values account for the different lengths of genes as well as for the different number of reads measured in two experiments. Reads mapping multiple times in the genome were assigned to the respective genes proportional to the expression level of a gene as measured by unique reads. Fold changes were computed as the ratio of the RPKM values for measured for a gene in two samples and the significance of differential gene expression was computed using the SAGEBetaBin method (24). Only genes with an absolute fold change >1.4 and a SAGEBetaBin significance score <0.01 were used for further steps. For the analysis of de-regulated genes measured by mRNA-seq, only genes with a minimal RPKM value of eight in at least one of the four conditions have been used.
Significant clusters of FoxP3 bound sequences in ChiP-seq data were identified using MACS (14) and a window size of 300 bases, requiring for a P < 10e−6. The minimal number of tags required for a cluster was determined from the input data. Significance values for the overlap between set of genes identified to be bound by FoxP3 in different species, i.e. mouse and human, were computed by the hypergeometric test.
Results were interpreted in the context of biologic processes and functions, as well as networks and pathways through the use of Ingenuity Pathway Analysis (Ingenuity Systems Inc., Redwood City, CA, USA).
RESULTS
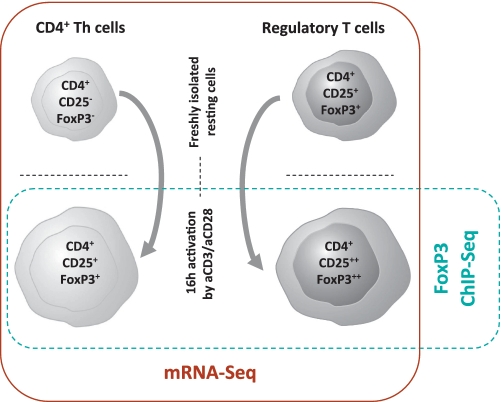
In the following, we will discuss the results of the expression profiling (mRNA-seq) and the analysis of FoxP3 binding-sites by chromatin-immunoprecipitation (ChiP-seq) experiments separately. The experimental setup is shown in Figure 1. Finally, the integration of both data sets allowed us to describe a more comprehensive understanding of the regulatory effects of FoxP3-binding in human activated Treg and CD4+ Th cells.
Figure 1.
Schematic overview of the experimental setup and the different human T cell populations used for RNA-seq and FoxP3 ChIP-seq analysis.
Expression profiling of human Treg and CD4+ Th cells by mRNA-seq
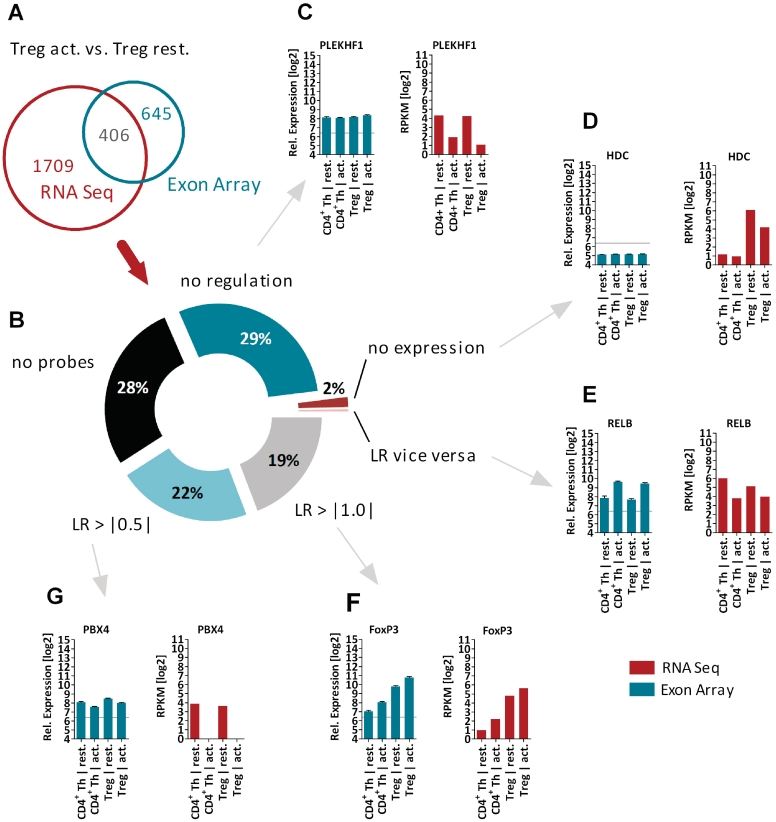
To analyze the transcriptomal landscape of primary human CD4+ Th and Treg cells, we sequenced the transcriptome of resting and activated Treg and CD4+ Th cells using Illumina's mRNA-seq method. Basic read mapping statistics are shown in Table 1. Expression profiling using short read data was performed as described in the ‘Materials and Methods’ section and an overview on the number of genes regulated in the different comparisons can be found in Table 2. Genes with a significant expression level (RPKM > 8) in at least one population and a log2 ratio (LR) of larger than one were taken into account. The differences between two populations in a state-specific manner have been compared, i.e. resting Treg versus resting CD4+ Th cells as well as activated Treg versus activated CD4+ Th cells. Finally, the transition from resting to the activated phenotype within a single cell type has been addressed. To qualify the confidence of the RNA-seq data, we proved the expression pattern of known T-cell markers, which have been described earlier as Treg-specific [e.g. FoxP3 (25,26), HPGD (27), CTLA4 (28), ZNFN1A2 (29,30)], as CD4+ Th cell specific [NKG7 (30), GZMB (30), CD40LG (31)] or as activation-specific [BIC (6), IRF4 (32), TNFRSF9 (30)]: all of them confirmed the expected expression pattern (data not shown). In addition, we compared the expression pattern of all 2115 genes (detected by RNA-seq to be regulated by comparing activated Tregs versus resting Treg cells) to the recently published mRNA expression profiling using Affymetrix Exon Arrays (E-TABM-779) (6). As the protocols for the purification of T-cell populations, the stimulation and the RNA extraction has been exactly the same, the differences between these two studies allowed us to address the differences based on the applied technologies. As shown Figure 2A, only 406 (19%) genes have been identified to be de-regulated by both technologies (cut-off LR > |1.0|, e.g. FoxP3, Figure 2F). We analyzed the remaining 1709 genes (81%) found to be regulated only by mRNA-seq and categorized the reasons for their absence (Figure 2B). 613 genes (29%) showed a significant expression level, but no regulation using the Affymetrix Exon Array technology (i.e. PLEKHF1, Figure 2C). 42 genes (2%) have not shown any significant expression levels using the Affymetrix Exon Arrays (i.e. HDC, Figure 2D). For only 8 genes a vice versa regulation has been observed (i.e. RELB, Figure 2E). 592 genes (28%) have not been measured on the microarray by probesets using the Affymetrix Exon Array core probeset annotation. By using the lower cuf-off of LR > |0.5|, additional 465 genes (22%) have been identified to be regulated also on the Exon Array platform (i.e. PBX4, Figure 2G).
Table 1.
Read mapping statistics for mRNA-seq
| Sample | All reads | Mapped reads | Exonic reads | Splice junction reads |
|---|---|---|---|---|
| Resting CD4+ Th | 15 277 248 | 13 055 740 | 9 998 139 | 551 483 |
| Activated CD4+ Th | 10 335 712 | 8 397 379 | 6 473 155 | 334 714 |
| Resting Treg | 13 655 820 | 11 567 471 | 8 817 608 | 474 661 |
| Activated Treg | 8 237 652 | 6 650 525 | 5 407 197 | 281 711 |
The table is showing the total number of single reads (paired end reads have been treated as two single reads), the number of reads mapping to the human genome, the number of exonic reads as well as the number of reads mapping to splice junctions.
Table 2.
Differentially expressed genes (DEGs) by comparing the RPKM values of separated T-cell populations
| Comparison | LR > 1.0 | LR < −1.0 | LR > |1.0| |
|---|---|---|---|
| Treg act./Treg rest. | 1056 | 1059 | 2115 |
| CD4+ Th act./CD4+ Th rest. | 924 | 926 | 1850 |
| Treg rest./CD4+ Th rest. | 171 | 64 | 235 |
| Treg act./CD4+ Th act. | 377 | 408 | 785 |
Shown are the numbers of genes which are either up-regulated (LR > 1.0) or down-regulated (LR < −1.0). The column LR > |1.0| shows the overall differentially expressed genes, LR = log2 ratio.
Figure 2.
Technology comparison of differentially expressed genes (Treg act. versus CD4+ Th cells). (A) Venn diagram for 2115 genes identified to be regulated by RNA-seq. Only 406 genes (19%) have been identified by the Affymetrix Exon Array technology, i.e. FoxP3 (F). (B) Analysis of reasons for not being detected by array technology. Of genes, 29% showed no regulation, i.e. PLEKHF1 (C); for 2% of genes no significant expression was detectable, i.e. HDC (D); only 42 genes showed a vice versa regulation pattern, i.e. RELB (E); 22% of genes the regulation could be confirmed by using a lower cuf-off for de-regulation (LR < |0.5|), i.e. PBX4 (G) and for 29% of genes no probeset was annotated at the Core data set using the Affymetrix Exon Arrays. The bar chart diagrams show in red for RNA-seq (in log2 RPKM values) and in green for Affymetrix Exon arrays (in log2 values of RMA normalized expression) the relative expression pattern for all profiled T cell populations. RPKM = reads per kilobase of exon per million mapped sequence reads, LR = Log2 ratio.
As recently described by Song et al. (33), also RNA-seq might miss transcripts at lower expression levels. Low expression and potentially insufficient transcript sampling combined with high RPKM threshold result in differences of differentially expressed genes, when compared to microarray experiments [645 genes are detected by arrays only (Figure 2A)]. The ‘Materials and Methods’ section states our very stringent threshold, that genes only with a RPKM > 8 in any of the four conditions have been included. To overcome the sensitivity issue in future experiments, a significant higher number of mapped reads will allow a decrease of the RPKM threshold. In summary, the gain of information achieved by applying RNA-seq weights out the loss of information by 2-fold in comparison to the Affymetrix Exon arrays.
Reasons for the discrepancies in results are mainly technology associated. The differences between microarrays and RNA-seq protocols in IVT amplification versus PCR amplification, in analogous versus digital signal detection, in knowledge of sequences versus unbiased sequencing, in regard to cross-hybridization versus distinguishing paralogous sequences, in spiked quantification versus absolute quantification of low-abundance transcripts have to be considered (17,34). In addition, the RNA-seq offers the identification of novel splice isoforms and the detection of SNPs. Still, for many applications, microarrays are the method of choice. Although the pricing for RNA-seq is reaching comparable levels, RNA-seq protocols still suffer from unknown biases such as those implied by the required ligation steps, and known biases where high abundance transcripts (housekeeping genes and/or ribosomal transcripts) make up the majority of sequencing data if not depleted (e.g. in some tissues 5% of the genes represent up to 75% of the reads sequenced).
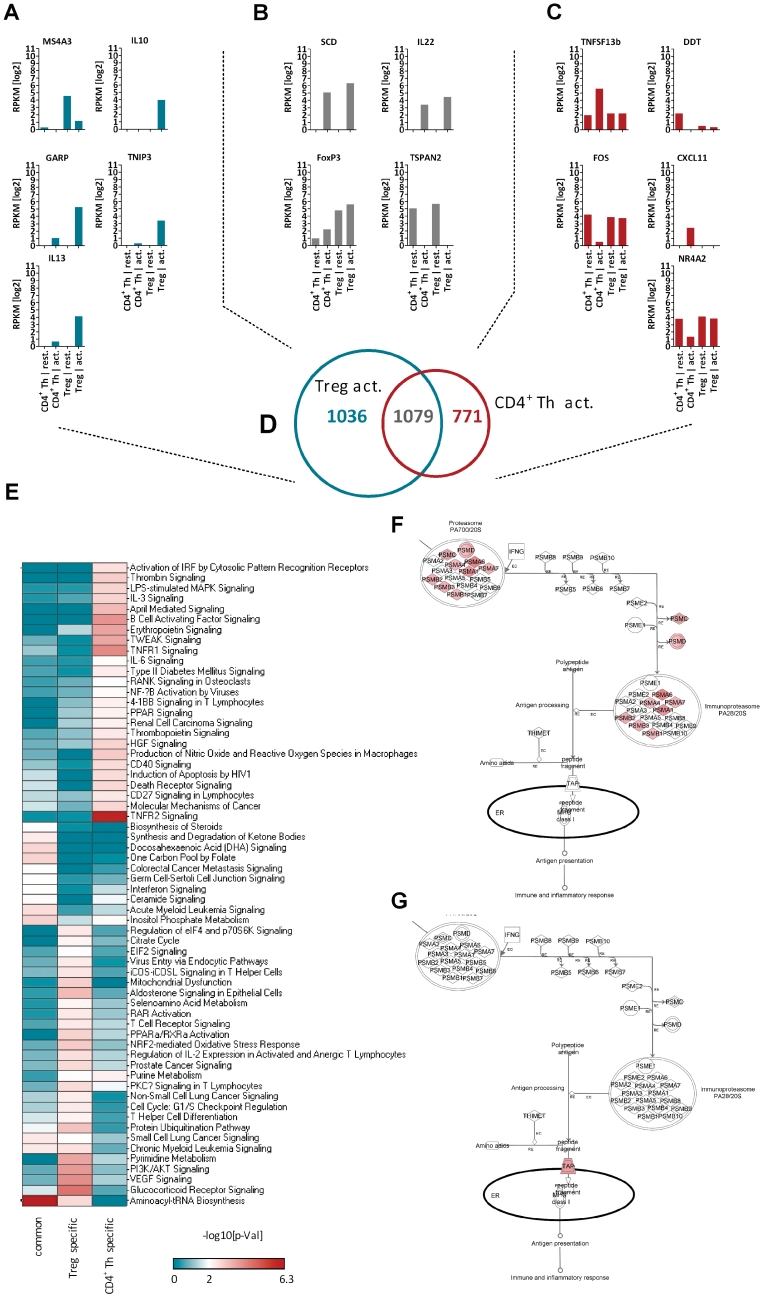
Next we compared the global gene expression pattern between activated Tregs and activated CD4+ Th cells. Therefore, we did not directly compare the gene expression levels of activated Tregs versus activated CD4+ Th cells. Instead of, we started to identify the genes which are regulated upon activation within the Treg populations (2115 genes) and CD4+ Th cell populations (1850 genes) independently. By comparing these two genesets, we were able to identify a geneset of 1036 genes, which are specifically regulated only in Treg cells (i.e. MS4A3, IL10, GARP (LRRC32), TNIP3 or IL-13; Figure 3A) and a geneset of 771 genes, which describe the genes specifically regulated in CD4+ Th cell activation (i.e. TNFSF13b, DDT, FOS, CXCL11 or NR4A2, Figure 3C). Finally, a common geneset of 1079 genes was defined, which contains genes regulated in Tregs as well as in CD4+ Th cells upon 16 h activation (i.e. SCD, IL22, FoxP3 or TSPAN2, Figure 3B). The three activation-specific genesets are shown as Venn diagram in Figure 3D and listed as Supplementary Table S1.
Figure 3.
Pathway Analysis of T cell activation. Shown are (A) genes specifically regulated upon activation of Treg cells. (B) Genes commonly regulated upon activation in both cell populations and (C) genes specifically regulated upon activation of CD4+ Th cells. (D) The Venn diagram shows the intersection of the activation signatures of CD4+ Th and Treg cells. (E) The three activation-specific genesets shown in Venn diagram have been used in Ingenuity Pathway Analysis to enrich influenced signaling pathways. Sixty two canonical signaling pathways scored with a significant −log10[P-value] > 2 for at least one of the three genesets. (F) The overlay of the epression data as LRs to the Protein Ubiquitination Pathway identified a couple members of the protease to be specifically up-regulated in activated Tregs, whereas no modulation of these has been observed after activation of CD4+ Th cells (G). The bar chart diagrams show the relative expression pattern for all in RNA-seq experiments profiled T cell populations in log2 RPKM values. RPKM = reads per kilobase of exon per million mapped sequence reads, LR = Log2 ratio.
The novel finding for Il-13 was proven by RT-PCR (not shown). Upon analyzing the RNA-seq data for additional CD4+ Th/CD4 memory cell marker gene expression (IFNg, IL4, CCR7, as well as CD62L), we are pretty confident that the IL-13 expression pattern did not originate from cell impurities, as it is expressed at that high levels in activated TREGs and did not show a major expression in activated Th cells.
Subsequently, Ingenuity Pathway Analysis was used to analyze the activation-specific genesets to enrich influenced signaling pathways. We found 62 canonical signaling pathways scored with a significant −log10[P-value] > 2 (Fisher's exact test) for at least one of the three genesets (Figure 3E). Upon activation, CD4+ Th cells significantly modulate IRF signaling, MAPK signaling, Tweak signaling, Il-6 signaling, NFκB activation, CD40 signaling, CD27 signaling and TNFR1 as well as TNFR2 signaling, whereas none of these canonical pathway were affected by activation of Tregs (Figure 3E). The other way around, Treg cells significantly modulate upon activation: Glucocorticoide receptor signaling, VEGF signaling, PI3K/AKT signaling, T-cell differentiation, PKCtheta signaling, NRF2 signaling, T-cell receptor signaling, ICOS signaling as well as the Protein Ubiquitination pathway (Figure 3E).
Each modulated signaling pathway was overlaid by the expression data to visualize the biologic significance, i.e. the Protein Ubiquitination Pathway with the LR (log2 expression changes) of Treg cells as well as with the LR of CD4+ Th cells as shown in Figure 3F or G. In CD4+ Th cells the proteasome is not affected by the activation thereof (Figure 3G). But specifically in activated Tregs a couple of the proteasome subunits are found to be strongly up-regulated (Figure 3F). The proteasome is a large multicatalytic complex which drives organized protein degradation. The proteasome plays a straightforward but critical role in the function of the adaptive immune system. Peptide antigens are displayed by the major histocompatibility complex class I (MHC) proteins on the surface of antigen-presenting cells. These peptides are products of proteasomal degradation of proteins originated by the invading pathogen. Although constitutively expressed proteasomes can participate in this process, a specialized complex composed of proteins whose expression is induced by interferon gamma produces peptides of the optimal size and composition for MHC binding. These proteins whose expression increases during the immune response include the 11S regulatory particle, whose main known biologic role is regulating the production of MHC ligands, and specialized β subunits called β1i, β2i and β5i with altered substrate specificity. The complex formed with the specialized β subunits is known as the immunoproteasome (35). The immunoproteasome seems to be up-regulated in both Treg and CD4+ Th cells upon activation, whereas the ‘standard’ proteasome is only up-regulated in Tregs upon activation (Figure 3F or G). This Treg specific regulation might explain the observation that long term culture of CD4+ Th cells in the presence of a small molecule inhibitor for LMP7 (also known as PSMB8, a catalytic subunit of the immunoproteasome) gave rise to a increased regulatory T-cell population (36).
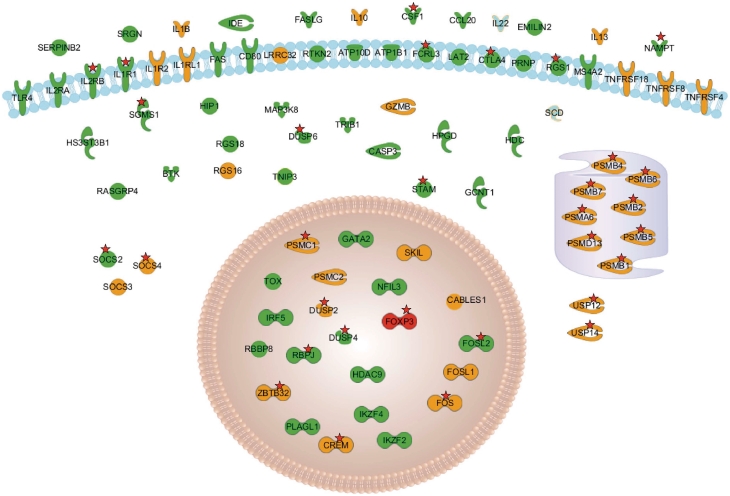
Figure 4 displays the some interesting marker genes distinguishing Treg from CD4+ Th cells. The complete picture can be found in Supplementary Table S1. Those include general Treg marker genes like FoxP3, CTLA4, TLR4, IL2RA and IL2RB as well as Treg specific activation markers [e.g. CABLES1, GARP (LRRC32) and TNFRSF18 (GITR)] that are found to be specifically up-regulated in activated Tregs only. Some of those genes correspond to newly identified lineage markers (i.e. HDC, BTK and GATA2) which have not been described before.
Figure 4.
Set of genes and their subcellular localization which are up-regulated in Treg cells compared to CD4+ Th cells. Genes shown in green are up-regulated in activated and resting Treg cells; orange genes are up-regulated upon activation of Treg cells but are not up-regulated upon CD4+ Th activation. The red stars mark genes which are bound by FoxP3 in activated Treg cells.
Finally, as shown in Figure 2F, FoxP3 is found to be expressed in all four samples. Low expression levels are found in CD4+ Th cells (resting expression level: 1.9 RPKM, activated expression level: 4.6 RPKM). In contrast, expression in Treg cells is >10-fold higher with an RPKM value of 27.4 in resting and 48.8 in activated Treg cells. Genes, whose promoters are bound by FoxP3 and the implications of FoxP3 binding on gene expression are discussed in detail below.
Identification of splice variant expression in mRNA-seq data
Alternative splicing plays important roles in many biologic processes (37) and has also been described to be of importance in the immune system (38) and also during T-cell activation (39). In addition to the detection in gene expression levels, the mRNA-seq technique allows also the analysis for splice variants. Dependent of the complexity of gene-specific splicing behavior, present or absent transcripts are detectable for each sample.
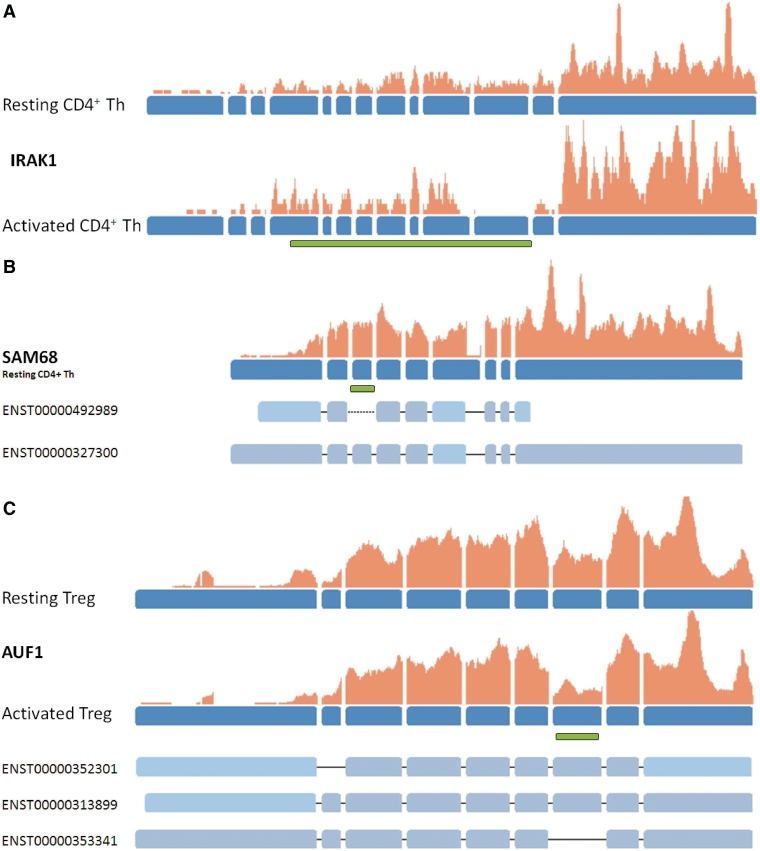
For IRAK1 (Interleukin receptor 1 associated kinase), a serine/threonine kinase, three different splice variants with opposing function are known (38,40). In our data set we find two splice variants (full length IRAK1 and IRAK1c) to be expressed. The full-length isoform of IRAK1 is detectable only in resting cell states of Tregs and CD4+ Th cells. Upon activation of these cells, exon 11 seems to be skipped: surprisingly, only the truncated isoform IRAK1c is detectable in activated CD4+ Th cells (Figure 5A). The skipped exon encodes to the C-terminal part of the kinase domain. This smaller variant isoform therefore likely lacks kinase activity, although it retains the ability to interact with many IRAK1-binding partners. It may function as a negative-feedback loop in which IRAK1 function is decreased in response to IL-1 stimulation. The presence and regulation of the truncated form of IRAK1 was proven indirectly by qRT–PCR using Roche's UPL technology (Supplementary Figure S1).
Figure 5.
Examples for the detection of splice variants in Treg and CD4+ Th cells. Exons of the genes are shown in blue, read coverage at a specific position is indicated by the orange bars. Examples for known transcript models of a gene are shown below the gene structures where solid lines indicate that a specific splice junction has been identified in the data while dotted lines indicate that a junction has not been found. Green bars indicate the location of specific protein domains. (A) displays changes in the expression of exon 11 between resting and activated CD4+ Th cells corresponding to the partial deletion of the kinase domain (shown by the green bar) of IRAK1. (B) For the SAM68 gene, exon skipping of exon 3 which represents a KH RNA binding domain (PF00013, indicated by the green bar) is only observed in resting Treg and CD4+ Th cells and never in activated states. (C) For AUF1 inclusion or exclusion of a Glycin-rich region (shown by the green bar) is found to occur in different levels in activated Treg and CD4+ Th cells compared to resting states.
SAM68 (shown in Figure 5B) is a protein which is recruited to the T-cell receptor and, once phosphorylated, functions as an adapter protein in signal transduction cascades by binding to SH2 and SH3 domain-containing proteins. In our data set, splicing events skipping the KH domain (PF00013), a domain known to be important for RNA binding, occurs only in resting CD4+ Th and Treg cells but is not observed in activated states. Variants lacking the KH domain have been shown to be expressed in growth-arrested cells only and are known to inhibit S phase (41).
For AUF1 (Heterogeneous nuclear ribonucleoprotein D0), shown in Figure 5C, different rates for the inclusion and exclusion of a Glycin-rich region are found. This region was shown to be important for the RNA-binding function (42). While in resting CD4+ Th and Treg cells both isoforms (with and/or without exon7) seem to be expressed at equal levels, a clear shift towards the isoform skipping exon7 is detectable in activated Treg cells. Evidence is shown in Figure 5 by the expression level of exon 7 and the number of junction reads voting for either variant (data not shown). The presence, majority and the regulation of the truncated form of AUF1 was confirmed by qRT–PCR using Roche's UPL technology (Supplementary Figure S2).
Two different splice variants have been described for FoxP3 in human (43). One isoform originates from an in-frame exon skipping event of exon 3 (protein coding exon 2) leading to a protein product which lacks 34 amino acids. Neither the forkhead nor the zinc-finger domains are affected by this event. Both variants have also been shown to be functional (43) such that their biologic role still has to be elucidated. In our mRNA-seq data, both variants are detected as being present in resting Treg cells. In activated Treg cells the number of reads mapping the 5′-end of the gene is very small and, likely due to missing data, no indication for the skipping event is found.
Other proteins for which differentially expressed splice variants are detected are IL2RA, LEF1 (both express mixtures of transcripts) and FYN. Specifically between resting and activated cell states for FYN different inclusion rates are found. Overall, this data displays the potential of mRNA-seq data to enable the analysis of state and cell specific splice variant expression and highlights the fact that interesting and important functions during CD4+ Th and Treg activation may be mediated and performed through differentially expressed isoforms.
Identification of Foxp3 binding sites in human T-cell populations and comparison to array-based measurements in mouse and human
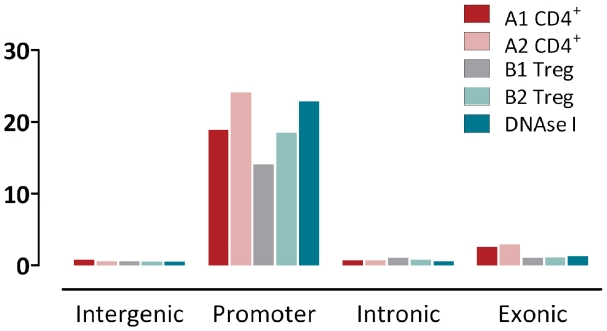
We performed ChiP-seq to identify genomic regions which are bound by FoxP3 in activated CD4+ Th and activated Treg cells of two human donors (biologic replicates). FoxP3-bound genomic DNA fragments from CD4+ Th and Treg cells were sequenced as described in the ‘Material and Methods’ section. Short sequence reads were mapped to the human genome and significant peaks, i.e. clusters of reads, were identified using MACS (14) and the cluster statistics are displayed in Table 3. In both cell populations and biologic replicates a large number of clusters were found, with many more clusters in Treg cells compared to CD4+ Th cells, likely due to higher expression levels of FoxP3. Clusters are mapped to intergenic, intronic, exonic or known promoter regions and, as shown in Figure 6, promoter regions appear to be highly enriched. They are mapped 14 to 24 times more often than expected by chance. As shown in Table 3, 32–51% of all clusters are mapped to known promoter sites (Ensembl 54). Genes detected in different biologic replicates overlap to a large extent. With respect to the smaller data set, 1264 (80.7%) of the CD4+ Th cell genes and 5627 (88.7%) of the Treg genes are identified in both biologic replicates indicating that the experiment in general returns highly reproducible results. 1170 genes (74% of the genes identified in sample A1) are detected in all four samples to be bound by FoxP3. Further, 1317 genes are found in A2 as well as B1 and B2. Those 2487 genes are likely to represent a set of genes which is bound by FoxP3 in both cell types. Only 26 genes are found in CD4+ Th cells only (samples A1 and A2). In contrast, 2897 genes are identified whose promoters are only bound by FoxP3 in Treg cells and which are candidates for a Treg specific regulatory effect of FoxP3.
Table 3.
FoxP3 cluster statistics for CD4+ Th and Treg cell populations obtained by Illumina ChiP-seq in two biologic replicates
| CD4+ Th (A1) | CD4+ Th (A2) | Treg (B1) | Treg (B2) | |
|---|---|---|---|---|
| Total number of clusters | 4429 | 7524 | 25 341 | 22 558 |
| Clusters overlapping with promoters | 1873 | 3876 | 8202 | 8976 |
| Clusters overlapping with DNAse I sites | 2930 | 5153 | 13 956 | 14 207 |
| Distinct genes mapped in promoter regions | 1566 | 3345 | 6341 | 7584 |
| Genes mapped in both biologic replicates | 1264 | 5627 | ||
Figure 6.
Cluster statistics for human FoxP3 clusters identified by MACS which shows the high enrichment of promoter elements overlapping with MACS clusters. Biologic replicates of isolated human CD4+ T-cells (A1 and A2) and human regulatory T-cells (B1 and B2).
To further evaluate the quality of the FoxP3 clusters obtained in our experiment we tested to what extent ChiP-seq clusters are associated to DNAse I hypersensitive sites from human primary CD4+ T-cells which have been published by Boyle et al. (44). DNAse I hypersensitive sites have been historically used to identify the location of regulatory regions, including enhancers, silencers, promoters and other locus control regions and can therefore provide additional evidence regarding data quality. The DNAse I sites were mapped onto the human genome and were subsequently clustered by MACS. As shown in Figure 6, DNAse I hypersensitive sites found in all four genomic categories but, just like FoxP3 clusters, they are highly enriched in promoter regions. The overlap between FoxP3 clusters and DNAse I clusters is even higher than the overlap of FoxP3 clusters with promoter regions. Between 55 and 68% of all FoxP3 clusters overlap with DNAse I clusters. Therefore, a larger fraction of FoxP3 binding sites which do not map to known promoter sites might be bound in other regions of regulatory relevance (i.e. cis- and/or trans-active regulatory elements like enhancer or silencers).
Using the Genomatix collection of transcription factor binding matrices, we searched for other, enriched transcription factor binding motifs in promoter regions bound by FoxP3. In both biologic replicates (B1 and B2) several transcription factor binding matrices are identified in FoxP3 bound regions more often than expected. Among the top scoring matrix families, four appear to be especially interesting in the biologic context. This includes the ETS-1 as well as the HAML (human acute myelogenous leukemia) factor family. Cooperative binding of members of the two families has been described in several cases in the context of T-cells including the cooperative binding of AML1 and Ets-1 to the TCR alpha enhancer (45) as well as composite binding of Ets-1 and CBF (another member of the HAML family) in the regulation of antigen receptor genes (46). Futhermore, binding sites for AP1F are overrepresented in FoxP3 bound regions. Members of this family are known to interact with both HAML and ETSF in various cases including the activation of GM-CSF in T-cells (47) and FoxP3 has been shown to suppress AP-1 transcriptional activity (48). Finally, interferon regulatory factor (IRFF) binding sites are overrepresented. A member of this family, IRF4, has been described to interact with multiple Ets factors to modulate CD68 expression in a cell type-specific manner (49). Also, it has only recently been shown (50) that in mouse Treg cells, high amounts of interferon regulatory factor-4 (IRF4) is dependent on Foxp3 expression. The authors proposed that IRF4 expression endows Treg cells with the ability to suppress TH2 responses a fact which might be confirmed by the over-representation of IRFF binding sites in FoxP3 bound genes such that collaborative binding might occur. The NFAT transcription factor motif which has been shown previously to interact with FoxP3 (51) is also overrepresented although less significantly than the four matrices described above.
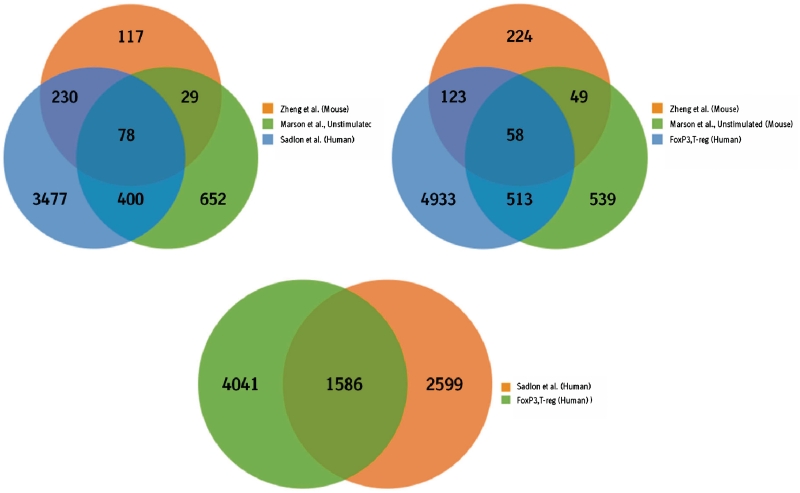
Next, we tested to what extent genes bound by FoxP3 in our human data set have also been identified in two studies by Zheng et al. (8) and Marson et al. (9) that analyzed FoxP3 occupancy in mouse and one recent study by Sadlon et al. (10) who identified human FoxP3 targets using ChiP-on-Chip experiments. In the Zheng data set, 1277 FoxP3 binding sites are annotated which correspond to 519 mouse genes in Ensembl 54. Out of those 454 can be mapped to a corresponding human ortholog and are used in subsequent analysis steps. In the Marson data set, 1335 binding sites in unstimulated and 1119 binding sites in stimulated hybridoma cells were identified (at a false discovery rate of 5%) which map to 1265 and 1055 distinct mouse genes. Out of those 1162 genes in unstimulated and 960 genes in stimulated cells have an annotated ortholog in the human genome. The 5579 genes identified by Sadlon et al. (10) in human correspond to 4185 human genes in Ensembl 54. The overlap between the genes in the three published data sets and the 5627 human FoxP3 bound genes in Treg cells is shown in Figure 7. It is interesting to note that the overlap of the two published mouse data sets is relatively small. Only 107 genes are detected in both data sets. Although this overlap is highly significant (P < 1.66e−42) it shows that even within the same species larger differences in genes detected to be bound by FoxP3 can be expected either due to the experimental design like cell lines or stimulation or due to genetic and biologic variance. Human FoxP3 bound genes in our data set significantly overlap with mouse genes. The human data set contains 181 genes detected in the Zheng study (P < 1.91e−14) and 561 genes from the Marson data set (P < 3.453e−77). 58 genes are found in all three studies and 743 genes are found in at least two data sets (49 of which are only found in the mouse). The largest and most significant overlap is found for the two human data sets. 1586 genes are consistently identified in both data sets as FoxP3 target genes (P < 1.65e−112) and are therefore consistent between two different experimental setups and two different technologies.
Figure 7.
Comparison of FoxP3 bound genes in human T-reg cells and three published data sets in mouse and human. FoxP3 target genes published by other groups have been identified using array based technologies (ChIP on Chip).
The effects of FoxP3 binding onto gene expression in human T-cells
As described above, 2897 genes are specifically bound by FoxP3 in activated Treg cells, 26 only in CD4+ Th cells and 2487 genes are bound in both cell types. We integrated this information with expression data from the mRNA-seq experiment of activated CD4+ Th and activated Treg cells, as discussed above. 3255 genes have been identified to be significantly regulated (absolute fold-change >1.4 and significance value <0.01) between activated CD4+ Th and Treg cells by mRNA-seq analysis. 1451 (44%) of those genes are also bound by FoxP3 either in Treg only (800 genes, 399 up-regulated, 401 down-regulated) or in Treg and CD4+ Th cells (647 genes, 362 upregulated, 285 downregulated). The fact that 44% of all regulated genes between activated Treg and CD4+ Th cells are bound by FoxP3 proofs its important role for activated Treg cells. The overlap is statistically highly significant (P < 4.55e−193). Marson et al. (9) described a highly suppressive function of FoxP3 in their data set where many more genes were down-regulated than up-regulated upon FoxP3 binding, a view which has also been taken by Ziegler et al. (2). In contrast to those findings and in correspondence with the Zheng study (8), our human data set suggests that activation and repression of genes by FoxP3 seem to be well balanced.
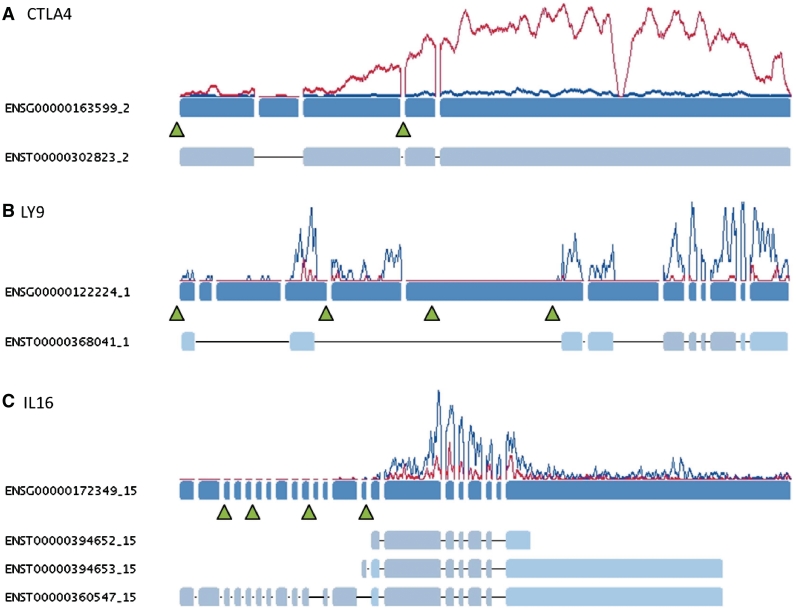
The gene which shows the highest up-regulation (17-fold) in Treg cells is CTLA4 which is bound by FoxP3 according to ChiP-seq data in two clusters as shown in Figure 8A. Other examples are IL1R1 (up-regulation: 13-fold), DUSP2, DUSP4 and DUSP6 (up to 8-fold) as well as CXCR6 and CCR4 (up to 4-fold). Interestingly, FoxP3 binds to its own promoter in Treg cells and may therefore regulate its own expression levels.
Figure 8.
Examples for the modulation of transcript expression by FoxP3. Exons of the genes are shown in blue, read coverage at a specific position is indicated by the red (activated Treg cells) and blue (activated CD4+ Th cells) lines. Examples for known transcript models of a gene are shown below the gene structure. Green arrows indicate positions of significant FoxP3 binding site peaks identified in activated Treg cells of both donors. (A) shows the up-regulation of CTLA4 likely mediated by two FoxP3 clusters, (B) LY9 is bound by FoxP3 at multiple sites in both cell lines but binding strength is 10× higher in Treg than in CD4+ TH cells, leading to the almost complete repression of the gene in Treg cells, (C) Short isoforms of the IL16 gene are known to be specifically expressed in T-cells. Expression and repression of the short variant of the gene may be mediated by FoxP3 which is bound to multiple sites of the gene including the promoter site of the short isoform in both cell types.
On the other hand, 401 genes bound by FoxP3 are significantly down-regulated in Treg cells. In our data set those genes include DPEP2 (60-fold down-regulated), ITGA4 (6-fold), TLR1 (7-fold), IFIT2 (8-fold), TRAT1 (3-fold) which is a positive regulator of the T-cell receptor signaling pathway as well as members of the TRIM (tripartite motif-containing) family (TRIM21, TRIM22, TRIM38, TRIM56). TRIM21 is known to increase IL2 production in T-cells (52).
2487 genes are bound by FoxP3 in activated CD4+ Th and Treg cells. Out of those, 647 (26%) genes appear to be significantly de-regulated in the comparison between the two cell types. Although there is no general correlation between binding strength (as measured by the number of reads found in a promoter cluster) and the resulting fold change in the gene expression measurement (see Supplementary Table S2), for 503 of those 647 genes (78%) binding strength is at least 2-fold higher in Treg cells compared to CD4+ Th cells. 362 genes are up-regulated in Treg cells while 285 genes are down-regulated. For example, IL16 and LY9 (lymphocyte antigen 9), which are both shown in Figure 8B and C, as well as IL7R appear to be down-regulated, while e.g. RGS1, IL2RB, FOS and FOSL2 (both part of the AP-1 transcription complex) are up-regulated.
Interestingly, the up-regulation of proteasome components in Treg cells discussed above clearly seems to be mediated by FoxP3 as many proteasome genes are bound by FoxP3 in their promoter regions (see also Figure 4). Also, as shown in Figure 8C for the example of IL16, FoxP3 may also be involved in the use of specific alternative transcription start sites in order to mediate expression of the T-cell specific isoform of the gene.
DISCUSSION
Treg cells play an essential role in the negative regulation of immune answers and FoxP3 has been shown to be one of the major regulator genes for developing the Treg gene signature. But despite many research in the field the mechanisms leading to the specific gene expression patterns are not completely understood. The last years have also shown that transcription factor binding may differ strongly from species to species such that results achieved in one model organism may not necessarily be transferred to others.
In this study we have analyzed for the effects of FoxP3 transcription factor binding on the development of the Treg gene signature in humans using the most recent and also most sensitive technological platforms available. We combined two approaches relying on next generation sequencing, namely mRNA-seq and ChiP-seq, which, in combination, allow for very detailed insights into the complex transcriptomal landscape of higher eukaryotes.
We have performed expression profiling of resting and activated Treg and CD4+ Th cells and identified gene signatures that are specifically de-regulated in the different cell types (Treg and CD4+ Th signature) or are regulated in both cell types upon activation (Activation signature). Signatures were compared to sets of genes identified in a similar study using Affymetrix Exon Arrays. In correspondence with other authors we find that while the key players are identified by both technologies, NGS can identify many more de-regulated genes likely to its higher sensitivity and the fact that any gene can be profilied without having to rely on pre-defined sets of probesets. Analysis of de-regulated pathways shows significant differences between Treg and CD4+ Th cells not only on the level of single genes.
Furthermore, using NGS data we can identify differences in splicing patterns like the expression of a kinase-dead isoform of the IRAK1 gene upon T-cell activation. Although it is not yet clear what function this kinase-dead isoform has in vivo, it is interesting to note that expression of the small isoform has been shown to be specific for activated Treg and CD4+ Th cells whereas both cell types express the full length isoform in resting states. Overexpression of IRAK1c in 293T or G292 cells suppressed NF-κB activation and blocked IL-1b-induced IL-6 as well as LPS and CpG-induced TNFα production. Mechanistically evidence was shown that IRAK1c functions as a dominant negative by failing to be phosphorylated by IRAK4, thus remaining associated with Tollip and blocking NF-κB activation (40). The presence of a regulated, alternative splice variant IRAK1c in activated T cells, that functions as a kinase-dead, dominant-negative protein adds further complexity to the variety of mechanisms that regulate TIR signaling and the subsequent inflammatory response.
By using ChiP-seq we identified genes bound by FoxP3 in activated Treg and CD4+ Th cells. By identifying promoter regions with significant FoxP3 peaks in short read data we obtained a set of more than 5000 genes which may be subject to a FoxP3 mediated regulation of the corresponding gene. Genes identified to be bound in human show a highly significant overlap with two published FoxP3 data sets in the mouse, although many more genes were identified in human. This is likely due to the higher sensitivity of ChiP-seq compared to array-based detection methods which again require a priori knowledge. Promoter regions bound by FoxP3 additionally show enrichment for other transcription factor binding sites like ETS-1, HAML, AP1 and IRFF (including IRF4) which are novel candidates for a cooperative function in controling T-cell development in human. Especially the correlation of FoxP3 binding sites with IRF4 appears to be interesting, as IRF4 has only recently been shown to be transcriptionally dependent on FoxP3.
In a last step, we combined both data sets in order to understand the regulatory effects of FoxP3 on the specific gene expression signature of Treg cells. More than 40% of the genes which appear to be de-regulated in activated Treg cells compared to activated CD4+ Th cells are bound by FoxP3 in their promoter regions showing the large importance of this transcription factor for developing the Treg signature. In contrast to some previous studies which proposed the major function of FoxP3 as a suppressor, we find that activation and repression of genes by FoxP3 are likely to be well balanced in human. Nobody before profiled that comprehensively human primary T cell populations. The functional relevance of the identified FoxP3-bound genes has been confirmed by earlier studies. In 2009, we published the functional relevance of FoxP3-bound miR-155 in sensititation CD4+ Th Cells for TREG mediated suppression (53). In addition, we (in human) and others (in mouse) could identify PDE3b as a FoxP3 target gene (54). PDE3b is involved in controlling the abundance of cyclic adenosine monophosphate (cAMP) and cAMP is known to suppress T cell function. Because Foxp3 blocks PDE3B expression, regulatory T cells may contain higher amounts of cAMP, which may play an important role in regulatory T cell function (54,55). Further examples for TREG-mediated T cell suppression of FoxP3 target genes have been described: DAB2 (56), CTLA4 (57) and EBI3 (IL-27/IL-35) (58). Beyond these selected examples, we did integrate the human FoxP3 occupancy and expression data with expression profiles derived from T cell populations of scurfy mice (FoxP3-deficient) (59,60). We downloaded the raw data (E-GEOD-11775), normalized, orthologous aligned (mouse to human) and integrated them with FoxP3-bound genes. A significant overlap of FoxP3-bound genes (ChIP-seq), which have been identified to be regulated upon T cell activation in human (RNA-seq), revealed no major regulation in activated T cell populations from scurfy mice (data not shown). Although there are differences in FoxP3 function and expression between human and mouse (61), by that kind of analysis we have been able to show indirect proof of functional relevance for the FoxP3-bound genes.
Taken together our study provides a comprehensive data set which allows analyzing the FoxP3 dependent development of the Treg gene signature as well as the differences between Treg cell and CD4+ Th cells in general. The use of next generation sequencing provides significantly deeper insights into the complex transcriptomal landscape of higher eukaryotes compared to array-based technologies and will help to further sharpen our understanding of transcriptional regulation of gene expression by transcription factors in the future.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online. The raw sequencing data have been uploaded to http://www.ncbi.nlm.nih.gov/sra under the accession number SRP006674.
FUNDING
The Boehringer Ingelheim Pharma GmbH & Co. KG. The funder was involved in study design and decision to publish the data and results (and to make them publicly available). The funder had no role in data collection and analysis or preparation of the article. Funding for open access charge: Boehringer Ingelheim Pharma GmbH & Co. KG will fully cover all publication charges. This study was part of the PhD project sponsored by Boehringer Ingelheim Pharma GmbH & Co. KG.
Conflict of interest statement. F.B., T.F., H.S., M.C.L., E.S., A.W. and T.H. are employees of Boehringer Ingelheim Pharma GmbH & Co. KG. DM is an employees of Boehringer Ingelheim Pharmaceuticals Inc. This does not alter the authors’ adherence to all the NAR policies on sharing data and materials.
Supplementary Material
ACKNOWLEDGEMENTS
We would like to thank Laurent Farinelli at Fasteris (Geneva, Switzerland) for discussions during the course of and performing of the RNA-seq work. We also thank Paul Labhart and Vassili Alexiadis at Genpathway Inc., (San Diego, USA) for DNA sample preparation and running the ChIP protocols. We are also grateful to Karsten Quast for bioinformatic assistance, Franz-Josef Schneider and Florian Gantner for their permanent support.
REFERENCES
- 1.Feuerer M, Hill JA, Mathis D, Benoist C. Foxp3+ regulatory T cells: differentiation, specification, subphenotypes. Nat. Immunol. 2009;10:689–695. doi: 10.1038/ni.1760. [DOI] [PubMed] [Google Scholar]
- 2.Ziegler SF. FOXP3: of mice and men. Annu. Rev. Immunol. 2006;24:209–226. doi: 10.1146/annurev.immunol.24.021605.090547. [DOI] [PubMed] [Google Scholar]
- 3.Lin W, Haribhai D, Relland LM, Truong N, Carlson MR, Williams CB, Chatila TA. Regulatory T cell development in the absence of functional Foxp3. Nat. Immunol. 2007;8:359–368. doi: 10.1038/ni1445. [DOI] [PubMed] [Google Scholar]
- 4.Sugimoto N, Oida T, Hirota K, Nakamura K, Nomura T, Uchiyama T, Sakaguchi S. Foxp3-dependent and -independent molecules specific for CD25+CD4+ natural regulatory T cells revealed by DNA microarray analysis. Int. Immunol. 2006;18:1197–1209. doi: 10.1093/intimm/dxl060. [DOI] [PubMed] [Google Scholar]
- 5.Williams LM, Rudensky AY. Maintenance of the Foxp3-dependent developmental program in mature regulatory T cells requires continued expression of Foxp3. Nat. Immunol. 2007;8:277–284. doi: 10.1038/ni1437. [DOI] [PubMed] [Google Scholar]
- 6.Stahl HF, Fauti T, Ullrich N, Bopp T, Kubach J, Rust W, Labhart P, Alexiadis V, Becker C, Hafner M, et al. miR-155 inhibition sensitizes CD4+ Th cells for TREG mediated suppression. PLoS ONE. 2009;4:e7158. doi: 10.1371/journal.pone.0007158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pfoertner S, Jeron A, Probst-Kepper M, Guzman CA, Hansen W, Westendorf AM, Toepfer T, Schrader AJ, Franzke A, Buer J, et al. Signatures of human regulatory T cells: an encounter with old friends and new players. Genome Biol. 2006;7:R54. doi: 10.1186/gb-2006-7-7-r54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zheng Y, Josefowicz SZ, Kas A, Chu TT, Gavin MA, Rudensky AY. Genome-wide analysis of Foxp3 target genes in developing and mature regulatory T cells. Nature. 2007;445:936–940. doi: 10.1038/nature05563. [DOI] [PubMed] [Google Scholar]
- 9.Marson A, Kretschmer K, Frampton GM, Jacobsen ES, Polansky JK, MacIsaac KD, Levine SS, Fraenkel E, von BH, Young RA. Foxp3 occupancy and regulation of key target genes during T-cell stimulation. Nature. 2007;445:931–935. doi: 10.1038/nature05478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sadlon TJ, Wilkinson BG, Pederson S, Brown CY, Bresatz S, Gargett T, Melville EL, Peng K, D'Andrea RJ, Glonek GG, et al. Genome-wide identification of human FOXP3 target genes in natural regulatory T cells. J. Immunol. 2010;185:1071–1081. doi: 10.4049/jimmunol.1000082. [DOI] [PubMed] [Google Scholar]
- 11.Kahvejian A, Quackenbush J, Thompson JF. What would you do if you could sequence everything? Nat. Biotechnol. 2008;26:1125–1133. doi: 10.1038/nbt1494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Roach JC, Glusman G, Smit AF, Huff CD, Hubley R, Shannon PT, Rowen L, Pant KP, Goodman N, Bamshad M, et al. Analysis of Genetic Inheritance in a Family Quartet by Whole-Genome Sequencing. Science. 2010;328:636–639. doi: 10.1126/science.1186802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lieberman-Aiden E, van Berkum NL, Williams L, Imakaev M, Ragoczy T, Telling A, Amit I, Lajoie BR, Sabo PJ, Dorschner MO, et al. Comprehensive mapping of long-range interactions reveals folding principles of the human genome. Science. 2009;326:289–293. doi: 10.1126/science.1181369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhang Y, Liu T, Meyer CA, Eeckhoute J, Johnson DS, Bernstein BE, Nussbaum C, Myers RM, Brown M, Li W, et al. Model-based analysis of ChIP-Seq (MACS) Genome Biol. 2008;9:R137. doi: 10.1186/gb-2008-9-9-r137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mardis ER. ChIP-seq: welcome to the new frontier. Nat. Methods. 2007;4:613–614. doi: 10.1038/nmeth0807-613. [DOI] [PubMed] [Google Scholar]
- 16.Wang ET, Sandberg R, Luo S, Khrebtukova I, Zhang L, Mayr C, Kingsmore SF, Schroth GP, Burge CB. Alternative isoform regulation in human tissue transcriptomes. Nature. 2008;456:470–476. doi: 10.1038/nature07509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tang F, Barbacioru C, Wang Y, Nordman E, Lee C, Xu N, Wang X, Bodeau J, Tuch BB, Siddiqui A, et al. mRNA-Seq whole-transcriptome analysis of a single cell. Nat. Methods. 2009;6:377–382. doi: 10.1038/nmeth.1315. [DOI] [PubMed] [Google Scholar]
- 18.Birzele F, Schaub J, Rust W, Clemens C, Baum P, Kaufmann H, Weith A, Schulz TW, Hildebrandt T. Into the unknown: expression profiling without genome sequence information in CHO by next generation sequencing. Nucleic Acids Res. 2010;38:3999–4010. doi: 10.1093/nar/gkq116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Becker C, Kubach J, Wijdenes J, Knop J, Jonuleit H. CD4-mediated functional activation of human CD4+CD25+ regulatory T cells. Eur. J. Immunol. 2007;37:1217–1223. doi: 10.1002/eji.200636480. [DOI] [PubMed] [Google Scholar]
- 20.Labhart P, Karmakar S, Salicru EM, Egan BS, Alexiadis V, O'Malley BW, Smith CL. Identification of target genes in breast cancer cells directly regulated by the SRC-3/AIB1 coactivator. Proc. Natl Acad. Sci. USA. 2005;102:1339–1344. doi: 10.1073/pnas.0409578102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sultan M, Schulz MH, Richard H, Magen A, Klingenhoff A, Scherf M, Seifert M, Borodina T, Soldatov A, Parkhomchuk D, et al. A global view of gene activity and alternative splicing by deep sequencing of the human transcriptome. Science. 2008;321:956–960. doi: 10.1126/science.1160342. [DOI] [PubMed] [Google Scholar]
- 22.Langmead B, Trapnell C, Pop M, Salzberg SL. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009;10:R25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mortazavi A, Williams BA, McCue K, Schaeffer L, Wold B. Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat. Methods. 2008;5:621–628. doi: 10.1038/nmeth.1226. [DOI] [PubMed] [Google Scholar]
- 24.Vencio RZ, Brentani H, Patrao DF, Pereira CA. Bayesian model accounting for within-class biological variability in Serial Analysis of Gene Expression (SAGE) BMC Bioinformatics. 2004;5:119. doi: 10.1186/1471-2105-5-119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hori S, Nomura T, Sakaguchi S. Control of regulatory T cell development by the transcription factor Foxp3. Science. 2003;299:1057–1061. [PubMed] [Google Scholar]
- 26.Sakaguchi S. Naturally arising Foxp3-expressing CD25+CD4+ regulatory T cells in immunological tolerance to self and non-self. Nat. Immunol. 2005;6:345–352. doi: 10.1038/ni1178. [DOI] [PubMed] [Google Scholar]
- 27.Eruslanov E, Kaliberov S, Daurkin I, Kaliberova L, Buchsbaum D, Vieweg J, Kusmartsev S. Altered expression of 15-hydroxyprostaglandin dehydrogenase in tumor-infiltrated CD11b myeloid cells: a mechanism for immune evasion in cancer. J. Immunol. 2009;182:7548–7557. doi: 10.4049/jimmunol.0802358. [DOI] [PubMed] [Google Scholar]
- 28.Sansom DM, Walker LS. The role of CD28 and cytotoxic T-lymphocyte antigen-4 (CTLA-4) in regulatory T-cell biology. Immunol. Rev. 2006;212:131–148. doi: 10.1111/j.0105-2896.2006.00419.x. [DOI] [PubMed] [Google Scholar]
- 29.Thornton AM, Korty PE, Tran DQ, Wohlfert EA, Murray PE, Belkaid Y, Shevach EM. Expression of Helios, an Ikaros transcription factor family member, differentiates thymic-derived from peripherally induced Foxp3+ T regulatory cells. J. Immunol. 2010;184:3433–3441. doi: 10.4049/jimmunol.0904028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lin W, Haribhai D, Relland LM, Truong N, Carlson MR, Williams CB, Chatila TA. Regulatory T cell development in the absence of functional Foxp3. Nat. Immunol. 2007;8:359–368. doi: 10.1038/ni1445. [DOI] [PubMed] [Google Scholar]
- 31.Jylhava J, Eklund C, Jylha M, Hervonen A, Hurme M. Expression profiling of immune-associated genes in peripheral blood mononuclear cells reveals baseline differences in co-stimulatory signalling between nonagenarians and younger controls: the vitality 90+ study. Biogerontology. 2010;11:671–677. doi: 10.1007/s10522-010-9274-7. [DOI] [PubMed] [Google Scholar]
- 32.Zheng Y, Chaudhry A, Kas A, deRoos P, Kim JM, Chu TT, Corcoran L, Treuting P, Klein U, Rudensky AY. Regulatory T-cell suppressor program co-opts transcription factor IRF4 to control T(H)2 responses. Nature. 2009;458:351–356. doi: 10.1038/nature07674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Liu S, Lin L, Jiang P, Wang D, Xing Y. A comparison of RNA-Seq and high-density exon array for detecting differential gene expression between closely related species. Nucleic Acids Res. 2011;39:578–588. doi: 10.1093/nar/gkq817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Shendure J. The beginning of the end for microarrays? Nat. Methods. 2008;5:585–587. doi: 10.1038/nmeth0708-585. [DOI] [PubMed] [Google Scholar]
- 35.Wang J, Maldonado MA. The ubiquitin-proteasome system and its role in inflammatory and autoimmune diseases. Cell Mol. Immunol. 2006;3:255–261. [PubMed] [Google Scholar]
- 36.Blanco B, Perez-Simon JA, Sanchez-Abarca LI, Caballero-Velazquez T, Gutierrez-Cossio S, Hernandez-Campo P, ez-Campelo M, Herrero-Sanchez C, Rodriguez-Serrano C, Santamaria C, et al. Treatment with bortezomib of human CD4+ T cells preserves natural regulatory T cells and allows the emergence of a distinct suppressor T-cell population. Haematologica. 2009;94:975–983. doi: 10.3324/haematol.2008.005017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Birzele F, Csaba G, Zimmer R. Alternative splicing and protein structure evolution. Nucleic Acids Res. 2008;36:550–558. doi: 10.1093/nar/gkm1054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lynch KW. Consequences of regulated pre-mRNA splicing in the immune system. Nat. Rev. Immunol. 2004;4:931–940. doi: 10.1038/nri1497. [DOI] [PubMed] [Google Scholar]
- 39.Ip JY, Tong A, Pan Q, Topp JD, Blencowe BJ, Lynch KW. Global analysis of alternative splicing during T-cell activation. RNA. 2007;13:563–572. doi: 10.1261/rna.457207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rao N, Nguyen S, Ngo K, Fung-Leung WP. A novel splice variant of interleukin-1 receptor (IL-1R)-associated kinase 1 plays a negative regulatory role in Toll/IL-1R-induced inflammatory signaling. Mol. Cell. Biol. 2005;25:6521–6532. doi: 10.1128/MCB.25.15.6521-6532.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Barlat I, Maurier F, Duchesne M, Guitard E, Tocque B, Schweighoffer F. A role for Sam68 in cell cycle progression antagonized by a spliced variant within the KH domain. J. Biol. Chem. 1997;272:3129–3132. doi: 10.1074/jbc.272.6.3129. [DOI] [PubMed] [Google Scholar]
- 42.McBride AE, Conboy AK, Brown SP, Ariyachet C, Rutledge KL. Specific sequences within arginine-glycine-rich domains affect mRNA-binding protein function. Nucleic Acids Res. 2009;37:4322–4330. doi: 10.1093/nar/gkp349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Smith EL, Finney HM, Nesbitt AM, Ramsdell F, Robinson MK. Splice variants of human FOXP3 are functional inhibitors of human CD4+ T-cell activation. Immunology. 2006;119:203–211. doi: 10.1111/j.1365-2567.2006.02425.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Boyle AP, Davis S, Shulha HP, Meltzer P, Margulies EH, Weng Z, Furey TS, Crawford GE. High-resolution mapping and characterization of open chromatin across the genome. Cell. 2008;132:311–322. doi: 10.1016/j.cell.2007.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mayall TP, Sheridan PL, Montminy MR, Jones KA. Distinct roles for P-CREB and LEF-1 in TCR alpha enhancer assembly and activation on chromatin templates in vitro. Genes Dev. 1997;11:887–899. doi: 10.1101/gad.11.7.887. [DOI] [PubMed] [Google Scholar]
- 46.Erman B, Cortes M, Nikolajczyk BS, Speck NA, Sen R. ETS-core binding factor: a common composite motif in antigen receptor gene enhancers. Mol. Cell. Biol. 1998;18:1322–1330. doi: 10.1128/mcb.18.3.1322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wang CY, Bassuk AG, Boise LH, Thompson CB, Bravo R, Leiden JM. Activation of the granulocyte-macrophage colony-stimulating factor promoter in T cells requires cooperative binding of Elf-1 and AP-1 transcription factors. Mol. Cell. Biol. 1994;14:1153–1159. doi: 10.1128/mcb.14.2.1153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lee SM, Gao B, Fang D. FoxP3 maintains Treg unresponsiveness by selectively inhibiting the promoter DNA-binding activity of AP-1. Blood. 2008;111:3599–3606. doi: 10.1182/blood-2007-09-115014. [DOI] [PubMed] [Google Scholar]
- 49.O'Reilly D, Quinn CM, El-Shanawany T, Gordon S, Greaves DR. Multiple Ets factors and interferon regulatory factor-4 modulate CD68 expression in a cell type-specific manner. J. Biol. Chem. 2003;278:21909–21919. doi: 10.1074/jbc.M212150200. [DOI] [PubMed] [Google Scholar]
- 50.Zheng Y, Chaudhry A, Kas A, deRoos P, Kim JM, Chu TT, Corcoran L, Treuting P, Klein U, Rudensky AY. Regulatory T-cell suppressor program co-opts transcription factor IRF4 to control T(H)2 responses. Nature. 2009;458:351–356. doi: 10.1038/nature07674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wu Y, Borde M, Heissmeyer V, Feuerer M, Lapan AD, Stroud JC, Bates DL, Guo L, Han A, Ziegler SF, et al. FOXP3 controls regulatory T cell function through cooperation with NFAT. Cell. 2006;126:375–387. doi: 10.1016/j.cell.2006.05.042. [DOI] [PubMed] [Google Scholar]
- 52.Ishii T, Ohnuma K, Murakami A, Takasawa N, Yamochi T, Iwata S, Uchiyama M, Dang NH, Tanaka H, Morimoto C. SS-A/Ro52, an autoantigen involved in CD28-mediated IL-2 production. J. Immunol. 2003;170:3653–3661. doi: 10.4049/jimmunol.170.7.3653. [DOI] [PubMed] [Google Scholar]
- 53.Stahl HF, Fauti T, Ullrich N, Bopp T, Kubach J, Rust W, Labhart P, Alexiadis V, Becker C, Hafner M, et al. miR-155 inhibition sensitizes CD4+ Th cells for TREG mediated suppression. PLoS ONE. 2009;4:e7158. doi: 10.1371/journal.pone.0007158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Gavin MA, Rasmussen JP, Fontenot JD, Vasta V, Manganiello VC, Beavo JA, Rudensky AY. Foxp3-dependent programme of regulatory T-cell differentiation. Nature. 2007;445:771–775. doi: 10.1038/nature05543. [DOI] [PubMed] [Google Scholar]
- 55.Bopp T, Becker C, Klein M, Klein-Hessling S, Palmetshofer A, Serfling E, Heib V, Becker M, Kubach J, Schmitt S, et al. Cyclic adenosine monophosphate is a key component of regulatory T cell-mediated suppression. J. Exp. Med. 2007;204:1303–1310. doi: 10.1084/jem.20062129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Jain N, Nguyen H, Friedline RH, Malhotra N, Brehm M, Koyanagi M, Bix M, Cooper JA, Chambers CA, Kang J. Cutting edge: Dab2 is a FOXP3 target gene required for regulatory T cell function. J. Immunol. 2009;183:4192–4196. doi: 10.4049/jimmunol.0902041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wing K, Onishi Y, Prieto-Martin P, Yamaguchi T, Miyara M, Fehervari Z, Nomura T, Sakaguchi S. CTLA-4 control over Foxp3+ regulatory T cell function. Science. 2008;322:271–275. doi: 10.1126/science.1160062. [DOI] [PubMed] [Google Scholar]
- 58.Collison LW, Workman CJ, Kuo TT, Boyd K, Wang Y, Vignali KM, Cross R, Sehy D, Blumberg RS, Vignali DA. The inhibitory cytokine IL-35 contributes to regulatory T-cell function. Nature. 2007;450:566–569. doi: 10.1038/nature06306. [DOI] [PubMed] [Google Scholar]
- 59.Kuczma M, Podolsky R, Garge N, Daniely D, Pacholczyk R, Ignatowicz L, Kraj P. Foxp3-deficient regulatory T cells do not revert into conventional effector CD4+ T cells but constitute a unique cell subset. J. Immunol. 2009;183:3731–3741. doi: 10.4049/jimmunol.0800601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kuczma M, Pawlikowska I, Kopij M, Podolsky R, Rempala GA, Kraj P. TCR repertoire and Foxp3 expression define functionally distinct subsets of CD4+ regulatory T cells. J. Immunol. 2009;183:3118–3129. doi: 10.4049/jimmunol.0900514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Ziegler SF. FOXP3: of mice and men. Annu. Rev. Immunol. 2006;24:209–226. doi: 10.1146/annurev.immunol.24.021605.090547. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.