Abstract
Heavy metal compounds have toxic and medicinal potential through capacity to form strong specific bonds with macromolecules, and the interaction of platinum drugs at the major groove nitrogen atom of guanine bases primarily underlies their therapeutic activity. By crystallographic analysis of transition metal–and in particular platinum compound–DNA site selectivity in the nucleosome core, we establish that steric accessibility, which is controlled by specific structural parameters of the double helix, modulates initial guanine–metal bond formation. Moreover, DNA conformational features can be linked to both similarities and distinctions in platinum drug adduct formation between the naked and nucleosomal DNA states. Notably, structures that facilitate initial platinum–guanine bond formation can oppose cross-link generation, rationalizing the occurrence of long-lived therapeutically ineffective monofunctional adducts. These findings illuminate DNA structure-dependent reactivity and provide a novel framework for understanding metal–double helix interactions, which should facilitate the development of improved chromatin-targeting medicinal agents.
INTRODUCTION
The pathologic and medicinal potential of heavy metal compounds relates closely to their ability to form strong bonding interactions with various biomolecules (1). Inside the cell, such bonding potential translates to the ability to elicit pronounced and long-lasting conformational changes in proteins and nucleic acids. The pharmacological effect of platinum-based chemotherapeutic agents (Supplementary Figure S1) is mediated through formation of DNA lesions, which interfere with genomic activities and ultimately trigger apoptosis (2,3). When these agents enter the low chloride anion environment within the cell, the chloride or carboxylate leaving groups can undergo aquation, generating reactive aqua-species. Initial attack on DNA coincides with formation of a single Pt–purine bond, corresponding to the monofunctional adduct (MFA), followed by potential chelation to yield a bifunctional adduct (cross-link). Drug reaction occurs at the N7 nitrogen atoms of purine bases, generating predominantly 1,2 intrastrand cross-links at GG and less frequently at AG dinucleotides, in addition to a minor fraction of GNG 1,3 intrastrand and other DNA adducts. The primary mechanism of action appears to relate to local dinucleotide kink distortions and double helix deformations that cause transcriptional arrest through stalling RNA polymerase (3).
In spite of their widespread application for decades in the treatment of specific cancers and the execution of many thousands of clinical and chemical studies, the principles dictating platinum drug site selectivity and cross-link formation are not well understood. In fact, the markedly non-uniform distribution of drug adducts observed for cellular DNA and for DNA in in vitro studies (4,5) indicate that site preference is governed by attributes that go beyond the guanine nucleotide distribution. Additionally, there is a lack of consensus on some of the basic mechanistic features (6,7), which likely arises from the fact that investigations have been based on one or several short DNA fragments of distinct sequence, yielding little overlap between related studies.
The fact that current platinum drugs elicit severe toxicity and resistance effects has prompted the search for safer and more effective agents (8). However, a full understanding of the mechanism of action—the weak points of cancer cells that are exploited—and the directed design of improved agents will depend on a detailed knowledge of drug adduct formation (9). Here, we have conducted a detailed biochemical, structural and analytical study with a variety of nucleosomal and naked DNA substrates to delineate the attributes underlying transition metal site preference and platinum compound adduct formation. Our findings shed light on the fundamental principles that govern platinum drug site selection and the generation of therapeutically active cross-links in the genome.
MATERIALS AND METHODS
Platinum compounds
Cisplatin (cisPt), carboplatin (carPt) and oxaliplatin (oxPt) were purchased from Sigma–Aldrich (Supplementary Figure S1). [(NH3)3PtCl]Cl (tamPt) and [(1,2-R,R-diaminocyclohexane)PtCl2] (clPt) were prepared according to established protocols (10,11). The purity of the commercially available compounds was 98–99% and that of the synthesized compounds was found to be >98% according to elemental analysis.
Nucleosome core particle generation and crystal preparation
Nucleosome core particle (NCP) constructs, NCP145, NCP146, NCP146b, NCP147 and NCP-TA, were prepared from recombinant Xenopus laevis histones and 145, 146 and 147 bp DNA fragments using established protocols (12−15). NCP crystals were grown as described previously and stabilized in a chloride-free substitute harvest buffer of 10 mM MnSO4, 50 mM K-cacodylate (pH 6.0), 24% (v/v) 2-methyl-2,4-pentanediol and 2% (w/v) trehalose (16).
Structural data reported here were obtained from NCP crystals derivatized by including Pt agent in the substitute harvest buffer at concentrations of 2 mM tamPt, 0.6 mM oxPt, 0.6 mM cisPt (1- to 2-day treatments) or 0.3 mM cisPt (4-day treatments). Crystals were subsequently allowed to incubate at room temperature for up to 4 days prior to data collection (17).
Structure solution and analysis
Single crystal X-ray diffraction data were recorded as described previously (13) at the Swiss Light Source (Paul Scherrer Institute, Villigen, Switzerland) using the PILATUS detector on beam line X06SA and a Mar225 CCD detector on beam line X06DA. The X-ray wavelength was tuned to the absorption edge of platinum (1.07 Å) for data collection. Data were processed with MOSFLM (18) and SCALA from the CCP4 suite (19). Structural refinement and model building were carried out with routines from the CCP4 suite. Structures for NCP145 and NCP146b in MnSO4 buffer, at 2.5 Å resolution and 2.55 Å resolution respectively, were solved by initial rigid body refinement of the earlier 2.65 Å NCP145 model (13) and the original 2.6 Å NCP146b model (12). Pt-derivatized NCP145 and NCP146b structures were solved by initial rigid body refinement of the (native) MnSO4 models. Pt occupancy and B-factor values were refined against the anomalous difference data using Mlphare (19). The initial occupancy values, on an arbitrary scale, were normalized with respect to the Pt atom having highest occupancy in the structure, which was assigned a value of 1.00.
Solvent accessible surface area (ASA) values for purine N7 atoms were obtained with the ArealMol routine of the CCP4 suite (19), using the default water molecule probe radius of 1.4 Å. In order to provide insight into the relationship between the initial DNA conformation, prior to soft metal coordination and the binding site preference for these metal species, we carried out the analyses such that Pt2+, Co2+ or Ni2+ crystallographic site occupancy was compared with the DNA structure of the respective native NCP crystals prepared in Mn2+ buffers. For the Pt compounds, the native MnSO4-models of NCP146b and NCP145 were utilized, which contain, respectively, a total of 60 and 58 guanine sites having an average guanine-N7-atom-ASA (ASAN7) of 10.7 ± 3.5 Å2 and 10.3 ± 4.1 Å2. For the 13- and 55-h tamPt and the 14-, 25- and 57-h oxPt treatments of NCP146b, 57, 55, 58, 57 and 58 guanine sites, respectively, are considered in the analysis. For the 13- and 55-h tamPt treatments of NCP145, 58 and 54 guanine sites, respectively, are considered in the analysis. The one to five guanine sites that were omitted from analysis in any case correspond to dynamic locations mostly near the DNA termini where Pt is observed to bind but displays a coordination distance ≥3 Å. In these instances, MFA formation is either preceded or accompanied by a substantial conformational change in the DNA, which cannot be differentiated.
In the analysis of DNA structure relative to Co2+/Ni2+ site occupancy, the comparison model was the 1.94 Å resolution NCP147 MnCl2 structure (12), which contains 60 guanine sites having an average ASAN7 of 9.5 ± 4.0 Å2. The analysis considered 105 bp of the 147 bp DNA, which excludes a central 42 bp section that undergoes substantial conformational changes from a Co2+/Ni2+-stabilized translation in the histone–DNA register arising from substitution of the intermediate-soft metals in place of Mn2+ in the crystals (20). The final guanine nucleotide set used for the analysis entailed 29 sites, whereby cross-linked GG dinucleotide sites (bifunctional coordination to both the 5′ G-N7 and 3′ G-O6 atoms), which involve a more complex reaction than MFA formation, were excluded from the analysis. The Mn2+ analysis was based on the metal occupancy values from the 2.75 Å resolution NCP147 MnSO4 data (21), collected at the manganese absorption edge, and the ASAN7 values from the 1.94 Å resolution NCP147 MnCl2 structure (12). The analysis included either the same set of 29 guanine sites for direct comparison with the Co2+/Ni2+ systems or the full complement of 56 sites (4 guanine nucleotides at GG sites involved in bifunctional Mn2+ coordination were omitted from the analysis).
DNA conformational analysis was conducted using 3DNA (22,23). Graphic figures were prepared with PyMOL (DeLano Scientific LLC, San Carlos, CA, USA). Principal component and statistical analyses were carried out with the XLSTAT routine of Microsoft Excel (Microsoft Corporation).
DNA footprinting and interstrand cross-linking analysis
Exonuclease III footprinting experiments for obtaining Pt adduct formation profiles, based on digest (3′–5′ direction) termination at DNA lesion sites, were conducted as described previously (16). 20 micromolar naked DNA or NCP in a buffer of 20 mM K-cacodylate (pH 6.0) were incubated with 5-, 5-, 5-, 20- or 200-fold molar excesses of cisPt, tamPt, clPt, oxPt or carPt, respectively, at room temperature for 48 h. For the low stoichiometry treatment shown in Supplementary Figure S12, only a 10-fold molar excess of oxPt was utilized. Samples were treated by heating at 55°C with the addition of 4 M NaCl to a final concentration of 2 M, followed by phenol–chloroform extraction and ethanol precipitation of the DNA, which was then 5′end-labelled with polynucleotide kinase. 675ng DNA samples were subjected to digestion with 250 U of exonuclease III for 1 h, which is a 10-fold excess over that minimally required for full digestion of the unplatinated DNA. The one exception corresponds to the ‘one-third’ digest shown in the left panel of Supplementary Figure S11, in which the tamPt samples were digested with one-half as much enzyme for only 40 min. After ethanol precipitation, digested DNA samples were subsequently deplatinated at 58°C by treatment with 1 M thiourea for 16 h, followed again by ethanol precipitation of the DNA. Modified Maxam–Gilbert purine sequencing standards were prepared initially as described (24), followed by 3′ dephosphorylation by treatment with 1 U of polynucleotide kinase per 20 ng DNA for 16 h. DNA fragments were resolved by denaturing PAGE (10% polyacrylamide, 8 M urea, 88 mM Tris–borate, 2 mM EDTA, pH 8.3), in which loaded quantities of Pt agent-treated DNA samples corresponded to approximately equal total radioactive counts visible on the gel.
For the interstrand cross-linking analysis, 20 µM naked DNA or NCP in a buffer of 20 mM K-cacodylate (pH 6.0) were incubated with 10-, 10-, 20- or 200-fold molar excesses of cisPt, tamPt, oxPt or carPt, respectively, at room temperature for 48 h (NCP146b and NCP145 samples). For the NCP-TA samples, 5-, 10-, 10- or 200-fold molar excesses of cisPt, clPt, oxPt or carPt, respectively, were utilized. Samples were treated by heating at 55°C with the addition of 4 M NaCl to a final concentration of 2 M, followed by phenol–chloroform extraction and ethanol precipitation of the DNA, which was then 5′end-labelled with polynucleotide kinase. DNA fragments were subsequently resolved by denaturing PAGE (10% polyacrylamide, 8 M urea, 88 mM Tris–borate, 2 mM EDTA, pH 8.3), with equivalent DNA quantities loaded for all samples.
Quantification of platinum adducts
The Pt agent:DNA molar stoichiometries of treatment used for the footprinting studies were chosen to achieve an approximately equal number of adducts per DNA molecule across the five different Pt compounds and between the naked and nucleosomal DNA samples. Samples were prepared by incubating 20 µM naked DNA or NCP in a buffer of 20 mM K-cacodylate (pH 6.0) with 5-, 5-, 5-, 20- or 200-fold molar excesses of cisPt, tamPt, clPt, oxPt or carPt, respectively, at room temperature for 48 h. Naked or nucleosomal DNA samples were subsequently prepared by heating either naked DNA or aliquots of NCP at 55°C with the addition of 4 M NaCl to a final concentration of 2 M, followed by phenol–chloroform extraction and ethanol precipitation of the DNA. DNA concentrations prior to Pt quantification were determined by spectrophotometric absorbance measurements at 260 nm. A Hitachi Z-2000 polarized Zeeman atomic absorption spectrophotometer was used to measure Pt content in the samples as described previously (25).
Measurements were conducted in duplicate over two independent trials and averaged between the five Pt compounds, yielding an average number of adducts per DNA molecule of 3.9 ± 0.7 and 3.5 ± 0.7 for the naked DNA and nucleosomal DNA states, respectively, for NCP146b samples. The corresponding naked and nucleosomal DNA values for NCP145 samples were 4.8 ± 1.3 and 4.1 ± 1.2, respectively. Adduct levels for the intact NCP were systematically higher, at 4.9 ± 1.2 and 5.1 ± 1.1, for NCP146b and NCP145, respectively. This is an apparent consequence of Pt agent reaction with histone methionine sulphur groups (16), causing more extensive overall adduct accumulation compared to DNA substrate alone.
RESULTS
Platinum adduct formation is modulated by solvent accessibility
We studied here the three most widely used platinum drugs, cisPt, carPt and oxPt, in addition to two other related compounds—tamPt and clPt (Supplementary Figure S1). In an earlier study on Pt drug-nucleosome site selectivity, we were able to observe adducts by X-ray crystallographic analysis of NCP crystals that had been treated with cisPt or oxPt, but this caused substantial reduction in diffraction quality from disordering of the crystals (16). In that initial study, we had utilized an NCP (NCP147) containing a 147 bp human α-satellite DNA fragment, which yielded roughly 3.5 Å resolution data sets. We subsequently experimented with different substrates and observed that crystals are profoundly more tolerant to Pt compound treatment when the NCP is composed of a 146 or 145 bp DNA. Disordering of these NCP146b and NCP145 crystals from platination is apparently much less severe than for NCP147 as a result of a ‘buffering’ effect from DNA stretching (see below).
It was possible to obtain up to 2.5 Å resolution data sets for NCP146b or NCP145 crystals heavily derivatized by tamPt, oxPt or cisPt (Supplementary Tables S1 and S2; the low reactivity of carPt and low solubility of clPt did not allow extensive derivatization of crystals on a reasonable time scale). By exploiting a special form of heavy atom X-ray scattering through tuning the X-ray wavelength to the absorption edge of platinum, we obtained exceptional profiles of Pt2+ ion location and site occupancy from the anomalous difference electron density maps (Figure 1 and Supplementary Figures S2−S4). With the exception of the nearly fully solvent accessible N7 atoms of the 5′ terminal adenine bases of NCP145 and NCP146b (average solvent accessible surface area = 23.2 ± 2.4 Å2)—the primary sites of adduct formation for oxPt—all other apparent sites of MFA formation on the DNA coincide with G-N7 groups. Aside from this DNA terminus artifact, the minor fraction of Pt adducts that initially appear to have formed at non-guanine sites or with unreasonable coordination geometry apparently relate to conformational changes in the double helix, which can be linked back to alterations in the DNA stretching configuration (Figure 2).
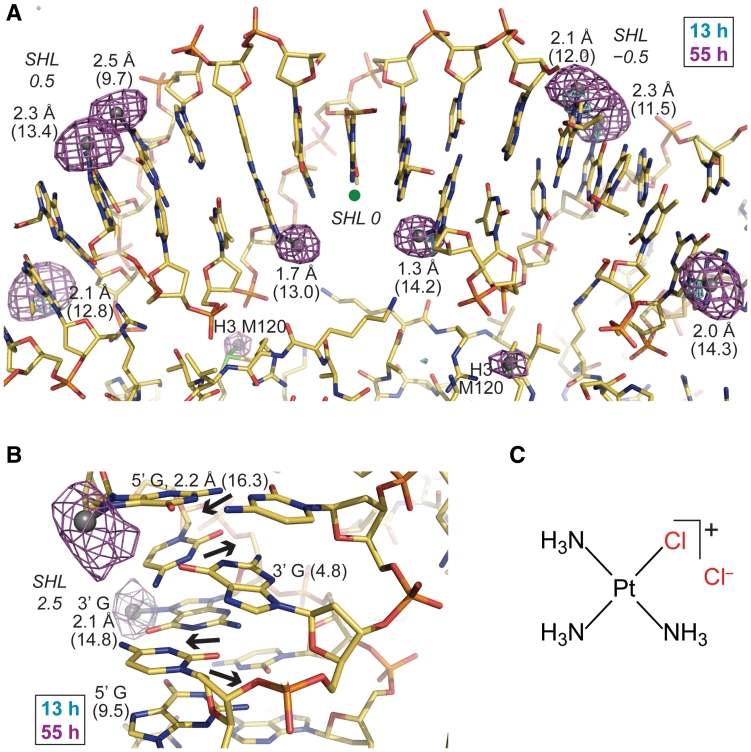
Figure 1.
Monofunctional Pt adduct formation in the nucleosome core. (A and B) Anomalous difference electron density maps (3.5σ-cyan, 4.5σ-magenta, A; 3.5σ, B) from 13- and 55-h tamPt treatments of NCP145 (A) and NCP146b (B) are superimposed on the respective refined 55-h models. Values in Å represent bond distances between Pt (silver spheres) and G-N7 [based on peak positions in the anomalous map; ideal bond length is ~2.0 Å (26)] and numbers in parentheses correspond to ASAN7 (Å2). A green dot designates the base pair at the nucleosome centre, through which the pseudo 2-fold axis of symmetry runs vertically (A). Alternating dinucleotide shift (displacement of base pairs into the major and minor grooves; arrows) causes pronounced fluctuation in consecutive ASAN7 values (B). (C) The chemical structure of the monofunctional agent, tamPt.
Figure 2.
Pt adduct formation in NCP crystals reveals a minor population with an alternate histone-DNA register. (A and B) Anomalous difference electron density maps (3.5σ, A; 4σ, B) from 13- and 55-h tamPt treatments of NCP146b are superimposed on the refined 55-h model (blue). A minor fraction of NCPs in the crystal have DNA stretching around SHL 5 as opposed to around SHL −5 (yellow; see Supplementary Figure S5), whereby the histone-DNA register relative to the major conformation (blue; stretching around SHL −5 and not around SHL 5) is translated by 1 bp (arrows). The minor conformation structure (yellow) corresponds to the opposing particle half of the refined model, which was superimposed via the histone fold regions. Values in Å represent bond distances between Pt (spheres) and G-N7 (A) or purine-N7 (B) and numbers in parentheses correspond to ASAN7 (Å2; B). Adduct-to-base correspondence is apparent from coordination geometry, base chemistry and anomalous map profile. Adducts associated with the minor stretching configuration are highlighted with a red asterisk.
DNA stretching is an intrinsic property of the nucleosome that is apparently confined to two unique locations where the histone binding configuration induces strain in the double helix (27); (Supplementary Figure S5). As such, the DNA is either compressed and undertwisted in the unstretched state or elongated and overtwisted in the stretched state. The stretched conformation is frequently accompanied by extreme kink distortions and is achieved through elevated dinucleotide twist and rise, which alters the histone-DNA register by increasing the ‘effective’ double helix length by 1 bp (13,28).
While the native NCP145 structure displays two incidences of stretching around SHL (SuperHelix Location, double helical turns from the nucleosome center) ± 2 and that of native NCP146b displays one incidence of stretching around SHL −5, NCP in solution and in the crystalline state can contain a mixture of stretching configurations (12,13,29–31); (Supplementary Figure S5). For NCP145 and NCP146b derivatization with cisPt, oxPt and tamPt, one observes adducts coinciding with at least two distinct stretching states for each NCP construct (Supplementary Figures S3 and S4). For tamPt and oxPt treatments of NCP146b, this appears to relate solely to two configurations; namely the predominant initial stretching at SHL −5 (only) and a secondary minor incidence where the stretch has relocalized to the opposing particle half (SHL 5). Thus, adduct formation is observed as a superimposition of these two NCP146b stretching conformations and the corresponding site selectivity can be derived by adduct assignment with respect to one or the other configuration (Figure 2).
As we had observed before for NCP147 (16), cisPt treatment of NCP145 and NCP146b gives rise to substantial disordering of the DNA, precluding comprehensive assignment of adduct formation profiles. Therefore, we utilized the data from the most extensive treatments of NCP146b with tamPt and oxPt and NCP145 with tamPt to analyse the relationship between DNA structure and adduct formation. Although there are several sites where some significant conformational change in the DNA must occur before or during adduct formation, the vast majority of sites coincide with the Pt2+ ion situated with roughly ideal bonding geometry with respect to the G-N7 atom (Figures 1 and 2, Supplementary Figures S3 and S4). In fact, these three heavily platinated structures are nearly identical to the corresponding native models (rmsd in DNA atomic positions of 0.3 Å in each case).
We had originally identified several DNA conformational features, which can influence the favourability or mode of divalent transition metal cation coordination at specific sites in the NCP (20,21,32). Here, we conducted a novel analysis uncovering a general principle that governs site selectivity—the ASAN7 (NCP146b average = 10.7 ± 3.5 Å2; NCP145 average = 10.3 ± 4.1 Å2) dictates the proclivity for MFA formation at these major groove sites. The correlations between NCP146b-Pt site occupancy and ASAN7 (Pearson’s r = 0.66, tamPt; r = 0.54, oxPt) (Supplementary Figures S6 and S7) are substantial, especially considering the general disordering effects of Pt adduct formation together with the large number of sites considered in the analyses (up to 58 guanine sites). Furthermore, the clustering of occupied and unoccupied tamPt and oxPt sites at high (13.1 ± 1.9 Å2, tamPt; 12.8 ± 2.7 Å2, oxPt) and low (8.3 ± 3.4 Å2, tamPt; 8.9 ± 3.3 Å2, oxPt) ASAN7 values is pronounced and the most accessible guanine sites tend to be the first to accumulate adducts. In addition, although localized disordering from oxPt treatment of NCP145 crystals does not allow comprehensive site assignment (Supplementary Figure S4), the same influence of ASAN7 can be seen for NCP145-tamPt site selectivity (r = 0.67; 12.9 ± 2.8 Å2, occupied; 7.5 ± 3.2 Å2, vacant) (Supplementary Figure S8). It is important to note that NCP145 is a distinct substrate, since it shares only 42% DNA sequence identity with NCP146b. As observed for MFA formation by the Pt compounds, we find that elevated ASAN7 also strongly promotes monofunctional binding of Mn2+, Ni2+ and Co2+ to NCP147 (Supplementary Figures S9 and S10, Supplementary Table S3).
Specific DNA conformational features dictate steric access
The stacking of bases in double helical DNA limits solvent accessibility within the major and minor grooves. Since ASAN7 modulates metal–DNA bond formation and varies dramatically over different locations, we wanted to determine if there are conformational features that primarily govern this parameter. We started by considering the change in ASAN7 with respect to changes in the six base pair step degrees of freedom for an idealized B-form DNA molecule (Supplementary Table S4). Importantly, the twist of the double helix renders the influence on ASAN7 of the 5′ base and the 3′ base flanking a particular base site distinct. Accordingly, this fundamental analysis shows that increases in the values of 5′ shift, tilt, slide and rise and 3′ tilt, twist and rise yield an increase in ASAN7, whereas, conversely, decreases in the values of 5′ roll and twist and 3′ shift, roll and slide yield increases in ASAN7 (Figure 3).
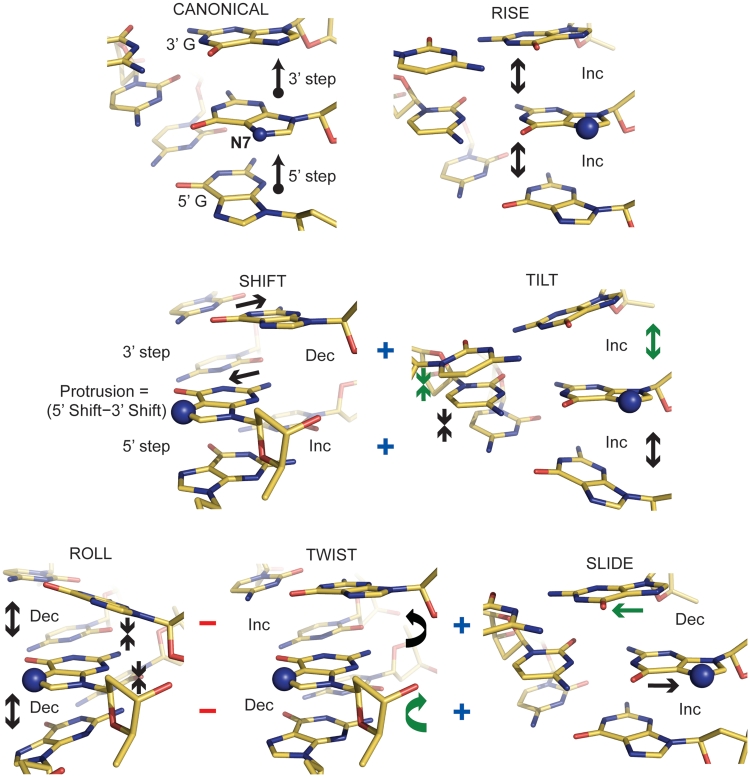
Figure 3.
Influence of DNA conformational elements on solvent accessibility within the major groove. The directional change (Inc, increase; Dec, decrease) for each of the six dinucleotide step parameters that yields an increased ASAN7 for the central guanine base is shown for idealized GGG trinucleotides (entailing a 5′ and a 3′ GG dinucleotide step). The ‘canonical’ reference point has values of 35° for twist, 3.3 Å for rise and 0 for the remaining four step parameters. The ‘+’ and ‘−’ symbols indicate the general conformational coupling that occurs in real DNA; shift and tilt are positively correlated, whereas roll is anti-correlated with twist and slide. With consideration of this coupling, parameters on either the 5′ or 3′ side that cannot work in a cooperative fashion with either shift or roll to elevate ASAN7 are indicated with arrows in green.
For assessing the empirical relationship between double helix conformation and ASAN7, we utilized the highest resolution native NCP model (NCP147; 12), as well as those of NCP146b and NCP145 (Supplementary Table S5). The main determinant of the ASAN7 is shift, which makes a far greater contribution than any of the other parameters apparently because it corresponds to direct displacement into the major groove solvent space. The relative contributions of 5′ versus 3′ shift differ somewhat, but we define a new parameter, protrusion, that equates to the sum displacement into the major groove with respect to both 5′ and 3′ base neighbours (Figure 3), and this parameter gives the highest single correlation with ASAN7.
From restraints imposed by the DNA backbone and base stacking interactions (in real DNA) alterations in roll are coupled to inverse changes of twist and slide and alterations in shift are coupled to changes in tilt (Supplementary Table S6). Therefore, although shift is clearly the driver of ASAN7 modulation, the contribution of 5′ shift is greater than that of 3′ shift (Supplementary Table S5), apparently because 5′ shift and tilt are in phase with each other and make a unified contribution to ASAN7, while the 3′ parameters are out of phase (Figure 3). Likewise, neither the 5′ nor 3′ individual contributions of roll, twist and slide towards ASAN7 have the appropriate phase relationships to provide a unified input, which presumably underlies their much weaker overall influence relative to shift.
Analysis of the platinated NCP146b and NCP145 models shows the predominance of shift in influencing tamPt and oxPt adduct formation (Supplementary Table S7). Moreover, the contribution of 5′ shift is substantially stronger than that of 3′ shift, which may relate to special steric demands imposed by the 5′ base. This is consistent with the fact that (for NCP146b) 5′ roll in the negative direction is also seen to significantly facilitate adduct formation—by increased steric clearance over the major groove edge accompanying bending into the minor groove (Figure 3). Such dominance by the 5′ side is likely a consequence of the fact that, due to the right-handed twist of the double helix, the 5′ flanking base is effectively rotated into the major groove relative to a G-N7 site, potentially blocking access from the 5′ direction—a circumstance opposite to that of the 3′ base, which is effectively recessed.
DNA structure modulates platinum drug cross-link formation
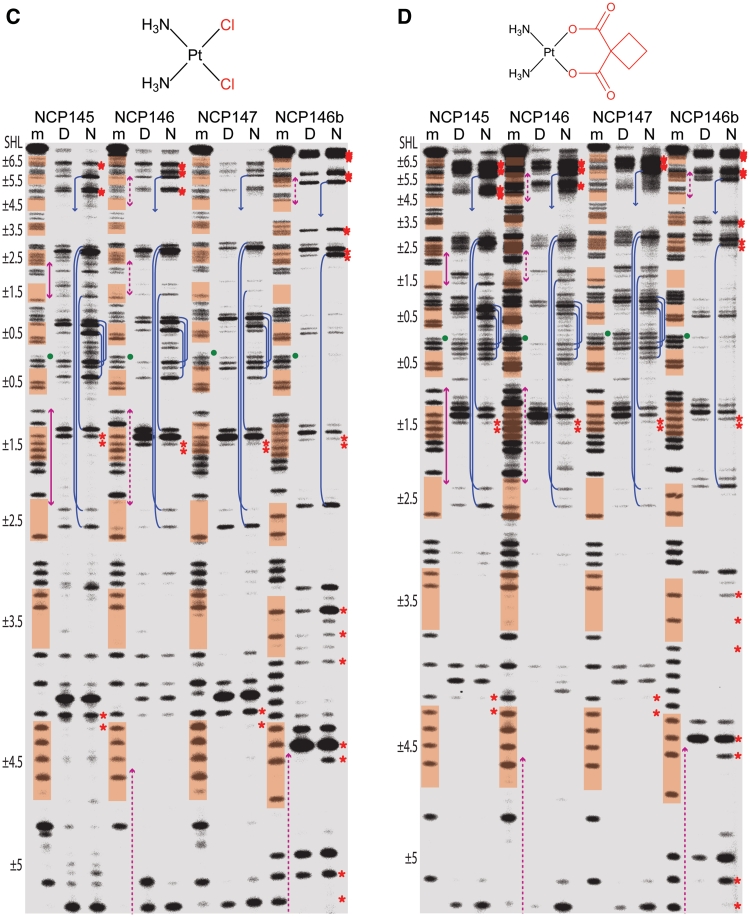
We had earlier discovered through footprinting analysis of naked and nucleosomal DNA substrates that histone octamer association can modulate cisPt and oxPt adduct formation profiles (16). This analysis was expanded here to include four NCP constructs and five Pt compounds (Figure 4, Supplementary Figures S11 and S12). We had originally assumed that exonuclease stop sites before isolated guanine nucleotides corresponded to MFAs. However, we find that our standard footprinting analysis of naked and nucleosomal templates treated with tamPt—which is capable of forming essentially only MFAs (Supplementary Figure S13)—yields almost no stop sites, indicating that MFAs are digested through much more readily than cross-links (Supplementary Figure S11). Accordingly, this shows that the footprints of the four bifunctional agents correspond effectively to cross-link formation profiles.
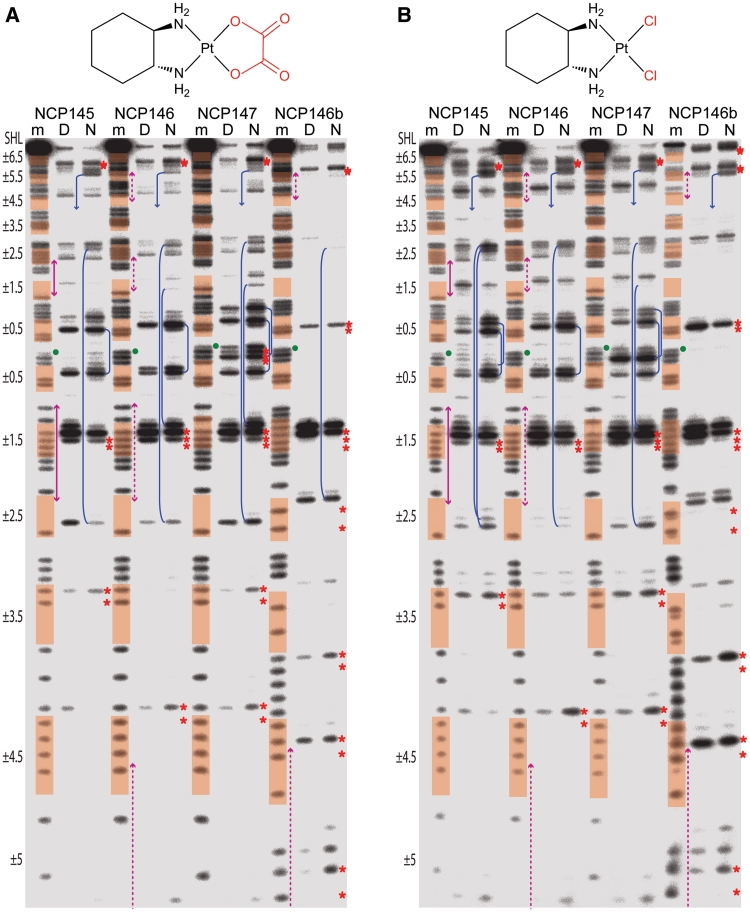
Figure 4.
Exonuclease digest analysis reveals Pt cross-link formation profiles. DNA samples, comprising four different NCP constructs, correspond to purine sequencing standard (m) or naked DNA (‘D’) and NCP (N) that were treated with oxPt (A), clPt (B), cisPt (C) or carPt (D). Minor groove inward-facing bases (highlighted in orange), regions of DNA stretching (magenta arrows) and the central nucleotide (green dot) are based on the native crystal structure assignments (Supplementary Figure S5; the palindromic DNA sequences read from 5′, bottom of gel, to 3′, top of gel). Prominent intrastrand cross-links at GG, AG, GA or GNG sites are indicated by red asterisks placed at the apparent purine nucleotides of the bifunctional adducts. Interstrand cross-links at GC sites within SHL ±0.5, ±1.5, ±2.5 and ±5.5 coincide with nuclease arrest upstream (3′) of guanine in either DNA strand (blue lines; multiple stop pairs are apparent in some cases). For clarity, cross-link features are illustrated only for the nucleosomal DNA (N) samples. See Supplementary Figure S12 for an illustration of cross-link assignment from stop site profiles.
The related 145, 146 and 147 DNA constructs each contain four unique GC dinucleotides while the 146b construct contains three such elements (Supplementary Figure S5), and we observed a substantial degree of interstrand cross-linking in both the naked and nucleosomal states (Supplementary Figure S13). With the knowledge that the Pt adduct footprints reveal cross-linking profiles, it is now apparent that many of the stop sites for the bifunctional agents correspond to GC interstrand cross-links (Figure 4). These stop sites arise as doublets, whereby nuclease digestion arrests upstream of guanine in either DNA strand. The overall intensity of the GC stops appear disproportionate considering that only a minor fraction of adducts correspond to interstrand cross-links. However, by extrapolation from the relative digestability of MFAs, cross-links that are more distorting or cumbersome would be expected to be especially resistant to nuclease digestion.
Some of the GC cross-links have enhanced prominence in the nucleosomal relative to the naked DNA samples, in particular at central locations (e.g. SHL ±0.5; Figure 4), and site-specific promotion of interstrand cross-linking by histone octamer association has been observed previously (33). Here, all of the GC sites are at locations where the minor groove faces inward and the centrally situated of these (SHL ±0.5, ±1.5 and ±2.5) are regions where minor groove bending has been found associated with alternating shift (34). This conformational pattern yields alternating high and low ASAN7, and indeed the former are found to be prominent sites of MFA formation in the crystal structures (Figure 1B and Supplementary Figure S14). In at least one instance, with a long-duration cisPt crystal treatment, we observe apparent interstrand cross-link formation at the SHL −0.5 and −1.5 GC sites of NCP145 (Supplementary Figure S15). Interestingly, the adduct position suggests the possibility of only a nominal conformational change associated with the cross-link, in stark contrast to the naked DNA crystal structure of the same cross-link type (35). Given their positions in the NCP constructs, the positive slide associated with the GC steps bent into the minor groove could facilitate interstrand cross-linking by bringing the N7 atoms into closer proximity.
The origin of DNA structure-based modulation of interstrand GC cross-linking apparently relates to a combination of ASAN7-based regulation of MFA formation and the influence of conformational features on the chelation step. In this regard, the main distinctions in the adduct formation profiles between the naked and nucleosomal states occur at locations where the two DNA forms have the greatest potential for differences: regions near the nucleosome centre where the histone-DNA contacts are strongest, one-half SHLs where DNA must bend (against its general tendency) into the minor groove and at locations of DNA stretching (12−14,28,34) (Figure 4).
Kinetics of platinum drug cross-link formation
Beyond the interstrand cross-links, the other main sites of cross-linking apparent in the footprinting analysis are also consistent with extensive adduct formation observed in the crystal structures (Figure 4, Supplementary Figures S3 and S4). In fact, we had previously shown that the footprint of NCP in solution compared to that in the crystalline state is very similar (16). For the especially clear oxPt crystal data, the most prominent cross-link formation occurs at the respective AGGG and AGGC elements within SHL 1.5 of NCP146b and NCP145 (Figure 5). This is in agreement with the cross-linking hotspot seen for oxPt and clPt in the footprinting analysis, but the cross-linking activity of cisPt and carPt towards this location is reduced as judged by the footprints (Figure 4). However, we do observe pronounced tamPt and cisPt MFA formation at SHL ±1.5 in crystals, even at early time points, which suggests that reduced cross-link formation may come about, at least in part, by inhibition of the chelation step (Supplementary Figure S14).
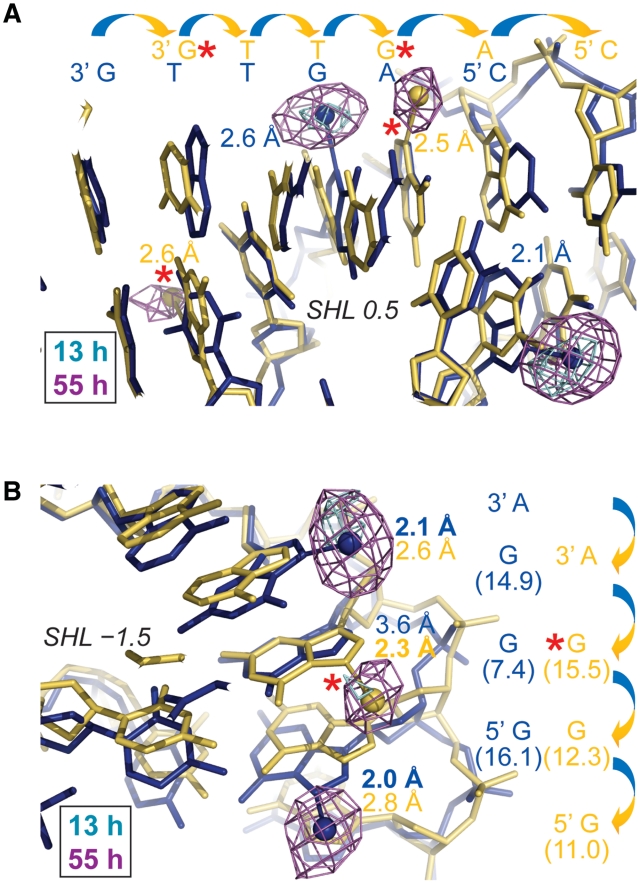
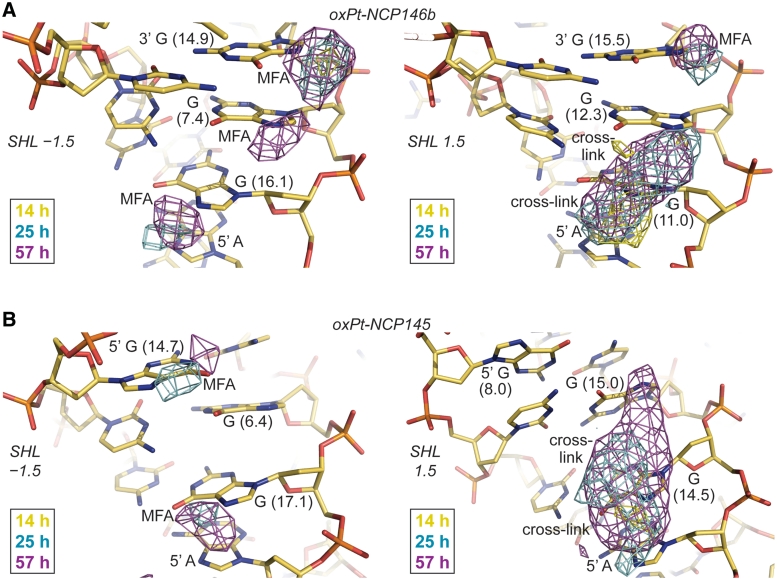
Figure 5.
Time lapse X-ray crystallography shows the evolution of Pt adducts. (A and B) Anomalous difference electron density maps (4σ, A; 3σ, B) from 14-, 25- and 57-h oxPt treatments of NCP146b and NCP145 are superimposed on the respective refined 25-h models. MFA formation occurs early at accessible G-N7 sites of SHL ±1.5 and apparent AG and GG cross-linking ensues later at SHL 1.5 (right). Differences in DNA structure and flexibility, as a consequence of distinct histone-DNA register (A) or stretching mode (B; note the massive kink into the minor groove at the SHL 1.5 GG of NCP145) between the two particle halves in the crystal, apparently account for the observed distinctions in chelation rate between SHL −1.5 and 1.5. Values in parentheses correspond to ASAN7 (Å2).
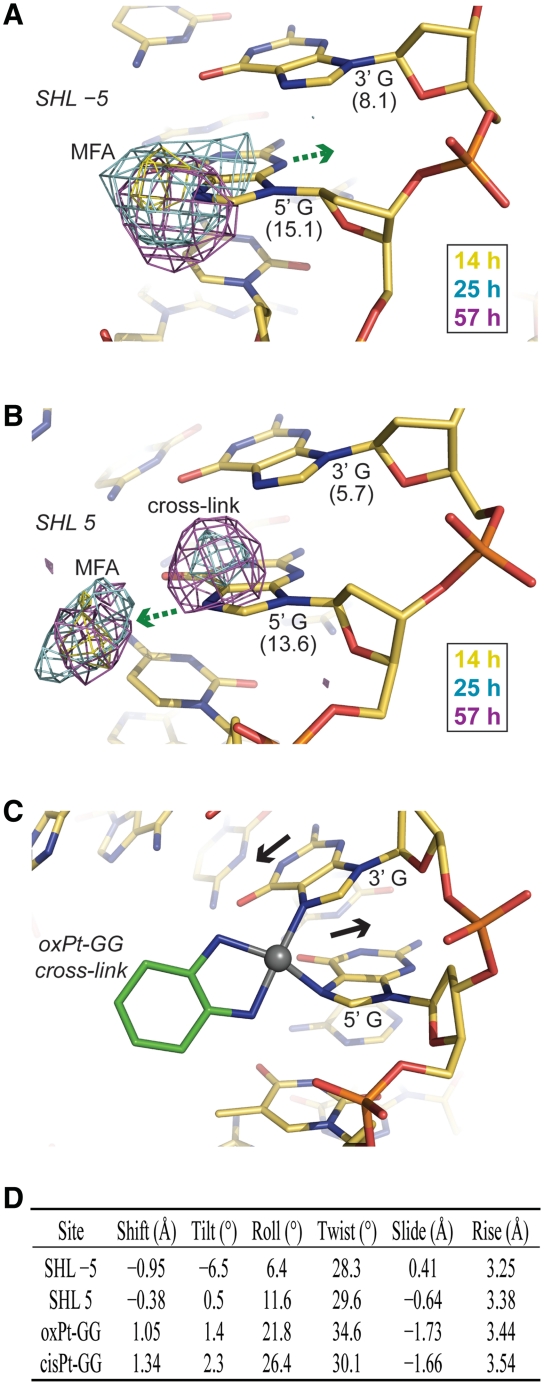
Differences in oxPt cross-link formation between SHL −1.5 and 1.5 in the crystals suggest that distinctions in DNA structure and flexibility modulate the intrastrand, as well as the interstrand, chelation rate (Figure 5). In fact, another location at the edge of SHL ±5 shows clear MFA formation at early time points, which can form chelates over the GG dinucleotide at later time points (Figure 6). Interestingly, extensive cross-link formation at this GG site is observed for the nucleosomal, but not the naked, footprint (Figure 4 and in particular with mild oxPt treatment, Supplementary Figure S12). This indicates nucleosome-based enhancement of cross-linking and importantly, ensuing cross-linking is observed crystallographically in only one of the particle halves, wherein the absence of DNA stretching gives a highly distinct initial conformation relative to the other symmetry-related site for which only pronounced MFA formation is seen (Figure 6). Comparison of the structural parameters of these two sites with those of the oligonucleotide crystal structure of an oxPt-GG cross-link (36) indicates that the pre-reaction conformation of the SHL 5 site that shows cross-link formation is much more similar to the oligonucleotide structure compared to that of SHL −5. Although the 5′ G-N7 is substantially more solvent accessible than the 3′ G-N7, which explains preferential attack at the 5′ site, conformational compatibility appears to account for a large difference in chelation rate. In this sense, given two sites of identical sequence, the site with a pre-cross-linking conformation closer to that of the cross-linked state should have a lower DNA distortion energy barrier associated with chelation.
Figure 6.
Time lapse X-ray crystallography reveals that DNA structure modulates Pt adduct chelation rate. (A and B) Anomalous difference electron density maps (3.5σ) from 14-, 25- and 57-h oxPt treatments of NCP146b are superimposed on the refined 25-h model. MFA formation at the 5′ G occurs early at both SHL ±5, but apparent GG cross-linking ensues only at SHL 5 (B). Values in parentheses correspond to ASAN7 (Å2). Movement of the 5′ G into the minor groove (A) or major groove (B; green arrows) must accompany MFA formation, indicating some degree of flexibility at this site. (C) Oligonucleotide crystal structure of an oxPt-GG cross-link (36) (base pair displacement into the major/minor grooves via shift, black arrows). (D) DNA structural parameters for the GG dinucleotide steps at SHL ±5 in the native NCP146b structure and in the oligonucleotide crystal structures of oxPt-GG (36) and cisPt-GG (26) cross-links.
DISCUSSION
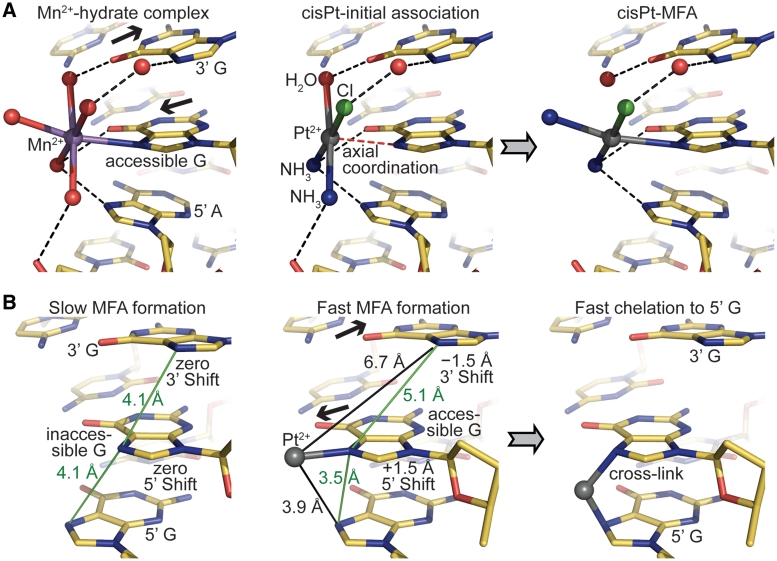
Mn2+, Ni2+ and Co2+ differ in coordination geometry compared to Pt2+ (octahedral versus square planar), but ligand substitution reactions for square planar compounds, such as MFA formation by Pt drugs, are initiated with weak axial coordination to the metal (37) (Figure 7A). Thus, the initial G-N7 associative species of Pt compounds to some extent resembles the coordination geometry for the other transition metals. In this respect, the different divalent metal species seemingly share a common mode of double helix bonding, which is regulated by solvent accessibility. For Pt compounds, sites that are sterically inaccessible disfavour MFA formation by preventing initial association and also possibly elevating the energetic barrier for the trigonal bipyramidal ligand exchange intermediate. Analogously, for the octahedral metals, steric clearance from the surrounding DNA bases is necessary for favourable water ligand interactions. Although steric access appears to be the dominant feature controlling guanine site selectivity for all the transition metal species we have studied here, there are clearly additional factors that influence binding (Supplementary Figures S6–S10). In particular, details of the DNA sequence and conformational context would affect association by dictating electrostatic interactions and hydrogen bonding capacity with respect to compound-specific metal centres and ligands (Figure 7A).
Figure 7.
Mechanistic features of Pt drug adduct formation. (A) The relationship between monofunctional association of octahedral transition metals (left) and MFA formation by square planar Pt compounds (centre and right). The model proposed to illustrate (monoaquated) cisPt reaction (centre to right) is identical to the structure of Mn2+-pentahydrate bound to an AGG element in NCP147 [left (32); atoms were only removed or recoloured]. The central guanine base (accessible G) has an elevated ASAN7 as a result of shift at the 5′ and 3′ sides (black arrows), which promotes metal bond formation. Hydrogen bonds appear as black dashed lines. (B) Model, based on idealized GGG trinucleotides, illustrating general factors controlling the kinetics of Pt adduct formation. Steric/solvent accessibility of the N7 atom, which is mainly governed by shift (black arrows), promotes MFA formation (centre). Consequently, a purine base to the 5′ side is in a much more favourable configuration, compared to a purine base to the 3′ side, for chelation to occur (right).
The cross-link formation profiles between the naked and nucleosomal states are overall alike and in fact more similar to each other than are the footprints between the different bifunctional agents (Figure 4). This indicates that cross-linking potential is primarily determined at the DNA sequence level and moreover implies that there is a considerable degree of similarity in sequence-dependent structure and flexibility between the two DNA states. Nonetheless, as we had observed in our previous study on NCP147 (16), at certain sites there are significant differences in the cross-linking profiles between the naked and nucleosomal states for the multiple substrates studied here. The tight systematic wrapping of the double helix produces regions where the DNA appears forced to deviate from its intrinsic preferences—including central locations, minor groove-inward positions and regions of stretching (12−14,28,34)—and these are the areas where one sees the greatest distinctions in cross-linking relative to naked DNA.
DNA conformation modulates Pt adduct formation and it appears that histone octamer association is capable of modifying the adduct formation profile by moreover altering DNA conformation/dynamics. This structure-based modulation can impact both MFA formation and chelation and an example of this are the prominent GC interstrand cross-links over the central nucleosome core region, for which the alternating shift and high slide values associated with smooth bending into the minor groove (34) are likely faciliatory. On the other hand, differences between the DNA-cross-link formation profiles of cisPt, carPt and oxPt are well known (3,4,16), and we also observe pronounced compound-specific differences in the footprints (Figure 4). With the substrates utilized in this study, one sees that the agents differing in only leaving groups (chloride versus dicarboxylate) give rise to more similar footprints than those that differ in only the carrier groups (NH3 versus DACH). Considering that the crystallographic data for tamPt and oxPt show that these two compounds, which differ markedly in both leaving and carrier groups, yield highly similar MFA profiles (r = 0.73; Supplementary Figure S16) suggests that—in addition to modulation of MFA formation by Pt species-specific steric and electrostatic factors—cross-linking distinctions also arise from differences in the kinetics of chelation.
The SHL ±1.5 DNA region is a general hotspot of adduct formation considering all of the data together, which is especially notable, since the corresponding histone binding motif has the greatest DNA distorting and positioning potential in the nucleosome (14). Conversely, well positioning DNAs will tend to have flexible sequence elements over this region, which most likely generally promotes cross-link formation. The adducts that form over the SHL ±1.5 AGGCA and AGGGA elements entail a combination of intrastrand AG and GG and interstrand GC cross-links depending on the agent, DNA state and construct. However, the extent of cross-linking at SHL ±1.5 in the nucleosomal relative to the naked state is notably stronger for oxPt/clPt compared to cisPt/carPt (Figure 4). Considering that we nonetheless observe pronounced MFA formation by tamPt and cisPt at these locations in the crystal suggests that the oxPt/clPt carrier group is more conducive to chelation, at this site, relative to the carrier groups of cisPt/carPt. In this sense, the reduced degree of kink distortion associated with an oxPt relative to a cisPt intrastrand cross-link, as gleaned from the oligonucleotide crystal structures (26,36; Figure 6D), may be more readily accommodated at this location. Moreover, as a minor groove-inward position, the DNA at SHL ±1.5 is bent in the opposite direction, making an oxPt cross-link effectively closer to the native conformation than a cisPt cross-link. Therefore, in a broad sense, as we have seen with the oxPt crystallographic data in comparing two other sites of identical sequence, MFA sites with pre-cross-linking conformations closer to that of the cross-linked state are likely to undergo chelation more rapidly (Figure 7B). This is also supported by the observation that DNA containing a preformed cisPt cross-link adopts major groove-inward positions on the histone octamer, which allows the major groove kink distortion to coincide with the native DNA bending direction (38). However, we have found no systematic bias towards cross-link generation at major versus minor groove-inward positions in the nucleosome across diverse sequence elements (other prominent cisPt and carPt cross-links can be seen at minor groove-inward locations; Figure 4), and this is apparently a consequence of an overall dominance of intrinsic DNA sequence-dependent behaviour in dictating cross-linking potential (16).
Understanding the kinetic factors that govern cross-link formation is important, since the MFAs of current Pt drugs are apparently therapeutically ineffective (2), and a key observation is that chelation is generally ~10-fold more rapid in the 5′ versus the 3′ direction for purine–purine dinucleotide sites (6,39). This underlies the occurrence of long-lived MFAs that are situated 5′ to a potential cross-linking base, which has been suggested to be a consequence of the Pt center being much closer to an N7 atom at the 5′ side (40). In addition, however, the fact that DNA conformations associated with an accessible guanine base are the most rapid sites of MFA formation means that potential cross-linking locations will tend to have a 5′ chelation site in an especially favourable configuration relative to the conversely highly unfavourable one associated with the 3′ direction (Figure 7B). Specifically, the pre-cross-linking shift configuration relative to attack in the 5′ direction is much more similar to cross-link structures (Figure 6D).
Our findings reveal how Pt drug-specific and DNA site-dependent influences can yield non-uniform adduct formation profiles not predicted from the purine distribution alone. In addition, the basic principles we have elucidated can provide a platform for developing more site selective metalloagents, possibly through exploitation of DNA structure-based modulation and additional features afforded by the proximal histone proteins in the nucleosome (9). For instance, carrier groups imposing specific steric challenges towards adduct formation, especially hindrance above or below the square Pt2+ plane, could potentially eliminate a host of counterproductive cellular targets. Finally, we note that the underlying structural principles governing base reactivity are likely relevant to many other DNA modifying species.
ACCESSION NUMBERS
Atomic coordinates and structure factors have been deposited in the Protein Data Bank under accession codes 3REH, 3REI, 3REJ, 3REK and 3REL.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Academic Research Council, the Ministry of Education, Singapore (grant 19/08); the Swiss National Science Foundation. Funding for open access charge: grant 19/08
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
We are grateful to C. Schulze-Briese, M. Wang, V. Olieric, A. Pauluhn, T. Tomizaki, M. Fuchs, R. Bingel-Erlenmeyer, G. Pompidor and C. Rajendran for exceptional support at the Swiss Light Source (Paul Scherrer Institute, Villigen, Switzerland).
REFERENCES
- 1.Glusker JP. Structural aspects of metal liganding to functional-groups in proteins. Adv. Protein Chem. 1991;42:1–76. doi: 10.1016/s0065-3233(08)60534-3. [DOI] [PubMed] [Google Scholar]
- 2.Jung YW, Lippard SJ. Direct cellular responses to platinum-induced DNA damage. Chem. Rev. 2007;107:1387–1407. doi: 10.1021/cr068207j. [DOI] [PubMed] [Google Scholar]
- 3.Todd RC, Lippard SJ. Inhibition of transcription by platinum antitumor compounds. Metallomics. 2009;1:280–291. doi: 10.1039/b907567d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Woynarowski JM, Chapman WG, Napier C, Herzig MC, Juniewicz P. Sequence- and region-specificity of oxaliplatin adducts in naked and cellular DNA. Mol. Pharmacol. 1998;54:770–777. doi: 10.1124/mol.54.5.770. [DOI] [PubMed] [Google Scholar]
- 5.Davies NP, Hardman LC, Murray V. The effect of chromatin structure on cisplatin damage in intact human cells. Nucleic Acids Res. 2000;28:2954–2958. doi: 10.1093/nar/28.15.2954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hambley TW. Platinum binding to DNA: structural controls and consequences. J. Chem. Soc. Dalton Trans. 2001:2711–2718. [Google Scholar]
- 7.Reedijk J. New clues for platinum antitumor chemistry: Kinetically controlled metal binding to DNA. Proc. Natl Acad. Sci. USA. 2003;100:3611–3616. doi: 10.1073/pnas.0737293100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kelland L. The resurgence of platinum-based cancer chemotherapy. Nat. Rev. Cancer. 2007;7:573–584. doi: 10.1038/nrc2167. [DOI] [PubMed] [Google Scholar]
- 9.Davey GE, Davey CA. Chromatin - a new, old drug target? Chem. Biol. Drug Des. 2008;72:165–170. doi: 10.1111/j.1747-0285.2008.00697.x. [DOI] [PubMed] [Google Scholar]
- 10.Morita H, Bailar JC., Jr Triamminechloroplatinum(II) chloride. Inorg. Syntheses. 1983;22:124–125. [Google Scholar]
- 11.Choi S, Vastag L, Larrabee YC, Personick ML, Schaberg KB, Fowler BJ, Sandwick RK, Rawji G. Importance of platinum(II)-assisted platinum(IV) substitution for the oxidation of guanosine derivatives by platinum(IV) complexes. Inorg. Chem. 2008;47:1352–1360. doi: 10.1021/ic701868b. [DOI] [PubMed] [Google Scholar]
- 12.Davey CA, Sargent DF, Luger K, Maeder AW, Richmond TJ. Solvent mediated interactions in the structure of the nucleosome core particle at 1.9 Å resolution. J. Mol. Biol. 2002;319:1097–1113. doi: 10.1016/S0022-2836(02)00386-8. [DOI] [PubMed] [Google Scholar]
- 13.Ong MS, Richmond TJ, Davey CA. DNA stretching and extreme kinking in the nucleosome core. J. Mol. Biol. 2007;368:1067–1074. doi: 10.1016/j.jmb.2007.02.062. [DOI] [PubMed] [Google Scholar]
- 14.Wu B, Mohideen K, Vasudevan D, Davey CA. Structural insight into the sequence dependence of nucleosome positioning. Structure. 2010;18:528–536. doi: 10.1016/j.str.2010.01.015. [DOI] [PubMed] [Google Scholar]
- 15.Dyer PN, Edayathumangalam RS, White CL, Bao Y, Chakravarthy S, Muthurajan UM, Luger K. Reconstitution of nucleosome core particles from recombinant histones and DNA. Methods Enzymol. 2004;375:23–44. doi: 10.1016/s0076-6879(03)75002-2. [DOI] [PubMed] [Google Scholar]
- 16.Wu B, Droge P, Davey CA. Site selectivity of platinum anticancer therapeutics. Nat. Chem. Biol. 2008;4:110–112. doi: 10.1038/nchembio.2007.58. [DOI] [PubMed] [Google Scholar]
- 17.Wu B, Ong MS, Groessl M, Adhireksan Z, Hartinger CG, Dyson PJ, Davey CA. A ruthenium antimetastasis agent forms specific histone protein adducts in the nucleosome core. Chemistry. 2011;17:3562–3566. doi: 10.1002/chem.201100298. [DOI] [PubMed] [Google Scholar]
- 18.Leslie AG. The integration of macromolecular diffraction data. Acta Crystallogr. D Biol. Crystallogr. 2006;62:48–57. doi: 10.1107/S0907444905039107. [DOI] [PubMed] [Google Scholar]
- 19.Bailey S. The CCP4 suite - programs for protein crystallography. Acta Crystallogr. D Biol. Crystallogr. 1994;50:760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- 20.Mohideen K, Muhammad R, Davey CA. Perturbations in nucleosome structure from heavy metal association. Nucleic Acids Res. 2010;38:6301–6311. doi: 10.1093/nar/gkq420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wu B, Davey CA. Using soft X-rays for a detailed picture of divalent metal binding in the nucleosome. J. Mol. Biol. 2010;398:633–640. doi: 10.1016/j.jmb.2010.03.038. [DOI] [PubMed] [Google Scholar]
- 22.Lu XJ, Olson WK. 3DNA: a software package for the analysis, rebuilding and visualization of three-dimensional nucleic acid structures. Nucleic Acids Res. 2003;31:5108–5121. doi: 10.1093/nar/gkg680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zheng GH, Lu XJ, Olson WK. Web 3DNA-a web server for the analysis, reconstruction, and visualization of three-dimensional nucleic-acid structures. Nucleic Acids Res. 2009;37:W240–W246. doi: 10.1093/nar/gkp358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Maxam AM, Gilbert W. Sequencing end-labeled DNA with base-specific chemical cleavages. Methods Enzymol. 1980;65:499–560. doi: 10.1016/s0076-6879(80)65059-9. [DOI] [PubMed] [Google Scholar]
- 25.Wu B, Davey CA. Platinum drug adduct formation in the nucleosome core alters nucleosome mobility but not positioning. Chem. Biol. 2008;15:1023–1028. doi: 10.1016/j.chembiol.2008.08.010. [DOI] [PubMed] [Google Scholar]
- 26.Todd RC, Lippard SJ. Structure of duplex DNA containing the cisplatin 1,2-{Pt(NH3)2}2+-d(GpG) cross-link at 1.77 Å resolution. J. Inorg. Biochem. 2010;104:902–908. doi: 10.1016/j.jinorgbio.2010.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tan S, Davey CA. Nucleosome structural studies. Curr. Opin. Struct. Biol. 2011;21:128–136. doi: 10.1016/j.sbi.2010.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Vasudevan D, Chua EY, Davey CA. Crystal structures of nucleosome core particles containing the ‘601’ strong positioning sequence. J. Mol. Biol. 2010;403:1–10. doi: 10.1016/j.jmb.2010.08.039. [DOI] [PubMed] [Google Scholar]
- 29.Tsunaka Y, Kajimura N, Tate S, Morikawa K. Alteration of the nucleosomal DNA path in the crystal structure of a human nucleosome core particle. Nucleic Acids Res. 2005;33:3424–3434. doi: 10.1093/nar/gki663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Edayathumangalam RS, Weyermann P, Dervan PB, Gottesfeld JM, Luger K. Nucleosomes in solution exist as a mixture of twist-defect states. J. Mol. Biol. 2005;345:103–114. doi: 10.1016/j.jmb.2004.10.012. [DOI] [PubMed] [Google Scholar]
- 31.Davey GE, Wu B, Dong Y, Surana U, Davey CA. DNA stretching in the nucleosome facilitates alkylation by an intercalating antitumour agent. Nucleic Acids Res. 2010;38:2081–2088. doi: 10.1093/nar/gkp1174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Davey CA, Richmond TJ. DNA-dependent divalent cation binding in the nucleosome core particle. Proc. Natl Acad. Sci. USA. 2002;99:11169–11174. doi: 10.1073/pnas.172271399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Millard JT, Wilkes EE. cis- and trans-Diamminedichloroplatinum(II) interstrand cross-linking of a defined sequence nucleosomal core particle. Biochemistry. 2000;39:16046–16055. doi: 10.1021/bi0022285. [DOI] [PubMed] [Google Scholar]
- 34.Richmond TJ, Davey CA. The structure of DNA in the nucleosome core. Nature. 2003;423:145–150. doi: 10.1038/nature01595. [DOI] [PubMed] [Google Scholar]
- 35.Coste F, Malinge JM, Serre L, Shepard W, Roth M, Leng M, Zelwer C. Crystal structure of a double-stranded DNA containing a cisplatin interstrand cross-link at 1.63 Å resolution: hydration at the platinated site. Nucleic Acids Res. 1999;27:1837–1846. doi: 10.1093/nar/27.8.1837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Spingler B, Whittington DA, Lippard SJ. 2.4 Å crystal structure of an oxaliplatin 1,2-d(GpG) intrastrand cross-link in a DNA dodecamer duplex. Inorg. Chem. 2001;40:5596–5602. doi: 10.1021/ic010790t. [DOI] [PubMed] [Google Scholar]
- 37.Helm L, Merbach AE. Inorganic and bioinorganic solvent exchange mechanisms. Chem. Rev. 2005;105:1923–1959. doi: 10.1021/cr030726o. [DOI] [PubMed] [Google Scholar]
- 38.Danford AJ, Wang D, Wang Q, Tullius TD, Lippard SJ. Platinum anticancer drug damage enforces a particular rotational setting of DNA in nucleosomes. Proc. Natl Acad. Sci. USA. 2005;102:12311–12316. doi: 10.1073/pnas.0506025102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Reeder F, Guo ZJ, Murdoch PD, Corazza A, Hambley TW, Berners-Price SJ, Chottard JC, Sadler PJ. Platination of a GG site on single-stranded and double-stranded forms of a 14-base oligonucleotide with diaqua cisplatin followed by NMR and HPLC - Influence of the platinum ligands and base sequence on 5′-G versus 3′-G platination selectivity. Eur. J. Bioch. 1997;249:370–382. doi: 10.1111/j.1432-1033.1997.00370.x. [DOI] [PubMed] [Google Scholar]
- 40.Dewan JC. Binding of the antitumor drug cis-[PtCl2(NH3)2] to crystalline ransfer RNAphe at 6-Å resolution. J. Am. Chem. Soc. 1984;106:7239–7244. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.