Abstract
MicroRNAs (miRNAs) are a class of noncoding small RNAs that act as negative regulators of gene expression. To identify miRNAs that may regulate human cell immortalization and carcinogenesis, we performed comparative miRNA array profiling of human normal and SV40-T antigen immortalized cells. We found that miR-296 was upregulated in immortalized cells that also had activation of telomerase. By an independent experiment on genomic analysis of cancer cells we found that chromosome region (20q13.32), where miR-296 is located, was amplified in 28/36 cell lines, and most of these showed enriched miR-296 expression. Overexpression of miR-296 in human cancer cells, with and without telomerase activity, had no effect on their telomerase function. Instead, it suppressed p53 function that is frequently downregulated during human cell immortalization and carcinogenesis. By monitoring the activity of a luciferase reporter connected to p53 and p21WAF1 (p21) untranslated regions (UTRs), we demonstrate that miR-296 interacts with the p21-3′UTR, and the Hu binding site of p21-3′UTR was identified as a potential miR-296 target site. We demonstrate for the first time that miR-296 is frequently upregulated during immortalization of human cells and contributes to carcinogenesis by downregulation of p53-p21WAF1 pathway.
INTRODUCTION
MicroRNAs (miRNAs) were first identified as regulators of development in Caenorhabditis elegans. They have subsequently emerged as a highly conserved class of noncoding RNAs that act as suppressors of messengers (mRNAs) by causing either their cleavage or translational inhibition. This recent appreciation of the regulatory role of small RNAs in mammalian systems has resulted in numerous studies to investigate their function in the normal biology of development and differentiation, and in disease phenotypes including cancers (1–6). miRNAs, 18–22-nucleotide (nt) in length, are transcribed as parts of longer molecules called pri-miRNAs that are processed in the cell nucleus into hairpin-structured pre-RNAs of 70–100 nt by the double-stranded RNA (dsRNA)-specific ribonuclease Drosha. Pre-miRNAs are exported from the nucleus to the cytoplasm by Exportin-5 where Dicer (a RNase III enzyme) cleaves them into 19 nt mature miRNAs with 1–4-nt 3′ overhangs at either end. One of the two strands of the mature miRNAs associates with an RNA-induced silencing complex (RISC) and become active miRNAs that suppress transcription of target genes (7,8). While many hundreds of mammalian miRNA genes have been identified and they are predicted to regulate about 30% of the human genome, little is known about their gene targets and functions (9,10).
It is a well accepted concept in carcinogenesis that nonrandom chromosomal abnormalities and accumulation of mutations in the open reading frames of oncogenes and tumor suppressors lead to loss of control of cell proliferation in cancer cells. Recent studies now suggest that the function of key genes may be altered by abnormal miRNA expression in oncogenesis, and it has been proposed that miRNA expression patterns may be used as diagnostic fingerprints and that miRNAs may be used as anticancer therapeutics (11–20). Indeed, many miRNA-encoding genes have been located at fragile sites in the genome, as well as in minimal regions of loss/gain associated with cancers (21). For example, miR-15a and miR-16-1 genes are located at chromosome 13q14, a region frequently deleted in pituitary tumors (22). miR-21 was found to be overexpressed in highly malignant human brain tumors, and was assigned functions including inhibition of apoptosis, increased angiogenesis and regulation of phosphatase and tensin homolog [PTEN (23–26)]. Inhibition of miR-21 in cultured hepatocellular carcinoma cells caused an upregulation of PTEN that was associated with decreased tumor cell proliferation, migration and invasion (26–28). In miRNA profiling studies, lung cancers were seen to have aberrant expression of about 45 miRNAs providing a specific miRNA signature (29). The let-7 miRNA family that negatively regulates oncogenic Ras is frequently downregulated, suggesting that let-7 miRNA has a tumor suppressor function (30–31). Renal cell carcinoma was associated with upregulation of a set of four miRNAs (miR-28, miR-185, miR-27 and let-7f-2); bladder cancers showed upregulation of 10 miRNAs (miR-223, miR-26b, miR-221, miR-103-1, miR-185, miR-23b, miR-203, miR-17-5p, miR-23a and miR-205) (32); colorectal cancers were found to have deregulated expression of miR-21, miR-31, miR-143 and miR-145 (28); and breast cancers were associated with increased miR-15a, miR-16, miR-17-5p, miR-21, miR-29b, miR-125b, miR-145, miR-155, miR-181b and let-7f (33). Furthermore, miR-29s was shown to regulate DNA methylation patterns. Enforced expression of miR-29s in lung cancer cell lines restored normal patterns of DNA methylation and induced re-expression of methylation-silenced tumor suppressor genes, such as FHIT and WWOX, and inhibited tumorigenicity in vitro and in vivo (34,35). The miR-34 family of miRNAs is downregulated in several types of cancers (36–38). These were shown to be induced by DNA damage and oncogenic stress in a p53-dependent manner, and to modulate p53-mediated apoptosis, cell cycle arrest and senescence by suppression of BCL2, MYCN, SIRT1 and E2F functions (39–44). p53 was also shown to regulate miRNA expression and processing several of them by interacting with Drosha (45–47), suggesting that there is a complex network of interactions in the regulation of tumor suppressors, their effectors and regulators.
In contrast to normal cells, cancer cells undergo continued proliferation and have mechanisms to maintain their telomeres, a most consistent manifestation of tumorigenesis in vitro and in vivo (48,49). The telomere maintenance mechanisms that get activated when cells escape from crisis and become immortalized include either the upregulation of telomerase activity or recombination-mediated alternative lengthening of telomeres (ALTs) (50). So far, no miRNAs have been shown to be involved in the events that allow cells to escape from crisis and become immortalized. In the present study, we used normal human fibroblasts and their SV40 T antigen-immortalized derivatives that showed telomerase-dependent (Tel) or -independent (ALT) maintenance of telomeres. miRNA array comparisons of the normal and telomerase positive cells showed upregulation of miR-296 in telomerase-positive immortal cells. Interestingly, miR-296 is located on chromosome 20q13.32 that was detected as a Gain-of-Locus in 28/36 of the cancer cell lines analyzed by Comparative Genomic Hybridization-Bacterial Artificial Chromosome (CGH-BAC) array. Molecular analyses of the effects of synthetic miR-296, its antagonist and an miR-296 expression plasmid in normal and immortalized cells (both Tel and ALT) led us to conclude that miR-296 had no direct impact on telomerase activity. Instead, it caused downregulation of the p53-p21WAF1 pathway (a major tumor suppressor pathway that is inactivated in the majority of cancers). We report that the Hu binding site of p21-3′UTR is a potential target of miR-296 and it therefore may promote carcinogenesis by downregulation of p21WAF1, in addition to its recently reported effects on angiogenesis (51).
MATERIALS AND METHODS
Cell lines and cell culture
All normal cell strains and cell lines were cultured in Dulbecco’s modified Eagle’s medium (DMEM; Gibco BRL, Grand Island, NY,USA) supplemented with 10% fetal bovine serum (Gibco BRL), penicillin (100 IU/ml) and streptomycin (50 µg/ml) in the presence of 5% CO2 at 37oC. Osteosarcoma (U-2 OS) cells were purchased from the American Type Culture Collection (ATCC, Manassas, VA, USA). Breast carcinoma cells (MCF 7) and normal human fibroblasts (TIG-1 and MRC5) were obtained from the Japanese Collection of Research Bioresources (JCRB, Japan). JFCF6 cells are normal jejunal fibroblasts from an individual with cystic fibrosis, and JFCF6/T22.5M, JFCF6/T1.J/1-3C and JFCF6/T1.J/6B are SV40-immortalized fibroblast lines derived from JFCF6 (A. Englezou, P. Bonnefin and R. Reddel, unpublished data). The latter two cell lines are referred to here as JFCF6-3C and JFCF6-6B.
Telomeric repeat amplification protocol assay
Telomerase was immunoaffinity purified from JFCF6 and JFCF6-6/T22.5M whole cell lysates (106 cells in a total volume of 200 µl) as described previously (52). This partially purified telomerase solution (2 µl) was used in a 50 µl polymerase chain reaction (PCR) reaction with 0.1 µg telomerase primer M2, and 0.05 µg reverse primer ACX (53,54). Telomerase extension of the primer was carried out at 25°C for 30 min, followed by PCR amplification of telomerase extension products using 95°C for 2 min and 30 cycles of 95°C for 10 s, 50°C for 25 s and 72°C for 30 s.
pCXbG or pCXbG-miR296 transfected cells (5 × 105) were collected and assayed for telomerase activity by Telo TTAGGG telomerase PCR enzyme-linked immunosorbent assay (ELISA; Roche, Mannhein, Germany). Serial dilutions of known concentrations of cell extract prepared from immortalized telomerase- expressing human kidney cell (HEK 293 cells) were used to establish a standard curve. U-2 OS cells were used as negative control.
miRNA array
Small RNAs <200 nt, including precursor and mature miRNAs, were extracted from JFCF6 p12 [PD11] and JFCF6/T22.5M p16 [PD45] cells with the mirVana miRNA isolation kit (Ambion, Austin, TX, USA) following the manufacturer’s protocol. Purified RNA was then labeled with Cy3 or Cy5 using the mirVana miRNA labeling kit (Ambion). Briefly, RNA was subjected to a tailing reaction with amine-modified nucleotide triphosphates by poly(A) polymerase, followed by amide formation using Cy dye ester. Labeled RNA was hybridized with oligonucleotides against a set of 229 human miRNAs arrayed on slides (Hokkaido-System Science, Japan), and detected by a scanner (Agilent Technologies, Santa Clara, CA, USA).
BAC array and CGH analysis
CGH analysis was performed on 24 breast cancer, seven gall bladder and four bile duct cell lines as described earlier (55,56). For CGH analysis, genomic DNA samples (500 ng) from the test (cancer cells) and the reference (human genomic DNA, Promega, USA) were labeled with cyanine5-dCTP and cyanine3-dCTP (Perkin Elmer; Fremont, CA, USA), respectively, in a random priming reaction using Bioprime DNA Labeling System (Invitrogen, Carlsbd, CA, USA) and Array Kit (Macrogen, Korea). Un-incorporated fluorescent nucleotides were removed by QIAquick PCR Purification Kit (Qiagen, Germany). Labeled test and reference DNAs were mixed in a solution (containing Cot−1 DNA, 100 µl and Yeast tRNA, 4 µl) (Macrogen) and were hybridized to the CGH array (4030 BAC clone DNAs in duplicate, covering the entire human genome with 1-Mbp resolution) provided by Macrogen (MAC ArrayTM KARYO 4000). The entire process from hybridization (37oC for 48–72 h) to washings [(i) 50% formamide/2X saline citrate at 46oC for 15 min; (ii) 0.1% sodium dodecyl sulfate (SDS)/2× SSC at 46oC for 30 min; (iii) PN buffer (0.1 MNa2PO4 and 0.1% NP40, Nakari Tesque, Japan) at 37°C for 15 min; and (iv) 2× SSC at 37oC for 5 min] was performed in a Hybri-station (Genomic Solutions, Ann Arbor, MI, USA). The array slides were scanned at 532 and 635 nm using GenePix4000A (Axon Instruments, Union, CA, USA) and analyzed by Mac Viewer software (Macrogen) as described earlier (55,56).
Cloning of miR-296, expression plasmid design and transfections
pCXGb-miR-296, a primary miR-296 region, was amplified from human genomic DNA by PCR using the following primers: 5′-aactcgaggaccacgtgagcccttgt-3′ and 5′-aagatatc cgggagactgtggcaga-3′. The PCR product was digested with SalI and EcoRV and introduced into pCXGb plasmid (generated from pCX-EGFP (57) between the XhoI and EcoRV sites (Figure 2A). The fragment encoding green fluorescent protein (GFP) fused with the blasticidin-resistance gene was amplified by PCR using the primers 5′-aaagaattcgccaccatggtgagcaaggg-3′, 5′-aaaaacaattgatatcactagtctcgagcggccgcctaggagccctcccacacataa-3′, and 5′-gacaaaggcttggcctggccatcgattttgtacagctcgtccatgc-3′ with pEGFP-C3 and pTracer-EF/bsd as the templates. The resultant product was digested with EcoRI and MfeI and inserted into the EcoRI sites of pCX-EGFP to generate pCXGb.
Figure 2.
miR-296 is amplified in cancer cell lines. (A) Gene locus for miR-296 on chromosome 20q13.32. (B) Summary of the comparative genomic hybridization (CGH) analysis of 36 cancer cell lines showing amplification (+) of 20q13.32. (C) Genomic PCR of miR-296 on normal and cancer cells showing amplification of miR-296 in cancer cells.
Transfections of pCXbG or pCXbG-miR296 plasmids were performed using Fugene (Roche Applied Science, Indianapolis, IN, USA). Typically, 3 µg of plasmid DNA was used per 6-cm dish of cells at 60–70% confluence. Transfected cells were detected by GFP fluorescence and the GFP-miR296 was amplified by PCR (denaturation: 95oC for 3 min followed by 35 cycles of denaturation: 95oC for 30 s, annealing: 60oC for 30 s and extension 72oC for 1 min), on the genomic DNA of the transfected cells using GFP (sense 5′-taccagacaaccattacctg-3′) and miR-296 (antisense 5′-gactcagcagctgacatgac-3′) primers.
Synthetic precursor miR-296 (PmiR-296) and its antagonist (AmiR-296)
Synthetic precursor miR-296 (PmiR-296) was purchased from Ambion. 25 nM of PmiR-296 was transfected into each cell type using Lipofectamine 2000 (Invitrogen). For the inhibition of miR-296, 25 nM miR-296 inhibitor oligonucleotide (AmiR-296, Ambion) that targeted miRNA-296 3p or control oligonucleotides were transfected. After 48 h, the cells were trypsinized, and harvested for further analysis.
Western blotting
Cells were lyzed in Nonidet P-40 lysis buffer in the presence of protease inhibitors. Protein concentration was determined using the BCA Protein Assay (Bio-Rad, Hercules, CA, USA). Each sample (20–50 μg) was separated by SDS–polyacrylamide gel electrophoresis (PAGE). The proteins were then electroblotted onto a polyvinylidene fluoride (PVDF) membrane. Membranes were blocked in TBS-T containing 5% dry milk. After blocking, membranes were incubated with anti-p53 (DO-1) (sc-126; Santa Cruz Biotech, Santa Cruz, CA, USA), and anti-p21 (C-19) (sc-397; Santa Cruz Biotech). Actin was used as an internal loading control (anti-actin antibody, Chemicon International, CA, USA). After probing with the first antibody as indicated, the blots were incubated with HRP (horse radish peroxidase)-conjugated secondary antibody (sc-2004 or sc-2005; Santa Cruz Biotech). Finally, the blots were developed using enhanced chemiluminescence (ECL) (RPN2132; GE Healthcare, Buckinghamshire, UK).
Immunostaining
Cells were fixed with prechilled (at −20°C) methanol/acetone (1:1) mixture for 10 min, rinsed thrice with phosphate-buffered saline (PBS), and blocked with 2% bovine serum albumin (BSA) for 1 h. Cells were incubated with appropriate primary antibodies at 4°C overnight. The anti-proliferating cell nuclear antigen (PCNA; 2568; Cell Signaling Technology) and p21 (sc-397; Santa Cruz Biotech) antibodies were recognized by Alexa Fluor 488 anti-mouse immunoglobulin G (IgG, Invitrogen) fluorophore-conjugated secondary antibodies. For 4′,6-diamidino-2-phenylindole (DAPI) staining, fixed cells were incubated in 1:500 of a stock solution of DAPI (5 mg/ml) at room temperature in dark for 30 min and were then washed with PBS. Actin filaments were labeled with Alexa Fluor-Phalloidin (Red). Finally, the stained cells were examined with a conventional fluorescence microscope or confocal scanning laser microscopy (Leica TCS SP2 AOBS microscope), using a 40× objective.
RNA isolation, reverse transcription-PCR and Quantitative PCR
For reverse transcription (RT)–PCR, total RNA was isolated from cells at 48 h post transfections using the RNeasy mini kit (M1705; Qiagen, Standford Valencia, CA, USA). Equal amounts of RNA (1 µg) were used in the RT reaction using oligo (dT)18 as the primer to generate the first-strand complementary DNA (cDNA), and M-MLV reverse transcriptase (A1250; Promega, Madison, WI, USA) was used to generate cDNA. The cDNA from each sample was then subjected to one round of PCR amplification consisting of an initial melting step (94°C for 5 min) followed by a number, optimized for each amplicon, of repetitive cycles (28–35) consisting of denaturation at 94°C for 30 s, annealing at 55°C for 1 min, and extension at 72°C for 30 s using primers specific to p53, p21, and GAPDH (internal control) mRNA. The primers used for the p21 RT–PCR were 5′-gacaccactggagggtgact-3′ (sense) and 5′-ggcgtttggagtggtagaaa-3′ (antisense), for p53 were 5′-aaggaaatctcaccccatcc-3′ (sense) and 5′-aaggctgcagtaagccaaga-3′ (antisense) and for GAPDH (internal control) were 5′-ACCACAGTCCATGCCATCAC-3′ (sense) and 5′-TCCACCACCCTGTTGCTGTA-3′ (antisense). To determine the quantitative-real-time PCR for p21 mRNA expression, total RNA was isolated from U-2 OS cells using TRIzol reagent. cDNA was synthesized using 5 μg of total RNA, oligo dT (2.5 mM) and Superscript Reverse Transcriptase II (200 U) in 20 μl using an RT kit (Invitrogen). Quantitative (Q)-PCR was performed using SYBR Green Master Mix (Applied Biosystems, Carlsbad, CA, USA). The following Q-PCR oligonucleotide primers sets were used: p21 sense (5′-cccgtgagcgat ggaact-3′), p21 antisense (5′-cgctcccaggcgaagtc-3′), GAPDH sense (5′-ccccttcattgacctcaactac-3′) glyceraldehyde 3-phosphate dehydrogenase (GAPDH) antisense (5′-tctcgctcctggaagatgg-3′).
Cell proliferation and viability assays
The effect of miR-296 on cell proliferation was determined by direct cell counting and by measuring the conversion of the tetrazolium salt MTT to formazan. Briefly, cells were treated with precursor miR-296 or anti miR296 (30 nM) in DMEM with 10% fetal bovine serum (FBS). After 2 days of incubation at 37oC, 200 µl of MTT (Sigma Chemical Corp., St. Louis, MO, USA) in PBS (2 µg/ml) was added to each well. After 4 h of incubation at 37°C, the supernatant was discarded and the precipitate was dissolved with 100 µl of dimethylsulfoxide (DMSO). Plates were then read on a microplate reader at 540 nm. Number of living cells was calculated from untreated cells cultured and treated with MTT in the same condition as in the experimental groups. Control and transfected cells were also stained for cell proliferation marker PCNA as described above.
Cloning of pGL4(EB)-p53-3′ untranslated region and pGL4(EB)-p21-3′UTR
pGL4(EB)-p53-3′ untranslated region (UTR); p53-3′UTR region was amplified by PCR using human genomic DNA with primers: 5′-aaaGAATTCtgacattctccacttcttgttccc-3′ and 5′-aaaAGATCTtggcagcaaagttttattgtaaaataagag-3′. The PCR product was digested with EcoRI and BglII and introduced into pGL4(EB) plasmid between EcoRI and BamHI sites. pGL4(EB)-p21-3′UTR; p21-3′UTR region was amplified by PCR using human genomic DNA with primers: 5′-aaaCAATTGTAAtccgcccacaggaagcctgc-3′ and 5′-aaaGGATCCtaaagtcactaagaatcatttattgagcacctg-3′. The PCR product was digested with MfeI and BamHI and introduced into pGL4(EB) plasmid between the EcoRI and BamHI sites.
Cloning of pGL4(EB)-HU and pGL4(EB)-p21-3′UTR
Nucleotide sequences in the p21 3′UTR mRNA (Hu binding site and two candidate CU rich regions (CU rich-1 and -2) were cloned into a reporter plasmid (58). The Hu binding site consisted of 28 nt (5′-AATTC TCTTAATTATTATTTGTGTTTTAATTTA G-3′ and 5′-GATCC TAAATTAAAACACAAATAATAATTAAGA G-3′). CU rich region-1 consisted of the following 32 nt (5′-AATTC CCTCATCCCGTGTTCTCCTTTTCCTCTCTCCC G-3′ and 5′-GATCC GGGAGAGAGGAAAAGGAGAACACGGGATGAGG G-3′) and CU rich region-2 consisted of 32 nt (5′-AATTC CTTCCTCATCCACCCCATCCCTCCCCAGTT G-3′ and 5′-GATCC AACTGGGGAGGGATGGGGTGGATGAGGAAG G-3′). Annealing of the single strands was performed by heating at 95°C (10 min) followed by slow cooling at room temperature. The double-stranded oligonucleotides were digested with EcoRI and BamHI, and cloned into pGL4(EB), generating luciferase-expressing vectors (pGL4(EB)-HU, pGL4(EB)-CU1 and pGL4(EB)-CU2) in which the luciferase open reading frame was followed by a 3′UTR containing the binding site of interest.
Luciferase reporter assay
U-2 OS or MCF 7 cells were cultured in 12-well plates and transfected with the luciferase constructs described above [pGL4, pGL4(EB)-p53-3′UTR, pGL4(EB)-p21-3′-UTR, pGL4(EB)-HU, pGL4(EB)-CU1 or pGL4(EB)-CU2 together with synthetic miR-296 (PmiR-296) or its antagonist (AmiR-296) using Lipofectamine (Gibco BRL)]. Luciferase assays were performed as described previously (59). Cells were transfected with 500 ng of various luciferase vectors using Lipofectamine (Gibco BRL). Luciferase activity was measured as described in the manufacturer's manual (Luciferase assay kit; Promega) with a luminometer and normalized to concentration of total protein. Protein content was measured with a BCA protein assay kit (Pierce Biotechnology, Rockford, IL, USA). Experiments were carried out in triplicate and repeated at least three times.
RESULTS AND DISCUSSION
Upregulation of miR-296 during human cell immortalization
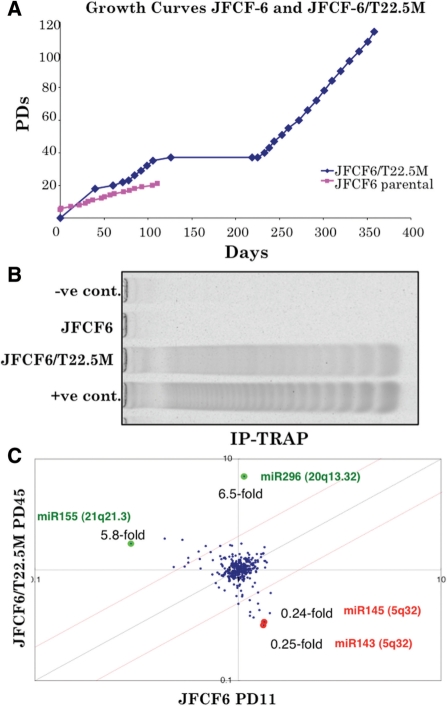
Growth curves of the parental JFCF6 cells and the SV40 T antigen transformed clone JFCF6/T22.5M are shown in Figure 1A. Whereas the parental JFCF6 cells underwent only 20 population doublings (PDs) before permanently ceasing proliferation, the JFCF6/T22.5M cells could be passaged for more than 100 PD, suggesting that they have undergone immortalization. They also showed activation of telomerase as demonstrated in Figure 1B. We performed miRNA array analysis of the parental JFCF6 cells at PD11 and telomerase-positive JFCF6/T22.5M derivatives at PD45, shortly after immortalization. As shown in Figure 1C, miRNA array analysis showed increased miR-296 (6.5-fold) and miR-155 (5.8-fold) expression in JFCF6/T22.5M cells. Two other miRNAs miR-143 and miR-145 were decreased in the immortalized JFCF6/T22.5M cells as compared to their parental counterpart JFCF6. We first examined the chromosomal localization of the genes encoding these miRNAs and investigated if any of these were located on chromosome regions commonly amplified in cancer cells by performing BAC-CGH array on 36 cancer cell lines (Figure 2). As shown, we found that the chromosome locus of miR-296 (20q13.32) (Figure 2A) was amplified in 28/36 cell lines (Figure 2B). By genomic PCR using miR-296 specific primers, amplification of miR-296 was confirmed in the cancer cell lines (Figure 2C). In some cancer cell lines, although amplification of the chromosome region 20q13.32 was not detected by BAC-CGH array, genomic PCR revealed amplification of miR-296 as compared to the control (Figure 2B and C). We presumed that this was due to the different resolution of BAC-CGH and PCR techniques. In order to verify that the gene amplification detected by PCR was not an artifact, we performed miR-296 PCR on the genomic DNA isolated from blood samples of 24 healthy individuals. In contrast to the cancer cell lines that showed amplification of miR-296 (3.08- to 7.2-fold as compared to the control; most cell lines showed 3-fold or higher amplification), none of the 24 normal genomic DNA samples showed miR-296 amplification (data not shown). Since miR-298 is found to be clustered with miR-296, we examined its status as well. As summarized in Table 1, miR-298 showed amplification in most of the cancer cell lines; but followed a different amplification pattern to that of the miR-296 (amplification ranged between 1.09- and 5.67-fold with many cell lines showing <3-fold increase) indicating that miR-296 and miR-298 might not be co-regulated. The data revealed that the copy number of miR-296 and miR-298 is increased in cancer cells, suggesting their involvement in carcinogenesis.
Figure 1.
miR-296 is upregulated in the SV40-immortalized telomerase-positive JFCF6 cells. (A) Growth curves of JFCF6 and its SV40 T antigen-immortalized derivative cell line JFCF-6/T22.5 M. (B) Telomerase activity in parental JFCF6 and its immortalized cell line JFCF6T/22.5 M. (C) miRNA array comparisons of the parental JFCF6 and a telomerase-positive immortalized derivative cell line, JFCF6/T22.5M, showing upregulation of miR-296 (6.5-fold) and miR-155 (5.8-fold), and downregulation of miR-145 (0.24-fold) and miR-143 (0.25-fold) in the latter.
Table 1.
miR-296 and miR-298 amplification in cancer cells
| Cell line | miR296 | miR298 |
|---|---|---|
| BT-474 | 3.10 | ND |
| HCC2218 | 3.08 | 4.00 |
| MDA-MB-231 | 3.18 | 1.09 |
| SKBR3 | 6.74 | 3.92 |
| MDA-MB-435S | 6.36 | 2.29 |
| AU565 | 4.62 | 1.45 |
| MDA-MB-468 | 7.24 | 1.73 |
| HCC1428 | 5.03 | 1.91 |
| HCC1954 | 3.29 | 5.69 |
| HS578T | 3.20 | 4.27 |
| HCC1008 | 3.78 | 3.18 |
| UACC812 | 5.85 | 5.15 |
| UACC893 | 5.36 | 1.18 |
| HCC1937 | 3.76 | 4.36 |
| MDA-MB-361 | 3.07 | 2.36 |
| ZR-75-1 | 4.08 | 2.98 |
| BT-483 | 3.91 | 4.00 |
| T47D | 2.40 | 5.09 |
| MDA-MB-175-V11 | 2.31 | 4.75 |
| MDA-MB-415 | 3.18 | 3.35 |
| HCC1806 | 3.23 | 1.55 |
| ZR-75-30 | 3.68 | 2.96 |
GAPDH was used as an internal control and the relative amplification (fold increase over the control, normal genomic DNA) in respective cell lines is shown. ND, not determined.
miR-296 suppresses the p53-p21WAF1 pathway
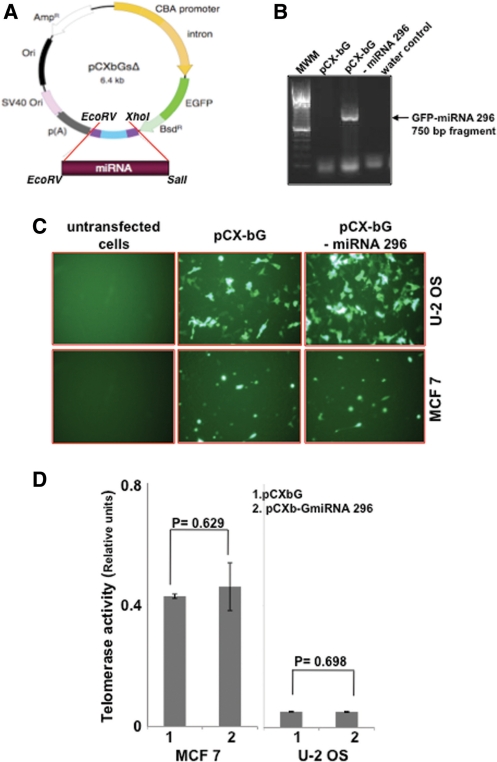
To examine the effect of miR-296 in cultured cells, we constructed a miR-296 overexpression vector. A 389-bp human genomic DNA fragment (encoding miR-296 precursor) was amplified by PCR on normal human genomic DNA and was expressed from the β-actin promoter in continuity with GFP in pCXbGsΔ vector, as illustrated in Figure 3A. We first confirmed the occurrence of GFP-miR-296 in transfected cells by PCR using GFP (sense) and miR-296 (antisense) primers. As shown in Figure 3B, expected size (750 bp) PCR product was detected in the pCXbGsΔ-miR-296 transfected cells only. Since miRNA array revealed an upregulation of miR-296 in telomerase-positive JFCF6-derived immortal cells (Figure 1C), we next investigated the relationship of miR-296 with the telomerase activity. We generated miR-296 overexpressing telomerase-positive (MCF 7 and JFCF6-6B) and telomerase-negative (U-2 OS and JFCF6-3C) cells. The transfected cells were selected in blasticidin (10 µg/ml)-supplemented medium and were seen to have bright GFP fluorescence (Figure 3C) with transfection efficiency of more than 80% both in telomerase-positive and -negative cells. Control and miR-296 transfected cells were assayed for telomerase activity by telomeric repeat amplification protocol (TRAP) assay. However, as shown in Figure 3D, there was no apparent difference in the level of telomerase activity in control and miR-296 transfected telomerase-positive or ALT cells.
Figure 3.
Exogenous miR-296 expression did not affect the telomerase activity. (A) Diagrammatic presentation of miR-296 expression vector, pCXGb-miR-296. The plasmid contained CMV enhancer/chicken β-actin promoter (CBA) driving the expression of the inserted miR-296 in continuity with enhanced GFP (EGFP) protein. Polyadenylation sequence p(A) was added to its 3′-end. The vector contained Simian Virus 40 (SV40) origin of replication (ori), blasticidin resistance (bsdR)- and ampicillin resistance (AmpR)- encoding sequences. (B) PCR amplication of GFP-miR-296 from control and miR-296 expressing vector transfected cells. (C) Exogenous expression of EGFP-miR-296 in human cancer cells with (MCF 7) and without (U-2 OS) telomerase activity. (D) Telomerase activity in control (1) and miR-296 transfected (2) cells showing no change in the telomerase activity in either the telomerase-positive (MCF 7) or -negative (U-2 OS) cells.
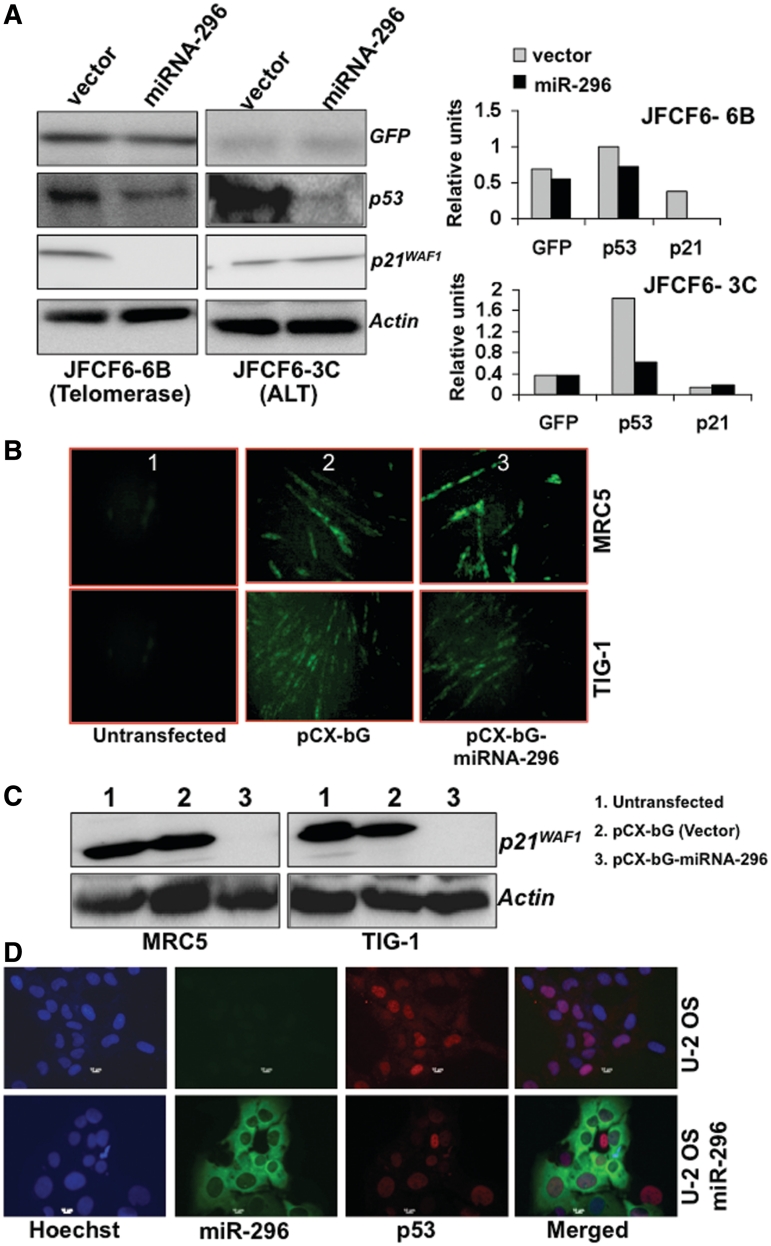
In human cells, immortalization and activation of telomerase are frequently associated with inactivation of tumor suppressor protein p53 function, raising the possibility of a link between these two pathways. It has also been shown that the overexpression of wild-type p53 downregulates the enzymatic activity of telomerase in various cancer cell lines through transcriptional repression of the gene encoding its catalytic subunit, human telomerase reverse transcriptase (hTERT) (60). We, therefore, investigated whether miR-296 had any impact on this pathway. As shown in Figure 4A, we found that the level of p53 protein decreased in cells overexpressing miR-296 and was independent of the telomerase activity status. Interestingly, p21WAF1, a major downstream effector of p53 was found to be strongly suppressed in the telomerase-positive line compared to the telomerase-negative (ALT) line derived from the same parental cells. In order to further investigate the role of miR-296 in regulation of the p53-p21WAF1 pathway and to exclude the effect of SV40 large T antigen, we used normal human fibroblasts with an active p53-p21WAF1 pathway. We found that miR-296 overexpression caused strong suppression of p21WAF1 expression in both TIG-1 and MRC5 cells (Figure 4B and C). We also examined the expression of p53 by immunostaining at a single cell level in control and miR-296 vector transfected U-2OS and MCF7 cells (both have wild-type p53). Whereas 75–80% of control cells showed bright p53 nuclear staining, only 3–5% of the miR-296 vector transfected cells (GFP fluorescence) showed such staining in both U-2 OS and MCF7 cells (Figure 4D and data not shown) demonstrating an inactivation of p53 by miR-296.
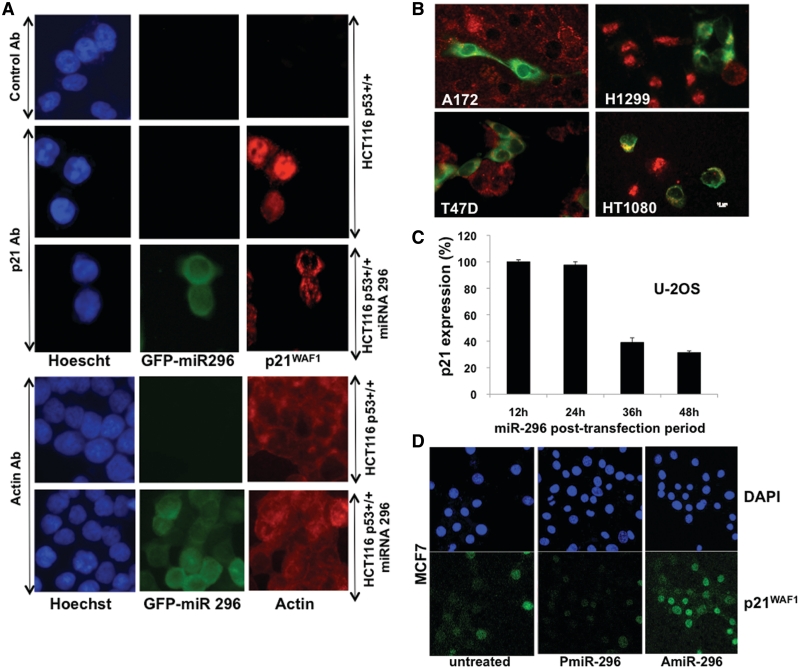
Figure 4.
Exogenous miR-296 expression inactivated the p53-21WAF1 pathway. (A) Immortal JFCF6-derived cells (6B, telomerase-positive and 3C, telomerase-negative) cells transfected with miR-296 showed decreased levels of p53. p21WAF1 showed decrease in telomerase-positive (JFCF6-6B) cells only. (B) Normal human fibroblasts transfected with miR-296 expression vector showing GFP fluorescence. (C) Normal human fibroblasts transfected with miR-296 showed a decrease in p21WAF1 protein. (D) Single-cell immunostaining for p53 in control and pCXbG-miR-296 transfected U-2 OS cells. Cells showing GFP fluorescence (GFP-miR-296) either lacked or showed faint nuclear staining for p53 (red).
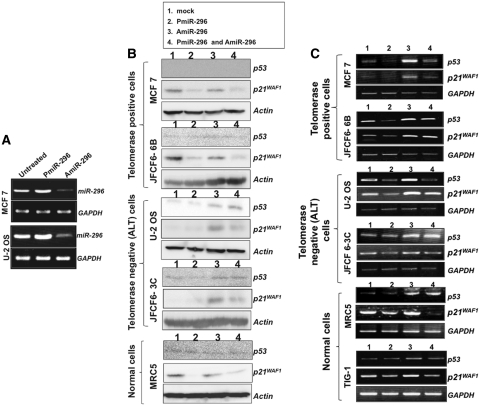
In order to confirm the above findings, we next used a synthetic precursor RNA for miR-296 (PmiR-296) and its inhibitor/antagonist (AmiR-296) that target miR-296 3p. At first, the effect of synthetic precursor and inhibitor was examined by RT–PCR on the transfected cells. As expected, the introduction of PmiR-296 resulted in an increase and of AmiR-296 in the decrease of miRNA-296 (Figure 5A). Furthermore, PmiR-296 transfected telomerase-positive (MCF 7 and JFCF6-6B) and normal (MRC5) cells showed strong suppression of p21WAF1 (Figure 5B). Co-transfection of PmiR-296 and its inhibitor (AmiR-296) partially canceled the suppressive effect of PmiR-296 on p21WAF1. The level of p21WAF1 in the untreated ALT cells (U-2 OS and JFCF6-3C) was lower than in the telomerase-positive cells, and remain unchanged in response to PmiR-296 treatment. However, the transfection of AmiR-296 caused an increase in both p53 and p21WAF1 levels. The data suggest that the p53-p21WAF1 pathway is regulated by miR-296. Of note, the effect of PmiR-296 on p53 (Figure 5B) was weak as compared to the miR-296 expressed from the vector (Figure 4). Since the protein levels are affected by their stability and interaction with other proteins (especially in case of wild-type p53 that is rapidly degraded), we examined the levels of p53 and p21WAF1 transcripts by RT–PCR. As shown in Figure 5C, PmiR-296 caused decrease in p53 and p21WAF1 transcripts, although to a different extent in different cell lines. Furthermore, a clear increase, both in p53 and p21WAF1 transcription with AmiR-296, and an antagonistic effect of PmiR-296 on AmiR-296 were detected (lanes 3 and 4, Figure 5C). These data confirmed that the miR-296 downregulates the p53-p21WAF1 pathway. We also examined the effect of PmiR-296 and AmiR-296 on cell proliferation. Transient transfection of the synthetic precursor of miR-296, PmiR-296, caused a 10–20% increase in cell number (observed at 48–60 h after the transfection); AmiR-296 led to a decrease in proliferation that was partially compensated by co-transfection of PmiR-296 (Figure 6A). Consistent with these data, (i) PmiR-296 resulted in an increase and AmiR-296 in decrease in viability of both MCF7 and U-2 OS cells (Figure 6B); (ii) PmiR-296 transfected OS cells showed increase in the level of cell proliferation marker PCNA (Figure 6C); and (iii) human normal cells (TIG-1) stably transfected with miR-296 showed increased proliferation (Figure 6D). Taken together, we report for the first time, that miR-296 downregulates the p53-p21WAF1 pathway and result in increased proliferation and viability of cells.
Figure 5.
miR-296 regulates p53-p21WAF1 pathway. (A) RT–PCR amplification of miR-296 from cells treated with either precursor PmiR-296 or antagonist AmiR-296. (B) p53 and p21WAF1 protein levels in cells transfected with synthetic precursor miR-296 (PmiR-296) and antagonist (AmiR-296) of miR-296. PmiR-296 transfected cells showed decreased levels of p53 and p21WAF1 proteins. The decrease was more pronounced in telomerase-positive (MCF 7 and JFCF6-6B) than in the telomerase-negative (U-2 OS and JFCF6-3C) cells. PmiR-296 transfected normal human fibroblasts showed a sharp decrease in p21WAF1. Cells transfected with miR-296 antagonist (AmiR-296, Lane 3) showed an increase in p21WAF1 and co-transfections of AmiR-296 with PmiR-296 (lane 4) partly antagonized the effect of PmiR-296 (lane 2). (C) RT–PCR analysis for miR-296 transcript in cells transfected with synthetic precursor miR-296 (PmiR-296) and antagonist (AmiR-296) of miR-296. PmiR-296 caused reduction in p53 and p21WAF1 transcripts (lane 2). AmiR-296 caused increase in miR-296 transcript (lane 3) and decrease in p53 and p21WAF1 obtained by PmiR-296 (lane 2) was compensated by co-transfection with Amir-296 (lane 4).
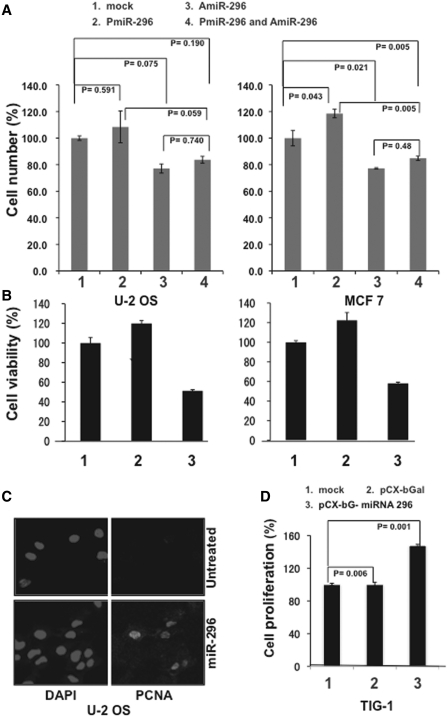
Figure 6.
miR-296 caused an increase in cell proliferation and cell viability. (A) Percent number of surviving U-2 OS and MCF 7 cells transfected with either PmiR-296 or AmiR-296 or the two together. Bars represent standard deviation calculated from three independent experiments. (B) Viability (MTT) assay of cells transfected with either PmiR-296 or AmiR-296. (C) PCNA staining of control and PmiR-296 treated cells. (D) Percent increase in number of TIG-1 cells transfected with control or miR-296 expression vector.
p21WAF-1 is a potential target for miR-296
In order to further investigate whether p53 and/or p21WAF1 are the targets of miRNA-296, we generated luciferase reporter plasmids having either the p53 3′UTR or the p21WAF1 3′UTR at the 3′ terminus of the luciferase open reading frame, as shown in Figure 7A. Cells were transfected with these reporters along with either the control vector or PmiR-296 expressing vector. We found that miR-296 had a weak but significant effect on the luciferase-p53-3′UTR. On the other hand, it caused robust suppression of luciferase-p21WAF1 3′UTR expression (Figure 7B). These data suggested that miR-296 targets both p53 and p21WAF1, the latter being a stronger target. Of note, it did not have any typical seed sequence for miR-296 in its 3′UTR. Several recent studies have reported that p21WAF1 mRNA levels are regulated not only at the transcriptional level, but also at the post-transcriptional level. The stability of p21WAF1 mRNA is controlled by Hu proteins that specifically bind to an AU-rich element in its 3′-UTR (58,61,62). Therefore, we subcloned the p21WAF1-3′UTR to the 3′UTR of luciferase as three fragments (HU region, CU rich region 1 and CU region 2) (Figure 7C and D) and investigated the effect of miR-296 on expression of these constructs. We found that the transfection of miR-296 decreased the expression of the construct containing the HU region (Figure 7E), indicating that miR-296 decreases p21WAF1 mRNA level via interaction with the HU region in its 3′UTR. Since this region also acts as a binding site for Hu protein that regulates the stability of p21WAF1, it is possible that miR-296 modulates the binding of the HU proteins, resulting in an unstable p21WAF1 mRNA. hWNK4 (human with-no-lysine kinase-4, a member of the serine-threonine protein kinase family associated with hypertension) has also been shown to be downregulated by miR-296 at the post-transcriptional level in a cell-specific pattern (63). Although we found that there is 40% homology between p21WAF1-3′UTR and hWNK4-3′UTR, the target sequence of miR-296 in the hWNK4-3′UTR (63) is different from the HU sequence defined in our study, suggesting that miR-296 may have multiple target sequences. The molecular details of the miR-296 binding to the HU region of p21WAF1-3′ UTR and mechanism of p21WAF1 suppression warrant further studies.
Figure 7.
miR-296 strongly targets p21WAF1 promoter. (A) Schematic representation of luciferase reporter in which p53- 3′UTR or p21WAF1-3′UTR were linked to the 3′-end of luciferase in a CMV promoter-driven expression plasmid. (B) Luciferase activity of Luc-p53-3′UTR or Luc-p21WAF1-3′UTR constructs. A weak but significant decrease in luciferase linked to p53-3′UTR was observed in miR-296 transfected cells. miR-296 caused sharp decline in luciferase expression from the Luc-p21WAF1-3′UTR construct. (C) Schematic representation of luciferase reporter in which p21WAF1-3′UTR was serially deleted to generate three constructs containing, HU binding site, CU rich region1 and CU rich region 2. (D) Sequence of HU binding site, CU rich region 1 and CU rich region 2. (E) Luciferase activity of the Luc-p21WAF1-3′UTR construct and its deletion mutants containing HU binding site, CU rich region 1 and CU rich region 2 showing that downregulation of luciferase expression is caused by interaction of miR-296 with the HU binding site.
We finally validated the effect of miR-296 on p21WAF1 expression in a variety of cancer cell lines. The cells transfected with miRNA expression plasmid were examined for p21WAF1 expression by immunostaining. As shown in Figure 8A, miR-296 overexpressing cells (green fluorescence) showed decreased p21WAF1 staining. Staining with the secondary antibody and an irrelevant control (actin) antibody were used as negative controls. Similar results were obtained in four other cell lines that showed either less or no p21WAF1 staining (red fluorescence) in miR-296 transfected cells confirming that miR-296 downregulates p21WAF1 (Figure 8B). We also performed Q-PCR to determine the half-life of p21 mRNA in miR-296 transfected cells. As shown in Figure 8C, p21 mRNA showed time-dependent decrease during 48 h post-transfection (about 70–80% decrease at 48 h). Furthermore, as seen by immuno-cytostaining, similar to the vector-driven expression of miR-296, synthetic PmiR-296 caused decrease in p21WAF1, whereas an increase was observed in AmiR-296 transfected cells (Figure 8D). The synthetic antagonist AmiR-296 was designed to target miR-296 3p, suggesting that the latter targets p21WAF1.
Figure 8.
miR-296 downregulates p21 expression. (A) p21WAF1 expression (red) in control and pCXbG-miR-296 transfected cells. Secondary antibody (control Ab) and actin staining were used as negative controls. (B) A variety of cancer cells examined for p21WAF1 expression after transfection of miR-296 expression construct (green) showed lack of red staining in green cells demonstrating that miR-296 downregulates p21WAF1 expression. (C) Quantitative-PCR for p21WAF1 in U2-O S cells treated with PmiR-296 showed time-dependent decrease after treatment with PmiR-296. (D) Cells treated with PmiR-296 showing decrease and with AmiR-296 showing increase in p21WAF1 staining. The data demonstrate that the miR-296 regulates p21WAF1.
Amplification of the 20q13.32 genomic region has been found in a wide variety of cancers (64–66). miR-296 has been categorized as an Angio-miR, along with miR-378 and miR-17-92 in the same gene cluster, due to its contribution to tumor angiogenesis (67,68). A study using co-culture of human U87 glioma and primary microvascular endothelial cells found that the elongated endothelial cells showing tubule formation and branching had upregulation of miR-296 (51). This was confirmed by culturing U87 glioma cells in angiogenic cocktail-supplemented medium. Feed-forward regulation was identified in which vascular endothelial growth factor (VEGF) induced miR-296 expression and that in turn targeted HGS, leading to increased levels of VEGFR2 and PDGFRb proteins and an increase response to VEGF. It was also induced by EGF and suggested a complex growth factor crosstalk mechanism leading to the development of angiogenesis. The expression of miR-296 was found to be upregulated in esophageal squamous cell cancer tissues and was associated with high mortality among the patients. Downregulation of miR-296 was shown to inhibit esophageal cancer cell progression by regulation of multiple signaling pathways including cyclin D1, p27, P-glycoprotein, Bcl-2, MDR1 and Bax, and was proposed as a potential target for intervention in this malignancy (69).
We have detected an upregulation of miR-296 in SV40 immortalized human cells, suggesting that it is an early event during cell transformation and carcinogenesis. We have shown, for the first time, that miR-296 is enriched in many cancer cell lines and suppresses p53-p21WAF1 pathway, an early event during immortalization of human cells. We provide evidence that miR-296 targets p21WAF-1 by interacting with its 3′ UTR. Since the p53-p21WAF1 pathway has several activities, including control of cell cycle, apoptosis, control of centrosome duplication and genomic stability, that are lost in human cancers, miR-296 may be a candidate therapeutic target for cancer therapy.
FUNDING
This work was supported by grants form the National Research Foundation of Korea (R15-2004-024-02001-0, 2010-0029220, 2009K001644) (to C-O. Yun); Australian Postgraduate Award, a Judith Hyam Memorial Trust Fund for Cancer Research Scholarship and a Research Scholar Award from Cancer Institute, New South Wales, Austrila (to Z.K.). Funding for open access charge: Lab budget.
Conflict of interest statement. None declared.
REFERENCES
- 1.Bartels CL, Tsongalis GJ. MicroRNAs: novel biomarkers for human cancer. Clin. Chem. 2009;55:623–631. doi: 10.1373/clinchem.2008.112805. [DOI] [PubMed] [Google Scholar]
- 2.Garzon R, Calin GA, Croce CM. MicroRNAs in cancer. Annu. Rev. Med. 2009;60:167–179. doi: 10.1146/annurev.med.59.053006.104707. [DOI] [PubMed] [Google Scholar]
- 3.Lee YS, Dutta A. MicroRNAs in cancer. Annu. Rev. Pathol. 2009;4:199–227. doi: 10.1146/annurev.pathol.4.110807.092222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Nimmo RA, Slack FJ. An elegant miRror: microRNAs in stem cells, developmental timing and cancer. Chromosoma. 2009;118:405–418. doi: 10.1007/s00412-009-0210-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Schickel R, Boyerinas B, Park SM, Peter ME. MicroRNAs: key players in the immune system, differentiation, tumorigenesis and cell death. Oncogene. 2008;27:5959–5974. doi: 10.1038/onc.2008.274. [DOI] [PubMed] [Google Scholar]
- 6.Shivdasani RA. MicroRNAs: regulators of gene expression and cell differentiation. Blood. 2006;108:3646–3653. doi: 10.1182/blood-2006-01-030015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Meltzer PS. Cancer genomics: small RNAs with big impacts. Nature. 2005;435:745–746. doi: 10.1038/435745a. [DOI] [PubMed] [Google Scholar]
- 8.Toledo F, Bardot B. Cancer: three birds with one stone. Nature. 2009;460:466–467. doi: 10.1038/460466a. [DOI] [PubMed] [Google Scholar]
- 9.Fabbri M, Croce CM, Calin GA. MicroRNAs. Cancer J. 2008;14:1–6. doi: 10.1097/PPO.0b013e318164145e. [DOI] [PubMed] [Google Scholar]
- 10.Zhang C. MicroRNomics: a newly emerging approach for disease biology. Physiol. Genomics. 2008;33:139–147. doi: 10.1152/physiolgenomics.00034.2008. [DOI] [PubMed] [Google Scholar]
- 11.Barbarotto E, Schmittgen TD, Calin GA. MicroRNAs and cancer: profile, profile, profile. Int. J. Cancer. 2008;122:969–977. doi: 10.1002/ijc.23343. [DOI] [PubMed] [Google Scholar]
- 12.Caldas C, Brenton JD. Sizing up miRNAs as cancer genes. Nat. Med. 2005;11:712–714. doi: 10.1038/nm0705-712. [DOI] [PubMed] [Google Scholar]
- 13.Cummins JM, He Y, Leary RJ, Pagliarini R, Diaz LA, Jr, Sjoblom T, Barad O, Bentwich Z, Szafranska AE, Labourier E, et al. The colorectal microRNAome. Proc. Natl Acad. Sci. USA. 2006;103:3687–3692. doi: 10.1073/pnas.0511155103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Folini M, Gandellini P, Longoni N, Profumo V, Callari M, Pennati M, Colecchia M, Supino R, Veneroni S, Salvioni R, et al. miR-21: an oncomir on strike in prostate cancer. Mol. Cancer. 2010;9:12. doi: 10.1186/1476-4598-9-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gandellini P, Folini M, Zaffaroni N. Towards the definition of prostate cancer-related microRNAs: where are we now? Trends Mol. Med. 2009;15:381–390. doi: 10.1016/j.molmed.2009.07.004. [DOI] [PubMed] [Google Scholar]
- 16.Hammond SM. MicroRNAs as oncogenes. Curr. Opin. Genet. Dev. 2006;16:4–9. doi: 10.1016/j.gde.2005.12.005. [DOI] [PubMed] [Google Scholar]
- 17.Lu J, Getz G, Miska EA, Alvarez-Saavedra E, Lamb J, Peck D, Sweet-Cordero A, Ebert BL, Mak RH, Ferrando AA, et al. MicroRNA expression profiles classify human cancers. Nature. 2005;435:834–838. doi: 10.1038/nature03702. [DOI] [PubMed] [Google Scholar]
- 18.Yanaihara N, Caplen N, Bowman E, Seike M, Kumamoto K, Yi M, Stephens RM, Okamoto A, Yokota J, Tanaka T, et al. Unique microRNA molecular profiles in lung cancer diagnosis and prognosis. Cancer Cell. 2006;9:189–198. doi: 10.1016/j.ccr.2006.01.025. [DOI] [PubMed] [Google Scholar]
- 19.Yang N, Coukos G, Zhang L. MicroRNA epigenetic alterations in human cancer: one step forward in diagnosis and treatment. Int. J. Cancer. 2008;122:963–968. doi: 10.1002/ijc.23325. [DOI] [PubMed] [Google Scholar]
- 20.Zhang L, Volinia S, Bonome T, Calin GA, Greshock J, Yang N, Liu CG, Giannakakis A, Alexiou P, Hasegawa K, et al. Genomic and epigenetic alterations deregulate microRNA expression in human epithelial ovarian cancer. Proc. Natl Acad. Sci. USA. 2008;105:7004–7009. doi: 10.1073/pnas.0801615105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Calin GA, Croce CM. MicroRNAs and chromosomal abnormalities in cancer cells. Oncogene. 2006;25:6202–6210. doi: 10.1038/sj.onc.1209910. [DOI] [PubMed] [Google Scholar]
- 22.Bottoni A, Piccin D, Tagliati F, Luchin A, Zatelli MC, degli Uberti EC. miR-15a and miR-16-1 downregulation in pituitary adenomas. J. Cell Physiol. 2005;204:280–285. doi: 10.1002/jcp.20282. [DOI] [PubMed] [Google Scholar]
- 23.Chan JA, Krichevsky AM, Kosik KS. MicroRNA-21 is an antiapoptotic factor in human glioblastoma cells. Cancer Res. 2005;65:6029–6033. doi: 10.1158/0008-5472.CAN-05-0137. [DOI] [PubMed] [Google Scholar]
- 24.Krichevsky AM, Gabriely G. miR-21: a small multi-faceted RNA. J. Cell Mol. Med. 2009;13:39–53. doi: 10.1111/j.1582-4934.2008.00556.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lu Z, Liu M, Stribinskis V, Klinge CM, Ramos KS, Colburn NH, Li Y. MicroRNA-21 promotes cell transformation by targeting the programmed cell death 4 gene. Oncogene. 2008;27:4373–4379. doi: 10.1038/onc.2008.72. [DOI] [PubMed] [Google Scholar]
- 26.Meng F, Henson R, Wehbe-Janek H, Ghoshal K, Jacob ST, Patel T. MicroRNA-21 regulates expression of the PTEN tumor suppressor gene in human hepatocellular cancer. Gastroenterol. 2007;133:647–658. doi: 10.1053/j.gastro.2007.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chang SS, Jiang WW, Smith I, Poeta LM, Begum S, Glazer C, Shan S, Westra W, Sidransky D, Califano JA. MicroRNA alterations in head and neck squamous cell carcinoma. Int. J. Cancer. 2008;123:2791–2797. doi: 10.1002/ijc.23831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Slaby O, Svoboda M, Fabian P, Smerdova T, Knoflickova D, Bednarikova M, Nenutil R, Vyzula R. Altered expression of miR-21, miR-31, miR-143 and miR-145 is related to clinicopathologic features of colorectal cancer. Oncology. 2007;72:397–402. doi: 10.1159/000113489. [DOI] [PubMed] [Google Scholar]
- 29.Ortholan C, Puissegur MP, Ilie M, Barbry P, Mari B, Hofman P. MicroRNAs and lung cancer: new oncogenes and tumor suppressors, new prognostic factors and potential therapeutic targets. Curr. Med. Chem. 2009;16:1047–1061. doi: 10.2174/092986709787581833. [DOI] [PubMed] [Google Scholar]
- 30.Jerome T, Laurie P, Louis B, Pierre C. Enjoy the silence: the story of let-7 microRNA and cancer. Curr. Genomics. 2007;8:229–233. doi: 10.2174/138920207781386933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Johnson SM, Grosshans H, Shingara J, Byrom M, Jarvis R, Cheng A, Labourier E, Reinert KL, Brown D, Slack FJ. RAS is regulated by the let-7 microRNA family. Cell. 2005;120:635–647. doi: 10.1016/j.cell.2005.01.014. [DOI] [PubMed] [Google Scholar]
- 32.Gottardo F, Liu CG, Ferracin M, Calin GA, Fassan M, Bassi P, Sevignani C, Byrne D, Negrini M, Pagano F, et al. Micro-RNA profiling in kidney and bladder cancers. Urol. Oncol. 2007;25:387–392. doi: 10.1016/j.urolonc.2007.01.019. [DOI] [PubMed] [Google Scholar]
- 33.Boggs RM, Wright ZM, Stickney MJ, Porter WW, Murphy KE. MicroRNA expression in canine mammary cancer. Mamm. Genome. 2008;19:561–569. doi: 10.1007/s00335-008-9128-7. [DOI] [PubMed] [Google Scholar]
- 34.Fabbri M, Garzon R, Cimmino A, Liu Z, Zanesi N, Callegari E, Liu S, Alder H, Costinean S, Fernandez-Cymering C, et al. MicroRNA-29 family reverts aberrant methylation in lung cancer by targeting DNA methyltransferases 3A and 3B. Proc. Natl Acad. Sci. USA. 2007;104:15805–15810. doi: 10.1073/pnas.0707628104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lujambio A, Esteller M. How epigenetics can explain human metastasis: a new role for microRNAs. Cell Cycle. 2009;8:377–382. doi: 10.4161/cc.8.3.7526. [DOI] [PubMed] [Google Scholar]
- 36.He L, He X, Lowe SW, Hannon GJ. microRNAs join the p53 network–another piece in the tumour-suppression puzzle. Nat. Rev. Cancer. 2007;7:819–822. doi: 10.1038/nrc2232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mraz M, Pospisilova S, Malinova K, Slapak I, Mayer J. MicroRNAs in chronic lymphocytic leukemia pathogenesis and disease subtypes. Leuk. Lymphoma. 2009;50:506–509. doi: 10.1080/10428190902763517. [DOI] [PubMed] [Google Scholar]
- 38.Bommer GT, Gerin I, Feng Y, Kaczorowski AJ, Kuick R, Love RE, Zhai Y, Giordano TJ, Qin ZS, Moore BB, et al. p53-mediated activation of miRNA34 candidate tumor-suppressor genes. Curr. Biol. 2007;17:1298–1307. doi: 10.1016/j.cub.2007.06.068. [DOI] [PubMed] [Google Scholar]
- 39.Chang TC, Wentzel EA, Kent OA, Ramachandran K, Mullendore M, Lee KH, Feldmann G, Yamakuchi M, Ferlito M, Lowenstein CJ, et al. Transactivation of miR-34a by p53 broadly influences gene expression and promotes apoptosis. Mol. Cell. 2007;26:745–752. doi: 10.1016/j.molcel.2007.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cole KA, Attiyeh EF, Mosse YP, Laquaglia MJ, Diskin SJ, Brodeur GM, Maris JM. A functional screen identifies miR-34a as a candidate neuroblastoma tumor suppressor gene. Mol. Cancer Res. 2008;6:735–742. doi: 10.1158/1541-7786.MCR-07-2102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hermeking H. The miR-34 family in cancer and apoptosis. Cell Death Differ. 2009;17:193–199. doi: 10.1038/cdd.2009.56. [DOI] [PubMed] [Google Scholar]
- 42.Raver-Shapira N, Marciano E, Meiri E, Spector Y, Rosenfeld N, Moskovits N, Bentwich Z, Oren M. Transcriptional activation of miR-34a contributes to p53-mediated apoptosis. Mol. Cell. 2007;26:731–743. doi: 10.1016/j.molcel.2007.05.017. [DOI] [PubMed] [Google Scholar]
- 43.Tazawa H, Tsuchiya N, Izumiya M, Nakagama H. Tumor-suppressive miR-34a induces senescence-like growth arrest through modulation of the E2F pathway in human colon cancer cells. Proc. Natl Acad. Sci. USA. 2007;104:15472–15477. doi: 10.1073/pnas.0707351104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yamakuchi M, Ferlito M, Lowenstein CJ. miR-34a repression of SIRT1 regulates apoptosis. Proc. Natl Acad. Sci. USA. 2008;105:13421–13426. doi: 10.1073/pnas.0801613105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Corney DC, Flesken-Nikitin A, Godwin AK, Wang W, Nikitin AY. MicroRNA-34b and MicroRNA-34c are targets of p53 and cooperate in control of cell proliferation and adhesion-independent growth. Cancer Res. 2007;67:8433–8438. doi: 10.1158/0008-5472.CAN-07-1585. [DOI] [PubMed] [Google Scholar]
- 46.Suzuki HI, Yamagata K, Sugimoto K, Iwamoto T, Kato S, Miyazono K. Modulation of microRNA processing by p53. Nature. 2009;460:529–533. doi: 10.1038/nature08199. [DOI] [PubMed] [Google Scholar]
- 47.Xi Y, Shalgi R, Fodstad O, Pilpel Y, Ju J. Differentially regulated micro-RNAs and actively translated messenger RNA transcripts by tumor suppressor p53 in colon cancer. Clin. Cancer Res. 2006;12:2014–2024. doi: 10.1158/1078-0432.CCR-05-1853. [DOI] [PubMed] [Google Scholar]
- 48.Aubert G, Lansdorp PM. Telomeres and aging. Physiol. Rev. 2008;88:557–579. doi: 10.1152/physrev.00026.2007. [DOI] [PubMed] [Google Scholar]
- 49.Cesare AJ, Reddel RR. Telomere uncapping and alternative lengthening of telomeres. Mech. Ageing Dev. 2008;129:99–108. doi: 10.1016/j.mad.2007.11.006. [DOI] [PubMed] [Google Scholar]
- 50.Reddel RR. The role of senescence and immortalization in carcinogenesis. Carcinogenesis. 2000;21:477–484. doi: 10.1093/carcin/21.3.477. [DOI] [PubMed] [Google Scholar]
- 51.Wurdinger T, Tannous BA, Saydam O, Skog J, Grau S, Soutschek J, Weissleder R, Breakefield XO, Krichevsky AM. miR-296 regulates growth factor receptor overexpression in angiogenic endothelial cells. Cancer Cell. 2008;14:382–393. doi: 10.1016/j.ccr.2008.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Cohen SB, Reddel RR. A sensitive direct human telomerase activity assay. Nat. Methods. 2008;5:355–360. doi: 10.1038/nmeth.f.209. [DOI] [PubMed] [Google Scholar]
- 53.Kim NW, Wu F. Advances in quantification and characterization of telomerase activity by the telomeric repeat amplification protocol (TRAP) Nucleic Acids Res. 1997;25:2595–2597. doi: 10.1093/nar/25.13.2595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wege H, Chui MS, Le HT, Tran JM, Zern MA. SYBR Green real-time telomeric repeat amplification protocol for the rapid quantification of telomerase activity. Nucleic Acids Res. 2003;31:E3–E3. doi: 10.1093/nar/gng003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Saito S, Ghosh M, Morita K, Hirano T, Miwa M, Todoroki T. The genetic differences between gallbladder and bile duct cancer cell lines. Oncol. Rep. 2006;16:949–956. [PubMed] [Google Scholar]
- 56.Saito S, Morita K, Hirano T. High frequency of common DNA copy number abnormalities detected by BAC array CGH in 24 cancer cell lines. Human Cell. 2009;22:1–10. doi: 10.1111/j.1749-0774.2008.00061.x. [DOI] [PubMed] [Google Scholar]
- 57.Okabe M, Ikawa M, Kominami K, Nakanishi T, Nishimune Y. Green mice' as a source of ubiquitous green cells. FEBS Lett. 1997;407:313–319. doi: 10.1016/s0014-5793(97)00313-x. [DOI] [PubMed] [Google Scholar]
- 58.Yano M, Okano HJ, Okano H. Involvement of Hu and heterogeneous nuclear ribonucleoprotein K in neuronal differentiation through p21 mRNA post-transcriptional regulation. J. Biol. Chem. 2005;280:12690–12699. doi: 10.1074/jbc.M411119200. [DOI] [PubMed] [Google Scholar]
- 59.Lee H, Choi JK, Li M, Kaye K, Kieff E, Jung JU. Role of cellular tumor necrosis factor receptor-associated factors in NF-kappaB activation and lymphocyte transformation by herpesvirus Saimiri STP. J. Virol. 1999;73:3913–3919. doi: 10.1128/jvi.73.5.3913-3919.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Shats I, Milyavsky M, Tang X, Stambolsky P, Erez N, Brosh R, Kogan I, Braunstein I, Tzukerman M, Ginsberg D, et al. p53-dependent down-regulation of telomerase is mediated by p21waf1. J. Biol. Chem. 2004;279:50976–50985. doi: 10.1074/jbc.M402502200. [DOI] [PubMed] [Google Scholar]
- 61.Gorospe M, Wang X, Holbrook NJ. p53-dependent elevation of p21Waf1 expression by UV light is mediated through mRNA stabilization and involves a vanadate-sensitive regulatory system. Mol. Cell Biol. 1998;18:1400–1407. doi: 10.1128/mcb.18.3.1400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Joseph B, Orlian M, Furneaux H. p21(waf1) mRNA contains a conserved element in its 3′-untranslated region that is bound by the Elav-like mRNA-stabilizing proteins. J. Biol. Chem. 1998;273:20511–20516. doi: 10.1074/jbc.273.32.20511. [DOI] [PubMed] [Google Scholar]
- 63.Mao J, Li C, Zhang Y, Li Y, Zhao Y. Human with-no-lysine kinase-4 3'-UTR acting as the enhancer and being targeted by miR-296. Int. J. Biochem. Cell Biol. 2010;41:872–878. doi: 10.1016/j.biocel.2010.06.006. [DOI] [PubMed] [Google Scholar]
- 64.Nishimura T. Total number of genome alterations in sporadic gastrointestinal cancer inferred from pooled analyses in the literature. Tumour Biol. 2008;29:343–350. doi: 10.1159/000176044. [DOI] [PubMed] [Google Scholar]
- 65.Staaf J, Jonsson G, Ringner M, Vallon-Christersson J, Grabau D, Arason A, Gunnarsson H, Agnarsson BA, Malmstrom PO, Johannsson OT, et al. High-resolution genomic and expression analyses of copy number alterations in HER2-amplified breast cancer. Breast Cancer Res. 2010;12:R25. doi: 10.1186/bcr2568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Uchida M, Tsukamoto Y, Uchida T, Ishikawa Y, Nagai T, Hijiya N, Nguyen LT, Nakada C, Kuroda A, Okimoto T, et al. Genomic profiling of gastric carcinoma in situ and adenomas by array-based comparative genomic hybridization. J. Pathol. 2010;221:96–105. doi: 10.1002/path.2686. [DOI] [PubMed] [Google Scholar]
- 67.Bonauer A, Boon RA, Dimmeler S. Vascular microRNAs. Curr. Drug Targets. 2010;11:943–949. doi: 10.2174/138945010791591313. [DOI] [PubMed] [Google Scholar]
- 68.Wang S, Olson EN. AngiomiRs – key regulators of angiogenesis. Curr. Opin. Genet. Dev. 2009;19:205–211. doi: 10.1016/j.gde.2009.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Hong L, Han Y, Zhang H, Li M, Gong T, Sun L, Wu K, Zhao Q, Fan D. The prognostic and chemotherapeutic value of miR-296 in esophageal squamous cell carcinoma. Ann. Surg. 2010;251:1056–1063. doi: 10.1097/SLA.0b013e3181dd4ea9. [DOI] [PubMed] [Google Scholar]