Abstract
Tra2β regulates a number of splicing switches including activation of the human testis-specific exon HIPK3-T in the Homeodomain Interacting Protein Kinase 3 gene. By testing HIPK3-T exons of different intrinsic strengths, we found Tra2β most efficiently activated splicing inclusion of intrinsically weak exons, although these were spliced at a lower overall level. Both the RRM and N-terminal RS-rich region of Tra2β were required for splicing activation. Bioinformatic searches for splicing enhancers and repressors mapped four physically distinct exonic splicing enhancers (ESEs) within HIPK3-T, each containing the known Tra2β AGAA-rich binding site. Surprisingly disruption of each single ESE prevented Tra2β-mediated activation, although single mutated exons could still bind Tra2β protein by gel shifts and functional splicing analyses. Titration experiments indicate an additive model of HIPK3-T splicing activation, requiring availability of an array of four distinct ESEs to enable splicing activation. To enable this efficient Tra2β-mediated splicing switch to operate, a closely adjacent downstream and potentially competitive stronger 5′-splice site is actively repressed. Our data indicate that a novel arrangement of multiple mono-specific AGAA-rich ESEs coupled to a weak 5′-splice site functions as a responsive gauge. This gauge monitors changes in the specific nuclear concentration of the RNA binding protein Tra2β, and co-ordinately regulates HIPK3-T exon splicing inclusion.
INTRODUCTION
In excess of 90% of human genes encode more than one mRNA isoform, with alternative splicing making an important contribution (1–3). Alternative splicing patterns of pre-mRNAs can be modulated in response to the concentration of specific nuclear RNA binding proteins within particular cells and tissues (4,5). The biological effect of splicing regulators is mediated through interpretation of an intrinsic splicing code embedded within pre-mRNA sequences. Individual splicing factors can both activate and repress exon inclusion, depending on where they bind in target pre-mRNAs (6,7). Although alternative splicing is found in every tissue, particularly high levels have been observed in the human brain and testes (8–11). The target sites for RNA binding proteins which make up the splicing code are being deciphered by experiments and bioinformatics (12), and must explain why some exons are spliced with wide anatomic distributions, while others have much more restricted tissue-specific patterns of splicing inclusion.
The SR family are an important class of splicing regulator proteins which contain domains enriched in arginine and serine residues (RS domains). Classical SR protein splicing regulators typically have one or two RNA recognition motifs (RRMs) required for binding to their target pre-mRNAs, and a C-terminal RS domain (13,14). The SR-like splicing regulator protein Tra2 is conserved between fruit flies and humans. Tra2 has an unusual modular structure compared with ‘classical’ SR proteins, with a central RRM flanked on the N-terminal side by a protein sequence enriched in arginine–serine (RS) dipeptides (19 RS dipeptides in humans), and on the C-terminal side by a hinge region and a smaller RS-rich region (seven RS dipeptides in humans). Tra2β protein binds most strongly to an AGAA target RNA sequence (15,16). A number of target exons respond to increasing nuclear concentrations of Tra2 (17–19). Amongst its known molecular targets, Drosophila Tra2 protein regulates a splicing switch in sex determination (20,21) and human Tra2β protein regulates a splicing switch in the tissue-specific exon HIPK3-T (19). The HIPK3-T exon is within the Homeodomain Interacting Protein Kinase 3 gene, which is implicated in apoptotic control. HIPK3-T is selected only in a single tissue, the testis, which has elevated Tra2β concentrations.
The mechanisms through which ubiquitously expressed factors like Tra2β can generate splicing switches in particular tissues are not well understood. To address this question, here we functionally dissected the sequences needed for splicing activation of HIPK3-T. We find that an array of exonic splicing enhancers responsive to Tra2β, coupled to a weak 5′-splice site act together as a gauge to enable splicing activation in response to increased nuclear concentrations of Tra2β. We show that this switch is delicately balanced by the interplay between Tra2β binding and 5′-splice site strength, allowing activation of alternative splicing within a narrow window, outside which the exon is either invisible to the spliceosome or activated constitutively.
MATERIALS AND METHODS
Computational analysis of putative ESE/ESS content of HIPK3-T and flanking introns
We used CisPlotter (unpublished), to compare and design HIPK3-T derivative sequences. Briefly, this program computes and plots the ESE or ESS score as shown in Figure 1A. We used the list of putative ESE and ESS octamers, chose the threshold z-score levels and dealt with overlapping motifs as described previously (22). This script accepts alternative nucleotides at specified positions and plots the resulting change in ESE or ESS scores in a different colour to facilitate mutagenesis design (Figure 3A).
Figure 1.
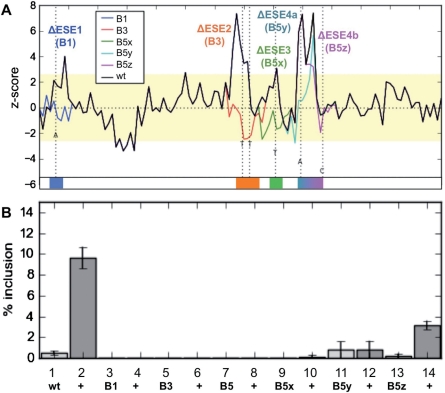
Modular organization of the HIPK3-T exon and activation by Tra2β through direct association with RNA. (A) z-score plot predicting the splicing control sequences in and around the HIPK3-T exon. Regions of the exon above the threshold for exonic splicing enhancers are labelled ESE1-4, and the region of the exon below the threshold for splicing silencers is labelled ESS. The architecture of the HIPK3-T exon and surrounding intron, and sequence of the HIPK3-T exon and flanking intron sequences are shown underneath. The strong 5′-splice site downstream of HIPK3-T is underlined, and its position in the sequence marked by an asterisk. (B) Schematic representation of Tra2β protein domains. (C) Analysis of the splicing response of wild-type HIPK3-T to full length and domain deleted Tra2β-GFP fusion proteins, assayed by RT–PCR and capillary gel electrophoresis, and presented as the percentage inclusion of HIPK3-T. All transfections and subsequent RNA analyses were performed at least in triplicate. (D) Example of RT–PCR from cells transfected with HIPK3-T and different expression constructs encoding Tra2β proteins, and analysed by capillary gel electrophoresis. +T = size of RT–PCR product including HIPK3-T exon. –T = size of RT–PCR product excluding HIPK3-T exon. (E) Expression levels of each of the different wild-type and domain deleted Tra2β-GFP fusion proteins, assayed by western blotting from transfected HEK293 cells in parallel with splicing assays. Levels of GFP fusion protein were measured using an α-GFP antibody, and levels of actin were measured on the same filter using an α-actin antibody to provide a loading control for each lane.
Figure 3.
Each of four Exonic Splicing Enhancer Regions (ESEs) are essential for Tra2β-mediated splicing activation. (A) The z-score plots of the wild-type HIPK3-T exon (black), superimposed with z-score plots for each of the point mutants which affected individual ESEs (shown as coloured lines, with the changed nucleotide indicated as a broken line). See also Table 1 for a complete description of all mutants used in this study. (B) The percentage exon inclusion obtained after transfection of each of these point mutants after transfection with GFP or Tra2β (+lane).
Constructs
Minigene splicing constructs were based on the wild-type HIPK3-T exon, and 269 and 267-nt upstream and downstream flanking intron sequence respectively, cloned into pXJ-41 as described previously (19). Site Directed Mutagenesis of HIPK3-T was carried out as previously described (22), mutants were verified by sequencing, and then subcloned back into the parent plasmid.
Deletion versions of human Tra2β protein were cloned in frame into pGFP3 using standard cloning techniques (19,23). Tra2βΔRS1 is missing amino acids 13–97, Tra2βΔRRM is missing amino acids 119–168, Tra2βΔRNP1 is missing amino acids 150–168 (Figure 2). For expression of full length Tra2β, its coding sequence was inserted in a pTT3 plasmid (24), in which a Flag tag was previously inserted in the Hind III site.
Figure 2.
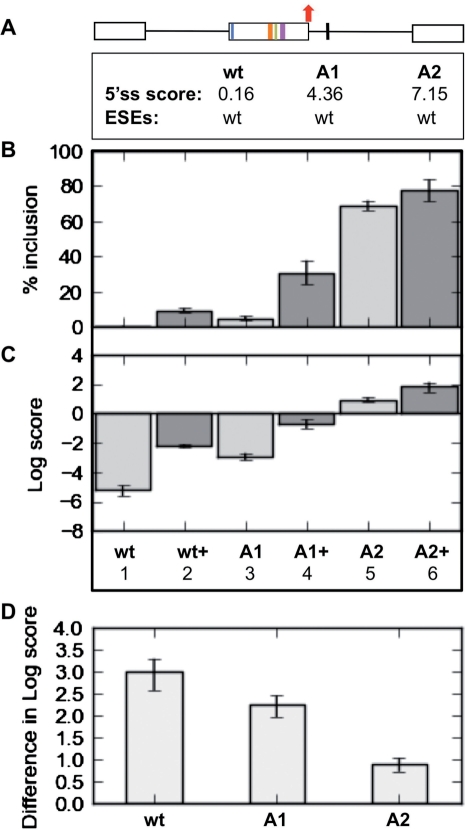
(A) The splicing response of HIPK3-T to Tra2β correlates with intrinsic exon strength defined by 5′-splice site. Correlation between 5′-splice site strength and splicing response of HIPK3-T in terms of percentage splicing inclusion in response to Tra2β (B), log-score plot for response to Tra2β (C), or difference in log scores (D). A1 and A2 variants have intermediate strength or strong natural 5′-splice site.
Minigene splicing analysis of HIPK3-T exon variants
Tissue culture, transfections of HEK293 cells, RNA purification and RT–PCR was carried out using primers and conditions as previously described (19) and the RT–PCR products were analysed by capillary gel electrophoresis. The HIPK3-T reporter construct (100 ng) was transfected with or without 300 ng of Tra2β expressing plasmid using GeneJammer (Agilent) under standard conditions for 24 h. To enable analysis of splicing patterns as well as monitoring the expression of transfected splicing factors, transfected cell cultures were split into two portions and analysed by western blotting as previously described (23).
Splicing was analysed as both a percentage splicing inclusion and a logarithmic splicing inclusion. We define log-score as the natural logarithm of the ratio of HIPK3 exon T inclusion to exclusion (logit transformation). Therefore there is a one-to-one mapping between percentage inclusion values and log-score values. A log-score of zero denotes 50% inclusion of the HIPK3 exon T. A positive log-score denotes >50% inclusion and a negative log-score denotes <50% exon inclusion. Note that the log-score values are unbounded and can theoretically range between +/– infinity. In reality they will be limited by the resolution capability of band detection, which here we have taken to be 0.1% which corresponds to a limit on the log-score value of −6.9.
To compare intrinsic splicing capability of two constructs, the difference of the two log-scores is taken. This procedure has several benefits over comparing fold change of the percentage inclusion values. Since the percentage inclusion values are bounded at 0 and 100%, fold change comparisons close to these boundaries are misleading. In contrast the log-score method has been designed to treat these two ends symmetrically. For example, percentage splicing inclusion changes from 2 to 5%; 95 to 98% or 40 to 64% each have a log-score difference of ~1, whereas the percentage inclusion fold change values would be 2.5, 1.03 and 1.6 respectively.
RNA affinity assay and EMSA
HIPK3-T exon variants or the downstream intron were cloned from the minigene splicing analysis constructs in pXJ41 vector into pBluescript, and then in vitro transcribed RNAs prepared as described (23). RNA affinity assays used in vitro transcribed RNAs attached to sepharose beads to pull down proteins from nuclear extracts as previously described (19). We tested for binding of candidate RNA binding proteins based on previous results (19) and on the availability of antisera. The sequences of RNAs used for electromobility shift assays (EMSAs) included the HIPK3-T exon and 11 nt of the upstream intron for wild-type or HIPK3-T exon derivatives B4, B5 and B6. Sequences are detailed in Table 1. Full length Tra2β was expressed at high level by transient transfection of suspension 293-EBNA cells (24) using FreeStyle 293-F cells, and FreeStyle expression medium according to the recommendations of Invitrogen. After 60 h transfection, 293-F cells were pelleted and washed in PBS. Extraction and purification of Tra2β was carried out according the protocol previously described (25), except that the Flag-Tra2β protein was immunopurified on an anti-Flag-M2 affinity gel and eluted by competition with FLAG peptide (SIGMA-ALDRICH). EMSAs were done as previously described (25).
Table 1.
HIPK3-T mutations used in this study
 |
The mutant name, systematic name, sequence effect of mutant are shown. Underneath the sequence of the HIPK3-T exon (uppercase) and downstream intron (lower case, downstream 5′-splice site is italicized). The ESEs are shown in coloured text consistent with Figure 1. Systematic name: e. denotes the exonic position of the mutation, e+ denotes downstream intron.
Antibodies
Proteins were detected by western blotting using antibodies specific for GFP (Clontech 632 381 Living Colours A v monoclonal antibody), tubulin (Sigma T5168 anti-α-tubulin monoclonal antibody), and Tra2β (Abcam polyclonal antibody ab31353), and others used previously (19,26).
RESULTS
Splicing activation of HIPK3-T is dependent on both the RRM and RS domains of Tra2β
The HIPK3-T exon has a weak 5′-splice site, although there is a potentially stronger downstream 5′-splice site that is not selected by the spliceosome (position indicated as an asterisk in Figure 1A). We used a pull down assay to monitor association of nuclear proteins with the HIPK3-T exon and with the downstream intronic region containing this strong 5′-splice site. Consistent with the observed physiological selection of the HIPK3-T exon, the exonic region strongly bound some SR proteins and the SR-like protein Tra2β (Figure 4C). In contrast, the downstream region did not associate with SR proteins, but did bind to hnRNPs, which are frequent splicing repressors (Figure 4C).
Figure 4.
ESE-depleted versions of HIPK3-T are still able to bind Tra2β. (A) Gel shift experiments (Electrophoretic Mobility Shift Assays) were carried out with either wild-type HIPK3-T RNA, or HIPK3-T variants with different ESE profiles. Radiolabelled RNAs were shifted with 0, 50 and 100 ng added Tra2β protein. The sequences of these RNAs are given in Table 1. (B) Purified full length Tra2β Flag tagged protein. (C) RNA binding proteins associated with the HIPK3-T exon and downstream intronic region are associated with a number of RNA splicing regulators. wt = wild-type HIPK3-T exon, DS = downstream RNA.
Tra2β is the only known splicing activator of HIPK3-T (19) and in preliminary experiments we found no other interacting RNA binding proteins significantly affected its splicing inclusion. To test if direct RNA binding was required for Tra2β-mediated splicing activation of HIPK3-T, we first inactivated the RRM with an independent deletion of RNP1 and deletion of the entire RRM (Figure 1B). We then tested the activity of the resulting mutants on splicing of the wild-type HIPK3-T exon by co-transfection experiments in HEK293 cells. In contrast to the wild-type Tra2β-GFP protein, mutants affecting the RRM (ΔRNP1 and ΔRRM) were unable to stimulate HIPK3-T exon inclusion (Figure 1C, D lanes 4 and 5 compared to lane 2) indicating direct binding of Tra2β-GFP to HIPK3-T target RNA is required for splicing activation. In several other SR protein splicing regulators, multiple domains can play partially redundant functions (27–30). We also deleted the N-terminal RS-rich region to test if the N- and C-terminal RS domains of Tra2β are functionally redundant, and found that Tra2β proteins lacking the N-terminal RS domain were unable to support splicing activation of HIPK3-T (Figure 1C and D lane 3). We observed equal levels of GFP-fusion proteins by western blotting (Figure 1E) for each of the full length and deletion derivatives of Tra2β, and these localized in the nucleus [data not shown, and (31)].
The amplitude of Tra2β-mediated splicing activation of HIPK3-T is dependent on intrinsic exon strength
HIPK3-T has a somewhat weak 5′-splice site (GAG|gttaaa), which has a score of 0.16 measured using a maximum entropy model (http://genes.mit.edu/burgelab, where a score of ~11 represents a perfect splice site). We tested whether this weak 5′-splice site normally contributes to exon skipping by mutating HIPK3-T to contain an intermediate strength (GAG|gttagg, score: 4.36 for construct A1) or strong 5′-splice site (GAG|gttagt, score: 7.15 for construct A2). Full details of all HIPK3-T mutants used in this study are given in Table 1. Both improved splice sites resulted in the HIPK3-T exon being more efficiently spliced even without the addition of exogenous Tra2β (compare Columns 1 with 3 and 5, Figure 2B). Therefore, the level of HIPK3-T exon inclusion shows a direct correlation to splice site strength, indicating that the weakness of the 5′-splice site is a major negative element restricting HIPK3-T exon splicing.
We next analysed the magnitude of the splicing response of the above 5′-splice site variants of HIPK3-T to Tra2β expression (Figure 2B, Columns 2, 4 and 6). To compare differences between splicing responses of different pre-mRNAs over the full dynamic range we monitored HIPK3-T splicing activation both as a percentage of HIPK3-T inclusion (showing overall levels of splicing inclusion, Figure 2B), and using a natural logarithm-score plot (Log-score, Figure 2C, see ‘Materials and Methods’ section). As expected, splicing of each of the HIPK3-T exons was activated by Tra2β, but with improved overall percentage splicing inclusion for the exons with strong (A2) or intermediate (A1) 5′-splice sites (Figure 3C). Importantly, plotting the difference in log-scores before and after co-transfection of Tra2β indicated a clear trend in splicing activation between pre-mRNAs: the splicing response of each of the pre-mRNAs to Tra2β was inversely correlated to 5′-splice site strength (Figure 2D). Hence weaker HIPK3-T exons respond more specifically to Tra2β-mediated splicing activation.
Analysis of the architecture of the HIPK3-T exon
To identify the splicing control elements that might mediate Tra2β-mediated HIPK3-T splicing activation we next analysed the entire exon sequence and flanking intron sequences for octamers predictive of splicing enhancers and silencers based on the bioinformatic approaches of Zhang and Chasin (32–34). Detailed analysis of the resulting octamer z-score plots indicated four distinct peaks in the HIPK3-T exon above the threshold value corresponding with putative Exonic Splicing Enhancers (ESEs), and a single Exonic Splicing Silencing Region (ESS) (Figure 1A). We name individual predicted ESEs here ESE 1–4 (Figure 1A). Each of the ESEs was GAA/AGA-rich (designated hereafter AGAA-rich), consistent with the known binding site for Tra2β (15,16,35)
Splicing activation of HIPK3-T by Tra2β requires the full complement of four ESEs
We next sought to precisely map sequences in the HIPK3-T exon that were responsive to Tra2β. Using the results of the HIPK3-T exon sequence analysis shown in Figure 1A, we systematically mutagenized each putative Tra2β responsive ESE with multiple single mutations (Figure 3 and Table 1). In each case site directed mutagenesis was designed using an in-house software based on the octamer z-scores, along with other matrices including HSF and ESE finder (36–38) to ensure that no predicted splicing silencer or enhancer elements were introduced into the HIPK3-T sequence (see ‘Materials and Methods’ section).
To our surprise, single mutation of any one of the four ESE regions was sufficient to completely prevent HIPK3-T splicing activation (Figure 3B). No HIPK3-T splicing inclusion was observed for pre-mRNAs made from constructs B1, B2 (disruption of ESE1), B3, B4 (disruption of ESE2), or B5 (disruption of ESE 3 and 4) (Figure 3B, Columns 1–8, Table 1 and data not shown). Further dissection of B5 into the individual point mutations B5x (disruption of ESE3), B5y (disruption of the first sub-peak of ESE4) and B5z (disruption of the second sub-peak of ESE4) indicated that only single point mutations in ESE3 (B5x) or in the first enhancer sub-peak of ESE4 (B5y) completely disabled Tra2β-mediated splicing activation (compare Columns 1 and 2 with Columns 9–12, Figure 3B). Disruption of the second sub-peak alone in ESE4 (B5z) only resulted in a partial loss of Tra2β-mediated splicing activation, most probably because the mutation preserves a AGAAG sequence in the first half of ESE4 (Columns 13, 14, Figure 3B).
HIPK3-T exons lacking single ESEs are still able to bind Tra2β protein
The above results indicate that ESEs 1–4 as defined by our approach are indeed true individual ESEs, and also each are absolutely required for Tra2β-mediated splicing activation of HIPK3-T. Each of the identified ESEs within HIPK3-T contains at least one AGAA motif, consistent with the known specific binding site of Tra2β protein (15,16). However, after each single ESE disruption there were three AGAA-containing ESEs remaining, so we predicted that such mutated HIPK3-T exons should still be able to bind to Tra2β protein even though splicing activation was lost.
To test this prediction we carried out gel shift experiments. In preliminary experiments we identified strong binding of bacterially expressed protein containing the RRM of Tra2β to the HIPK3-T exon and no binding to the downstream intronic RNA (data not shown). This indicated that the RRM was important for interaction with HIPK3-T in vitro. Since Tra2β protein can multimerize via its RS domains (31), full length Tra2β protein was then expressed and purified from mammalian cells (Figure 4B). Purified protein was then used to test in vitro association with radiolabelled RNAs comprising either wild-type HIPK3-T, HIPK3-T exon with a single ESE disruption (B4), HIPK3-T exon with a double ESE disruption (B5), and HIPK3-T exon with a triple ESE disruption (B6).
Strong and almost complete Tra2β binding was observed to wild-type HIPK3-T exon even at the lowest concentration of added Tra2β (Figure 4A, lanes 1–5 and 16–20). Additional band shifts were obtained with increasing concentrations of Tra2β, most likely due to binding of several Tra2β molecules to the T exon (Figure 4, lanes 1–5). Three major sizes of shift were observed for both wild-type and mutant B4 HIPK3-T exons. Very slightly less strong shifts at low Tra2β concentrations were observed after single ESE disruption (lanes 6–10), indicating that HIPK3-T exons lacking single ESEs are still able to bind Tra2β efficiently. Consistent with expectations from the AGAA-rich ESE elements, less RNA was shifted for probes containing double (B5, Figure 4A, lanes 21–25) and particularly triple ESE mutations (B6, Figure 4A, lanes 11–15). Indeed, at the lower concentrations of Tra2β protein tested (10 and 20 ng) a significant part of the B5 and B6 RNA remained un-shifted, and the highest molecular weight band shift was not observed in the B6 mutant even with the highest concentration of Tra2β (lane 15 compared to 5).
We also compared the binding of wild-type HIPK3-T to mutant RNAs with double ESE disruptions (Figure 4C, wt and B5) by pull down from nuclear extracts. No difference in binding of Tra2β relative to ASF/SF2 was observed for any of these RNAs. Taken together these data indicate that each of the identified putative ESEs are able to interact with Tra2β protein, and mutation of one (B4) or two (B5) ESEs does not dramatically change the interaction properties of the exon. However, while B5 and wild-type RNAs form similar sized protein–RNA complexes, in vivo only the wild-type RNA sequence supports splicing activation by Tra2β.
Splicing of HIPK3-T exon variants with stronger splice sites need fewer ESEs for splicing activation, but activate with lower amplitude
Taken together the above experiments indicated that while in vivo splicing activation of the HIPK3-T exon is severely compromised after loss of only a single ESE sequence, Tra2β protein is still able to bind to HIPK3-T RNA in vitro efficiently after single ESE disruption. Hence although Tra2β binding might take place to a reduced complement of ESEs, four functional ESEs are the minimum required to enable splicing activation of the wild-type HIPK3-T exon by Tra2β.
We further tested whether a combination of three ESEs, although non-functional in the wild-type context of exon HIPK3-T, might retain some splicing enhancer activity by analysing splicing of engineered versions of HIPK3-T with stronger splice sites. ESE2 was mutated in the context of both the intermediate (to give C1) and strong 5′-splice site (to give C2) derivatives of HIPK3-T. We then tested each of these engineered HIPK3-T exon variants for splicing activation by Tra2β (Figure 5). Consistent with Tra2β still binding to ESE-disrupted HIPK3-T exons, ESE2 removal reduced rather than prevented Tra2β-mediated splicing in the context of an improved 5′-splice site (Figure 5B and C, Columns 3–6). As previously observed, pre-mRNAs made from B3 were not activated for splicing by Tra2β (Figure 5B, Columns 1–2).
Figure 5.
HIPK3-T exon variants with stronger splice sites have a reduced ESE content requirement for splicing activation, but are activated with a lower amplitude. (A) Organisation of pre-mRNAs used in this experiment. Splicing responses of HIPK3-T exon variants B3, C1 and C2, described fully in Table 1, are shown in terms of percentage splicing inclusion in response to Tra2β (B), Log-score plot for response to Tra2β (C), or difference in log scores (D).
Analysis of this data by plotting the difference in the log-score of splicing activation before and after co-transfection with Tra2β in Figure 5D again showed a clear correlation between the quantitative response to Tra2β and the overall intrinsic strength of the exon. The pre-mRNA with the intermediate 5′-splice site (C1) showed a strongly increased responsiveness to Tra2β after ESE2 mutation (compare the log score plot for C1 in Figure 5 to wt and A1 in Figure 2), although splicing inclusion levels were reduced comparatively to the wild-type HIPK3-T substrate. This was exactly the trend described in the previous section for the wild-type combination of four ESEs. HIPK3-T variants with the strong 5′-splice site that either contained an ESE2 mutation (C2), or not (A2) showed very similar behaviour showing these pre-mRNAs are less dependent on Tra2β for splicing inclusion (compare A2 and C2 in Figures 2 and 5 middle and bottom panels). Identical results were also obtained on combining another single ESE disruption mutant (B4) with the intermediate and strong 5′-splice site mutants (mutations C3 and C4, Table 1, and data not shown).
The response of HIPK3-T to Tra2β is most consistent with an additive mode of splicing activation
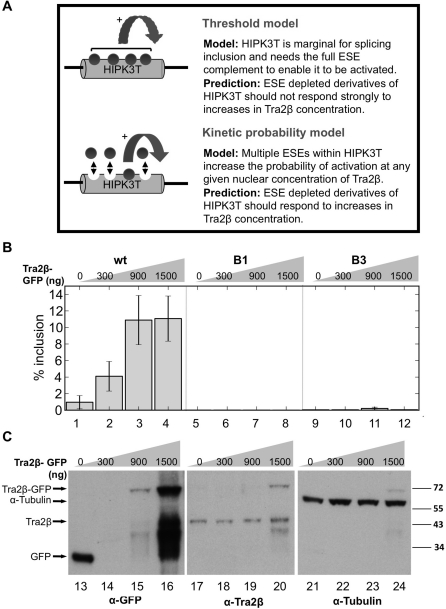
The results shown in Figures 2 and 5 indicate that only the combination of the four ESEs and the weak 5′-splice site present in the wild-type HIPK3-T exon are able to induce a specific and efficient on/off splicing switch in response to Tra2β expression. These results are consistent with two conceptually different models of HIPK3-T splicing regulation (Figure 6A). The first ‘splicing threshold’ model predicts that concomitant occupancy of each of a minimum number of four ESE sites would be essential for HIPK3-T splicing activation. According this model, decreasing the ESE number would totally incapacitate the exon with regards to splicing activation and increasing Tra2β concentration within the cell should not be able to rescue splicing.
Figure 6.
Splicing of ESE depleted versions of HIPK3-T is not rescued by higher concentrations of nuclear Tra2β protein. (A) Two possible models of HIPK3-T splicing inclusion in response to Tra2β. (B) Graphs of dose responses of wild-type HIPK3-T and mutant HIPK3-T B1 and B3 to increasing concentrations of Tra2β (transfections of 300, 900 and 1500 ng of Tra2β expressing plasmid). (C) Levels of GFP-containing proteins were detected by α-GFP (left panel); Tra2β protein detected using α-Tra2β (middle panel); and α-tubulin (right panel) measured in parallel in transfected cells.
The second ‘kinetic probability’ model is based on the probability of Tra2β occupancy of the HIPK3-T exon, through analogy with the model for Tra2 protein regulation of Drosophila Dsx splicing (17). In this model, multiple ESE sites would increase the probability of co-residency of Tra2β and U1 snRNP on the HIPK3-T exon, and ensure splicing activation on nascent RNA before the transcription of downstream competing exons (Model 2 in Figure 6A). In this case binding of Tra2β to only one ESE may be sufficient for activation. Thus increasing the nuclear concentration of Tra2β might compensate for the decreased ESE content by increasing the probability of ESE occupancy.
In order to test between the predictions of these two models, we carried out transfections with a gradient of different Tra2β concentrations. Splicing of the wild-type HIPK3-T exon was efficiently activated in response to increasing Tra2β-GFP. Although B1 or B3, with three of four ESEs remaining, showed a minimal response to Tra2β, only very low levels of exon inclusion are achieved (Figure 6), and thus even high levels of Tra2β cannot compensate for an incomplete set of ESEs even in very high concentrations of Tra2β. Western analysis of transfected cells is shown in Figure 6C, which shows the increase in Tra2β-GFP along the concentration gradient, and also the levels of endogenous Tra2β protein and alpha-tubulin in the same cells.
Splicing repression of a strong downstream 5′-splice site enables the HIPK3-T splicing switch to operate
The above data indicate that a combination of four ESEs coupled to a weak 5′-splice site enable an efficient on/off splicing switch in response to increasing concentrations of Tra2β. However, proper operation of the HIPK3-T splicing switch system would require either any nearby competing stronger 5′-splice sites not dependent on Tra2β activation to be actively repressed, or for the Tra2β protein to activate splicing at the upstream 5′-splice site.
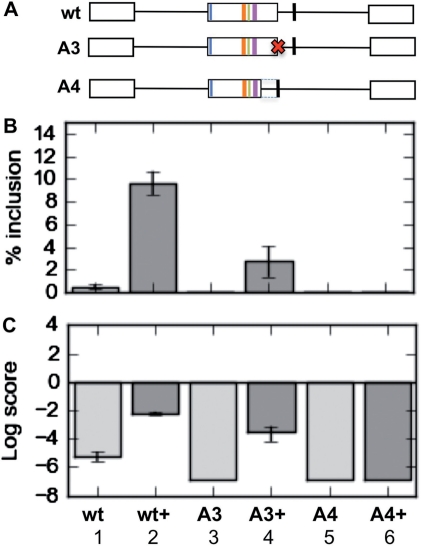
Sequence analysis indicated that immediately downstream of HIPK3-T is a strong 5′-splice site consensus sequence that is in fact not selected by the spliceosome in vivo (19). This sequence has a score of 6.25 compared to the natural 5′-splice site which only measures 0.16 using a maximum entropy model (http://genes.mit.edu/burgelab). To test whether this downstream splice site is actively repressed, or whether Tra2β preferentially activates the upstream 5′-splice site irrespective of intrinsic splice site strength, we first removed the weak upstream natural 5′-splice site (mutant A3). In this case we only observed very weak splicing at the downstream site (Figure 7, Columns 3 and 4). This indicates that the upstream natural 5′-splice site is indeed preferentially activated by the Tra2β sites in the exon, but the downstream splice site is functional at some level since it is activated by Tra2β when the upstream weaker site is absent.
Figure 7.
The stronger 5′-splice site downstream of the HIPK3-T exon is repressed. (A) Organization of the different pre-mRNAs used in this experiment. (B) Percentage of splicing inclusion of each of the engineered HIPK3-T exons at the downstream splice site in response to co-transfection of either GFP or Tra2β-GFP. (C) Splicing inclusion of each of the HIPK3-T exon derivativesmonitored by a log-score plot.
To test for active repression of the downstream site we then replaced the natural 5′-splice site with the downstream stronger 5′-splice site, by simply deleting 37 nt off the end of the exon in place of the weak natural 5′-splice site (mutant A4). In this case we observed no splicing inclusion even with transfected Tra2β (Figure 7, Columns 5 and 6). This confirms that this downstream 5′-splice site region is also actively repressed, and moving the repressive elements closer to the HIPK3-T exon may repress splicing inclusion. Consistent with this model, the downstream intronic region binds a high concentration of hnRNPs, but not SR proteins (Figure 4C).
DISCUSSION
Alternative splice site selection is regulated by ubiquitously expressed splicing factors that act on sequences located within or in the vicinity of exons. This gives rise to at least two variables for each exon, namely, the complement of splicing factors available in the target tissue, which is often controlled within very narrow concentration ranges; and secondly, the intrinsic strength of the exon reflected by its splice site strengths and complement of positive and negative regulatory elements. The recognition of an exon is based on a finely regulated balance between these two variables.
It has been a conceptual challenge in the field to explain how individual exons are regulated in a tissue or cell specific manner in response to ubiquitously expressed factors. We present here evidence for an interesting and novel splicing switch mechanism, that relies on a balance between the number of binding sites for a single splicing factor coupled to 5′-splice site strength. In the HIPK3-T system, a very weak exon which is not recognized in any tissue type tested other than testis contains an array of AGAA-rich ESEs and is activated by Tra2β. Although the HIPK3-T exon binds in nuclear extracts to a number of splicing regulator proteins in addition to Tra2β (19), none of these other RNA binding proteins, particularly ASF/SF2 which may also bind to a GAA-rich target sequence (23,39), could activate or repress this exon in in vivo minigene experiments (data not shown). In contrast to transfected cells, in vitro splicing substrates containing the HIPK3-T splicing enhancer do require addition of ASF/SF2 to enable splicing activation of a 5′-splice site by Tra2β (19). This in vitro requirement is consistent with the need for exogenous SR proteins to support splicing of other pre-mRNA substrates in S100 extracts, since Tra2β cannot provide this additional activity.
Interestingly, loss of even a single ESE sequence resulted in complete loss of exon activation which could not be rescued by elevating levels of Tra2β. An array of ESEs responsive to a single splicing factor where disruption of just a single ESE results in loss of exon recognition is novel and consistent with a model in which exon strength is finely balanced with ESE content to enable an efficient splicing switch to take place in the testis in response to specifically to Tra2β concentration. Such gauges may be used by exons to measure the nuclear concentration of specific splicing regulators that are ubiquitously expressed but at varying tissue-specific levels, and thus establish cell type-specific splicing responses. Importantly, to enable these gauges to work properly potentially stronger consensus downstream 5′-splice sites would need to be actively repressed, or else these stronger sites would be constitutively selected by the spliceosome. We have experimentally confirmed repression of a potentially competing 5′-splice site for HIPK3-T. Similar repression of downstream 5′-splice sites has also been observed for other regulated exons with weak 5′-splice sites (40). While HIPK3-T contains multiple similar ESEs all responding to Tra2β concentrations, splicing of other exons have been reported to be individually dependent on multiple different functional ESEs responsive to different RNA binding proteins (34). This suggests that a model of splicing control in which multiple ESEs respond to the nuclear concentration of specific RNA binding proteins might extend also to combinatorial control. The number of Tra2β binding sites required within an exon to establish a splicing response is likely to depend on splice site strength and other splicing signals. For example, the inclusion of another Tra2β-responsive exon such as SMN2 exon 7, which has a single Tra2β binding site, additionally depends on binding of hnRNPG and SRp30c (15).
Our analysis here of the human testis specific HIPK3-T exon makes interesting comparison to the mammalian SFRS10 exon 2 which is also regulated by Tra2β. The SFRS10 gene encodes Tra2β protein, and exon 2 is a ubiquitously spliced poison exon which enables autoregulation in response to increasing levels of Tra2β protein (41). Similarly to HIPK3-T, SFRS10 exon 2 has 4 AGAA rich ESEs distributed all along the exon. However, SFRS10 exon 2 has a moderate 5′-splice site rather than the weak 5′-splice site found in HIPK3-T. In contrast to the HIPK3-T exon, inactivation of one ESE in the SFRS10 exon 2 does not completely abolish Tra2β activation, likely due to the associated moderately strong 5′-splice site. Hence the HIPK3-T and SFRS10 exons represent two prototypical examples of alternative exons regulated by the generally expressed splicing regulator protein Tra2β. Splicing inclusion can be either highly tissue-specific as for HIPK3-T or differentially regulated in various tissues as in SFRS10 exon 2. A general principle suggested from these two examples is that the anatomic spectrum of different Tra2β-responsive exons may be established in response to Tra2β concentration through an inter-play between the number of Tra2β-responsive ESEs and associated splice site strengths and other local splicing signals. Tra2β-responsive exons with stronger 5′-splice sites might be activated more ubiquitously than weaker exons like HIPK3-T with weaker 5′-splice sites. With these weaker exons, splicing inclusion would be switched on in specific tissues like the testis with elevated Tra2β concentrations and/or down-regulation of some splicing repressors (42). It is also possible that HIPK3-T is under regulation by trans-acting factors like PP1 which regulate Tra2β activity (43).
We propose an array of Tra2β binding sites within the HIPK3-T exon are deciphered to give tissue specific patterns of splicing activation in response to the expression levels of Tra2β. Very similar mechanisms of Tra2 splicing regulation have been shown to take place in Drosophila, both on the 3′-splice site (44) and on 5′-splice site regions (45). Activation of the Doublesex female-specific 3′-splice site is modulated in response to the number of multi-site enhancers for Tra2, and activation increases linearly rather than synergistically in response to this ESE number (17). In flies, increases in numbers of Tra2-responsive ESEs have been suggested to increase the probability of assembly of a splicing activator complex on Doublesex pre-mRNA, rather than enabling cooperative assembly of multiple binding splicing complexes containing Tra2. In this case the number of enhancer binding sites may influence the probability of U1 snRNP binding to the alternatively spliced exon. However, despite potential subtle mechanistic differences, human and fly orthologues of Tra2 protein seem to regulate splicing switches in a somewhat analogous fashion, both requiring assembly of Tra2 proteins on a responsive gauge. Regulated splice variants of other Drosophila mRNAs are similarly made in response to different threshold values of Tra2 concentrations in different tissues (18).
In this study we also found that Tra2β has a magnified activating effect on intrinsically weak exons. This magnified effect makes the HIPK3-T splicing switch more efficient. Since the testis has an increased Tra2β concentration (19), this observed splicing activation effect of Tra2β on weakly defined exons is also likely to apply to other intrinsically weak exons with Tra2β binding sites, and thus might contribute to the high overall levels of exon inclusion observed in the testis (8–11).
FUNDING
BBSRC (grant number BB/D013917/1) and Wellcome Trust (grant number WT080368MA) awarded to D.J.E.; a Royal Society Joint International Grant (awarded to D.J.E. and J.S.); EURASNET NoE (European Commission FP6) (to J.S.). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript. Funding for open access charge: Wellcome Trust.
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
The authors thank Dr Y. Durocher for the gift of the plasmid pTT3, and Dr F. Mauxion and B. Muller for their help for culture and transfection of FreeStyle 293-F cells.
REFERENCES
- 1.Pan Q, Shai O, Lee LJ, Frey BJ, Blencowe BJ. Deep surveying of alternative splicing complexity in the human transcriptome by high-throughput sequencing. Nat. Genet. 2008;40:1413–1415. doi: 10.1038/ng.259. [DOI] [PubMed] [Google Scholar]
- 2.Wang ET, Sandberg R, Luo S, Khrebtukova I, Zhang L, Mayr C, Kingsmore SF, Schroth GP, Burge CB. Alternative isoform regulation in human tissue transcriptomes. Nature. 2008;456:470–476. doi: 10.1038/nature07509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Nilsen TW, Graveley BR. Expansion of the eukaryotic proteome by alternative splicing. Nature. 2010;463:457–463. doi: 10.1038/nature08909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fu XD. Towards a splicing code. Cell. 2004;119:736–738. doi: 10.1016/j.cell.2004.11.039. [DOI] [PubMed] [Google Scholar]
- 5.Wang Z, Burge CB. Splicing regulation: from a parts list of regulatory elements to an integrated splicing code. RNA. 2008;14:802–813. doi: 10.1261/rna.876308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Llorian M, Schwartz S, Clark TA, Hollander D, Tan LY, Spellman R, Gordon A, Schweitzer AC, de la Grange P, Ast G, et al. Position-dependent alternative splicing activity revealed by global profiling of alternative splicing events regulated by PTB. Nat. Struct. Mol. Biol. 2010;17:1114–1123. doi: 10.1038/nsmb.1881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ule J, Stefani G, Mele A, Ruggiu M, Wang X, Taneri B, Gaasterland T, Blencowe BJ, Darnell RB. An RNA map predicting Nova-dependent splicing regulation. Nature. 2006;444:580–586. doi: 10.1038/nature05304. [DOI] [PubMed] [Google Scholar]
- 8.Castle JC, Zhang C, Shah JK, Kulkarni AV, Kalsotra A, Cooper TA, Johnson JM. Expression of 24,426 human alternative splicing events and predicted cis regulation in 48 tissues and cell lines. Nat. Genet. 2008;40:1416–1425. doi: 10.1038/ng.264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Clark TA, Schweitzer AC, Chen TX, Staples MK, Lu G, Wang H, Williams A, Blume JE. Discovery of tissue-specific exons using comprehensive human exon microarrays. Genome Biol. 2007;8:R64. doi: 10.1186/gb-2007-8-4-r64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yeo G, Holste D, Kreiman G, Burge CB. Variation in alternative splicing across human tissues. Genome Biol. 2004;5:R74. doi: 10.1186/gb-2004-5-10-r74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.de la Grange P, Gratadou L, Delord M, Dutertre M, Auboeuf D. Splicing factor and exon profiling across human tissues. Nucleic Acids Res. 2010;38:2825–2838. doi: 10.1093/nar/gkq008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Barash Y, Calarco JA, Gao W, Pan Q, Wang X, Shai O, Blencowe BJ, Frey BJ. Deciphering the splicing code. Nature. 2010;465:53–59. doi: 10.1038/nature09000. [DOI] [PubMed] [Google Scholar]
- 13.Long JC, Caceres JF. The SR protein family of splicing factors: master regulators of gene expression. Biochem. J. 2009;417:15–27. doi: 10.1042/BJ20081501. [DOI] [PubMed] [Google Scholar]
- 14.Bourgeois CF, Lejeune F, Stevenin J. Broad specificity of SR (serine/arginine) proteins in the regulation of alternative splicing of pre-messenger RNA. Prog. Nucleic Acid Res. Mol. Biol. 2004;78:37–88. doi: 10.1016/S0079-6603(04)78002-2. [DOI] [PubMed] [Google Scholar]
- 15.Clery A, Jayne S, Benderska N, Dominguez C, Stamm S, Allain FH. Molecular basis of purine-rich RNA recognition by the human SR-like protein Tra2-beta1. Nat. Struct. Mol. Biol. 2011;18:443–450. doi: 10.1038/nsmb.2001. [DOI] [PubMed] [Google Scholar]
- 16.Tsuda K, Someya T, Kuwasako K, Takahashi M, He F, Unzai S, Inoue M, Harada T, Watanabe S, Terada T, et al. Structural basis for the dual RNA-recognition modes of human Tra2-{beta} RRM. Nucleic Acids Res. 2010;39:1538–1553. doi: 10.1093/nar/gkq854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hertel KJ, Maniatis T. The function of multisite splicing enhancers. Mol. Cell. 1998;1:449–455. doi: 10.1016/s1097-2765(00)80045-3. [DOI] [PubMed] [Google Scholar]
- 18.Qi J, Su S, McGuffin ME, Mattox W. Concentration dependent selection of targets by an SR splicing regulator results in tissue-specific RNA processing. Nucleic Acids Res. 2006;34:6256–6263. doi: 10.1093/nar/gkl755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Venables JP, Bourgeois CF, Dalgliesh C, Kister L, Stevenin J, Elliott DJ. Up-regulation of the ubiquitous alternative splicing factor Tra2beta causes inclusion of a germ cell-specific exon. Hum. Mol. Genet. 2005;14:2289–2303. doi: 10.1093/hmg/ddi233. [DOI] [PubMed] [Google Scholar]
- 20.Baker BS. Sex in flies: the splice of life. Nature. 1989;340:521–524. doi: 10.1038/340521a0. [DOI] [PubMed] [Google Scholar]
- 21.Hoshijima K, Inoue K, Higuchi I, Sakamoto H, Shimura Y. Control of doublesex alternative splicing by transformer and transformer-2 in Drosophila. Science. 1991;252:833–836. doi: 10.1126/science.1902987. [DOI] [PubMed] [Google Scholar]
- 22.Grellscheid SN, Smith CW. An apparent pseudo-exon acts both as an alternative exon that leads to nonsense-mediated decay and as a zero-length exon. Mol. Cell. Biol. 2006;26:2237–2246. doi: 10.1128/MCB.26.6.2237-2246.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Liu Y, Bourgeois CF, Pang S, Kudla M, Dreumont N, Kister L, Sun YH, Stevenin J, Elliott DJ. The germ cell nuclear proteins hnRNP G-T and RBMY activate a testis-specific exon. PLoS Genet. 2009;5:e1000707. doi: 10.1371/journal.pgen.1000707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Durocher Y, Perret S, Kamen A. High-level and high-throughput recombinant protein production by transient transfection of suspension-growing human 293-EBNA1 cells. Nucleic Acids Res. 2002;30:E9. doi: 10.1093/nar/30.2.e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cavaloc Y, Bourgeois CF, Kister L, Stevenin J. The splicing factors 9G8 and SRp20 transactivate splicing through different and specific enhancers. RNA. 1999;5:468–483. doi: 10.1017/s1355838299981967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dreumont N, Hardy S, Behm-Ansmant I, Kister L, Branlant C, Stevenin J, Bourgeois CF. Antagonistic factors control the unproductive splicing of SC35 terminal intron. Nucleic Acids Res. 2010;38:1353–1366. doi: 10.1093/nar/gkp1086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Caceres JF, Krainer AR. Functional analysis of pre-mRNA splicing factor SF2/ASF structural domains. EMBO J. 1993;12:4715–4726. doi: 10.1002/j.1460-2075.1993.tb06160.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shaw SD, Chakrabarti S, Ghosh G, Krainer AR. Deletion of the N-terminus of SF2/ASF permits RS-domain-independent pre-mRNA splicing. PLoS One. 2007;2:e854. doi: 10.1371/journal.pone.0000854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhu J, Krainer AR. Pre-mRNA splicing in the absence of an SR protein RS domain. Genes Dev. 2000;14:3166–3178. doi: 10.1101/gad.189500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zuo P, Manley JL. Functional domains of the human splicing factor ASF/SF2. EMBO J. 1993;12:4727–4737. doi: 10.1002/j.1460-2075.1993.tb06161.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nayler O, Cap C, Stamm S. Human transformer-2-beta gene (SFRS10): complete nucleotide sequence, chromosomal localization, and generation of a tissue-specific isoform. Genomics. 1998;53:191–202. doi: 10.1006/geno.1998.5471. [DOI] [PubMed] [Google Scholar]
- 32.Coles JL, Hallegger M, Smith CW. A nonsense exon in the Tpm1 gene is silenced by hnRNP H and F. RNA. 2009;15:33–43. doi: 10.1261/rna.1225209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhang XH, Chasin LA. Computational definition of sequence motifs governing constitutive exon splicing. Genes Dev. 2004;18:1241–1250. doi: 10.1101/gad.1195304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhang XH, Kangsamaksin T, Chao MS, Banerjee JK, Chasin LA. Exon inclusion is dependent on predictable exonic splicing enhancers. Mol. Cell. Biol. 2005;25:7323–7332. doi: 10.1128/MCB.25.16.7323-7332.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tacke R, Tohyama M, Ogawa S, Manley JL. Human Tra2 proteins are sequence-specific activators of pre-mRNA splicing. Cell. 1998;93:139–148. doi: 10.1016/s0092-8674(00)81153-8. [DOI] [PubMed] [Google Scholar]
- 36.Cartegni L, Wang J, Zhu Z, Zhang MQ, Krainer AR. ESEfinder: a web resource to identify exonic splicing enhancers. Nucleic Acids Res. 2003;31:3568–3571. doi: 10.1093/nar/gkg616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Desmet FO, Hamroun D, Lalande M, Collod-Beroud G, Claustres M, Beroud C. Human Splicing Finder: an online bioinformatics tool to predict splicing signals. Nucleic Acids Res. 2009;37:e67. doi: 10.1093/nar/gkp215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Smith PJ, Zhang C, Wang J, Chew SL, Zhang MQ, Krainer AR. An increased specificity score matrix for the prediction of SF2/ASF-specific exonic splicing enhancers. Hum. Mol. Genet. 2006;15:2490–2508. doi: 10.1093/hmg/ddl171. [DOI] [PubMed] [Google Scholar]
- 39.Sanford JR, Wang X, Mort M, Vanduyn N, Cooper DN, Mooney SD, Edenberg HJ, Liu Y. Splicing factor SFRS1 recognizes a functionally diverse landscape of RNA transcripts. Genome Res. 2009;19:381–394. doi: 10.1101/gr.082503.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yu Y, Maroney PA, Denker JA, Zhang XH, Dybkov O, Luhrmann R, Jankowsky E, Chasin LA, Nilsen TW. Dynamic regulation of alternative splicing by silencers that modulate 5′ splice site competition. Cell. 2008;135:1224–1236. doi: 10.1016/j.cell.2008.10.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Stoilov P, Daoud R, Nayler O, Stamm S. Human tra2-beta1 autoregulates its protein concentration by influencing alternative splicing of its pre-mRNA. Hum. Mol. Genet. 2004;13:509–524. doi: 10.1093/hmg/ddh051. [DOI] [PubMed] [Google Scholar]
- 42.Kamma H, Portman DS, Dreyfuss G. Cell type-specific expression of hnRNP proteins. Exp. Cell. Res. 1995;221:187–196. doi: 10.1006/excr.1995.1366. [DOI] [PubMed] [Google Scholar]
- 43.Novoyatleva T, Heinrich B, Tang Y, Benderska N, Butchbach ME, Lorson CL, Lorson MA, Ben-Dov C, Fehlbaum P, Bracco L, et al. Protein phosphatase 1 binds to the RNA recognition motif of several splicing factors and regulates alternative pre-mRNA processing. Hum. Mol. Genet. 2008;17:52–70. doi: 10.1093/hmg/ddm284. [DOI] [PubMed] [Google Scholar]
- 44.Lynch KW, Maniatis T. Assembly of specific SR protein complexes on distinct regulatory elements of the Drosophila doublesex splicing enhancer. Genes Dev. 1996;10:2089–2101. doi: 10.1101/gad.10.16.2089. [DOI] [PubMed] [Google Scholar]
- 45.Lam BJ, Bakshi A, Ekinci FY, Webb J, Graveley BR, Hertel KJ. Enhancer-dependent 5′-splice site control of fruitless pre-mRNA splicing. J. Biol. Chem. 2003;278:22740–22747. doi: 10.1074/jbc.M301036200. [DOI] [PMC free article] [PubMed] [Google Scholar]