Abstract
In isolated nucleoli from Tetrahymena thermophila, low concentrations of the intercalating agent proflavine inhibit both transcription termination and splicing of the rRNA precursor. Proflavine also exerts an in vivo effect on the process of transcription termination under conditions, where the growth rate is only slightly reduced. Thus, approximately 40% of the rRNA precursor molecules, accumulated in nucleoli during 60 min of treatment with the drug, are longer than the normal 35S rRNA precursor. R-Loop mapping of these longer precursor molecules isolated after 30 and 60 min of incubation demonstrates that the RNA polymerases have a 50 fold lower elongation rate in the spacer region than in the coding region. Proflavine in the given concentration is found to have no significant effect on the splicing of properly terminated precursor molecules. In contrast, none of the longer non-terminated molecules are found to be spliced. These results indicate that proflavine primarily affects the process of transcription termination and that the splicing event is inhibited due to the improper termination of the precursor molecule.
Full text
PDF





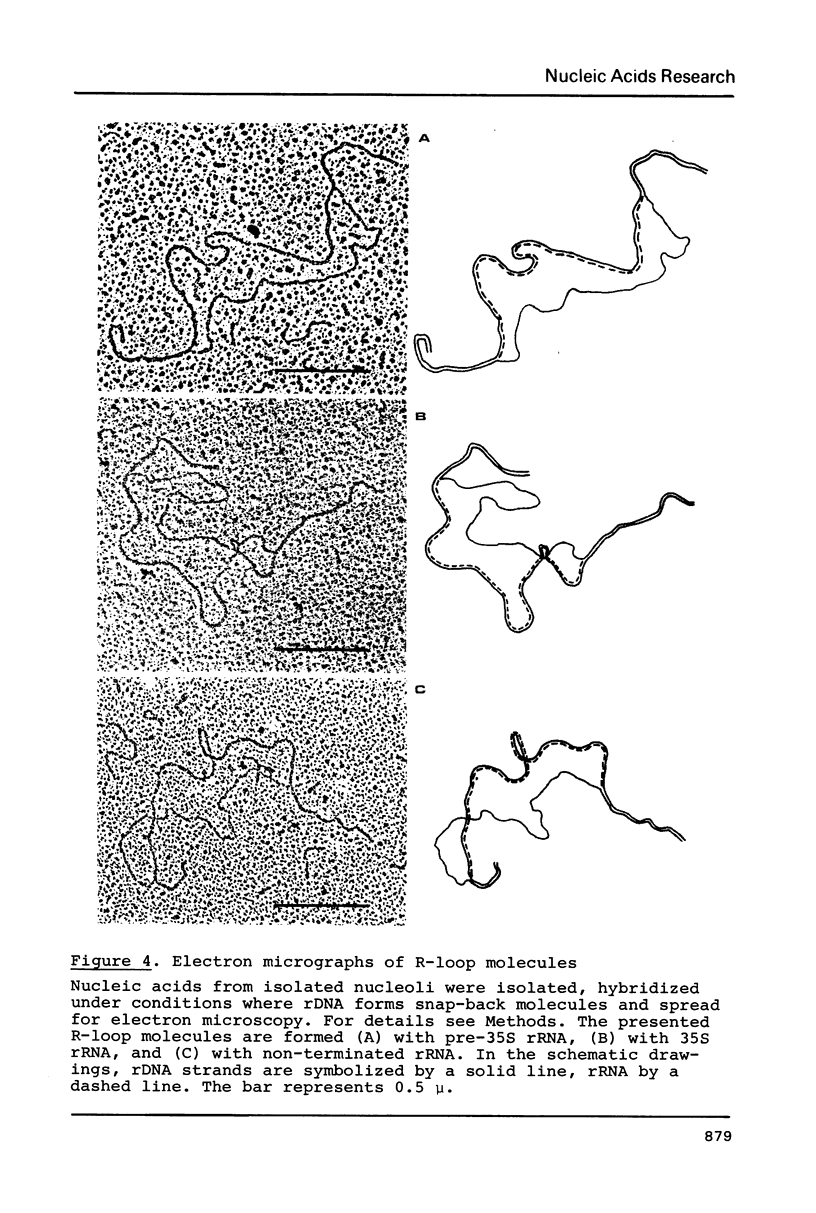

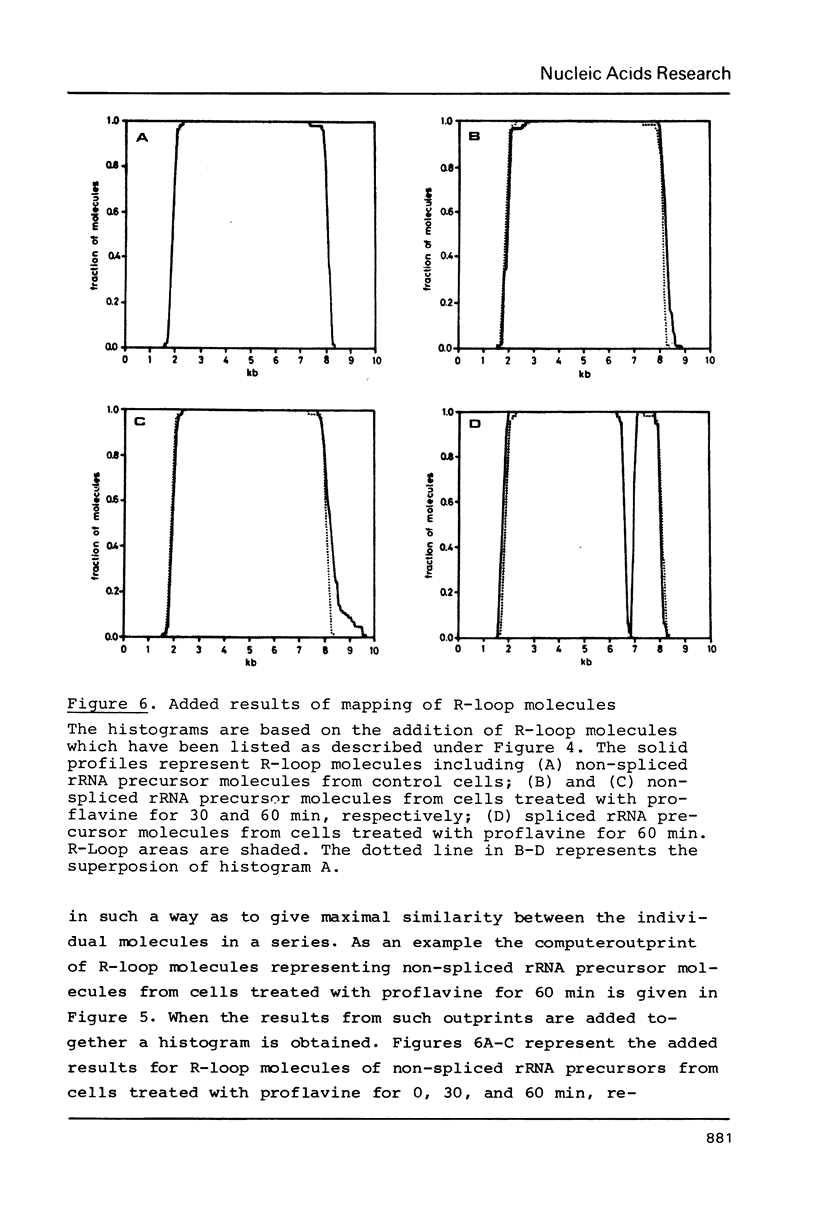





Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Abelson J. RNA processing and the intervening sequence problem. Annu Rev Biochem. 1979;48:1035–1069. doi: 10.1146/annurev.bi.48.070179.005131. [DOI] [PubMed] [Google Scholar]
- Borchsenius S., Bonven B., Leer J. C., Westergaard O. Nuclease-sensitive regions on the extrachromosomal r-chromatin from Tetrahymena pyriformis. Eur J Biochem. 1981 Jul;117(2):245–250. doi: 10.1111/j.1432-1033.1981.tb06329.x. [DOI] [PubMed] [Google Scholar]
- Brehm S. L., Cech T. R. Fate of an intervening sequence ribonucleic acid: excision and cyclization of the Tetrahymena ribosomal ribonucleic acid intervening sequence in vivo. Biochemistry. 1983 May 10;22(10):2390–2397. doi: 10.1021/bi00279a014. [DOI] [PubMed] [Google Scholar]
- Brimacombe R. Secondary structure and evolution of ribosomal RNA. Nature. 1981 Nov 19;294(5838):209–210. doi: 10.1038/294209a0. [DOI] [PubMed] [Google Scholar]
- Brinker J. M., Madore H. P., Bello L. J. Stabilization of heterogeneous nuclear RNA by intercalating drugs. Biochem Biophys Res Commun. 1973 Jun 8;52(3):928–934. doi: 10.1016/0006-291x(73)91026-7. [DOI] [PubMed] [Google Scholar]
- Carin M., Jensen B. F., Jentsch K. D., Leer J. C., Nielsen O. F., Westergaard O. In vitro splicing of the ribosomal RNA precursor in isolated nucleoli from Tetrahymena. Nucleic Acids Res. 1980 Dec 11;8(23):5551–5566. doi: 10.1093/nar/8.23.5551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cech T. R., Tanner N. K., Tinoco I., Jr, Weir B. R., Zuker M., Perlman P. S. Secondary structure of the Tetrahymena ribosomal RNA intervening sequence: structural homology with fungal mitochondrial intervening sequences. Proc Natl Acad Sci U S A. 1983 Jul;80(13):3903–3907. doi: 10.1073/pnas.80.13.3903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiu N. H., Bruszewski W. B., Salzman N. P. Evidence for the role of double-helical structures in the maturation of simian virus-40 messenger RNA. Nucleic Acids Res. 1980 Jan 11;8(1):153–168. doi: 10.1093/nar/8.1.153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Din N., Engberg J., Gall J. G. The nucleotide sequence at the transcription termination site of the ribosomal RNA gene in Tetrahymena thermophila. Nucleic Acids Res. 1982 Mar 11;10(5):1503–1513. doi: 10.1093/nar/10.5.1503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dudov K. P., Dabeva M. D., Hadjiolov A. A. Simple agar--urea-gel electrophoretic fractionation of high molecular weight ribonucleic acids. Anal Biochem. 1976 Nov;76(50):250–258. doi: 10.1016/0003-2697(76)90283-9. [DOI] [PubMed] [Google Scholar]
- Engberg J., Nasir-ud-Din, Eckert W. A., Kaffenberger W., Pearlman R. E. Detailed transcription map of the extrachromosomal ribosomal RNA genes in Tetrahymena thermophila. J Mol Biol. 1980 Sep 25;142(3):289–313. doi: 10.1016/0022-2836(80)90274-0. [DOI] [PubMed] [Google Scholar]
- Engberg J., Nasir-ud-Din, Eckert W. A., Kaffenberger W., Pearlman R. E. Detailed transcription map of the extrachromosomal ribosomal RNA genes in Tetrahymena thermophila. J Mol Biol. 1980 Sep 25;142(3):289–313. doi: 10.1016/0022-2836(80)90274-0. [DOI] [PubMed] [Google Scholar]
- Gocke E., Leer J. C., Nielsen O. F., Westergaard O. Transcriptional properties of nucleoli isolated from Tetrahymena. Nucleic Acids Res. 1978 Nov;5(11):3993–4006. doi: 10.1093/nar/5.11.3993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groudine M., Eisenman R., Weintraub H. Chromatin structure of endogenous retroviral genes and activation by an inhibitor of DNA methylation. Nature. 1981 Jul 23;292(5821):311–317. doi: 10.1038/292311a0. [DOI] [PubMed] [Google Scholar]
- Jelinek W., Darnell J. E. Double-stranded regions in heterogeneous nuclear RNA from Hela cells. Proc Natl Acad Sci U S A. 1972 Sep;69(9):2537–2541. doi: 10.1073/pnas.69.9.2537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaback D. B., Angerer L. M., Davidson N. Improved methods for the formation and stabilization of R-loops. Nucleic Acids Res. 1979 Jun 11;6(7):2499–2317. doi: 10.1093/nar/6.7.2499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimball P. C., Duesberg P. H. Virus interference by cellular double-stranded ribonucleic acid. J Virol. 1971 Jun;7(6):697–706. doi: 10.1128/jvi.7.6.697-706.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kruger K., Grabowski P. J., Zaug A. J., Sands J., Gottschling D. E., Cech T. R. Self-splicing RNA: autoexcision and autocyclization of the ribosomal RNA intervening sequence of Tetrahymena. Cell. 1982 Nov;31(1):147–157. doi: 10.1016/0092-8674(82)90414-7. [DOI] [PubMed] [Google Scholar]
- Leer J. C., Tiryaki D., Westergaard O. Termination of transcription in nucleoli isolated from Tetrahymena. Proc Natl Acad Sci U S A. 1979 Nov;76(11):5563–5566. doi: 10.1073/pnas.76.11.5563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robertson H. D., Dickson E., Jelinek W. Determination of nucleotide sequences from double-stranded regions of HeLa cell nuclear RNA. J Mol Biol. 1977 Oct 5;115(4):571–589. doi: 10.1016/0022-2836(77)90103-6. [DOI] [PubMed] [Google Scholar]
- Snyder A. L., Kann H. E., Jr, Kohn K. W. Inhibition of the processing of ribosomal precursor RNA by intercalating agents. J Mol Biol. 1971 Jun 14;58(2):555–565. doi: 10.1016/0022-2836(71)90371-8. [DOI] [PubMed] [Google Scholar]
- Southern E. M. Detection of specific sequences among DNA fragments separated by gel electrophoresis. J Mol Biol. 1975 Nov 5;98(3):503–517. doi: 10.1016/s0022-2836(75)80083-0. [DOI] [PubMed] [Google Scholar]
- Stern R., Friedman R. M. Double-stranded RNA synthesized in animal cells in the presence of actinomycin D. Nature. 1970 May 16;226(5246):612–616. doi: 10.1038/226612a0. [DOI] [PubMed] [Google Scholar]
- Westergaard O., Gocke E., Nielsen O. F., Leer J. C. Effect of lucanthone (miracil D) on transcription of ribosomal RNA genes from Tetrahymena in vivo and in vitro. Nucleic Acids Res. 1979 Jun 11;6(7):2391–2402. doi: 10.1093/nar/6.7.2391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White R. L., Hogness D. S. R loop mapping of the 18S and 28S sequences in the long and short repeating units of Drosophila melanogaster rDNA. Cell. 1977 Feb;10(2):177–192. doi: 10.1016/0092-8674(77)90213-6. [DOI] [PubMed] [Google Scholar]
- Yannarell A., Niemann M., Schumm D. E., Webb T. E. Proflavine sensitivity of RNA processing in isolated nuclei. Nucleic Acids Res. 1977 Mar;4(3):503–511. doi: 10.1093/nar/4.3.503. [DOI] [PMC free article] [PubMed] [Google Scholar]