Abstract
The 90S pre-ribosome, also known as the small subunit (SSU) processome, is a large multisubunit particle required for the production of the 18S rRNA from a pre-rRNA precursor. Recently, it has been shown that the formation of this particle entails the initial association of the tUTP subunit with the nascent pre-RNA and, subsequently, the binding of Rrp5/UTP-C and U3 snoRNP/UTP-B subunits in two independent assembly branches. However, the mode of assembly of other 90S pre-ribosome components remains obscure as yet. In this study, we have investigated the assembly of three proteins (Utp20, Imp4 and Bms1) previously regarded as potential nucleating factors of the 90S particle. Here, we demonstrate that the loading of those three proteins onto the pre-rRNA takes place independently of Rrp5/UTP-C and, instead, occurs downstream of the tUTP and U3/UTP-B subcomplexes. We also demonstrate that Bms1 and Utp20 are required for the recruitment of a subset of proteins to nascent pre-ribosomes. Finally, we show that proteins associated through secondary steps condition the stability of the two assembly branches in partially assembled pre-ribosomes. These results provide new information about the functional relationships among 90S particle components and the events that are required for their stepwise incorporation onto the primary pre-rRNA.
INTRODUCTION
The formation of eukaryotic ribosomes requires the production and assembly of four rRNAs and ~80 ribosomal proteins. In Saccharomyces cerevisiae, this process begins with the RNA polymerase I-dependent transcription of the polycistronic 35S rRNA precursor in the nucleolus (for a scheme, see Supplementary Figure S1). This pre-RNA undergoes a series of modifications and cleavage steps to yield three of the four rRNAs present in mature ribosomes (1). The initial cleavage steps take place at the A0, A1, A2 and A3 sites of the 35S rRNA (1–3). The first three cleavages generate the 20S pre-rRNA, a precursor that is exported to the cytosol and cleaved to render the 18S rRNA of the small 40S ribosomal subunit. The cleavage at the A3 site generates the 27SA3 pre-rRNA that, upon further maturation, will yield the 5.8S and 25S rRNAs that will form part of the large 60S ribosomal subunit (Supplementary Figure S1). These two processing pathways are not mutually dependent, since cleavage at A3 can precede cuts at A0, A1 and A2 sites (Supplementary Figure S1). Concomitant to the cleavage events, the rRNAs assemble with ribosomal proteins and the 5S rRNA, a 60S subunit component that is independently transcribed by RNA polymerase III.
All those processing and assembly reactions require numerous non-ribosomal factors that associate with pre-rRNAs in complexes known as pre-ribosomal particles (4–11). One of them is the 90S pre-ribosomal particle, also known as the small subunit (SSU) processome. In addition to the primary pre-rRNA, the U3 snoRNP and early assembled ribosomal proteins, this particle harbors ≃50 non-ribosomal factors that play structural, regulatory and cleavage roles (1,6,10,12–17). Recent work has shed light on the recruitment mechanism employed to assemble some of those factors onto the nascent pre-rRNA. Thus, it has been shown that different subsets of those proteins are grouped together in structurally autonomous subunits that pre-exist before the synthesis of the 35S pre-RNA and the formation of the 90S particle (14,15,17–24). These building blocks include the subunits known as tUTP (U3 protein complex required for transcription), UTP-B, UTP-C, Mpp10/Imp3/Imp4 and Bms1/Rcl1. tUTP (also known as UTP-A) is composed of seven proteins, and UTP-B and UTP-C both contain six proteins. Mmp10/Imp3/Imp4 and Bms1/Rcl1, as indicated by their names, are composed of three and two components, respectively. It has also been shown that some of those subunits, together with snoRNPs, associate with the pre-rRNA in a hierarchical and stepwise manner (Figure 1A) (18,25). One of the first steps in the formation of the 90S pre-ribosome is the binding of the tUTP subunit to the nascent pre-rRNA, an event that is a condition sine qua non for the subsequent binding of other pre-ribosomal subunits and proteins. A large number of 90S particle components bind to this nucleation core following two mutually independent assembly branches. One branch involves the recruitment of Rrp5 and the subsequent incorporation of the UTP-C subunit in an Rrp5-dependent manner. In the second branch, the UTP-B subunit and the U3 snoRNP co-assemble onto the nucleation core to form a highly stable 90S particle intermediary that also contains the Mpp10/Imp3/Imp4 subcomplex, the GTPase Bms1 and other pre-ribosomal proteins (18). However, it is not known as yet whether those latter components associate to the pre-rRNA through assembly steps that are concurrent or downstream to the loading of the U3 snoRNP and the UTP-B subunits onto the 35S pre-rRNA.
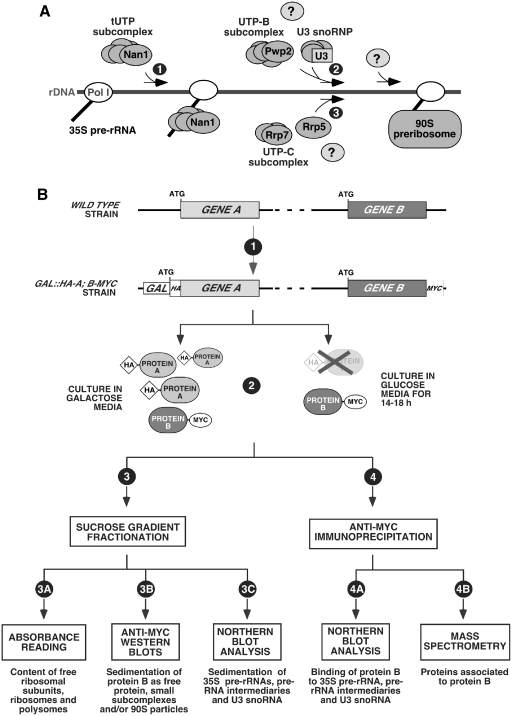
Figure 1.
(A) Diagram of the steps required for the assembly of the primary subunits of the 90S pre-ribosome onto the 35 pre-rRNA. The initial binding of the tUTP subunit, which occurs at an early and independent step (step 1), is required for the recruitment of other components and subunits through two independent assembly branches. One of these branches requires the UTP-B and U3 snoRNP subcomplexes (step 2). The other branch involves the stepwise binding of Rrp5 and the UTP-C subunit (step 3). The assembly of other factors through each one of these two branches might be concurrent or subsequent to the association of the primary building subunits (question marks). Nan1, Pwp2, Rrp5 and Rrp7 are the proteins of the 90S particle that have been epitope tagged or conditionally depleted in different experiments of this study. (B) Schematic outline of the experimental strategy used in this work. The aim of this study was to analyze the assembly behavior of specific 90S pre-ribosome components in the presence and absence of other 90S pre-ribosome proteins. Details about the purpose of the techniques and steps outlined in this Figure are given in the text at the beginning of the ‘Results’ section.
To get further insights into the assembly mechanism of the 90S pre-ribosomal particle, we decided to investigate in the present work the hierarchy of assembly of three 90S pre-ribosomal particle components, Imp4, Bms1 and Utp20. Imp4 is a RNA binding factor that forms a stable heterotrimeric subcomplex with Imp3 and Mpp10 (20,21). This subunit binds directly to the U3 snoRNA and is able to induce, in vitro, structural rearrangements in the U3 snoRNA that alter its base pairing with the pre-rRNA (26,27). Bms1 is another stable component of 90S pre-ribosomes that forms a small subcomplex with Rcl1 (14,15,22,28,29). Bms1 is a GTPase that has been proposed to deliver Rcl1 to pre-ribosomes when bound to GTP (30,31). However, the timing and purpose of this regulated delivery is still poorly characterized. Finally, Utp20 is a HEAT-repeat protein present in both 90S and pre-40S particles (32). Although its function is unknown, we have previously hypothesized that it could be important for the building of early 90S particle intermediates because, according to proteomic and bioinformatics analyses, Utp20 seems to be heavily interconnected and in close physical proximity to components of the tUTP, UTP-B and UTP-C subunits (18).
The results presented here clarify the mechanism of assembly of those three proteins onto nascent pre-ribosomes, their role as ancillary assembly factors for the incorporation of additional layers of pre-ribosomal proteins onto the 90S particle and shed light on their intrinsic functions in the context of pre-rRNA biogenesis.
MATERIALS AND METHODS
Yeast strains and genetic methods
Yeast strains used in this study are listed in Supplementary Table S1. Conditional mutant strains for RRP5, NAN1, PWP2, UTP20, IMP4 and BMS1 under the control of the GAL1 promoter were generated by the one-step polymerase chain reaction (PCR) strategy. This rendered in-frame fusions of a KANMX6-GAL1-HA cassette upstream of the ATG of the corresponding gene. These strains are referred to in the text as GAL::HA-RRP5, GAL::HA-NAN1, GAL::HA-PWP2, GAL::HA-UTP20, GAL::HA-IMP4 and GAL::HA-BMS1. MYC carboxyl-terminal tagged alleles for UTP20, IMP4, BMS1, PWP2 and RRP7 were generated also by one-step integration of PCR cassettes in the corresponding locus. The MYC-tagged versions are the only source of protein in the cell and their expression is under the gene endogenous promoters. These alleles are referred to in the main text as UTP20-MYC, IMP4-MYC, BMS1-MYC, PWP2-MYC and RRP7-MYC. All MYC-tagged alleles used in this study are fully functional. Wild-type cells modified to express any of those alleles presented normal growth rates and showed no detectable alterations in the content of rRNAs, levels of ribosomal subunits and polysomes, or production and sedimentation of pre-rRNA precursors. Strains carrying the conditional GAL1-driven constructs were cultured at 30°C in either galactose (YP-Gal, 0.4% yeast extract, 0.8% peptone, 0.1 mM adenine, 2% galactose) or glucose (YPD, 0.4% yeast extract, 0.8% peptone, 0.1 mM adenine, 2% glucose). For protein depletion, the incubation times in glucose were as follows: 18 h in glucose for GAL::HA-PWP2 cells; 16 h in glucose for GAL::HA-NAN1, GAL::HA-UTP20, GAL::HA-IMP4 and GAL::HA-BMS1 cells; and 14 h in glucose for GAL::HA-RRP5 cells.
Sucrose density-gradient analysis
Polysome analysis and fractionation of lysates through 7–50% sucrose gradients were performed as described (19). Extract equivalents to 15 absorption units at 260 nm (A260) were layered per gradient. Fractions from each gradient were analyzed by western and northern blot. Protein samples (40 µl) of each fraction were analyzed directly on 8% SDS–polyacrylamide gels. Total RNA was extracted from 100 µl aliquots of each fraction by the hot-phenol method and analyzed on 1.2% agarose–formaldehyde gels.
Northern blot analysis
RNAs from total cellular lysates, gradient fractionations or co-immunoprecipitations were prepared by the hot-phenol method as described (18). The oligonucleotides used for analyzing pre-rRNA intermediaries are the following: region D-A2 of 35S pre-rRNA: 5′-TTAAGCGCAGGCCCGGCT-3′; region A2–A3 of 35S pre-rRNA: 5′-TGTTACCTCTGGGCC-3′. The oligonucleotide probe to analyze the U3 snoRNA levels was 5′-GGATTGCGGACCAAGCTAA-3′. Oligonucleotide labeling, northern blotting and hybridization were performed essentially as described (19).
Co-immunoprecipitation experiments
Cell cultures were grown to A600 between 0.8 and 1.0. Extract equivalents to 15 A600 units, prepared as for polysome sucrose gradient analysis (19), were taken to 250 µl of PP buffer and mixed with 0.5 ml of IP buffer (20 mM Tris–HCl (pH 7.5), 5 mM MgCl2, 150 mM potassium acetate, 1 mM dithiothreitol, 0.2% Triton X-100) containing a mixture of protease inhibitors (Complete™, Roche), 600 U/ml RNasin (Promega) and 2 µg of anti-MYC 9E10 monoclonal antibody (Roche), and incubated at 4°C for 2 h under gentle rotation. Following incubation with Gammabind™ Sepharose beads (GE Healthcare), immunoprecipitates were washed at 4°C five times for 5 min with IP buffer. For protein analyses, one-fifth of the immunoprecipitated material was resuspended in 200 µl of SDS loading buffer, boiled and analyzed by western blot. For RNA analyses, the rest of the immunoprecipitated material was resuspended in 400 µl of 50 mM sodium acetate, 10 mM EDTA (pH 5.2), 1% SDS and processed for RNA extraction using the hot-phenol procedure (33). After ethanol precipitation, RNAs were resuspended in formaldehyde loading buffer, separated by electrophoresis on 1.2% agarose–formaldehyde and analyzed by northern blot using as probes the oligonucleotides described above.
Purification of pre-ribosomal complexes and mass spectrometry analysis
Purifications of Utp20p-MYC-, Imp4p-MYC-, Bms1-MYC-, Pwp2-MYC- or Rrp7p-MYC-containing complexes in conditional mutant strains were performed using a large-scale anti-MYC co-immunoprecipitation approach. Preparation of lysates, chromatographic procedures and protein identification by mass spectrometry were carried out essentially as described (18). Complexes were always purified in several independent experiments and the associated proteins to each bait protein, in each experimental condition, were found to be the same every time. A protein was considered to be associated with a given bait protein when it was detected as a strongly stained band in the corresponding gels of the complex purifications performed with that bait. If so, it was included in Figures 4 and 7. A protein was considered as ‘not present’ in a particular purified complex when: (i) it was not detected in the silver-stained SDS–PAGE gels. (ii) The electrophoretic band identified by mass spectrometry did not correspond to such protein. (iii) The relative amount of that protein was drastically reduced in the bait purifications when compared to the amounts that co-purified with other baits. In addition, we also detected some strongly stained protein bands that could not be identified by mass spectrometry (8% of all stained bands analyzed) and possibly corresponded to mixtures of protein degradation products or denatured IgG chains released from the anti-MYC beads after the boiling step. Finally, we identified proteins that had reported functions not related to ribosomal biogenesis. These proteins, representing ≃11% of all stained bands, were found associated to the chosen bait regardless of the experimental conditions used, suggesting that they likely represent unspecific products obtained along the purification process. The identity of these bands can be provided upon request. All mass spectrometry analyses were performed in the Genomics and Proteomics Unit of the Centro de Investigacion del Cancer of Salamanca.
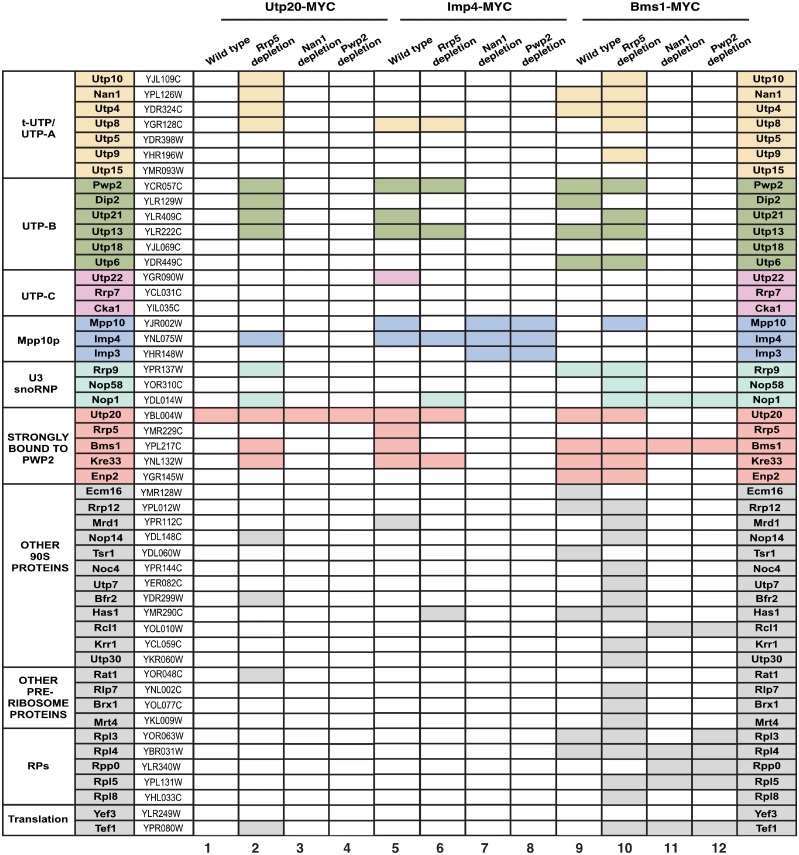
Figure 4.
Protein composition analysis of the complexes formed by Utp20, Imp4 and Bms1 in wild type and Rrp5-, Nan1- or Pwp2-depleted cells. The MYC-tagged proteins used as baits and the proteins depleted before complex purifications are indicated on the top. The proteins co-purifying with the baits that were identified in these analysis, the designations of the corresponding open reading frames, and the subcomplex they belong to are indicated in the three first columns and in the last column. The five proteins highlighted in red are not components of the known primary UTP subunits, but consistently co-purify with Pwp2 in our purifications from both wild-type and Rrp5-depleted cells. A shaded rectangle indicates that the protein was found associated to the bait.
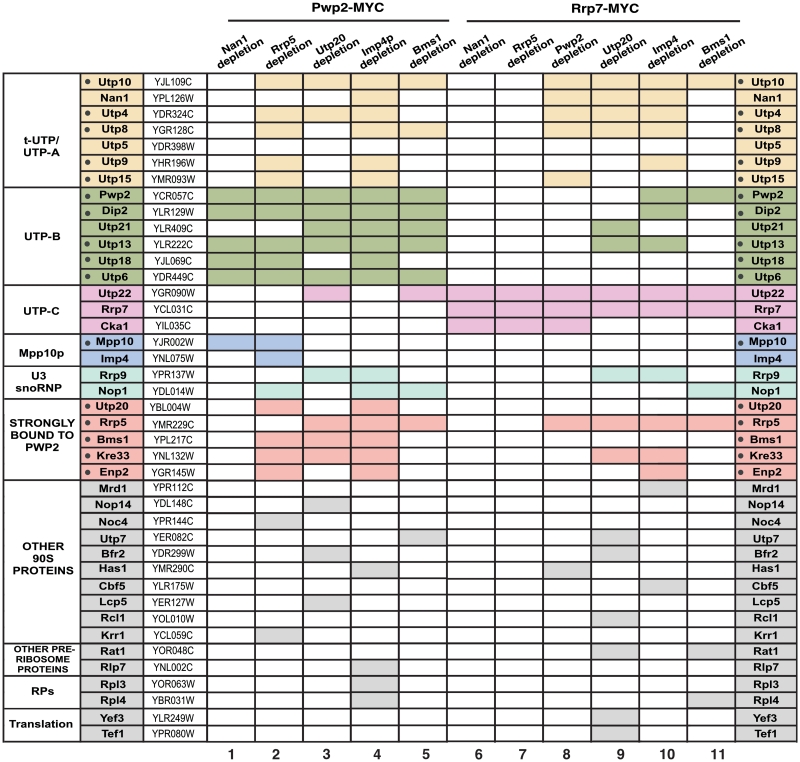
Figure 7.
Protein composition analysis of pre-ribosomal complexes in cells depleted of Nan1, Rrp5, Utp20, Imp4 or Bms1. The MYC-tagged proteins used as baits, and the proteins depleted before complex purifications are indicated on the top. The proteins co-purifying with the baits that were identified in these analyses, the designations of the corresponding open reading frames and the subcomplex they belong to are indicated in the three first columns and in the last column. A subset of 16 components of the 90S pre-ribosome that consistently co-purify with Pwp2 in our analysis, both in wild-type and Rrp5-depleted cells, are dot-marked in the second column on the left. A shaded rectangle indicates that the protein was found associated to the bait.
RESULTS
Experimental design
We used a similar strategy to that used before in the elucidation of the hierarchy of assembly of the U3 snoRNP, tUTP, UTP-B and UTP-C subunits in the 90S pre-ribosomal particle (18). To this end, we carried out a systematic analysis of the assembly behavior of each of the three proteins under study in the presence or absence of: (i) each of the other two proteins under study in this work; (ii) each of the previously characterized 90S primary subunits (tUTP, UTP-B and Rrp5) (Figure 1B). To achieve this goal, we first generated yeast strains expressing two types of epitope-tagged proteins: in the case of proteins to be depleted, their loci were modified using standard homologous recombination techniques to encode HA-tagged proteins under the regulation of the inducible GAL1 promoter (Figure 1B, step 1). This strategy enabled us to trigger either the overexpression or the repression of the protein under analysis using galactose or glucose in the culture medium, respectively. To monitor the behavior of other 90S particle factors in those mutant strains, the corresponding endogenous loci were modified to express MYC-tagged versions of the proteins under their own promoters (Figure 1B, step 1). Once generated, these cells were maintained in galactose media and, when needed, shifted to glucose media for 14–18 h to track down the behavior of the MYC-tagged protein in the presence or absence of the HA-tagged protein (Figure 1B, step 2). Extracts from each experimental condition were subsequently analyzed using a number of independent techniques. First, lysates were fractionated on sucrose gradients to determine: (i) the generation of 40S and 60S ribosomal subunits, of 80S ribosomes and polysomes by means of the continuous reading of A254 of the collected gradient fractions after the ultracentrifugation step (Figure 1B, step 3A). (ii) The sedimentation pattern of the MYC-tagged protein in the presence/absence of the selected HA-tagged protein using anti-MYC western blot analyses in aliquots obtained from gradient fractions (Figure 1B, step 3B). (iii) The production/sedimentation pattern of pre-rRNA precursors and the U3 snoRNA using northern blot analysis with appropriate 32P-labeled probes (Figure 1B, step 3C). Second, lysates were subjected to analytical anti-MYC immunoprecipitations to detect the interaction of the MYC-tagged protein under study with: (i) the 35S pre-rRNA and downstream biosynthetic products (Figure 1B, step 4A). (ii) The U3 snoRNA (Figure 1B, step 4A). Third, lysates were used in preparative anti-MYC immunoprecipitations and mass spectrometry analyses to identify the proteins and/or 90S pre-ribosomal subunits associated with the MYC-tagged protein (Figure 1B, step 4B). By repeating those experiments with all the possible combinations of the proteins under study, this experimental strategy allowed us to get a view of the behavior of these factors in terms of their order of assembly onto the 90S pre-ribosomal particle and their participation in the formation of partially assembled intermediaries of the 90S particle.
The incorporation of Utp20, Imp4 and Bms1 into nascent pre-ribosomes takes place independently of the Rrp5/UTP-C assembly branch
Utp20, Imp4 and Bms1 were previously detected in proteomics experiments complexed with both tUTP and UTP-B subunit components in the absence of Rrp5 (18), suggesting that they are loaded onto the 90S pre-ribosomal core following the U3 snoRNP/UTP-B assembly route. To verify this possibility, we investigated the behavior of Utp20-MYC, Imp4-MYC and Bms1-MYC proteins in cells expressing or lacking Rrp5. In the presence of Rrp5, Utp20-MYC (Figure 2A, top panel on the left), Imp4-MYC (Figure 2C, top panel on the left) and Bms1-MYC (Figure 2E, top panel on the left) were detected in high molecular weight ≃90S complexes that co-sedimented with both the 35S pre-rRNA (Figure 2A, C and E; middle panels on the left, fractions 9–12) and the U3 snoRNA (Figure 2A, C and E; bottom panels on the left, fractions 9–12). In addition, we observed that those three proteins were present in fractions outside the 90S sedimentation range. Thus, Utp20-MYC was detected at high levels in ≃40S–60S fractions (Figure 2A, top panel on the left, fractions 5–7). This sedimentation profile was similar to that observed in cells expressing wild-type levels of Rrp5 (Supplementary Figure S2), and was consistent with previous data indicating that Utp20 is present in both 90S and pre-40S pre-ribosomes (32). Imp4 was detected at the top fractions of the gradient (Figure 2C, top panel on the left, fractions 2–4; Supplementary Figure S2), the region that presumably contains free Mpp10–Imp3–Imp4 subunits. In the case of Bms1, we had to restrict our analyses to its presence in high molecular weight complexes. This protein was routinely degraded in the top fractions of the sucrose gradients and, therefore, we could not obtain a reliable quantification of its presence in small size complexes (Figure 2E, top panel on the left). However, Bms1 was much more stable in the intermediate and bottom regions of the gradient, being consistently detected in the ≃90S (Figure 2E, top panel on the left, fractions 10–12) and ≃40S fractions (Figure 2E, top panel on the left, fractions 6–8). This sedimentation profile, which is similar to the one observed in wild-type cells (Supplementary Figure S2), is consistent with the published studies indicating that Bms1 is a component of both 90S and 40S pre-ribosomes (22). Interestingly, we observed that the depletion of Rrp5 elicited no significant change in the sedimentation profile of Utp20–MYC (Figure 2A, top panel on the right, fractions 10–12), Imp4–MYC (Figure 2C, top panel on the right, fractions 10–11) and Bms1–MYC (Figure 2E, top panel on the right, fractions 10–12) in large molecular weight complexes, suggesting that they assemble onto the 90S pre-ribosomal particle in an Rrp5-independent manner. Northern blot analyses confirmed that the 35S pre-rRNA (Figure 2A, C and E; middle panels) and the U3 snoRNA (Figure 2A, C and E; bottom panels) displayed the expected sedimentation profiles in the three GAL-HA-RRP5 strains used in the study both under Rrp5 overexpression (Figure 2A, C and E; left panels) and depletion (Figure 2A, C and E; right panels) conditions. To verify that the assembly of those three proteins onto the 90S pre-ribosomal particle was Rrp5 independent, we carried out co-immunoprecipitation experiments coupled with northern blot analyses to monitor their interaction with the 35S pre-rRNA and the U3 snoRNA. These experiments indicated that Utp20–MYC and Imp4–MYC could associate with both the 35S pre-rRNA (Figure 2B and D, middle panels) and the U3 snoRNA (Figure 2B and D, bottom panels) regardless of the presence or not of Rrp5 in the yeast lysates. We also detected the association of Bms1 with the 35S pre-rRNA (Figure 2F, middle panels) and the U3 snoRNA (Figure 2F, bottom panels) in both Rrp5 positive and negative cells. However, in this case, those interactions took place much less efficiently than in the case of the co-immunoprecipitations made with Utp20–MYC and Imp4–MYC. This is possibly due to the deficient recovery of intact Bms1–MYC in Rrp5-depleted cells (Figure 2F, upper panel on the right, compare lines 5 and 8). Utp20–MYC (Figure 2B, third panel from the top on the right, lane 8), Imp4–MYC (Figure 2D, third panel from the top on the right, lane 8) and Bms1–MYC (Figure 2F, third panel from the top on the right, lane 8) could also associate stably with the 24S pre-rRNA, an aberrant rRNA species typically generated in Rrp5-depleted cells (34). The presence of the MYC-tagged proteins in the appropriate total cellular lysates (Figure 2B, D and F; top panels on the left) and immunoprecipitates (Figure 2B, D and F; top panels on the right) was confirmed by immunoblot analysis with anti-MYC antibodies. A similar approach using anti-HA antibodies was used to monitor the expression levels (Figure 2B, D and F; second panels from top on the left) or co-immunoprecipitation (Figure 2B, D and F; second panels from top on the right) of HA-tagged Rrp5 in these experiments. As a further control for the efficient elimination of Rrp5 in cells grown in glucose media, we verified that its elimination led to the expected blockage in the cleavage of the 35S pre-rRNA at the A0–A3 sites. Consistent with such blockage, we confirmed the disappearance of the 23S pre-rRNA and the concomitant accumulation of the aberrant 24S pre-rRNA (Figure 2B, D and F; middle panels), the formation of partially assembled pre-ribosomal particles containing tUTP and UTP-B subunit elements but not UTP-C proteins (18) (data not shown, see below), and the expected loss of 40S and 60S subunits (data not shown). These results, combined with previous evidence from proteomic analyses (18), indicate that Utp20, Imp4 and Bms1 assemble onto the primary pre-rRNA in an Rrp5-independent manner.
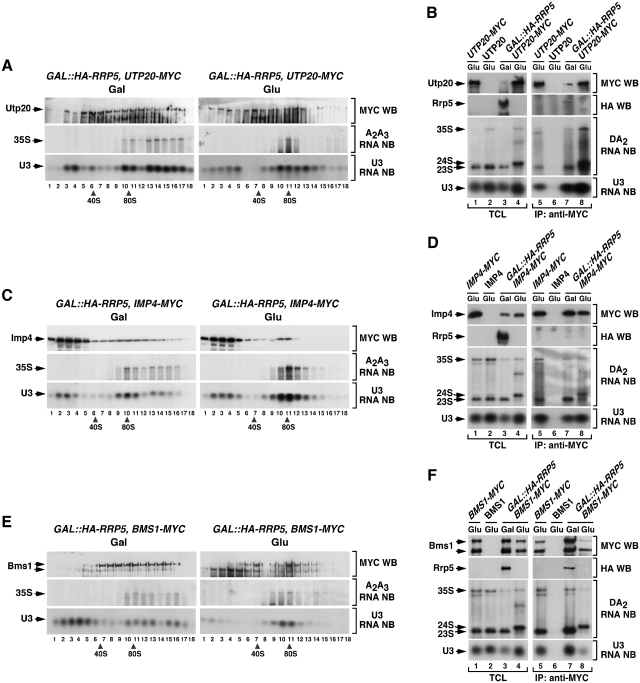
Figure 2.
The interaction of Utp20, Imp4 and Bms1 with the 35S pre-rRNA is independent of Rrp5. (A, C and E) Sedimentation behavior in sucrose gradients of Utp20 (A), Imp4 (C) and Bms1 (E) in the presence and absence of Rrp5. Cellular extracts prepared from the indicated yeast strains (top) grown in medium containing either galactose (left panels) or glucose (right panels) were resolved in 7–50% linear sucrose gradients. After ultracentrifugation, 0.5 ml fractions were collected from the top of the tube. The content of Utp20–MYC (A), Imp4–MYC (C) and Bms1–MYC (E) in each fraction was analyzed by anti-MYC immunoblotting (first panels from top). In parallel, total RNAs were prepared from each fraction and analyzed by northern blot with either an oligonucleotide probe to the 35S A2–A3 region (second panels from top) or to the U3 snoRNA (bottom panels). The number of each fraction and the sedimentation positions of 40S and 80S particles are indicated at the bottom. Gal, galactose; Glu, glucose; WB, western blot; NB, northern blot. (B, D and F) Co-immunoprecipitation of Utp20 (B), Imp4 (D) and Bms1 (F) with pre-RNAs and U3 snoRNA in the presence and absence of Rrp5. Total cellular lysates (lanes 1–4) and anti-MYC immunoprecipitates (lanes 5–8) prepared from the indicated yeast strains and growth conditions (top) were analyzed by anti-MYC and anti-HA immunoblotting (first and second panels from top), and northern blot analysis with either an oligonucleotide probe to the 35S D-A2 region (third panels from top) or to the U3 snoRNA (bottom panels). TCL, total cellular lysates; IP, immunoprecipitation.
The recruitment of Utp20 to early pre-ribosomes takes place in a U3 snoRNP/UTP-B-dependent manner
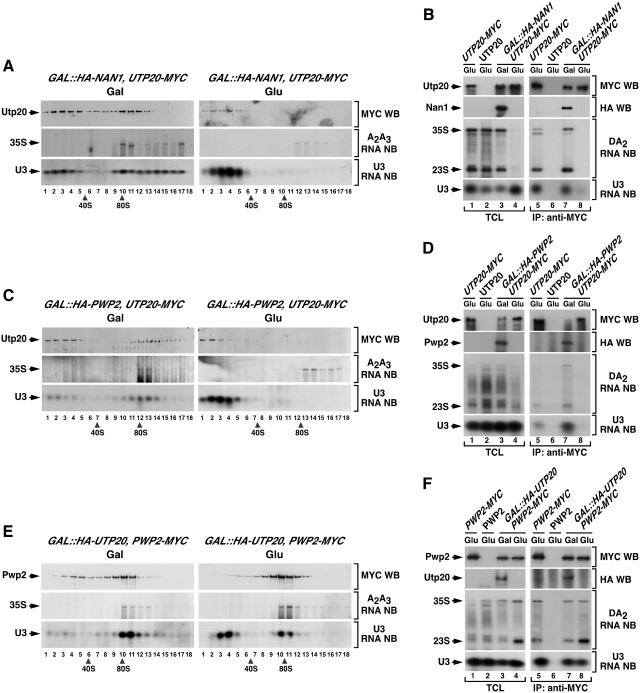
It has been previously found that the binding of the tUTP subunit to the primary pre-rRNA is required for the subsequent docking of several 90S particle components (Figure 1A) (18,25). To determine if that is also the case for Utp20, we examined its behavior in the presence and absence of Nan1, a known component of the tUTP complex (25). For that purpose, we generated a conditional GAL::HA-NAN1 strain expressing constitutively Utp20–MYC. Upon depletion of Nan1, we observed that Utp20–MYC disappeared from the 90S gradient fractions (Figure 3A, top panel on the right), suggesting that its ability to dock onto pre-ribosomal complexes is Nan1- and tUTP dependent. As expected, we observed that Nan1-depleted cells showed reduced 35S pre-rRNA levels (Figure 3A, middle panel on the right) as well as the accumulation of the U3 snoRNP in gradient fractions corresponding to free U3 snoRNPs that are not complexed with the 35S pre-rRNA (Figure 3A, bottom panel on the right). Confirming the gradient results, we could not detect any co-immunoprecipitation of Utp20 with the 35S pre-rRNA (Figure 3B, third panel from top), the 23S pre-rRNA (Figure 3B, third panel from top) and the U3 snoRNA (Figure 3B, bottom panel) when Nan1 was absent from the lysates. These results indicate that the assembly of Utp20 requires the prior incorporation of the tUTP subunit.
Figure 3.
The recruitment of Utp20 to early pre-ribosomes is secondary to the assembly of UTP-B/U3. (A, C and E) Sedimentation behavior in sucrose gradients of Utp20 (A and C) and Pwp2 (E) in the presence and absence of Nan1 (A), Pwp2 (C) and Utp20 (E). Cellular extracts prepared from the indicated yeast strains (top) grown in medium containing either galactose (left panels) or glucose (right panels) were resolved in 7–50% linear sucrose gradients. After ultracentrifugation, 0.5 ml fractions were collected from the top of the tube. The content of Utp20–MYC (A and C) and Pwp2–MYC (E) in each fraction was analyzed by anti-MYC immunoblotting (first panels from top). In parallel, total RNAs were prepared from each fraction and analyzed by northern blot as indicated in Figure 2. The number of each fraction and the sedimentation points of 40S and 80S particles are indicated at the bottom. Gal, galactose; Glu, glucose; WB, western blot; NB, northern blot. (B, D and F) Co-immunoprecipitation of Utp20 (B, D) and Pwp2 (F) with pre-RNAs and U3 snoRNA in the presence and absence of Nan1 (B), Pwp2 (D) and Utp20 (F). Total cellular lysates (lanes 1–4) and anti-MYC immunoprecipitates (lanes 5–8) prepared from the indicated yeast strains and growth conditions (top) were analyzed by anti-MYC and anti-HA immunoblotting (upper panels), and northern blot analysis as described in Figure 2. TCL, total cellular lysates; IP, immunoprecipitation.
Next, we decided to ascertain the assembly hierarchy between Utp20 and the UTP-B subunit. To this end, we used a GAL::HA-PWP2;UTP20-MYC strain to monitor the docking of Utp20 onto the pre-rRNA in the presence or absence of Pwp2, a known component of the UTP-B subunit (14,15,19). We observed that the depletion of Pwp2 induced a shift in the sedimentation of Utp20–MYC from 35S pre-rRNA enriched fractions (Figure 3C, top panel on the left, fractions 13–14) to gradient positions consistent with uncomplexed proteins (Figure 3C, top panel on the right, fractions 1–3). A change in the sedimentation pattern was also observed for the U3 snoRNA (Figure 3C, compare bottom panels). Consistent with those results, we observed using co-immunoprecipitation and northern blot analysis that Utp20–MYC could not associate with the 35S pre-rRNA (Figure 3D, third panel from top), the 23S pre-rRNA (Figure 3D, third panel from top) and the U3 snoRNAs (Figure 3D, bottom panel) when Pwp2 was absent from the yeast lysates. The presence of Utp20–MYC and HA-Pwp2 in the appropriate total cellular lysates (Figure 3D, first and second panels on the left) and immunoprecipitates (Figure 3D, first and second panels on the right) was confirmed by immunoblot analysis with anti-MYC and anti-HA antibodies, respectively. Taken together, these results indicate that the assembly of Utp20 onto the 90S pre-ribosomal particle requires the presence of the UTP-B complex.
Two possible scenarios could explain this UTP-B dependency. Thus, it is plausible that Utp20 co-assembles with, and is required for, the docking of both UTP-B and U3 snoRNP onto the tUTP-primed 35S pre-rRNA. Alternatively, it could be possible that Utp20 is part of a secondary assembly wave that takes place only upon the docking of UTP-B and U3 snoRNP on the nascent pre-ribosomal particle. If the former hypothesis were correct, we would expect that the assembly of both UTP-B and U3 snoRNP had to be Utp20 dependent. In contrast, if the latter hypothesis were correct, such assembly step has to be Utp20 independent. To distinguish those two possibilities, we generated a GAL::HA-UTP20;PWP2-MYC strain to monitor the assembly of the UTP-B component Pwp2 and the U3 snoRNA onto the 35S pre-rRNA in the presence and absence of Utp20. Using sucrose gradient sedimentation analyses, we found that the co-sedimentation profiles of Pwp2-MYC (Figure 3E, compare top left and right panels, fractions 10–11) and the U3 snoRNA (Figure 3E, compare middle left and right panels, fractions 10–11) with the 35S pre-rRNA were wild type like in the absence of Utp20. Likewise, we observed using co-immunoprecipitation and northern blot analyses that the efficiency of the interaction of Pwp2-MYC with the 35S pre-rRNA (Figure 3F, third panel from the top on the right), the 23S pre-rRNA (Figure 3F, third panel from the top on the right) and the U3 snoRNA (Figure 3F, bottom panel on the right) did not change in the absence of Utp20. This was not due to deficient repression of Utp20, because northern blot analyses showed that Utp20-deficient cells displayed the expected accumulation of 35S and 23S pre-rRNAs (Figure 3F, third panel from the top on the left), the loss of 40S subunits (data not shown) and the formation of defective 90S pre-ribosomes (data not shown and see below). These results indicate that the recruitment of Utp20 takes place after the incorporation of the UTP-B/U3 snoRNP complex onto the tUTP-primed 35S pre-rRNA.
To gather information on the physical interactions established by Utp20 with other 90S particle factors and on its presence in pre-assembly or post-cleavage subcomplexes, we conducted proteomic analyses to identify factors associated to Utp20 under the different experimental conditions described above. In wild-type cells, Utp20–MYC was detected mostly as an isolated, non-complexed protein (Figure 4, column 1; and Supplementary Figure S3A). This finding indicates that Utp20 does not form part of an additional pre-ribosomal subunit and, in addition, that it is weakly bound to other pre-ribosomal particle components. Instead, we found somewhat surprisingly that Utp20–MYC does form a more stable association with intrinsic components of the tUTP, UTP-B and Mpp10 subunits in Rrp5-depleted cells than in wild-type cells (Figure 4, column 2; Supplementary Figure S3B). Such factors were previously found to be present in partially assembled pre-ribosomes produced under the same conditions (18). These results are fully consistent with our experiments showing that Utp20 assembles onto the primary pre-rRNA in an Rrp5-independent manner and, in addition, suggest that this protein becomes either stably bound or trapped into the structural assembly intermediaries that are formed in Rrp5-deficient cells. Consistent with the observations indicating that the assembly of Utp20 is UTP-B dependent, we observed that those interactions were lost in cells lacking either Nan1 (a tUTP subunit component; Figure 4, column 3; Supplementary Figure S3C) or Pwp2 (an UTP-B subunit element; Figure 4, column 4; Supplementary Figure S3D). Collectively, our results indicate that Utp20 incorporates individually onto nascent pre-ribosomes in a second assembly wave that takes place upon the initial incorporation of tUTP and UTP-B/U3 snoRNP complexes. Instead, its assembly does not require the prior docking onto the tUTP-primed 35S pre-rRNA of the Rrp5 and UTP-C complexes.
The recruitment of Imp4 to early pre-ribosomes occurs downstream of the assembly of the UTP-B/U3 snoRNP complex
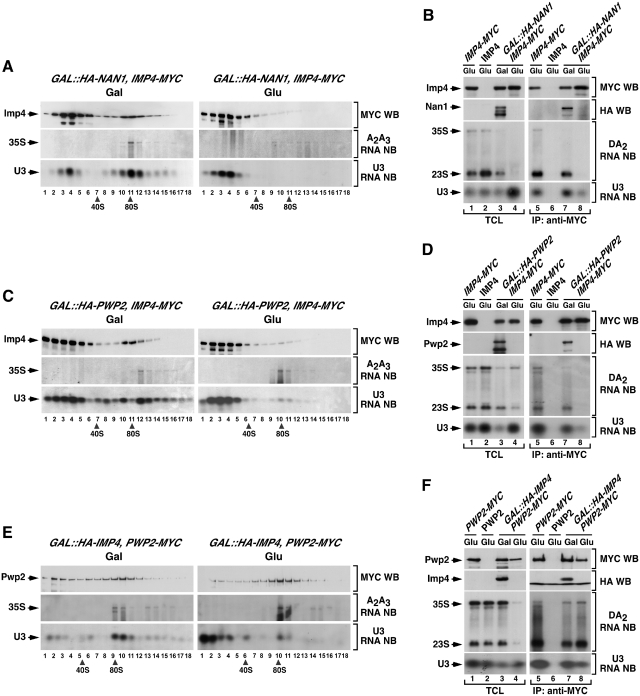
To study the order of assembly of Imp4, we first analyzed whether its incorporation onto the 35S pre-rRNA took place concomitantly or upon the docking of the tUTP or UTP-B/U3 snoRNP complexes. In the absence of Nan1, a structural component of the tUTP subunit, we observed that Imp4–MYC displayed ≃15–20S sedimentation coefficients, the expected size for free Mpp10–Imp4–Imp3 complexes (Figure 5A, compare upper panels on the right and left). In agreement with this, we observed that the Nan1 deficiency induced the dissociation of Imp4–MYC from 35S pre-rRNA (Figure 5B, third panel from the top on the right), the 23S pre-rRNA (Figure 5B, third panel from the top on the right) and the U3 snoRNA (Figure 5B, bottom panel on the right). Identical results were observed in cells lacking Pwp2, an UTP-B component (Figure 5C and D), indicating that the assembly of Imp4–MYC onto the 90S pre-ribosomal particle is both tUTP- and UTP-B dependent. This is consistent with previous results indicating that Mpp10, another component of the Mpp10–Imp3–Imp4 subunit, binds the primary pre-rRNA in a Pwp2- and U3-snoRNA-dependent manner (19).
Figure 5.
The recruitment of Imp4 to early pre-ribosomes is secondary to the assembly of UTP-B/U3. (A, C and E) Sedimentation behavior in sucrose gradients of Imp4 (A and C) and Pwp2 (E) in the presence and absence of Nan1 (A), Pwp2 (C) and Imp4 (E). Cellular extracts prepared from the indicated yeast strains (top) grown in medium containing either galactose (left panels) or glucose (right panels) were resolved in 7–50% linear sucrose gradients. After ultracentrifugation, 0.5 ml fractions were collected from the top of the tube. The content of Imp4–MYC (A and C) and Pwp2–MYC (E) in each fraction was analyzed by anti-MYC immunoblotting (first panels from top). In parallel, total RNAs were prepared from each fraction and analyzed by northern blot as indicated in Figure 2. The number of each fraction and the sedimentation points of 40S and 80S particles are indicated at the bottom. Gal, galactose; Glu, glucose; WB, western blot; NB, northern blot. (B, D and F) Co-immunoprecipitation of Imp4 (B and D) and Pwp2 (F) with pre-RNAs and U3 snoRNA in the presence and absence of Nan1 (B), Pwp2 (D) and Imp4 (F). Total cellular lysates (lanes 1–4) and anti-MYC immunoprecipitates (lanes 5–8) prepared from the indicated yeast strains and growth conditions (top) were analyzed by anti-MYC and anti-HA immunoblotting (upper panels), and northern blot analysis as described in Figure 2. TCL, total cellular lysates; IP, immunoprecipitation.
We next investigated whether Imp4 was required for the assembly of the UTP-B subunit onto the 35S pre-rRNA using a GAL::HA-IMP4;PWP2-MYC strain. In our sucrose gradient sedimentation and co-immunoprecipitation analysis, we observed that Pwp2 kept wild-type-like sedimentation profiles (Figure 5E, compare upper left and right panels) and could associate with the 35S and 23S pre-rRNAs (Figure 5F, third panel from top on the right) in the absence of Imp4. Confirming the effective depletion of Imp4 in these experiments, additional controls demonstrated that Imp4-depleted cells exhibited the previously reported defects in A0–A2 pre-rRNA processing and in 40S subunit production (data not shown). These results indicate that the recruitment of the UTP-B subunit onto the pre-rRNA is independent of Imp4. In contrast to this, our sucrose gradient fractionation studies indicated that the loss of Imp4 induced a partial elimination of the U3 snoRNA from 90S fractions (Figure 5E, compare bottom panels, fractions 9–11) and a concomitant accumulation of this snoRNA in the top fractions of the sucrose gradients (Figure 5E, compare bottom panels, fractions 1–2). Likewise, we observed using co-immunoprecipitation and northern blot analyses that the levels of association of Pwp2 with the U3 snoRNA were lower in Imp4-deficient than in control cells (Figure 5F, bottom panel on the right). Because the assembly of the UTP-B component Pwp2, which depends on the U3 snoRNP (19), is not affected in Imp4-depleted cells (Figure 5E and F), we surmise that the reduced Pwp2/U3 snoRNA association is not due to the inefficient incorporation of the U3 snoRNP onto the 90S pre-ribosomal particle. Instead, it must derive from a reduction in the affinity between U3 snoRNA and UTP-B when they are in the context of a partially assembled pre-ribosome that lacks Imp4 (see ‘Discussion’ section).
The comparison of the Imp4-associated proteome in wild type, Rrp5-, Nan1- and Pwp2-depleted cells confirmed the assembly hierarchy of Imp4 relative to the aforementioned proteins and, at the same time, allowed the identification of its closest interacting neighbors both inside and outside the pre-ribosomal particles. We detected the association of Imp4–MYC with many 90S particle proteins, including components of the tUTP and UTP-B subunits in both wild type and Rrp5-deficient cells (Figure 4; columns 5 and 6, respectively). However, we found that Imp4–MYC associated exclusively with the other two components of its subunit, Mpp10 and Imp3, in Nan1- (Figure 4, column 7; Supplementary Figure S3G) and Pwp2-deficient (Figure 4, column 8; Supplementary Figure S3H) cells. Based on all these findings, we conclude that Imp4 is recruited to the nascent pre-ribosomal particle in the context of the Mpp10–Imp3–Imp4 complex after the UTP-B subunit has already been docked onto the particle.
The recruitment of Bms1 to early pre-ribosomes occurs downstream of the assembly of the UTP-B/U3 snoRNP complex
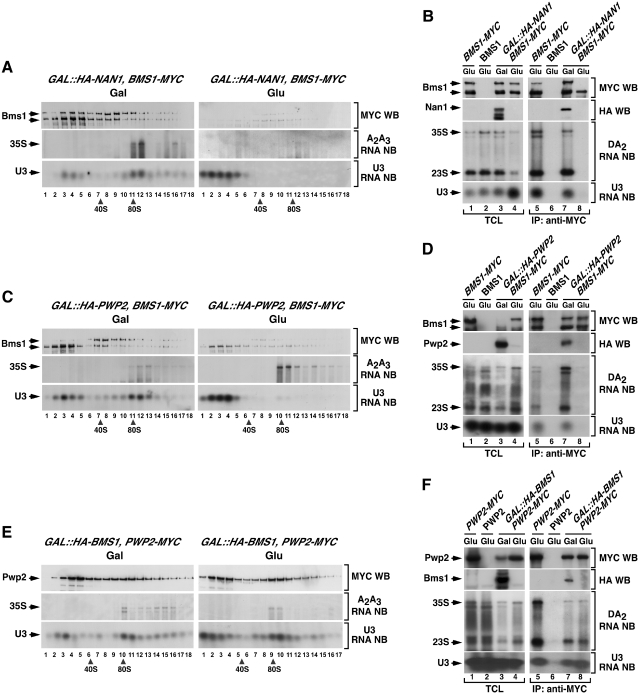
We next investigated the assembly mechanism of the GTPase Bms1 onto nascent pre-ribosomes. Using the same strategy used with Utp20 and Imp4, we found that the depletion of either Nan1 or Pwp2 induced a loss of Bms1–MYC from the high molecular weight ≃90S fractions of sucrose gradients (Figure 6A and C). The disappearance of Bms1–MYC from large complexes was not accompanied by an accumulation of the protein in the upper regions of the gradients (Figure 6A and C). This was probably due to the low stability of free Bms1 in the gradient sedimentation experiments. In the case of Nan1-depleted cells, the loss of Bms1–MYC from pre-ribosomal particles could not be confirmed with co-immunoprecipitation analysis, because the recovery of intact protein by immunoprecipitation was very low (Figure 6B, lane 8). In the case of Pwp2-depleted cells, the co-immunoprecipitation assays did confirm the results from the sucrose gradient analyses and showed that Bms1–MYC is not stably associated with the 35S pre-rRNA and the U3 snoRNA in the absence of Pwp2 (Figure 6D). Together, these findings indicate that Bms1 depends on tUTP and UTP-B for its docking onto 90S pre-ribosomes. Our experiments also indicated that Bms1 is totally dispensable for the assembly of both the UTP-B subnit and the U3 snoRNP onto the nascent pre-ribosomal particle (Figure 6E and F). Consistent with the above assembly hierarchy, proteomics analyses showed that Bms1–MYC could form stable complexes in wild-type and Rrp5-deficient cells with a wide collection of 90S particle proteins, including U3 snoRNP components as well as tUTP, UTP-B and Mpp10–Imp3–Imp4 subunit elements (Figure 4, columns 9 and 10, respectively). In contrast, such interactions were not detected when Bms1–MYC was purified from cell lysates derived from Nan1- and Pwp2-deficient cells (Figure 4, columns 11 and 12, respectively). Under the latter purification conditions, we could only detect a stable interaction of the Bms1–MYC bait with Rcl1, Nop1 and a reduced number of ribosomal proteins. These results suggest that, as previously described in wild-type cells (22), there is a pool of free Bms1 protein that is complexed to Rcl1.
Figure 6.
The recruitment of Bms1 to nascent pre-ribosomes is secondary to the assembly of UTP-B/U3. (A, C and E) Sedimentation behavior in sucrose gradients of Bms1 (A and C) and Pwp2 (E) in the presence and absence of Nan1 (A), Pwp2 (C) and Bms1 (E). Cellular extracts prepared from the indicated yeast strains (top) grown in medium containing either galactose (left panels) or glucose (right panels) were resolved in 7–50% linear sucrose gradients. After ultracentrifugation, 0.5 ml fractions were collected from the top of the tube. The content of Bms1–MYC (A, C) and Pwp2–MYC (E) in each fraction was analyzed by anti-MYC immunoblotting (first panels from top). In parallel, total RNAs were prepared from each fraction and analyzed by northern blot as indicated in Figure 2. The number of each fraction and the sedimentation points of 40S and 80S particles are indicated at the bottom. Gal, galactose; Glu, glucose; WB, western blot; NB, northern blot. (B, D and F) Co-immunoprecipitation of Bms1 (B and D) and Pwp2 (F) with pre-RNAs and U3 snoRNA in the presence and absence of Nan1 (B), Pwp2 (D) and Bms1 (F). Total cellular lysates (lanes 1–4) and anti-MYC immunoprecipitates (lanes 5–8) prepared from the indicated yeast strains and growth conditions (top) were analyzed by anti-MYC immunoblotting (upper panels), and northern blot analysis as described in Figure 2. TCL, total cellular lysates; IP, immunoprecipitation.
Bms1, but not Utp20 or Imp4, plays a relevant role in the overall building of the mature 90S pre-ribosome
In order to further characterize the role of these proteins in the context of the formation of the 90S pre-ribosomal particle, we decided to investigate whether this second assembly wave entailed interdependencies among Utp20, Imp4 and Bms1. We hypothesized that if their assembly was interdependent, the protein composition of the pre-ribosomal complexes formed in the absence of each of those three proteins had to be very similar. In contrast, such composition should be different if they were assembling in either an independent- or hierarchically determined manner. To investigate this issue, we compared the interactome of Pwp2–MYC, a UTP-B subunit element, in wild-type, Utp20-, Imp4- and Bms1-deficient cells. For comparison, we also characterized the Pwp2 interactome of Rrp5-depleted cells. We chose Pwp2 for these analyses because previous studies have shown that this protein is an excellent bait for the efficient isolation of 90S pre-ribosome structural intermediates (14,18). Since the yield of recovery of some proteins in pre-ribosome intermediates is quite variable, we decided to focus our attention on the detection of a subset of 90S particle components that are tightly associated to Pwp2 in both wild-type and Rrp5-depleted cells (18). Those proteins, which will be referred to hereafter as the ‘Pwp2 signature’, include five components of the tUTP subunit (Utp10, Utp4, Utp8, Utp9, Utp15), four components of the UTP-B subunit (Dip2, Utp13, Utp18, Utp6), a component of the Mpp10 subcomplex (Mpp10) and the proteins Rrp5, Utp20, Bms1, Kre33 and Enp2 (Figure 7, column 2).
Using this experimental strategy, we observed that the Pwp2 interactome exhibited variations in its complexity depending on the presence or absence of the three proteins under study. In the case of Imp4-depleted cells, the Pwp2 interactome contained 14 out of the 15 elements of the Pwp2 signature (Figure 7, column 4; Supplementary Figure S4B). In Bms1-depleted cells, the Pwp2 interactome was significantly reduced, since it kept all UTP subunit elements but lost all the components of the Pwp2 signature that did not belong to UTP subunits with the only exception of Rrp5 (Figure 7, compare column 5 with columns 2 and 3; Supplementary Figure S4A–C). In Utp20-deficient cells, the Pwp2 interactome was composed of UTB subunit elements and some (Rrp5, Bms1, Kre33), but not all, of the proteins that are not stably associated to such subunits.
To further confirm these observations, we purified pre-ribosome complexes using as bait Rrp7, an UTP-C subunit protein. As in the case of the experiments with Pwp2, we observed that the Rrp7 interactome was significantly simpler in Bms1-depleted cells (Figure 7, column 11; Supplementary Figure S4F) than in Utp20- (Figure 7, column 10; Supplementary Figure S4E) or Imp4-deficient (Figure 7, column 11; Supplementary Figure S4D) cells. In fact, Rrp7 was quite inefficient in pulling down tUTP and UTP-B components in the absence of Bms1.
Importantly, these experiments also revealed that the assembly of Bms1 onto the pre-ribosomal particle does not depend on Utp20 or Imp4 for its binding to the pre-rRNA, as demonstrated by the detection of Bms1 in the Pwp2–MYC complexes obtained from both Utp20- (Figure 7, column 3) and Imp4-deficient (Figure 7, column 4) cells. Instead, Utp20 and Mpp10–Imp3–Imp4 subunit proteins do depend on Bms1 for their stable interaction with Pwp2–MYC (Figure 7, column 5).
Taken together, these results indicate that: (i) Utp20, Imp4 and Bms1 are not recruited to nascent pre-ribosomes through a common cooperative step. (ii) Bms1, but not Utp20 or Imp4, is required for the assembly of a significant number of proteins onto the initial pre-ribosomal core. (iii) The docking of Bms1 onto nascent pre-ribosomes takes place before, and is a conditio sine qua non for the incorporation of Utp20 and the Mpp10–Imp3–Imp4 subcomplex. (iv) Bms1 contributes to the stability of the UTP-B/UTP-C interaction within the nascent pre-ribosomal particle.
DISCUSSION
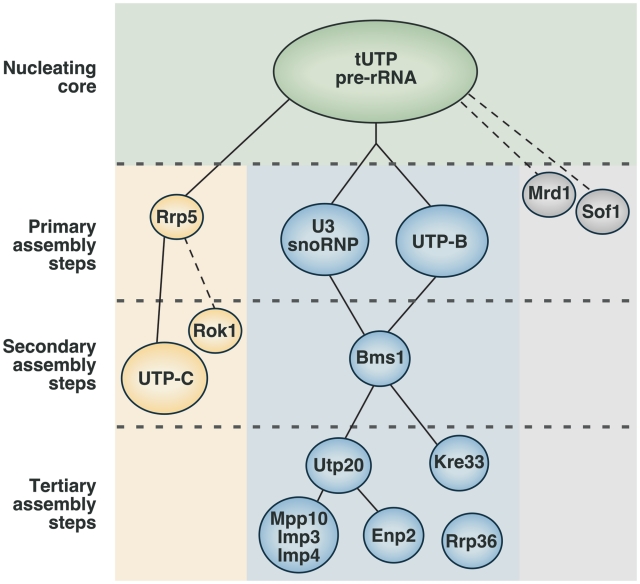
The results presented here, in combination with those previously published by us and other groups (15,18,19,25), are consistent with a model for the construction of the 90S pre-ribosomal particle that relies on the addition of UTP subunits and satellite proteins in a highly hierarchical manner (Figure 8). According to this model, the formation of the 90S pre-ribosomal particle is initiated by the incorporation of the tUTP subunit onto the 35S pre-RNA, and the subsequent, and tUTP-dependent, docking of additional proteins and subunits in two different assembly branches. The growth of one of these assembly branches requires the sequential incorporation of Rrp5 and the UTP-C subunit. The growth of the other assembly branch takes place as the UTP-B subunit and the U3 snoRNP are co-assembled onto the nascent pre-ribosomal particle. A number of experimental observations indicate that this multiunit structure, formed by the initial pre-rRNA, tUTP, U3snoRNP, UTP-B and Rrp5, is the primary structural core from which the 90S pre-ribosomal particle is built upon. Thus, we have shown that those subunits can stably bind to the 35S pre-rRNA in the context of intermediate pre-ribosomal assemblies (18). Furthermore, the presence of those units is essential for the recruitment of a large plethora of 90S particle constituents. In contrast, the elimination of other components of the mature 90S pre-ribosomal particle such as Utp20, Imp4 or Bms1 does not cause a deleterious effect in the docking of the primary subunits onto the 35S pre-rRNA.
Figure 8.
Assembly hierarchy model of the 90S pre-ribosome. The binding of the tUTP subunit initiates the formation of the 90S particle and is required for the subsequent loading of the rest of the components. Two separate, and mutually independent, primary assembly steps have been identified. One of the steps involves the assembly of the U3 snoRNP and UTP-B subunits, and is required for the recruitment of at least 20 components of the particle, including the GTPase Bms1. The other primary assembly step is the binding of Rrp5, which is required for the subsequent incorporation of the UTP-C subunit. Bms1 is required for a secondary assembly step that drives the assembly of several proteins, including Utp20, Enp2, Kre33 and the Mpp10 subcomplex. This hierarchy of interactions has been established from this work and previous studies that analyzed the interdependence between factors for their binding to the 35S pre-rRNA, and the composition of pre-ribosomal complexes formed in the absence of specific proteins (18,19,25). In addition, available evidence indicates that Mrd1 and Sof1 might be primary assembly factors (37,38), and that Rok1 and Rrp36 are recruited through the Rrp5- and the UTP-B/U3-dependent branches, respectively (39,40). Rcl1, a protein that forms a subcomplex with Bms1, might be recruited to early pre-ribosomes together with Bms1. However, there is still not enough evidence to place the Rcl1–Bms1 subunit in this hierarchical model. Dashed lines indicate the possibility of intermediate assembly steps. See ‘Discussion’ section for more details about this model.
Furthermore, our data indicate that the maturation of the 90S pre-ribosomal particle takes place in a rather asymmetric manner (Figure 8). Consistent with this, we have shown that a large number of the 90S particle components (≃20) are incorporated only through the U3 snoRNP/UTP-B subunit assembly branch and independently of the Rrp5/UTP-C branch. In addition, we have demonstrated that proteins that do not belong to the primary subunits incorporate to the nascent pre-ribosomal particle through secondary and tertiary assembly steps that are influenced by specific factors. We have shown that a second-order nucleator is the GTPase Bms1, since its presence within the growing pre-ribosomal particle is critical to ensure the incorporation of a considerable number of pre-ribosomal proteins including the Mpp10–Imp3–Imp4 subcomplex, Utp20, Kre33 and Enp2. Likewise, Utp20 may play a similar role in a third wave of assembly, because we have seen that the pre-ribosomal particle lacks some of the satellite proteins (Mpp10–Imp3–Imp4, Enp2) in Utp20-deficient cells. With the exception of the Mpp10–Imp3–Imp4 complex, it seems that most of the proteins that do not belong to the primary scaffold incorporate into the particle as single proteins or very small heteromolecular complexes rather than as UTP-like subunits.
Our data also suggest that the incorporation of new proteins is accompanied by significant changes in the structure of nascent pre-ribosomal particles that greatly modify the interactions between their components. For example, we have observed that Imp4 (and therefore, the Mpp10–Imp3–Imp4 subunit) is required for enhancing the stability of the U3 snoRNP/Pwp2 complex within the 90S particle core. Likewise, the incorporation of the second Bms1-dependent assembly layer results in an increased stability of the association between the UTP-B and UTP-C subunits within the particle. We have also seen in our proteomics experiments that the strength of the association of Utp20 with other 90S pre-ribosomal proteins decreases significantly between the intermediate steps of the assembly and the fully mature particle. These results indicate that some of the more peripheral proteins/subunits might induce conformational rearrangements that alter the interactions between the two assembly branches to form a particle that is competent for cleaving the pre-rRNA at sites A0–A2.
Our results have also shed some light on the still obscure properties of the proteins under study in this work. Based on in vitro observations indicating that the Mpp10–Imp3–Imp4 subunit can induce structural rearrangements in the U3 snoRNA that alter its base pairing with the 35S pre-rRNA (26,27), it was previously hypothesized that this subunit could act as a regulatory element that controls the formation or maintenance of U3 snoRNA/pre-rRNA interactions. Despite those data, direct evidence for the actual function of this subcomplex was lacking, and it was unclear whether Mpp10–Imp3–Imp4 was actually important for the initial docking of the U3 snoRNP onto the pre-rRNA or, alternatively, whether it played roles in the ensuing U3 molecular rearrangements that take place upon its annealing to the pre-rRNA precursor. Our results have shown that the presence of the Mpp10–Imp3–Imp4 subunit is not required for the efficient building of large and stable pre-ribosomal complexes that contain U3 snoRNP, tUTP proteins, UTP-B elements and many other known 90S particle components such as Rrp5, Utp20, Bms1, Kre33, Enp2, Has1, Cbf5 and Rlp7. These data indicate that the Mpp10–Imp3–Imp4 subcomplex does not participate in the recruitment of the bulk of 90S particle transacting factors, including U3 snoRNP, to the 35S pre-rRNA. Therefore, if the Mpp10–Imp3–Imp4 subunit exerts an influence on the U3-pre-rRNA interaction, it must do so once the U3 snoRNP and the rest of proteins of the ‘core’ and the downstream Bms1-dependent layer of proteins have already assembled onto the pre-rRNA. In agreement with this mechanistic model, we have demonstrated here that the Mpp10–Imp3–Imp4 subunit is recruited at a hierarchically late stage during the formation of the 90S particle. Our observation that the U3 snoRNP has a higher tendency to dissociate from the pre-ribosomal complexes formed in the absence of Mpp10–Imp3–Imp4 than from smaller less-complex pre-ribosome assemblies, such as those formed in the absence of Bms1 or Utp20, also suggests that Mpp10–Imp3–Imp4 influences the U3–pre-RNA interaction at a late assembly step. Previous studies in human cells showing that the binding of Mpp10 to U3 is dispensable for its docking onto the pre-rRNA are also fully consistent with this model (35).
Bms1 is the only GTPase present in 90S pre-ribosomes and, therefore, it has been considered an obvious candidate to regulate their assembly or activation. In this regard, previous studies have shown that Bms1 can form a small complex with Rcl1, a protein structurally similar to RNA 3′ cyclases that is essential for the cleavage of the 35S pre-rRNA precursor at the A0, A1 and A2 sites (22,29,36). Furthermore, it was proposed that Bms1 could be implicated in the delivery of Rcl1 to pre-ribosomes in a GTP-dependent manner (30,31). Despite this evidence, the actual role of Bms1 and Rcl1 within the 90S particle has remained unclear. We observed that Bms1 is loaded onto the 35S pre-rRNA only after the incorporation of the ‘core’ components but before the docking of other satellite components such as the Mpp10–Imp3–Imp4 subunit, Utp20, Kre33 and Enp2. These findings strongly indicate that instead of initiating the assembly of 90S pre-ribosomes, Bms1 must be implicated in a secondary assembly step required for the recruitment of a significant number of 90S particle components. Interestingly, electron microscopy analysis of chromatin spreads have previously revealed the initial formation of small 5′ terminal knobs on nascent pre-rRNA transcripts that then undergo a one-step collapse into a large particle, presumably mature 90S pre-ribosomes, when transcription of the 18S rRNA is almost complete (2). Whether such event involves the secondary Bms1-mediated assembly step described here is an interesting possibility to be addressed in the future. It also remains to be characterized the specific role that Rcl1 might play in the whole assembly process of the 90S particle.
Although we have not mapped the assembly hierarchy of the whole proteome of the 90S particle, we can use evidence derived from other publications to make inferences about the mode of recruitment of some other components not studied in our work. For example, it has been shown that Mrd1 and Sof1 dock onto the 35S pre-rRNA in both an Rrp5- and U3 snoRNP-independent manner (37,38), suggesting that they might be recruited through other primary assembly steps (Figure 8). Likewise, it is known that the Rrp36 and the Rok1 helicase are recruited to pre-ribosomal complexes through the UTP-B/U3 snoRNP and the Rrp5/UTP-C assembly route, respectively (39,40) (Figure 8). Some information about the possible order of recruitment of many other 90S particle factors can be inferred from their presence or absence in different partially assembled complexes. However, detailed validation studies, like the one described here, will be required to establish at which stage they actually incorporate onto nascent pre-ribosomal complexes involved in the biogenesis of the small ribosomal subunits.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Spanish Ministry of Science and Innovation (MICINN) (BFU-2008-02729 and RD06/0020/0001); Castilla-León Autonomous Government (GR95); Samuel Solórzano Barruso Foundation (FS/2-2009). MICINN funding is co-sponsored by the European Union FEDER Program. Funding for open access charge: Spanish Ministry of Science and Innovation (MICINN) (BFU-2008-02729).
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
We thank M.Blázquez for expert technical assistance and the personnel of CIC Proteomics Facility for their excellent work in protein complex characterization. We would like to specially thank Xosé R. Bustelo for fruitful discussions and support.
REFERENCES
- 1.Venema J, Tollervey D. Ribosome synthesis in Saccharomyces cerevisiae. Annu. Rev. Genet. 1999;33:261–311. doi: 10.1146/annurev.genet.33.1.261. [DOI] [PubMed] [Google Scholar]
- 2.Osheim YN, French SL, Keck KM, Champion EA, Spasov K, Dragon F, Baserga SJ, Beyer AL. Pre-18S ribosomal RNA is structurally compacted into the SSU processome prior to being cleaved from nascent transcripts in Saccharomyces cerevisiae. Mol. Cell. 2004;16:943–954. doi: 10.1016/j.molcel.2004.11.031. [DOI] [PubMed] [Google Scholar]
- 3.Kos M, Tollervey D. Yeast pre-rRNA processing and modification occur cotranscriptionally. Mol. Cell. 37:809–820. doi: 10.1016/j.molcel.2010.02.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fatica A, Tollervey D. Making ribosomes. Curr. Opin. Cell Biol. 2002;14:313–318. doi: 10.1016/s0955-0674(02)00336-8. [DOI] [PubMed] [Google Scholar]
- 5.Tschochner H, Hurt E. Pre-ribosomes on the road from the nucleolus to the cytoplasm. Trends Cell Biol. 2003;13:255–263. doi: 10.1016/s0962-8924(03)00054-0. [DOI] [PubMed] [Google Scholar]
- 6.Fromont-Racine M, Senger B, Saveanu C, Fasiolo F. Ribosome assembly in eukaryotes. Gene. 2003;313:17–42. doi: 10.1016/s0378-1119(03)00629-2. [DOI] [PubMed] [Google Scholar]
- 7.Henras AK, Soudet J, Gerus M, Lebaron S, Caizergues-Ferrer M, Mougin A, Henry Y. The post-transcriptional steps of eukaryotic ribosome biogenesis. Cell Mol. Life Sci. 2008;65:2334–2359. doi: 10.1007/s00018-008-8027-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Staley JP, Woolford JL., Jr Assembly of ribosomes and spliceosomes: complex ribonucleoprotein machines. Curr. Opin. Cell Biol. 2009;21:109–118. doi: 10.1016/j.ceb.2009.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kressler D, Hurt E, Bassler J. Driving ribosome assembly. Biochim. Biophys. Acta. 2010;1803:673–683. doi: 10.1016/j.bbamcr.2009.10.009. [DOI] [PubMed] [Google Scholar]
- 10.Phipps KR, Charette JM, Baserga SJ. The SSU processome in ribosome biogenesis - progress and prospects. WIREs RNA. 2:1–21. doi: 10.1002/wrna.57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Panse VG, Johnson AW. Maturation of eukaryotic ribosomes: acquisition of functionality. Trends Biochem. Sci. 35:260–266. doi: 10.1016/j.tibs.2010.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bernstein KA, Gallagher JE, Mitchell BM, Granneman S, Baserga SJ. The small-subunit processome is a ribosome assembly intermediate. Eukaryot. Cell. 2004;3:1619–1626. doi: 10.1128/EC.3.6.1619-1626.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dragon F, Gallagher JE, Compagnone-Post PA, Mitchell BM, Porwancher KA, Wehner KA, Wormsley S, Settlage RE, Shabanowitz J, Osheim Y, et al. A large nucleolar U3 ribonucleoprotein required for 18S ribosomal RNA biogenesis. Nature. 2002;417:967–970. doi: 10.1038/nature00769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Grandi P, Rybin V, Bassler J, Petfalski E, Strauss D, Marzioch M, Schafer T, Kuster B, Tschochner H, Tollervey D, et al. 90S pre-ribosomes include the 35S pre-rRNA, the U3 snoRNP, and 40S subunit processing factors but predominantly lack 60S synthesis factors. Mol. Cell. 2002;10:105–115. doi: 10.1016/s1097-2765(02)00579-8. [DOI] [PubMed] [Google Scholar]
- 15.Krogan NJ, Peng WT, Cagney G, Robinson MD, Haw R, Zhong G, Guo X, Zhang X, Canadien V, Richards DP, et al. High-definition macromolecular composition of yeast RNA-processing complexes. Mol. Cell. 2004;13:225–239. doi: 10.1016/s1097-2765(04)00003-6. [DOI] [PubMed] [Google Scholar]
- 16.Schafer T, Strauss D, Petfalski E, Tollervey D, Hurt E. The path from nucleolar 90S to cytoplasmic 40S pre-ribosomes. EMBO J. 2003;22:1370–1380. doi: 10.1093/emboj/cdg121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Watkins NJ, Segault V, Charpentier B, Nottrott S, Fabrizio P, Bachi A, Wilm M, Rosbash M, Branlant C, Luhrmann R. A common core RNP structure shared between the small nucleoar box C/D RNPs and the spliceosomal U4 snRNP. Cell. 2000;103:457–466. doi: 10.1016/s0092-8674(00)00137-9. [DOI] [PubMed] [Google Scholar]
- 18.Perez-Fernandez J, Roman A, De Las Rivas J, Bustelo XR, Dosil M. The 90S preribosome is a multimodular structure that is assembled through a hierarchical mechanism. Mol. Cell. Biol. 2007;27:5414–5429. doi: 10.1128/MCB.00380-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dosil M, Bustelo XR. Functional characterization of Pwp2, a WD family protein essential for the assembly of the 90 S pre-ribosomal particle. J. Biol. Chem. 2004;279:37385–37397. doi: 10.1074/jbc.M404909200. [DOI] [PubMed] [Google Scholar]
- 20.Wehner KA, Gallagher JE, Baserga SJ. Components of an interdependent unit within the SSU processome regulate and mediate its activity. Mol. Cell. Biol. 2002;22:7258–7267. doi: 10.1128/MCB.22.20.7258-7267.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Granneman S, Gallagher JE, Vogelzangs J, Horstman W, van Venrooij WJ, Baserga SJ, Pruijn GJ. The human Imp3 and Imp4 proteins form a ternary complex with hMpp10, which only interacts with the U3 snoRNA in 60-80S ribonucleoprotein complexes. Nucleic Acids Res. 2003;31:1877–1887. doi: 10.1093/nar/gkg300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wegierski T, Billy E, Nasr F, Filipowicz W. Bms1p, a G-domain-containing protein, associates with Rcl1p and is required for 18S rRNA biogenesis in yeast. RNA. 2001;7:1254–1267. doi: 10.1017/s1355838201012079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rudra D, Mallick J, Zhao Y, Warner JR. Potential interface between ribosomal protein production and pre-rRNA processing. Mol. Cell. Biol. 2007;27:4815–4824. doi: 10.1128/MCB.02062-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kuhn H, Hierlmeier T, Merl J, Jakob S, Aguissa-Toure AH, Milkereit P, Tschochner H. The Noc-domain containing C-terminus of Noc4p mediates both formation of the Noc4p-Nop14p submodule and its incorporation into the SSU processome. PloS One. 2009;4:e8370. doi: 10.1371/journal.pone.0008370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gallagher JE, Dunbar DA, Granneman S, Mitchell BM, Osheim Y, Beyer AL, Baserga SJ. RNA polymerase I transcription and pre-rRNA processing are linked by specific SSU processome components. Genes Dev. 2004;18:2506–2517. doi: 10.1101/gad.1226604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gerczei T, Shah BN, Manzo AJ, Walter NG, Correll CC. RNA chaperones stimulate formation and yield of the U3 snoRNA-Pre-rRNA duplexes needed for eukaryotic ribosome biogenesis. J. Mol. Biol. 2009;390:991–1006. doi: 10.1016/j.jmb.2009.05.072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gerczei T, Correll CC. Imp3p and Imp4p mediate formation of essential U3-precursor rRNA (pre-rRNA) duplexes, possibly to recruit the small subunit processome to the pre-rRNA. Proc. Natl Acad. Sci. USA. 2004;101:15301–15306. doi: 10.1073/pnas.0406819101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gelperin D, Horton L, Beckman J, Hensold J, Lemmon SK. Bms1p, a novel GTP-binding protein, and the related Tsr1p are required for distinct steps of 40S ribosome biogenesis in yeast. RNA. 2001;7:1268–1283. doi: 10.1017/s1355838201013073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tanaka N, Smith P, Shuman S. Crystal structure of Rcl1, an essential component of the eukaryal pre-rRNA processosome implicated in 18s rRNA biogenesis. RNA. 17:595–602. doi: 10.1261/rna.2571811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Karbstein K, Jonas S, Doudna JA. An essential GTPase promotes assembly of preribosomal RNA processing complexes. Mol. Cell. 2005;20:633–643. doi: 10.1016/j.molcel.2005.09.017. [DOI] [PubMed] [Google Scholar]
- 31.Karbstein K, Doudna JA. GTP-dependent formation of a ribonucleoprotein subcomplex required for ribosome biogenesis. J. Mol. Biol. 2006;356:432–443. doi: 10.1016/j.jmb.2005.11.052. [DOI] [PubMed] [Google Scholar]
- 32.Dez C, Dlakic M, Tollervey D. Roles of the HEAT repeat proteins Utp10 and Utp20 in 40S ribosome maturation. RNA. 2007;13:1516–1527. doi: 10.1261/rna.609807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ausubel FM, Brent R, Kingston RE, Moore DD, Seidman JG, Smith JA, Struhl K. Current Protocols in Molecular Biology. New York: John Wiley & Sons, Inc.; 1994. [Google Scholar]
- 34.Venema J, Tollervey D. RRP5 is required for formation of both 18S and 5.8S rRNA in yeast. EMBO J. 1996;15:5701–5714. [PMC free article] [PubMed] [Google Scholar]
- 35.Granneman S, Vogelzangs J, Luhrmann R, van Venrooij WJ, Pruijn GJ, Watkins NJ. Role of pre-rRNA base pairing and 80S complex formation in subnucleolar localization of the U3 snoRNP. Mol. Cell. Biol. 2004;24:8600–8610. doi: 10.1128/MCB.24.19.8600-8610.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Billy E, Wegierski T, Nasr F, Filipowicz W. Rcl1p, the yeast protein similar to the RNA 3′-phosphate cyclase, associates with U3 snoRNP and is required for 18S rRNA biogenesis. EMBO J. 2000;19:2115–2126. doi: 10.1093/emboj/19.9.2115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bax R, Vos HR, Raue HA, Vos JC. Saccharomyces cerevisiae Sof1p associates with 35S Pre-rRNA independent from U3 snoRNA and Rrp5p. Eukaryot. Cell. 2006;5:427–434. doi: 10.1128/EC.5.3.427-434.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Segerstolpe A, Lundkvist P, Osheim YN, Beyer AL, Wieslander L. Mrd1p binds to pre-rRNA early during transcription independent of U3 snoRNA and is required for compaction of the pre-rRNA into small subunit processomes. Nucleic Acids Res. 2008;36:4364–4380. doi: 10.1093/nar/gkn384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Vos HR, Bax R, Faber AW, Vos JC, Raue HA. U3 snoRNP and Rrp5p associate independently with Saccharomyces cerevisiae 35S pre-rRNA, but Rrp5p is essential for association of Rok1p. Nucleic Acids Res. 2004;32:5827–5833. doi: 10.1093/nar/gkh904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gerus M, Bonnart C, Caizergues-Ferrer M, Henry Y, Henras AK. Evolutionarily conserved function of RRP36 in early cleavages of the pre-rRNA and production of the 40S ribosomal subunit. Mol. Cell. Biol. 30:1130–1144. doi: 10.1128/MCB.00999-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.