Abstract
Pentatricopeptide repeat (PPR) proteins are particularly numerous in plant mitochondria and chloroplasts, where they are involved in different steps of RNA metabolism, probably due to the repeated 35 amino acid PPR motifs that are thought to mediate interactions with RNA. In non-photosynthetic eukaryotes only a handful of PPR proteins exist, for example the human LRPPRC, which is involved in a mitochondrial disease. We have conducted a systematic study of the PPR proteins in the fission yeast Schizosaccharomyces pombe and identified, in addition to the mitochondrial RNA polymerase, eight proteins all of which localized to the mitochondria, and showed some association with the membrane. The absence of all but one of these PPR proteins leads to a respiratory deficiency and modified patterns of steady state mt-mRNAs or newly synthesized mitochondrial proteins. Some cause a general defect, whereas others affect specific mitochondrial RNAs, either coding or non-coding: cox1, cox2, cox3, 15S rRNA, atp9 or atp6, sometimes leading to secondary defects. Interestingly, the two possible homologs of LRPPRC, ppr4 and ppr5, play opposite roles in the expression of the cox1 mt-mRNA, ppr4 being the first mRNA-specific translational activator identified in S. pombe, whereas ppr5 appears to be a general negative regulator of mitochondrial translation.
INTRODUCTION
The pentatricopeptide repeat (PPR) protein family is characterized by repeats of a degenerate 35 amino acid sequence, related to the tetratricopeptide repeat (TPR), which is a 34 amino acid motif involved in protein–protein interactions (1). The PPR family is one of the largest protein families known in plants, with over 450 members in Arabidopsis thaliana and even more in other land plants. However, PPR proteins are found in limited numbers in animals and fungi, and are extremely rare in bacteria, they have only been found in a few bacterial pathogens of eukaryotes. It is particularly striking that the bacterial ancestors of mitochondria and chloroplasts lack PPR proteins, because the available experimental data and protein localization predictions suggest that PPR proteins are almost exclusively located in organelles (2). This implies that the current PPR proteins are the result of rapid evolution. Nearly 70% of plant PPR proteins are predicted to be mitochondrial (2), but the majority of PPR proteins studied so far have been chloroplastic, possibly because mitochondrial PPR mutants in plants are often embryo-lethal.
All PPR proteins are thought to bind RNA via the PPR motif. Models of this motif based on the TPR anti-parallel helical structure, suggest that contrary to the TPR motif, the side chains in the central groove of the PPR repeats tend to be hydrophilic and positively charged (1), consistent with an interaction with RNA molecules rather than proteins. In some cases, RNA binding has now been directly demonstrated, and the sequences of the cis-elements recognized by the PPR motif are known (3). However, with the exception of some proteins that contain additional domains involved in RNA editing, PPR proteins do not carry an enzymatic activity and probably act as adaptors that mediate the interaction between their target RNA and other proteins (4). This gives PPR proteins great functional flexibility, and explains how they can participate in all the steps of RNA metabolism from transcription to degradation, affecting diverse processes such as development, organelle biogenesis, energetic regulation and cytoplasmic male sterility [for a review see (5)].
Concerning mitochondrial PPR proteins, one, the human RNA polymerase POLRMT, is clearly required for transcription, although the function of its single PPR motif is unknown (6). Other PPR proteins are involved in post-transcriptional steps. In plants a large number of these proteins are involved in mitochondrial transcript editing [for review see (7) and references therein]. Mitochondrial PPR proteins are also involved in RNA processing: tRNA excision, precursor mRNA cleavage and transcript end maturation e.g. (8–10). This can lead to RNA degradation, for example the targeted degradation of the human mitochondrial leucine tRNA by the PPR protein PTCD1 (11). Many mitochondrial PPR proteins in fungi and animals appear to stabilize both coding and non-coding RNA, e.g. the yeast ATP6/8 co-transcript which is protected by Aep3 (12), and the 12S or 15S rRNAs which are protected from degradation by the human PTCD3 or yeast Dmr1 (13,14). The relationships between transcript stabilization and translation are often very tight, and several factors appear to affect both, but in reality it is often difficult to distinguish between the two. For example in budding yeast, Rmd9 is thought to convey mRNAs to their site of translation and also loosely interacts with the ribosome (15,16), while Pet309 stabilizes the intron-containing cox1 transcript, and activates translation of the mature cox1 mRNA (17). In the case of Pet309 it has been shown that the PPR motifs are not required for the stability function (18). Several other mitochondrial PPR proteins are involved in general translation, and in Trypanosoma, numerous PPR proteins are bona fide ribosomal proteins (19,20).
Given the variety of RNA-linked functions mediated by PPR proteins, it is not surprising that a PPR protein, LRPPRC/LRP130, has been implicated in a human disease. Mutations in LRPPRC are responsible for a French-Canadian Leigh syndrome; however, the precise role of LRPPRC is far from clear. It has been proposed to regulate mitochondrial transcription (21), the stability and/or translation of the cox1 and cox3 mRNAs (22–24) and to be a general regulator of the stability and handling of all human mitochondrial mRNAs (25,26). In addition LRPPRC is present in the nucleus where it seems to play a role in the regulation of gluconeogenesis and mitochondrial biogenesis (27,28), thus contributing to the coordination of nucleo–mitochondrial interactions.
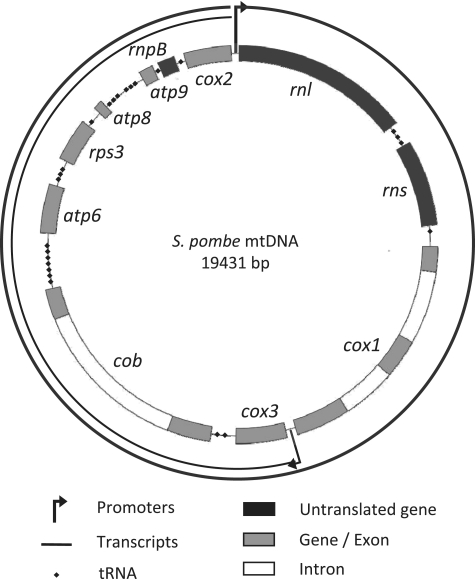
To further our understanding of the functions of the PPR protein family, we have used classical blast searches and the fungal specific SCIPHER algorithm developed by the Golik laboratory (29) to identify all the PPR proteins in Schizosaccharomyces pombe. Apart from the mitochondrial RNA polymerase, we have identified eight proteins, containing three to sixteen predicted PPR motifs, and we have undertaken a study of these proteins. The fission yeast, S. pombe, has several particularities that make it a very attractive model for the study of mitochondrial PPR proteins. First, it is a petite-negative yeast, so most mutants that compromise mitochondrial gene expression maintain their mitochondrial genome, unlike budding yeast, where mutants deficient in mitochondrial gene expression often lose their mitochondrial DNA (mtDNA), making any further study of their function very difficult. Second, the mtDNA of S. pombe resembles that of mammalian cells as it is very compact and uses similar transcription and processing features. Three of the respiratory complexes (III, IV and V) are of dual genetic origin, both nuclear and mitochondrial. In S. pombe, the mitochondrial genome contains eight major protein coding genes (cox1, 2, 3, cytb, atp6, 8, 9, rps3), the two rRNAs rnl and rns, the predicted RNAse P RNA rnpB, and 25 tRNAs (Figure 1). In most strains, only cytb and cox1 contain introns (one and two, respectively), and an intron-less strain has been constructed (30). Efficient transcription of the S. pombe mtDNA requires both the mitochondrial transcription factor (Mft1) and RNA polymerase (Rpo41) (31). The mtDNA contains two principal promoters of different strengths, yielding two overlapping primary transcripts that are generally processed by tRNA excision and cleavage downstream of a CC-rich motif to generate the rRNAs and mRNAs that contain short flanking regions (32). Consequently, S. pombe mitochondrial mRNAs resemble those of human mitochondria, which mostly lack untranslated regions (33), suggesting that they might be stabilized and translated using similar mechanisms and factors.
Figure 1.
Map of the S. pombe mtDNA. The promoter located in front of the large rRNA gene rnl is the major promoter that yields a 19.4 kb transcript corresponding to the full mtDNA. A second minor promoter is located between the cox1 and cox3 genes and drives the transcription of a 10.5 kb RNA containing the genes from cox3 to cox2. The 5′-end of mature RNAs are processed through tRNA punctuation, except for rnl and cox3 whose 5′-ends are determined by the start of transcription. The 3′-end processing occurs at a C-rich motif located a few nucleotides after each open reading frame. However the 3′-end of the RNAse P RNA (rnpB) is produced by tRNA processing and the processing signal of rnl is unknown (32). Genes coding RNAs are depicted in black, protein encoding genes are in gray and introns in white. In our intron-containing strain, cox1 contains two introns and cytb one intron.
Here we present a genome-wide study of all the PPR proteins of a single organism, S. pombe. Previously, nothing was known about the function of these eight PPR proteins. We have shown that they are all mitochondrial proteins and they are involved in mitochondrial biogenesis, although they target different steps in RNA metabolism and have different substrates.
MATERIALS AND METHODS
Strains, plasmids, media and genetic methods
All strains are described in Supplementary Table S1 and were grown at 28 or 36°C as indicated. The different wild-type strains used were NB205-6A (h− ade6-M216 ura4-D18 his3Δ leu1-32 rho+ [three mitochondrial introns], (34), P3 (h+ ade7-50 rho+ [no mitochondrial introns], (30), and the Δpku70 and Δpku80 strains FY14145 and FY14191 (a gift from the National BioResource Program in Japan). Plasmids used or constructed during this work were derivatives of pGEM-T-easy (Promega), pDUAL-FFH1, pDUAL-YFH1 (35) and pTG1754/Not (36). Media and genetic methods were as described in (36,37). Schizosaccharomyces pombe asci were microdissected directly from the mixture of haploid, diploid and sporulating cells. Schizosaccharomyces pombe transformation (38) was improved by (i) using single stranded salmon sperm DNA as carrier, (ii) regenerating cells in complete liquid medium overnight, (iii) plating onto 5% glucose selective medium as described in (39). Yeast genomic DNA was extracted according to (40).
Deletion of the PPR genes
Gene deletions were constructed by the PCR method (41) using pFA6a-kanMX6 (carrying the kanR gene that confers G418 resistance) (42), or pAG25 (carrying the natR gene that confers Nourseotricin resistance) (43). PCR fragments containing the resistance gene were generated using hybrid oligonucleotides containing 75–80 bases of homology with the recipient locus flanking the gene of interest and transformed into NB205-6A, P3 and/or FY14145/FY14191. Transformants, able to grow in the presence of the appropriate drug, were streaked on selective medium, and the genomic DNA of single colonies was analyzed by PCR to look both for the correct insertion of the deletion cassette and the absence of the wild-type sequence. Colonies carrying the expected deletion were back-crossed to a wild-type strain to verify the cosegregation of the resistance marker with the gene deletion and in some cases to segregate away the pku70 or pku80 mutation.
Epitope tagging of the ppr genes
Plasmids containing the different tagged ppr genes under the control of the nmt1 promoter (44) were purchased from the RIKEN consortium and tested for their ability to complement the corresponding mutants; with the exception of ppr1, all tagged genes complemented the gene deletion (data not shown). For the seven ppr plasmids that showed a good complementation, the plasmids were cut by NotI and transformed into the corresponding Δppr mutants to integrate the tagged version into the leu1 locus (44). Sequence analysis of the ppr1 insert obtained from the RIKEN showed that it contained three mutations. Thus the wild-type ppr1 gene together with some other ppr genes were epitope tagged at their chromosomal locus using a PCR strategy (41), and the plasmids pFA6a-3HA-kanMX6, pFA6a-13Myc-KanMX6 (45), and pBS7 (YFP-Venus, NCRR Yeast Resource Centre, University of Washington). After integration, these constructions were verified by PCR amplification of the fusion site and epitope tag, followed by sequencing.
Northern blot analyses
Schizosaccharomyces pombe cells were grown to exponential phase (100 Klett units or 1 OD600) in complete glucose medium, total RNAs were extracted using the hot phenol protocol (46) and run on a formaldehyde gel before transfer onto Hybond-C extra membranes. The blots were successively hybridized with various probes at 65°C under standard saline conditions. After overnight hybridization, the blots were washed briefly several times with 6xSSPE before exposure (from a few hours up to 1 week at −70°C). Mitochondrial probes were PCR or restriction fragments labeled with [α32P]dATP and a random priming kit (Invitrogen).
35S labeling of mitochondrial proteins
Schizosaccharomyces pombe cells were grown to early exponential phase in minimal, or complete, 5% raffinose medium containing 0.1% glucose. Mitochondrial proteins were labeled at 30°C by a 3 h incubation of whole cells with 35S methionine and cysteine (Bioactif-Hartmann) in the presence of cycloheximide (10 mg/ml), which specifically blocks cytoplasmic translation. Proteins were extracted as described in (47) and samples were run on 16% acrylamide—0.5% bisacrylamide SDS gels, after drying, the gel was exposed to a film for 1 day, or up to several weeks, or onto a phosphorimager screen at room temperature.
Purification of mitochondria, alkali treatment and western blotting
Mitochondria were purified from S. pombe cells grown in complete glucose medium as described previously (39). Alkali treatment of isolated membrane and soluble mitochondrial fractions was performed as described in (48). Samples were run on 10 or 12% SDS–PAGE before western blotting. Primary antibodies were: anti-human Hsp60, 1/1000, (Sigma H3524); anti-Saccharomyces cerevisiae Arg8: 1/4000, a gift from T.D. Fox (49); anti-S. pombe Cox2, 1/2500 (50); anti-S. pombe Cytb, 1/1000 (I. Kühl and N. Bonnefoy, unpublished data); anti-Flag, 1/1000 (Sigma F185); anti-HA 1/1000 (Sigma H6908); anti-cMyc 1/5000 (Covance PRB-150C); Secondary antibodies were diluted 1/10 000-fold.
Cytochrome spectra
Low temperature cytochrome spectra of S. pombe cell paste were recorded using a Cary 400 spectrophotometer after addition of sodium dithionite to fully reduce the cytochromes (51). The absorption maxima were 603, 560, 554 and 548 nm for cytochromes aa3, b, c1 and c, respectively. The S. pombe, the cytochrome c peak always shows a 544 nm shoulder that disappears in a cytochrome c mutant (N. Bonnefoy, unpublished data).
RESULTS AND DISCUSSION
Homology searches for S. pombe PPR proteins uncover a nine-member family
In the nucleus, the regulation of transcription is an important factor in the overall control of gene expression; in contrast, the regulation of mitochondrial gene expression appears to occur principally at the post-transcriptional level. Thus it is perhaps surprising that only a few motifs typical of RNA manipulating proteins, such as the PPR motif, have been identified in mitochondrial proteins (2). We have conducted a systematic genome-wide analysis to identify and partially characterize the fission yeast proteins containing PPR motifs.
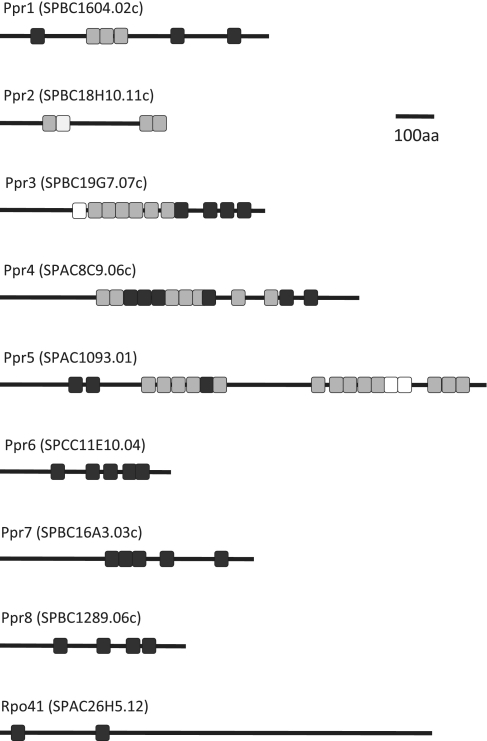
Our initial survey for S. pombe PPR proteins used blast comparisons starting with known fungal or animal PPR proteins, such as Pet309 (17) or LRPPRC (22). Searches were then repeated using the newly identified S. pombe PPR proteins as baits. In this way, we were able to detect eight PPR proteins, which we have named Ppr1 to 8 (Figure 2). In parallel, the SCIPHER algorithm, which was specifically designed by the Golik Laboratory to find fungal PPR motifs (29), was run on the complete S. pombe genome. This identified the same 8 PPR proteins and the mitochondrial RNA polymerase, Rpo41, which like its human counterpart POLRMT, is a PPR protein (6,29) (Figure 2). Thus, as in other fungi and in animals, we found that the fission yeast PPR family is rather small: nine PPR proteins have been identified, amongst which four had not been annotated as PPR proteins in databases such as Uniprot. According to the SCIPHER algorithm, the S. pombe proteins contain between 2 (Rpo41) and 16 (Ppr5) PPR motifs and are between 432 (Ppr2) and 1261 (Ppr5) amino acids long (Table 1).
Figure 2.
The PPR proteins of S. pombe. Gray boxes show the PPR motifs annotated in Uniprot and detected by the SCIPHER algorithm, black boxes show PPR motifs that were only found with the SCIPHER algorithm and white boxes are PPR motifs that are annotated in the UniProt database but were not found using the SCIPHER algorithm. In Ppr4, the boundaries of the predicted motifs eight and nine are unclear, and they may correspond to a single motif. The ninth S. pombe PPR protein is the mitochondrial RNA polymerase.
Table 1.
Summary of the S. pombe Ppr protein features and mutant phenotypes
| Ppr |
|||||||||
|---|---|---|---|---|---|---|---|---|---|
| 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | ||
| Protein features | |||||||||
| Length (amino acid) | 697 | 432 | 687 | 931 | 1261 | 443 | 658 | 481 | |
| No. of PPR motifs | 6 | ≥3 | ≥10 | 13 | ≥16 | 5 | 5 | 4 | |
| Membrane (%) | 30 | 80 | 100 | 100 | 100 | 100 | 50 | 40 | |
| Possible homologs | |||||||||
| Saccharomyces cerevisiae | Dmr1 | Pet309 | a | Aep2 | |||||
| Homo sapiens | PTCD3 | ||||||||
|
Δppr |
|||||||||
|---|---|---|---|---|---|---|---|---|---|
| Δ1 | Δ2 | Δ3 | Δ4 | Δ5 | Δ6 | Δ7 | Δ8 | wt | |
| Phenotype of Δppr mutants | |||||||||
| Growth on galactose | − | ± | − | − | ++++ | − | − | ++ | +++ |
| Growth on antimycin | +++ | + | ++ | + | +++ | − | − | + | + |
| Spectra | |||||||||
| Cytochrome aa3 | − | − | − | − | + | − | − | ± | + |
| Cytochrome bc1 | ± | ± | ± | ± | + | ± | ± | + | + |
| RNAs most affected | cox2 | None | 15S | cox1 | None | atp9 | atp6 | cox1 | None |
| cox3 | cytb | if Σib | if Σib | ||||||
| Proteins most affected | Cox2 | All | All | Cox1 | All up | All esp | All | Cox1 | None |
| Cox3 | Atp9 | ||||||||
| Mt protein level | |||||||||
| Cox2 | − | − | − | − | ++++ | ± | ± | + | +++ |
| Cytb | ± | − | − | ± | ++++ | + | + | +++ | +++ |
aPet309 is identified by Blast searches using Ppr5 as a bait, however from our study it does not appear to be a functional homolog.
bΣi: mitochondrial genome where cox1 contains two introns; ‘esp’: especially.
The mutant phenotypes are described qualitatively by comparison to the wild-type, the number of + signs indicates the strength of the phenotype, +/− just detectable and - not detected.
Five of the nine S. pombe proteins shared clear structural homologies with S. cerevisiae proteins (Table 1), all of which have mitochondrial functions: Ppr3 with Dmr1, involved in 15S rRNA stability (14), Ppr4 and Ppr5 with Pet309, required for translation of the cox1 mRNA (18), Ppr6 with Atp13/Aep2, necessary for the stability of the atp9 mRNA (52,53), and Rpo41 with the other mtRNA polymerases. The four other S. pombe PPR proteins did not present any significant homology to proteins in other organisms outside the PPR motif. As the aim of our study of the S. pombe PPR proteins was to characterize new factors involved in mitochondrial gene expression, we decided to exclude Rpo41 and focus on the eight new PPR proteins.
All predicted PPR proteins are mitochondrial proteins
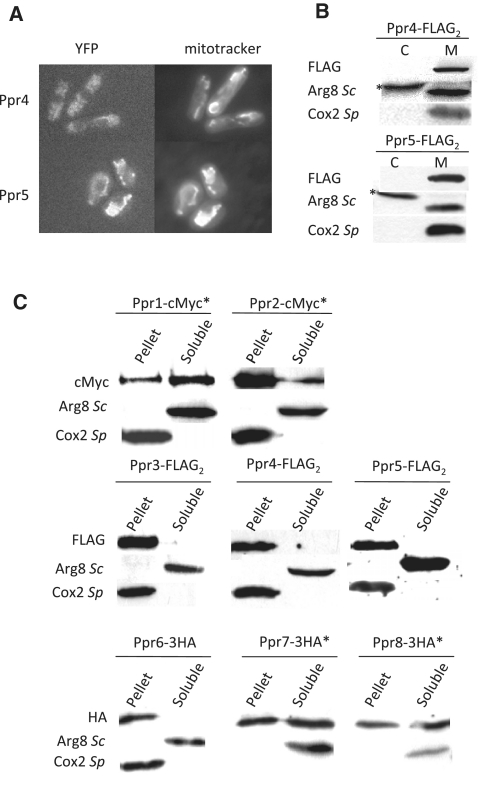
The previously characterized PPR proteins are almost exclusively specific to organelles (2). Consistent with this observation, six of the eight S. pombe PPR proteins were clearly proposed to be mitochondrial, based on the analysis of YFP gene fusions under the control of the thiamine sensitive promoter nmt1 that were constructed and analyzed in the framework of the S. pombe proteome localization program (44). The two exceptions were Ppr4 and Ppr5, where the data were unclear; we tagged the corresponding genes with YFP at their chromosomal locus and saw an overlap with the mitochondrial marker mitotracker for Ppr5 (Figure 3A). However, for Ppr4 the signal was weak, leading to high background probably due to auto-fluorescence of vacuoles. FLAG-versions of both Ppr4 and Ppr5 were then generated and the tagged strains were subjected to cell fractionation and western blot (Figure 3B) that showed a clear localization into the mitochondrial fraction. Thus all eight S. pombe PPR proteins are mitochondrial.
Figure 3.
Cellular localization of the S. pombe Ppr proteins. (A) Fluorescent imaging of Ppr4-YFP and Ppr5-YFP. The ppr4 and ppr5 genes were fused to YFP-Venus. Cells were grown on YPGA 2% glucose with an excess of adenine, and observed with a Zeiss Axioplan 2 microscope linked to a Cool Snap camera (Princeton Instruments). Mitotracker was obtained from Molecular Probes. The signal for Ppr4-YFP was considerably weaker than for Ppr5-YFP. As a consequence the Ppr4-YFP image also shows autofluorescence from the vacuoles. (B) Purification of mitochondria from Ppr4-FLAG2 and Ppr5-FLAG2. Cells producing FLAG-tagged Ppr4 or Ppr5 proteins were fractionated to separate the mitochondria (M) from the cytosolic fraction (C). Protein samples were subjected to western blot analysis with the anti-FLAG antibody and with two control antibodies: anti-Cox2Sp, that labels the inner mitochondrial membrane protein Cox2, and anti-Arg8Sc, that labels the soluble S. pombe mitochondrial protein Arg1 as well as a cross-reacting cytosolic protein (marked with an asterisk). An equal amount of protein was loaded in each lane. (C) Subcellular localization of all Ppr proteins. Mitochondria containing cMyc, FLAG or 3HA-tagged versions of the different Ppr proteins were alkali-treated to separate the soluble and membrane fraction, the total quantity of protein in each fraction was loaded on a 10 or 12% gel and analyzed with the appropriate anti-tag antibodies. Control antibodies are as in panel B anti-Arg8Sc for the soluble fraction and anti-Cox2Sp for the membrane fraction. The proteins marked with an asterisk were tagged at their chromosomal locus. FLAG versions were integrated at the leu1 locus in a background where the endogenous ppr gene was deleted.
To validate the mitochondrial localization of the different proteins, ppr2-ppr8 were FLAG-tagged at the leu1 locus under the control of the nmt1 promoter using the Riken consortium plasmid resources (44) and ppr1 was tagged with a cMyc13 epitope. Mitochondria were purified by cell fractionation and in all cases, a clear signal was obtained for the tag in the mitochondrial fraction and not in the post-mitochondrial supernatant (data not shown), thus confirming that all eight PPR proteins from S. pombe are located in the mitochondria.
To further refine the mitochondrial localization of the PPR proteins, carbonate extractions were performed on purified mitochondria to test the association of each of the eight PPR proteins with the mitochondrial membranes (Figure 3C). In our initial experiments, Ppr3, 4 and 5 remained fully associated with the mitochondrial membranes upon carbonate extraction, like the Cox2 control, showing that they are integral membrane proteins. The other proteins were seen to partition between the soluble and membrane fraction. As all these constructions except ppr1 were expressed from the nmt1 promoter, we were concerned that the partition between the soluble and membrane fractions might be due to the over-expression of the proteins. To overcome this problem the carbonate fractionation was repeated using 3′HA3 or cMyc13 ppr fusions integrated at the chromosomal locus under the control of the endogenous promoter. In these experiments Ppr6 remained fully associated with the mitochondrial membranes while Ppr1, 2, 7 and 8 were found in both the soluble and membrane fractions, suggesting that they might be peripheral proteins.
Thus all S. pombe PPR proteins are mitochondrial and interact more or less strongly with the mitochondrial membranes. This is consistent with the idea that all RNA metabolism and translation events in mitochondria occur in close proximity to the membrane, the final destination of the mitochondrially-encoded proteins.
A phenotypic and molecular analysis of the Δppr mutants identifies two functional classes
To investigate the function of the different PPR proteins, each gene was disrupted in a mitochondrial intron-containing and intron-less background; this latter background facilitates the interpretation of some experiments and also allows the detection of RNA splicing defects. For reasons of clarity, the analysis of each mutant will be presented individually, although all mutants were studied in parallel using the following techniques.
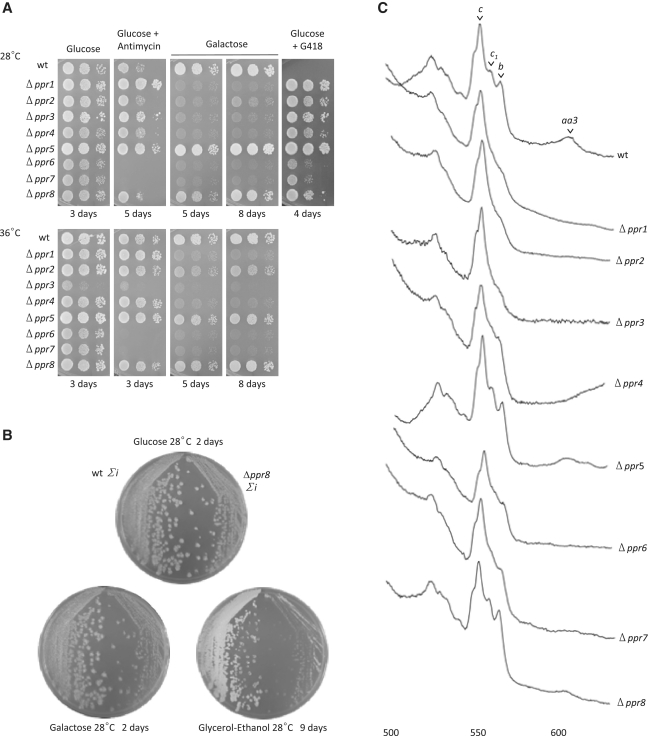
Growth characteristics were tested on various media, including galactose, and glucose containing antimycin (Figure 4A and B). In S. pombe, an inability to grow on galactose is indicative of a strong defect in the mitochondrial respiratory chain (39) whereas sensitivity to antimycin under fermentation reflects an ATP synthase deficiency (37). When electron transfer is blocked by antimycin A, ATP synthase activity becomes essential for viability by generating a membrane potential. Antimycin sensitivity could only be tested in the intron-containing background because the intron-less strain is antimycin resistant (30). Cytochrome spectra were recorded to determine whether the heme containing respiratory enzymes (complex III, containing cytochrome b and c1, and complex IV, containing cytochrome aa3), were affected in the mutants (Figure 4C).
Figure 4.
(A) Growth phenotypes of the Δppr mutants. Ten-fold serial dilutions of freshly grown Δppr mutants in the intron-containing background were spotted onto complete medium containing either galactose, which is used as a non-fermentable carbon source in S. pombe, glucose or glucose supplemented with G418 (indicative of the deletion) or antimycin A (to test for a possible complex V deficiency). Plates were incubated at 28 or 36°C for the indicated time. (B) Phenotype of the deletion of ppr8 in both intronic backgrounds. Intron-containing (Σi) Δppr8 (Δ8) cells and the isogenic wild-type NB205-6A were streaked on complete glucose medium and replica-plated onto complete medium containing 2% glucose, 2% galactose + 0.1% glucose, or 3% glycerol, 3% ethanol and incubated as indicated before taking photographs. (C) Cytochrome spectra of Δppr mutants. Cells of the Δppr mutants in the intron-containing background were grown for 2–3 days on glucose medium and low temperature cytochrome spectra were recorded after the addition of dithionite to fully reduce the cytochromes. The peaks corresponding to cytochromes aa3, b, c1 and c are indicated.
In addition, molecular studies were undertaken to analyze mitochondrial gene expression, because PPR proteins are expected to act on organelle RNAs to modulate transcriptional, post-transcriptional and/or translational steps. Experiments were performed both in the mitochondrial intron-less and intron-containing backgrounds to measure the steady state levels of all mitochondrial rRNAs and mRNAs in the Δppr mutants (Figure 5), as well as the in vivo labeling of newly synthesized mitochondrial proteins with 35S amino acids, in presence of cycloheximide that blocks cytoplasmic translation (Figure 6A). For reasons that are not clear, in vivo labeling experiments are much less efficient in S. pombe than in S. cerevisiae; therefore, experiments were repeated three to six times. Qualitatively, the results appeared similar whatever the mitochondrial intron content, although labeling in the intron-less context was generally more efficient and thus more reliable. Thus a representative result in the intron-less background comparing all eight mutants is presented (Figure 6A). Western blotting with the few antibodies available against mitochondrially-encoded proteins from S. pombe (i.e. Cox2 and Cytb) was also performed to analyze the steady state levels of these mitochondrial proteins (Figure 6B).
Figure 5.
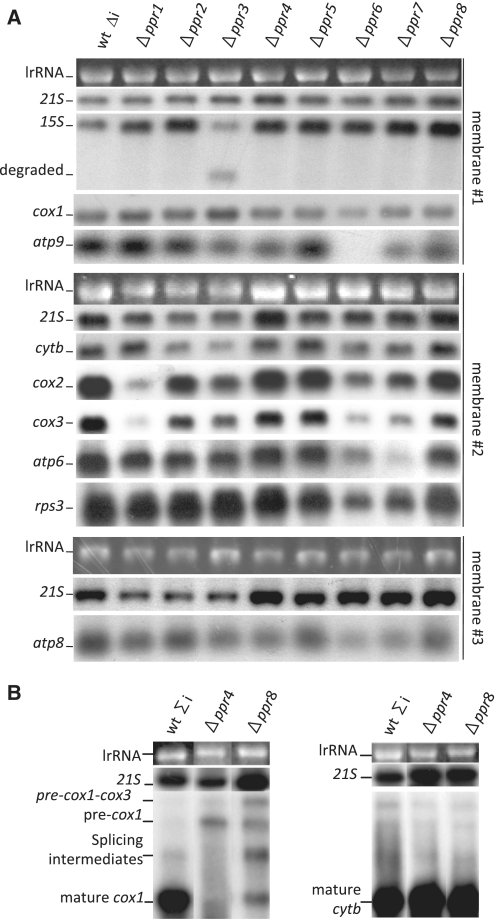
(A) Analysis of the steady state mtRNA levels in the Δppr mutants in the intron-less background. Total RNA from the intron-less P3 wild-type and isogenic Δppr mutants were hybridized to different probes as indicated. Three representative membranes are presented with the UV photograph of the cytoplasmic large rRNA and a control hybridization with the 21S large mitochondrial RNA. All hybridizations have been repeated two or three times and only the changes consistently observed in all experiments are described in the text. For unknown reasons, there is slightly less 21S RNA in the mutants Δppr1, 2 and 3. The same RNA sample was used in preparation for each membrane, but in membrane 1, the wild-type sample appears to have transferred less efficiently. (B) Northern blot of the mosaic genes cox1 and cytb in the Δppr4 and Δppr8 mutants in an intron-containing background. Total RNA from the intron-containing NB205-6A wild-type and isogenic Δppr4 and Δppr8 mutants and hybridized to the intron-containing cox1 and cytb probes as indicated. Precursor and mature RNAs are marked. The membranes are presented with the UV photograph of the cytoplasmic large rRNAs and a control hybridization with the 21S large mitochondrial RNA.
Figure 6.
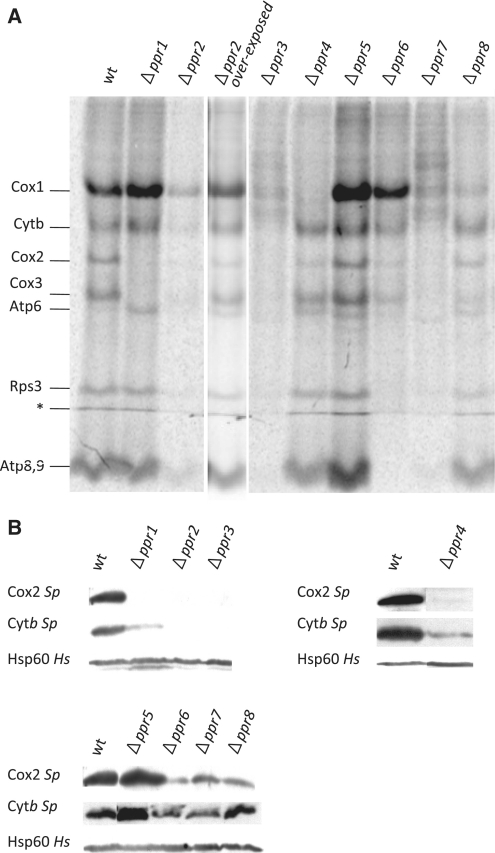
(A) In vivo 35S labeling of Δppr mutants. The various Δppr mutants were labeled in vivo with 35S methionine/cysteine in the presence of cycloheximide, which blocks cytoplasmic translation, and similar amounts of total proteins were loaded on an SDS–PAGE gel, as verified by coomassie blue staining (not shown). A typical result in the intron-less background is presented after a 3 day phosphorimager exposure, however, similar patterns have been obtained whatever the mitochondrial intron content. For the Δppr2 strain that showed a very weak signal, a 3-month exposure of the gel onto a film is also shown. The band marked with the asterisk is a migration artifact. For reasons that are unknown, Atp6 is often more efficiently labeled in respiratory mutants (especially cytochrome oxidase mutants) compared to the wild-type. (B) Steady state level of mitochondrial proteins in the various Δppr mutants. Purified mitochondria from the eight Δppr mutants in the intron-less background were analyzed by SDS–PAGE and western blot. Transferred proteins were hybridized to S. pombe antibodies raised against Cox2 and Cytb. The loading control is Hsp60 that recognizes a mitochondrial matrix protein.
We found that none of the Δppr strains showed a general decrease in all the transcripts belonging to one or both of the two transcription units of the S. pombe mitochondrial genome. Thus apart from the polymerase Rpo41, none of the PPR proteins appear to be involved in general transcription. Similarly, none of the PPR proteins are involved only in mitochondrial intron splicing, as the intron-less genome does not restore growth on non-fermentable medium to any of the respiratory-deficient mutants. In addition, processing does not seem to be affected since none of mutants accumulated aberrant RNA intermediates, contrary to the observations of Hoffmann et al. (54) who examined the processing mutants pah1 and par1. In fine, the Δppr mutants appear to fall in two classes: those that seem to primarily affect mRNA stability (Ppr1, 3, 6, 7), and those that seem to act downstream, at a translational step (Ppr2, 4, 5, 8), for a summary see Table 1.
Ppr1 and Ppr3 appear to stabilize a limited number of mitochondrial RNAs
Among the PPR proteins playing an RNA stability role, Ppr1 and Ppr3 appeared to be the most specific. Δppr1 cells could not grow on galactose (Figure 4A), and lacked cytochrome aa3 in spectra, whilst cytochrome b and c1 were significantly decreased. Conversely, the mutant grew better than the wild-type on medium containing antimycin, suggesting that the ATP synthase functioned very efficiently. At the molecular level, deletion of ppr1 led to a strong reduction in the levels of the cox2 and cox3 mRNAs (Figure 5A), as a consequence, Cox2 and Cox3 were absent in the in vivo labeling experiments (Figure 6A) and Cox2 was lacking in western blot (Figure 6B). These data show that Ppr1 increases the stability of both the cox2 and cox3 mRNAs, which explains the lack of Cox2 and Cox3 proteins. However, the stability function of Ppr1 does not exclude a supplementary role in mRNA translation. In maize chloroplasts, PPR10 stabilizes the atpH mRNA by binding 5′-UTR and blocking the action of exoribonucleases, but the binding of PPR10 also uncovers a ribosome binding site allowing translation of the atpH mRNA (55,56). The cox1 mRNA is stable in the absence of Ppr1, suggesting that the effects on cox2 and cox3 are specific and not part of a general defect in all the subunits of complex IV. In addition, the decrease of steady state Cytb is probably due to a partial destabilization of complex III due to the absence of complex IV.
The Δppr3 mutant could not grow on galactose, and it also showed a clear thermosensitivity on glucose containing media (Figure 4A). In addition cytochrome aa3, b and c1 were strongly decreased (Figure 4C). Deletion of ppr3 led to a strong reduction in the 15S-rRNA associated with the appearance of shorter RNAs that are probably degradation products. There is also a reduction in cytb mRNA (Figure 5A). In addition there was a general decrease in the synthesis of all the mitochondrial proteins and the tested proteins could not be detected by western blot.
These results suggest that Ppr3 is primarily required for the stability of the small rRNA, and that the severe reduction in the level of the small rRNA in the Δppr3 mutant will perturb protein synthesis, leading to a general mitochondrial translation defect. Ppr3 is structurally homologous to Dmr1 from S. cerevisiae (14) and the human PTCD3 (13), both of which are involved in the maintenance of the small mitochondrial rRNA. However, it is possible that the S. pombe protein has acquired another function, as the deletion of ppr3 also leads to a reduction in the level of cytb mRNA and apart from Ppr3, no PPR protein specifically targeting the cytb mRNA has been identified so far in S. pombe.
Ppr6 and 7 appear to target primarily the atp9 and 6 mRNAs, respectively
The Δppr6 and Δppr7 strains, could not grow on galactose, but also showed low viability on glucose containing G418 and were strongly sensitive to antimycin A on glucose medium (Figure 4A), thus revealing a defect in the ATP synthase (complex V). In addition, the peaks for cytochromes aa3, b and c1 were decreased although not absent (Figure 4C). The deletion of ppr6 and ppr7 caused a more or less pronounced reduction in the levels of all the protein coding RNAs, the most dramatic being the absence of atp9 in Δppr6 and the very strong decrease of atp6 in Δppr7 (Figure 5A). As expected, Atp9 and Atp6 appeared to be absent from Δppr6 and Δppr7, respectively, and there was a generalized reduction in mitochondrial protein synthesis, which was more pronounced for Δppr7. In the in vivo labeling experiments, a band below the level of Atp6 could be seen in Δppr7 cells, but the origin of this band is unknown. However, the steady state levels of Cox2 and Cytb appeared to be normal; unfortunately no antibodies are available to test mitochondrially-encoded subunits of the ATP synthase.
The Δppr6 and Δppr7 mutants seem to have a general effect on the stability of many mRNAs as well as translation. However, the most specific consequence of the Δppr6 and Δppr7 mutations appears to be a strong decrease in the level of the atp9 and atp6 mRNAs, respectively, this is consistent with the ATP synthase defect that is indicated by the antimycin sensitivity. It is possible that the more general problems in translation and complex assembly could be secondary effects of the ATP synthase F0 defect. In S. cerevisiae, complex V mutants often have a long-range effect by destabilizing the mtDNA, impairing mitochondrial translation and leading to wider pleiotropic effects [for review (57)]. However, we cannot exclude that Ppr6 and Ppr7 could be general mRNA stability factors with multiple targets, some such as atp9 and atp6 being more sensitive. This could be similar to LRPPRC that has multiple RNA targets, amongst which COX1 and COX3 are more sensitive to degradation when LRPPRC is mutated or silenced (25,26). Ppr6 is a sequence homolog of Atp13/Aep2 from S. cerevisiae, which is specifically required for the stability of atp9 (52,53), and was also recently identified as a PPR protein (29). Our results show that Ppr6 is similar to Aep2, but may have acquired other functions.
Ppr4 is a specific translational activator with a single target
Amongst the PPR proteins that affect translation, Ppr4 is the most specific. Cells devoid of Ppr4 could not grow on galactose (Figure 4A) and cytochrome spectra showed that cytochrome aa3 was lacking whilst cytochrome b and c1 were somewhat decreased (Figure 4C). In the intron-less mitochondrial background (Figure 5A), deletion of ppr4 did not seem to cause an obvious change in the steady state levels of the mitochondrial RNAs, however in an intron-containing background, we could not detect any mature cox1 mRNA and there was an accumulation of the unspliced pre-mRNA containing the two cox1 introns, these introns encode maturases that are necessary for their own excision (Figure 5B). Significantly, the expression of the intron-containing cytb mRNA was not affected. In accordance, the Δppr4 strain showed a specific absence of Cox1 synthesis and a reduction in Cox2 synthesis (Figure 6A) and Cox2 was barely detectable in the steady state (Figure 6B).
These observations strongly suggest that the only function of Ppr4 is to specifically activate the translation of the cox1 mRNA, and that a deficiency in the synthesis of the maturases encoded by the cox1 introns is responsible for the splicing defect rather than an additional function of Ppr4 as a splicing factor. The reduced level in the Cox2 protein labeling despite normal cox2 mRNA levels in Δppr4 cells is probably a secondary defect of the absence of the Cox1 protein. In S. pombe, in vivo protein synthesis experiments require rather long incorporation times (3 h as opposed to 5–15 min in S. cerevisiae), so the result is a combination of synthesis and stability and in S. cerevisiae, it is known that the absence of Cox1, Cox2 is particularly unstable (58).
Ppr4 shares significant similarities with Pet309 from S. cerevisiae (17), which specifically activates the translation of the cox1 mRNA and helps to stabilize intron containing cox1 transcripts. It is difficult to ascertain whether Ppr4 is also important for the stability of the intron-containing cox1 transcript, as S. pombe has fewer introns in cox1 than S. cerevisiae. Thus, Ppr4 is the first mRNA-specific translational activator discovered in fission yeast. It is striking that cox1 translational activators have been found in S. cerevisiae, Pet309 and Mss51 [(18,59,60) and references therein], in humans, Taco1 (61) and now in S. pombe. This emphasizes the crucial role played by cox1 translation in the regulation of mitochondrial biogenesis, as demonstrated by the feedback regulatory loop mediated by Mss51 and other factors in S. cerevisiae (62–64). It will be of great interest to determine the precise target sequence of Ppr4 on the cox1 mRNA.
Ppr8 modestly affects mitochondrial translation
In contrast to Δppr4, Δppr8 cells only showed a slight growth defect on galactose medium compared to the wild-type whatever the temperature (Figure 4A), but exhibited a clear growth arrest on glycerol medium (Figure 4B). The cytochrome spectrum of Δppr8 cells was only slightly affected, with less cytochrome aa3 (Figure 4C). In the intron-less mitochondrial background (Figure 5A), Δppr8 did not seem to cause an obvious change in the steady state levels of the mitochondrial RNAs. In the intron-containing background, the deletion of ppr8 led to a reduction in the level of the mature cox1 mRNA and an accumulation of all the splicing intermediates and the pre-cox1-cox3 intermediate; again cytb was not affected (Figure 5B). Finally, there was a general but limited reduction in mitochondrial protein synthesis with a specific drop in the synthesis of Cox1 (Figure 6A), but for the proteins tested, the steady state levels seem normal (Cytb) or only slightly decreased (Cox2) (Figure 6B). Thus the absence of Ppr8 seems to cause a general but rather limited decrease in translation, which appears more pronounced for Cox1; however, it is not possible to conclude if it has a single, or multiple targets.
Ppr2 is a general translation factor
Cells deleted for ppr2 were defective for growth on galactose (Figure 3A) although some weak growth was visible upon longer incubation, and in cytochrome spectra, cytochromes aa3, b and c1 were strongly decreased. The steady state of mitochondrial RNA was wild-type, except for a modest reduction in the cytb mRNA. However there was a large generalized reduction in the synthesis of all the mitochondrial proteins, although all bands were visible after a long exposure, and the tested proteins could not be detected in western blots. This shows that the deletion of ppr2 leads to a drastic reduction in the synthesis of all the mitochondrially synthesized proteins and an absence of steady state Cytb and Cox2, possibly because the level of synthesis is too low to allow efficient assembly of the complexes and the subunits are degraded. Thus Ppr2 appears to be a general translation factor.
Ppr5 is a negative regulator of mitochondrial gene expression
Finally, the results in Figure 4A (intron-containing mitochondrial background), showed that deletion of ppr5 did not affect growth under these conditions, indeed the Δppr5 strain appears to grow even better than the wild-type, especially on glucose medium. In accordance with the growth data, the Δppr5 cytochrome spectrum was wild-type (Figure 4C) and there was no obvious change in the steady state levels of the mitochondrial RNAs (Figure 5A). The most striking result is that the deletion of ppr5 led to a clear increase in the synthesis, but not the steady state level, of the mitochondrial proteins (compare Figure 6A and B). Thus, Ppr5 is the only S. pombe PPR protein whose absence seems to stimulate, rather than impair, mitochondrial biogenesis and especially mitochondrial protein synthesis. This suggests that Ppr5 could be a general negative regulator of translation. In addition, a YFP tagged version of Ppr5 seemed more highly expressed during growth on glucose rather than galactose, which would be consistent with a role as a repressor of mitochondrial biogenesis during fermentation (data not shown).
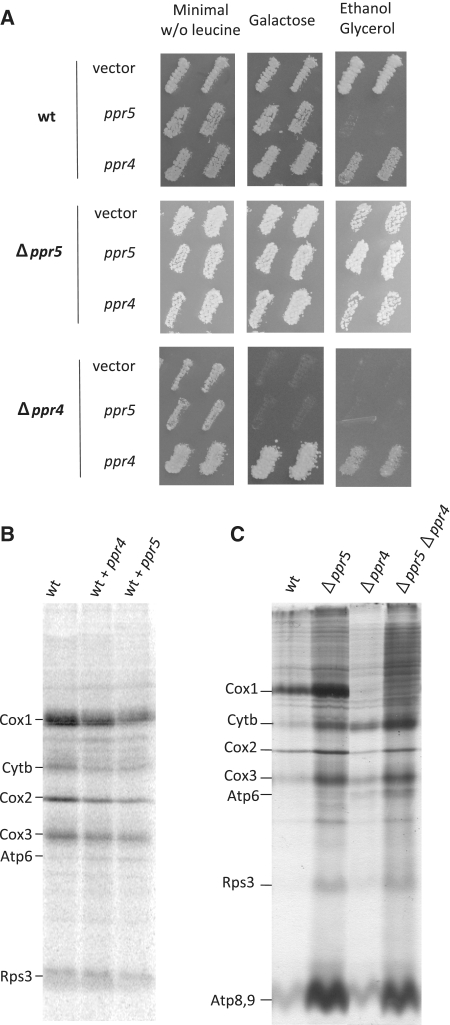
To further analyze the function of Ppr5, we determined the effect of ppr5 over-expression on growth and mitochondrial translation. A FLAG version of ppr5 under the control of the inducible nmt1 promotor was integrated at the leu1 locus in the wild-type NB205-6A and isogenic Δppr5 and Δppr4 (which does not synthesize Cox1) strains. Controls were either integrations of the vector or of a linearized plasmid producing Ppr4-FLAG in the same three strains. The transformants were tested for growth on non-fermentable medium (Figure 7A). Whereas the FLAG versions of Ppr5 or Ppr4 complemented the corresponding mutants they compromised growth of the wild-type on ethanol-glycerol medium, Ppr5 over-expression was particularly deleterious. Next, we looked at in vivo labeling of mitochondrial proteins in these strains (Figure 7B). Whereas over-expression of Ppr4 had a mild effect, the labeling of mitochondrially-synthesized proteins was decreased when Ppr5 was over-produced, consistent with Ppr5 being a negative regulator of translation.
Figure 7.
Effect of ppr4 and ppr5 over-expression or deletion in different strain backgrounds. (A) Effect of ppr4 and ppr5 over-expression on growth. Wild-type (NB205-6A), Δppr4 or Δppr5 cells were transformed with integrative leu1 plasmids expressing functional tagged versions of ppr4 or ppr5 under the control of the nmt1 promoter (from the Riken consortium), or with the control vector. [Leu+] transformants were patched on minimal glucose medium lacking leucine, replica-plated onto complete galactose or ethanol glycerol medium and incubated at 28°C for 3–7 days. (B) 35S labeling of mitochondrial proteins in cells overexpressing ppr4 or ppr5. Wild-type strains transformed with the control plasmid or functional tagged versions of ppr4 or ppr5 under the control of the nmt1 promoter from the upper part of panel A were labeled with 35S amino acids in presence of cycloheximide. Similar amounts of protein were loaded in each lane as verified by coomassie blue staining (data not shown). (C) 35S labeling of mitochondrial proteins in Δppr single and double mutants. Mitochondrial proteins of the wild-type (NB205-6A) and of single or double Δppr4 and 5 mutants in the mitochondrial intron containing background were labeled as in panel B. Similar amounts of protein were loaded in each lane as verified by coomassie blue staining (data not shown).
If the deletion of Ppr5 relieves an inhibition of translation, we would predict that it might be able to counteract a mutation that decreases Cox1 synthesis, unless the mutated component is essential for cox1 translation. To test this hypothesis, ppr5 was deleted in the Δppr4 mutant that specifically abolishes Cox1 synthesis. In the Δppr4 Δppr5 double mutant, Cox1 is not synthesized as in the Δppr4 single mutant; however, other mitochondrial proteins such as Cytb, Cox3 and Atp9 are more strongly labeled as in the single Δppr5 mutant (Figure 7C). Thus the deletion of ppr5 increases the general translation efficiency but cannot overcome the cox1 translation defect in a Δppr4 mutant, showing that the deletion of ppr4 causes a specific block in cox1 translation that is epistatic to Δppr5.
These data are consistent with Ppr5 being a general negative regulator of mitochondrial translation, and confirm that Ppr4 plays an essential role in cox1 translation. In humans, another PPR protein PTCD1, has been proposed to inhibit mitochondrial translation by decreasing the stability of the mitochondrial leucine tRNAs (11). We have probed several mitochondrial tRNAs from S. pombe including the two leucine tRNAs but did not detect any increase in the steady state levels in cells lacking Ppr5 (data not shown). Thus even though Ppr5 might play a similar role to PTCD1 in humans, the mechanism by which it represses translation is probably different.
CONCLUSION
In summary, this study is a systematic functional analysis of all the identified PPR proteins in a given organism. The fact that the PPR motif is highly degenerate raises the question of whether we have indeed identified all the members of the family present in the S. pombe genome. We expected to find a PPR protein involved in S. pombe tRNA processing, since in both the yeast S. cerevisiae (29) and Man (8), one subunit of RNAse P contains PPR motifs, irrespective of whether an RNA component is present (S. cerevisiae) or not (human). However no sequence homolog can be found in S. pombe, suggesting that either RNAse P is too divergent to be detected by sequence comparison, or that tRNA processing occurs via a different system despite the presence of a possible gene for an RNAse P RNA in the S. pombe mtDNA. Possible candidates could be the Suv3 and Dss1 homologs Pah1 and Par1, which have been implicated in the 3′-end processing of fission yeast mRNAs (54).
While it is clear that S. pombe PPR proteins act at different levels and that some have general effects, whereas others have very specific effects on individual RNAs, no correlation could be made between the obvious structural characteristics of the proteins and these two general classes of function, RNA stability or translation. Both contain PPR proteins that are 100% membrane bound and others that partition between the membrane and the mitochondrial matrix when extracted with carbonate. Although the number of PPR motifs in a given protein varies considerably in both groups, there is a tendency for the specificity to increase with the number of PPR motifs. For example, ppr4, which appears to have the most specific phenotype when deleted, has 13 PPR motifs, whilst ppr2, which gives a general phenotype when deleted, only has three. If this is the case, the negative regulator Ppr5 does not seem to follow this scheme as it contains the highest number of PPR motifs but has a general effect; however, at present the mode of action of Ppr5 is unknown and its target could be highly specific, yet result in a general reduction in translation. Thus further studies will be necessary to define the structure function relationship of the different PPR proteins, analyze their interaction with their target RNAs and clarify the different roles of this fascinating family of proteins in mitochondrial biogenesis.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
The Agence Nationale pour la Recherche (ANR) (JCJC06-0163 to N.B. and I.K.); the Fondation pour la Recherche Médicale (FDT20091217787 to N.B. and I.K.). L.D. is recipient of a PhD fellowship from the Ministère de l'Education Nationale, de la Recherche et la Technologie. Funding for open access charge: ANR.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
We are indebted to Pawel Golik for providing unpublished results of the bioinformatic identification of S. pombe PPR proteins. We thank Geneviève Dujardin and Thomas D. Fox both for fruitful discussions and for the gift of antibodies and G.D. for critical reading of the article. Laurence Maréchal-Drouard gave advice on tRNA hybridizations and B. Schäfer provided the P3 S. pombe intron-less strain. The Δku strains were gifts from the National Bio Resource Program in Japan. FLAG-tagged versions of the ppr genes were distributed by the RIKEN Bioresource Center.
REFERENCES
- 1.Small ID, Peeters N. The PPR motif - a TPR-related motif prevalent in plant organellar proteins. Trends Biochem. Sci. 2000;25:46–47. doi: 10.1016/s0968-0004(99)01520-0. [DOI] [PubMed] [Google Scholar]
- 2.Lurin C, Andres C, Aubourg S, Bellaoui M, Bitton F, Bruyere C, Caboche M, Debast C, Gualberto J, Hoffmann B, et al. Genome-wide analysis of Arabidopsis pentatricopeptide repeat proteins reveals their essential role in organelle biogenesis. Plant Cell. 2004;16:2089–2103. doi: 10.1105/tpc.104.022236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hayes ML, Mulligan RM. Pentatricopeptide repeat proteins constrain genome evolution in chloroplasts. Mol. Biol. Evol. 2011;7:2029–2039. doi: 10.1093/molbev/msr023. [DOI] [PubMed] [Google Scholar]
- 4.Delannoy E, Stanley WA, Bond CS, Small ID. Pentatricopeptide repeat (PPR) proteins as sequence-specificity factors in post-transcriptional processes in organelles. Biochem. Soc. Trans. 2007;35:1643–1647. doi: 10.1042/BST0351643. [DOI] [PubMed] [Google Scholar]
- 5.Schmitz-Linneweber C, Small I. Pentatricopeptide repeat proteins: a socket set for organelle gene expression. Trends Plant Sci. 2008;13:663–670. doi: 10.1016/j.tplants.2008.10.001. [DOI] [PubMed] [Google Scholar]
- 6.Lightowlers RN, Chrzanowska-Lightowlers ZM. PPR (pentatricopeptide repeat) proteins in mammals: important aids to mitochondrial gene expression. Biochem. J. 2008;416:e5–e6. doi: 10.1042/BJ20081942. [DOI] [PubMed] [Google Scholar]
- 7.Zehrmann A, Verbitskiy D, Hartel B, Brennicke A, Takenaka M. PPR proteins network as site-specific RNA editing factors in plant organelles. RNA Biol. 2011;8:67–70. doi: 10.4161/rna.8.1.14298. [DOI] [PubMed] [Google Scholar]
- 8.Holzmann J, Frank P, Loffler E, Bennett KL, Gerner C, Rossmanith W. RNase P without RNA: identification and functional reconstitution of the human mitochondrial tRNA processing enzyme. Cell. 2008;135:462–474. doi: 10.1016/j.cell.2008.09.013. [DOI] [PubMed] [Google Scholar]
- 9.Xu F, Ackerley C, Maj MC, Addis JB, Levandovskiy V, Lee J, Mackay N, Cameron JM, Robinson BH. Disruption of a mitochondrial RNA-binding protein gene results in decreased cytochrome b expression and a marked reduction in ubiquinol-cytochrome c reductase activity in mouse heart mitochondria. Biochem. J. 2008;416:15–26. doi: 10.1042/BJ20080847. [DOI] [PubMed] [Google Scholar]
- 10.Hattori M, Sugita M. A moss pentatricopeptide repeat protein binds to the 3′ end of plastid clpP pre-mRNA and assists with mRNA maturation. FEBS J. 2009;276:5860–5869. doi: 10.1111/j.1742-4658.2009.07267.x. [DOI] [PubMed] [Google Scholar]
- 11.Rackham O, Davies SM, Shearwood AM, Hamilton KL, Whelan J, Filipovska A. Pentatricopeptide repeat domain protein 1 lowers the levels of mitochondrial leucine tRNAs in cells. Nucleic Acids Res. 2009;37:5859–5867. doi: 10.1093/nar/gkp627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ellis TP, Helfenbein KG, Tzagoloff A, Dieckmann CL. Aep3p stabilizes the mitochondrial bicistronic mRNA encoding subunits 6 and 8 of the H+-translocating ATP synthase of Saccharomyces cerevisiae. J. Biol. Chem. 2004;279:15728–15733. doi: 10.1074/jbc.M314162200. [DOI] [PubMed] [Google Scholar]
- 13.Davies SM, Rackham O, Shearwood AM, Hamilton KL, Narsai R, Whelan J, Filipovska A. Pentatricopeptide repeat domain protein 3 associates with the mitochondrial small ribosomal subunit and regulates translation. FEBS Lett. 2009;583:1853–1858. doi: 10.1016/j.febslet.2009.04.048. [DOI] [PubMed] [Google Scholar]
- 14.Puchta O, Lubas M, Lipinski KA, Piatkowski J, Malecki M, Golik P. DMR1 (CCM1/YGR150C) of Saccharomyces cerevisiae encodes an RNA-binding protein from the pentatricopeptide repeat family required for the maintenance of the mitochondrial 15S ribosomal RNA. Genetics. 2010;184:959–973. doi: 10.1534/genetics.110.113969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nouet C, Bourens M, Hlavacek O, Marsy S, Lemaire C, Dujardin G. Rmd9p controls the processing/stability of mitochondrial mRNAs and its overexpression compensates for a partial deficiency of Oxa1p in Saccharomyces cerevisiae. Genetics. 2007;175:1105–1115. doi: 10.1534/genetics.106.063883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Williams EH, Butler CA, Bonnefoy N, Fox TD. Translation initiation in Saccharomyces cerevisiae mitochondria: functional interactions among mitochondrial ribosomal protein Rsm28p, initiation factor 2, methionyl-tRNA-formyltransferase and novel protein Rmd9p. Genetics. 2007;175:1117–1126. doi: 10.1534/genetics.106.064576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Manthey GM, McEwen JE. The product of the nuclear gene PET309 is required for translation of mature mRNA and stability or production of intron-containing RNAs derived from the mitochondrial COX1 locus of Saccharomyces cerevisiae. EMBO J. 1995;14:4031–4043. doi: 10.1002/j.1460-2075.1995.tb00074.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tavares-Carreon F, Camacho-Villasana Y, Zamudio-Ochoa A, Shingu-Vazquez M, Torres-Larios A, Perez-Martinez X. The pentatricopeptide repeats present in Pet309 are necessary for translation but not for stability of the mitochondrial COX1 mRNA in yeast. J. Biol. Chem. 2008;283:1472–1479. doi: 10.1074/jbc.M708437200. [DOI] [PubMed] [Google Scholar]
- 19.Mingler MK, Hingst AM, Clement SL, Yu LE, Reifur L, Koslowsky DJ. Identification of pentatricopeptide repeat proteins in Trypanosoma brucei. Mol. Biochem. Parasitol. 2006;150:37–45. doi: 10.1016/j.molbiopara.2006.06.006. [DOI] [PubMed] [Google Scholar]
- 20.Pusnik M, Small I, Read LK, Fabbro T, Schneider A. Pentatricopeptide repeat proteins in Trypanosoma brucei function in mitochondrial ribosomes. Mol. Cell. Biol. 2007;27:6876–6888. doi: 10.1128/MCB.00708-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sondheimer N, Fang JK, Polyak E, Falk MJ, Avadhani NG. Leucine-rich pentatricopeptide-repeat containing protein regulates mitochondrial transcription. Biochemistry. 2010;49:7467–7473. doi: 10.1021/bi1008479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mootha VK, Lepage P, Miller K, Bunkenborg J, Reich M, Hjerrild M, Delmonte T, Villeneuve A, Sladek R, Xu F, et al. Identification of a gene causing human cytochrome c oxidase deficiency by integrative genomics. Proc. Natl Acad. Sci. USA. 2003;100:605–610. doi: 10.1073/pnas.242716699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Xu F, Morin C, Mitchell G, Ackerley C, Robinson BH. The role of the LRPPRC (leucine-rich pentatricopeptide repeat cassette) gene in cytochrome oxidase assembly: mutation causes lowered levels of COX (cytochrome c oxidase) I and COX III mRNA. Biochem J. 2004;382:331–336. doi: 10.1042/BJ20040469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Debray FG, Morin C, Janvier A, Villeneuve J, Maranda B, Laframboise R, Lacroix J, Decarie JC, Robitaille Y, Lambert M, et al. LRPPRC mutations cause a phenotypically distinct form of Leigh syndrome with cytochrome c oxidase deficiency. J. Med. Genet. 2011;48:183–189. doi: 10.1136/jmg.2010.081976. [DOI] [PubMed] [Google Scholar]
- 25.Gohil VM, Nilsson R, Belcher-Timme CA, Luo B, Root DE, Mootha VK. Mitochondrial and nuclear genomic responses to loss of LRPPRC expression. J. Biol. Chem. 2010;285:13742–13747. doi: 10.1074/jbc.M109.098400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sasarman F, Brunel-Guitton C, Antonicka H, Wai T, Shoubridge EA. LRPPRC and SLIRP interact in a ribonucleoprotein complex that regulates posttranscriptional gene expression in mitochondria. Mol. Biol. Cell. 2010;21:1315–1323. doi: 10.1091/mbc.E10-01-0047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cooper MP, Uldry M, Kajimura S, Arany Z, Spiegelman BM. Modulation of PGC-1 coactivator pathways in brown fat differentiation through LRP130. J. Biol. Chem. 2008;283:31960–31967. doi: 10.1074/jbc.M805431200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Topisirovic I, Siddiqui N, Lapointe VL, Trost M, Thibault P, Bangeranye C, Pinol-Roma S, Borden KL. Molecular dissection of the eukaryotic initiation factor 4E (eIF4E) export-competent RNP. EMBO J. 2009;28:1087–1098. doi: 10.1038/emboj.2009.53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lipinski KA, Puchta O, Surandranath V, Kudla M, Golik P. Revisiting the yeast PPR proteins - application of an iterative hidden markov model algorithm reveals new members of the rapidly evolving family. Mol. Biol. Evol. 2011 doi: 10.1093/molbev/msr120. (in press) [DOI] [PubMed] [Google Scholar]
- 30.Schäfer B, Merlos-Lange AM, Anderl C, Welser F, Zimmer M, Wolf K. The mitochondrial genome of fission yeast: inability of all introns to splice autocatalytically, and construction and characterization of an intronless genome. Mol. Gen. Genet. 1991;225:158–167. doi: 10.1007/BF00282654. [DOI] [PubMed] [Google Scholar]
- 31.Jiang H, Sun W, Wang Z, Zhang J, Chen D, Murchie AI. Identification and characterization of the mitochondrial RNA polymerase and transcription factor in the fission yeast Schizosaccharomyces pombe. Nucleic Acids Res. 2011 doi: 10.1093/nar/gkr103. (in press) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schäfer B. RNA maturation in mitochondria of S. cerevisiae and S. pombe. Gene. 2005;354:80–85. doi: 10.1016/j.gene.2005.03.032. [DOI] [PubMed] [Google Scholar]
- 33.Temperley RJ, Wydro M, Lightowlers RN, Chrzanowska-Lightowlers ZM. Human mitochondrial mRNAs–like members of all families, similar but different. Biochim. Biophys. Acta. 2010;1797:1081–1085. doi: 10.1016/j.bbabio.2010.02.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chiron S, Suleau A, Bonnefoy N. Mitochondrial translation: elongation factor tu is essential in fission yeast and depends on an exchange factor conserved in humans but not in budding yeast. Genetics. 2005;169:1891–1901. doi: 10.1534/genetics.104.037473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Matsuyama A, Shirai A, Yashiroda Y, Kamata A, Horinouchi S, Yoshida M. pDUAL, a multipurpose, multicopy vector capable of chromosomal integration in fission yeast. Yeast. 2004;21:1289–1305. doi: 10.1002/yea.1181. [DOI] [PubMed] [Google Scholar]
- 36.Bonnefoy N, Kermorgant M, Brivet-Chevillotte P, Dujardin G. Cloning by functional complementation, and inactivation, of the Schizosaccharomyces pombe homologue of the Saccharomyces cerevisiae gene ABC1. Mol. Gen. Genet. 1996;251:204–210. doi: 10.1007/BF02172919. [DOI] [PubMed] [Google Scholar]
- 37.Bonnefoy N, Kermorgant M, Groudinsky O, Dujardin G. The respiratory gene OXA1 has two fission yeast orthologues which together encode a function essential for cellular viability. Mol. Microbiol. 2000;35:1135–1145. doi: 10.1046/j.1365-2958.2000.01781.x. [DOI] [PubMed] [Google Scholar]
- 38.Okazaki K, Okazaki N, Kume K, Jinno S, Tanaka K, Okayama H. High-frequency transformation method and library transducing vectors for cloning mammalian cDNAs by trans-complementation of Schizosaccharomyces pombe. Nucleic Acids Res. 1990;18:6485–6489. doi: 10.1093/nar/18.22.6485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chiron S, Gaisne M, Guillou E, Belenguer P, Clark-Walker GD, Bonnefoy N. Studying mitochondria in an attractive model: Schizosaccharomyces pombe. Methods Mol. Biol. 2007;372:91–105. doi: 10.1007/978-1-59745-365-3_7. [DOI] [PubMed] [Google Scholar]
- 40.Hoffman CS, Winston F. A ten-minute DNA preparation from yeast efficiently releases autonomous plasmids for transformation of Escherichia coli. Gene. 1987;57:267–272. doi: 10.1016/0378-1119(87)90131-4. [DOI] [PubMed] [Google Scholar]
- 41.Wach A. PCR-synthesis of marker cassettes with long flanking homology regions for gene disruptions in S. cerevisiae. Yeast. 1996;12:259–265. doi: 10.1002/(SICI)1097-0061(19960315)12:3%3C259::AID-YEA901%3E3.0.CO;2-C. [DOI] [PubMed] [Google Scholar]
- 42.Wach A, Brachat A, Pohlmann R, Philippsen P. New heterologous modules for classical or PCR-based gene disruptions in Saccharomyces cerevisiae. Yeast. 1994;10:1793–1808. doi: 10.1002/yea.320101310. [DOI] [PubMed] [Google Scholar]
- 43.Goldstein AL, McCusker JH. Three new dominant drug resistance cassettes for gene disruption in Saccharomyces cerevisiae. Yeast. 1999;15:1541–1553. doi: 10.1002/(SICI)1097-0061(199910)15:14<1541::AID-YEA476>3.0.CO;2-K. [DOI] [PubMed] [Google Scholar]
- 44.Matsuyama A, Arai R, Yashiroda Y, Shirai A, Kamata A, Sekido S, Kobayashi Y, Hashimoto A, Hamamoto M, Hiraoka Y, et al. ORFeome cloning and global analysis of protein localization in the fission yeast Schizosaccharomyces pombe. Nat. Biotechnol. 2006;24:841–847. doi: 10.1038/nbt1222. [DOI] [PubMed] [Google Scholar]
- 45.Petracek ME, Longtine MS. PCR-based engineering of yeast genome. Methods Enzymol. 2002;350:445–469. doi: 10.1016/s0076-6879(02)50978-2. [DOI] [PubMed] [Google Scholar]
- 46.Ausubel FM, Brent R, Kingston RE, Moore DD, Seidman JG, Struhl K. Current protocols in Molecular Biology. Boston, Mass: John Wiley & sons; 1993. [Google Scholar]
- 47.Gouget K, Verde F, Barrientos A. In vivo labeling and analysis of mitochondrial translation products in budding and in fission yeasts. Methods Mol. Biol. 2008;457:113–124. doi: 10.1007/978-1-59745-261-8_8. [DOI] [PubMed] [Google Scholar]
- 48.Lemaire C, Dujardin G. Preparation of respiratory chain complexes from Saccharomyces cerevisiae wild-type and mutant mitochondria: activity measurement and subunit composition analysis. Methods Mol. Biol. 2008;432:65–81. doi: 10.1007/978-1-59745-028-7_5. [DOI] [PubMed] [Google Scholar]
- 49.Steele DF, Butler CA, Fox TD. Expression of a recoded nuclear gene inserted into yeast mitochondrial DNA is limited by mRNA-specific translational activation. Proc. Natl Acad. Sci. USA. 1996;93:5253–5257. doi: 10.1073/pnas.93.11.5253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Gaisne M, Bonnefoy N. The COX18 gene, involved in mitochondrial biogenesis, is functionally conserved and tightly regulated in humans and fission yeast. FEMS Yeast Res. 2006;6:869–882. doi: 10.1111/j.1567-1364.2006.00083.x. [DOI] [PubMed] [Google Scholar]
- 51.Claisse ML, Pere-Aubert GA, Clavilier LP, Slonimski PP. Method for the determination of cytochrome concentrations in whole yeast cells. Eur. J. Biochem. 1970;16:430–438. doi: 10.1111/j.1432-1033.1970.tb01098.x. [DOI] [PubMed] [Google Scholar]
- 52.Ackerman SH, Gatti DL, Gellefors P, Douglas MG, Tzagoloff A. ATP13, a nuclear gene of Saccharomyces cerevisiae essential for the expression of subunit 9 of the mitochondrial ATPase. FEBS Lett. 1991;278:234–238. doi: 10.1016/0014-5793(91)80124-l. [DOI] [PubMed] [Google Scholar]
- 53.Ellis TP, Lukins HB, Nagley P, Corner BE. Suppression of a nuclear aep2 mutation in Saccharomyces cerevisiae by a base substitution in the 5′-untranslated region of the mitochondrial oli1 gene encoding subunit 9 of ATP synthase. Genetics. 1999;151:1353–1363. doi: 10.1093/genetics/151.4.1353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hoffmann B, Nickel J, Speer F, Schafer B. The 3′ ends of mature transcripts are generated by a processosome complex in fission yeast mitochondria. J. Mol. Biol. 2008;377:1024–1037. doi: 10.1016/j.jmb.2008.01.038. [DOI] [PubMed] [Google Scholar]
- 55.Prikryl J, Rojas M, Schuster G, Barkan A. Mechanism of RNA stabilization and translational activation by a pentatricopeptide repeat protein. Proc. Natl Acad. Sci. USA. 2011;108:415–420. doi: 10.1073/pnas.1012076108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Barkan A. Expression of plastid genes: organelle-specific elaborations on a prokaryotic scaffold. Plant physiology. 2011;155:1520–1532. doi: 10.1104/pp.110.171231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ackerman SH, Tzagoloff A. Function, structure, and biogenesis of mitochondrial ATP synthase. Prog. Nucleic Acid Res. Mol. Biol. 2005;80:95–133. doi: 10.1016/S0079-6603(05)80003-0. [DOI] [PubMed] [Google Scholar]
- 58.Lemaire C, Robineau S, Netter P. Molecular and biochemical analysis of Saccharomyces cerevisiae cox1 mutants. Curr. Genet. 1998;34:138–145. doi: 10.1007/s002940050378. [DOI] [PubMed] [Google Scholar]
- 59.Perez-Martinez X, Broadley SA, Fox TD. Mss51p promotes mitochondrial Cox1p synthesis and interacts with newly synthesized Cox1p. EMBO J. 2003;22:5951–5961. doi: 10.1093/emboj/cdg566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Zambrano A, Fontanesi F, Solans A, de Oliveira RL, Fox TD, Tzagoloff A, Barrientos A. Aberrant translation of cytochrome c oxidase subunit 1 mRNA species in the absence of Mss51p in the yeast Saccharomyces cerevisiae. Mol. Biol. Cell. 2007;18:523–535. doi: 10.1091/mbc.E06-09-0803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Weraarpachai W, Antonicka H, Sasarman F, Seeger J, Schrank B, Kolesar JE, Lochmuller H, Chevrette M, Kaufman BA, Horvath R, et al. Mutation in TACO1, encoding a translational activator of COXI, results in cytochrome c oxidase deficiency and late-onset Leigh syndrome. Nat. Genet. 2009;41:833–837. doi: 10.1038/ng.390. [DOI] [PubMed] [Google Scholar]
- 62.Pierrel F, Bestwick ML, Cobine PA, Khalimonchuk O, Cricco JA, Winge DR. Coa1 links the Mss51 post-translational function to Cox1 cofactor insertion in cytochrome c oxidase assembly. EMBO J. 2007;26:4335–4346. doi: 10.1038/sj.emboj.7601861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Fontanesi F, Soto IC, Horn D, Barrientos A. Mss51 and Ssc1 facilitate translational regulation of cytochrome c oxidase biogenesis. Mol. Cell Biol. 2010;30:245–259. doi: 10.1128/MCB.00983-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Perez-Martinez X, Butler CA, Shingu-Vazquez M, Fox TD. Dual functions of Mss51 couple synthesis of Cox1 to assembly of cytochrome c oxidase in Saccharomyces cerevisiae mitochondria. Mol. Biol. Cell. 2009;20:4371–4380. doi: 10.1091/mbc.E09-06-0522. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.