Abstract
The difference in melting temperature of a double-stranded (ds) DNA molecule in the absence and presence of bound ligands can provide experimental information about the stabilization brought about by ligand binding. By simulating the dynamic behaviour of a duplex of sequence 5′-d(TAATAACGGATTATT)·5′-d(AATAATCCGTTATTA) in 0.1 M NaCl aqueous solution at 400 K, we have characterized in atomic detail its complete thermal denaturation profile in <200 ns. A striking asymmetry was observed on both sides of the central CGG triplet and the strand separation process was shown to be strongly affected by bonding in the minor groove of the prototypical interstrand crosslinker mitomycin C or the monofunctional tetrahydroisoquinolines trabectedin (Yondelis®), Zalypsis® and PM01183®. Progressive helix unzipping was clearly interspersed with some reannealing events, which were most noticeable in the oligonucleotides containing the monoadducts, which maintained an average of 6 bp in the central region at the end of the simulations. These significant differences attest to the demonstrated ability of these drugs to stabilize dsDNA, stall replication and transcription forks, and recruit DNA repair proteins. This stabilization, quantified here in terms of undisrupted base pairs, supports the view that these monoadducts can functionally mimic a DNA interstrand crosslink.
INTRODUCTION
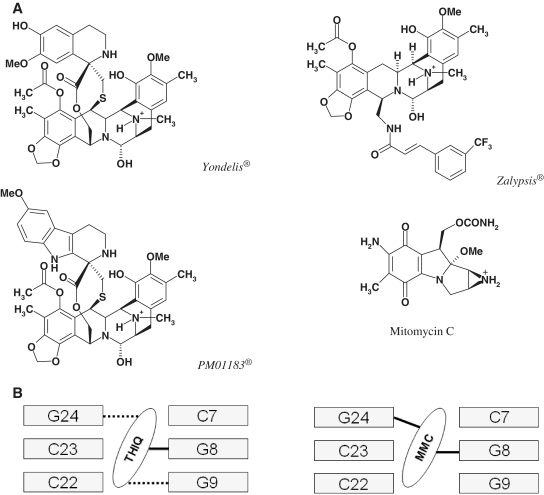
The stability of a DNA double helix containing two complementary polynucleotide chains interacting with each other via Watson–Crick (WC) hydrogen bonds largely depends on its length, base composition, sequence and presence of mismatches, but it can also be strongly perturbed either globally or locally by alterations in pH, temperature and solvent (1) as well as by the binding of proteins such as helicases, polymerases or topoisomerases in living cells (2). In fact, local melting leading to strand separation is necessary for such vital processes as replication, transcription, recombination or repair (3) and can be affected by the presence of non-covalently bound [e.g. mono- and bis-intercalators (4)] or covalently bonded drugs [e.g. cisplatin and nitrogen mustards (5)]. Within the latter category we have focused on several anti-tumour agents (Figure 1) that form a covalent bond with the exocyclic N2 of select guanines in the DNA minor groove: (i) on the one hand, the well-known mitomycin C (MMC), which upon reductive activation is capable of physically connecting the two strands by simultaneously bonding to the two guanines present at a CpG step in a double-stranded (ds) DNA molecule (6), and (ii) on the other hand, the tetrahydroisoquinoline derivatives (7) Yondelis®, Zalypsis® and PM01183®, which form monoadducts in the minor groove that have been shown in fluorescence-based assays to increase substantially the melting temperature of selected DNA oligonucleotides (8–10). An interstrand crosslink (ICL) is considered a highly damaging lesion in DNA and clinically relevant for anti-neoplastic action because it represents a complex challenge to the DNA repair machinery (11).
Figure 1.
(A) Chemical structures of the N-protonated forms of the drugs used in the melting simulations. In the case of the tetrahydroisoquinolines, dehydration of the hemiaminal (carbinolamine) yields the reactive iminium intermediate that reacts with the 2-amino group of a guanine (7). In the case of MMC, two-electron reduction of the quinone ring facilitates methoxide elimination, formation of a leuco-aziridinomitosene and opening of the aziridine ring to provide the initial alkylating agent that leads to a DNA monoadduct with the 2-amino group of a guanine; the second alkylating centre is the iminium ion that forms upon the reverse Michael elimination of carbamic acid from the monoadduct and undergoes a nucleophilic attack by the 2-amino group of the guanine in the opposite strand at a CpG step (6). (B) Schematic of the binding modes of the tetrahydroisoquinolines Yondelis®, Zalypsis® and PM-01183® (THIQ, left) and MMC (right) to the central CGG triplet of the DNA 15-mer studied. Covalent and hydrogen bonds are shown, respectively, as solid and dotted lines. Base numbers refer to their position in the 15-mer: strand bearing the adduct = 1–15; complementary strand = 16–30.
Advances in computer power, force field development (12) and simulation methodologies applied to the study of nucleic acid structures have led to a maturity in the field that enables theoretical results to fill in some of the missing features that the experimental tools cannot provide (13,14). Thus simulations describing the thermal melting properties of several DNA duplexes have been carried out using both simplified coarse-grained models in the absence of ions and solvent molecules (15–18) and atomistic models in either a continuum solvent model (19) or explicit water (20–22). A remaining limitation in modelling this process is that the time scale usually covered in most MD simulations (tens or hundreds of nanoseconds) is much shorter, with some notable recent exceptions (19), than the time over which melting actually takes place. The workaround has been either to study very short DNA oligomers [e.g. CGC (20) or CGCG (18)] or to carry out the simulations under conditions that favour WC hydrogen bond disruption and strand separation, as for example, increased salt concentration and elevated temperature (21) or the presence of miscible pyridine in the aqueous solvent (19). Another less explored alternative has been to use a combination of replica exchange and metadynamics to simulate the helix to coil transition of three different hexanucleotides (23). Interestingly, by comparing results from melting simulations of Dickerson’s dodecamer in the presence of only water or a water–pyridine mixture, it was concluded that strand separation is actually triggered by thermal energy and water molecules, with pyridine’s major role being to stabilize partially disrupted microstates through stacking with the solvent-exposed extrahelical nucleobases (19).
Recent work from our labs (24) showed that a bead-immobilized DNA template containing a central CGG triplet surrounded by A:T pairs on both sides was weakly sensitive to XPF, a structure-specific endonuclease that preferentially cleaves one strand of duplex DNA at the 5′ side of a junction with single-stranded (ss) DNA (25). Our finding that the cleavage intensity increased, in a concentration-dependent manner, if the polynucleotide was pre-treated with either Yondelis® or Zalypsis®, led us to think that XPF, which is known to play a key role in multiple steps of ICL repair (26) and cuts the strand having the unpaired 3′ flap, was probably recognizing a ds/ss discontinuity brought about by the particular choice of trinucleotide repeats (TAA and ATT) on each side of the drug-targeted CGG triplet. Our reasoning was that the enhanced XPF endonucleolytic activity observed in the presence of the drug adducts was likely to arise from increased stabilization of the A:T-rich region in which the central CGG triplet was embedded.
With the aim of gaining further insight into the effects of a covalently bonded drug on the process of DNA-strand separation, we have used all-atom molecular dynamics (MD) simulations in saline solutions at high temperature to examine and compare the thermal denaturation of an A:T-rich DNA 15-mer containing a central CGG triplet in the absence and presence of a drug covalently bonded to the underlined guanine. Complete loss of WC base pairing was observed for the drug-free oligonucleotide after 190 ns of simulation, whereas the central regions of the drug-bonded duplexes remained hydrogen bonded to different extents at the final time of 200 ns. Our findings thus support the view that the three tetrahydroisoquinoline adducts studied functionally mimic an ICL, despite the fact that these drugs are covalently bonded to only one strand. Moreover, several of the DNA conformational states that were characterized during the melting transitions were unique to the MMC bis-adduct, and this observation might account for some of the differences observed experimentally regarding the repair of these lesions in mammalian cells (8).
MATERIALS AND METHODS
Construction of the deoxyoligonucleotides and force field parameters for the non-standard residues
An initial model for the drug-free 15-mer of sequence 5′-d(TAATAACGGATTATT)·5′-d(AATAATCCGTTATTA) was built using the nucgen module in the AMBER suite of programs (http://ambermd.org/) and optimized parameters for B-DNA (27). The structures of Yondelis® (8), Zalypsis® (9) and PM01183® (10) covalently bonded to the central guanine in the CGG triplet were built as previously reported. The N2(G)-MMC-N2(G) bisadduct with 1″-α stereochemistry (6) was modelled by changing the connectivity and subsequent energy refinement of the MMC-N2(G) monoadduct structure deposited in the Protein Data Bank [PDB (28)] with code 199D and then transferring the ICL to the 15-mer by best-fit superposition on the central CpG step. Electrostatic potential-derived (ESP) point charges for the atoms making up the ICL were assigned from the wavefunction calculated using a 6–31 G(d) basis set, as implemented in the ab initio quantum chemistry program Gaussian03 (29), and subsequent fitting employing the RESP methodology (30). Parameters derived for drug atoms were consistent with those present for polynucleotides and proteins in the AMBER force field (31), currently incorporating corrections (parmbsc0) for an improved description of DNA conformations on a multinanosecond time scale (11).
MD simulations
Each molecular system was immersed in a truncated octahedron of TIP3P water molecules (32) that extended 15 Å away from any solute atom so that the resulting volume was enough to accommodate the separation of the strands upon DNA melting. Incorporation of the appropriate number of sodium ions (33) at random locations ensured electrical neutrality. A salt concentration of 0.1 M NaCl was then achieved by addition to each complex of equivalent amounts of Na+ and Cl− ions. The cut-off distance for the non-bonded interactions was 9 Å and periodic boundary conditions were used. Electrostatic interactions were represented using the smooth particle-mesh Ewald method (34) with a grid spacing of 1 Å. The SHAKE algorithm (35) was applied to all bonds involving hydrogens, and an integration step of 2.0 fs was used throughout. The simulation protocol made use of the pmemd module in AMBER 10 (36) and consisted of a number of steps. First, solvent molecules and counterions were relaxed by energy minimization and allowed to redistribute around the positionally restrained solute (25 kcal·mol−1·Å−2) during 50 ps of MD at constant temperature (300 K) and pressure (1 atom) essentially as described previously (37). These initial harmonic restraints were gradually reduced in a series of progressive energy minimizations until they were completely removed. The resulting systems were then heated from 100 to 400 K during 25 ps, equilibrated at 400 K for 1 ns in the absence of any restraints and further simulated under the same conditions up to a total time of 200 ns during which system coordinates were collected every 20 ps for further analysis. Each complete simulation took ~2000 h of CPU time running in parallel on 32 IBM Power PC 970MP processors at the Barcelona Supercomputer Centre (MareNostrum).
Analysis of the MD trajectories
Three-dimensional structures and trajectories were visually inspected using the computer graphics program PyMOL (38). Interatomic or centroid–centroid distances, angles and root mean square deviations (rmsd) from a given structure were monitored using the ptraj module in AMBER. Our criteria for WC hydrogen-bonded pairs were (i) a distance between H-bond donor and acceptor atoms shorter than 3.2 Å, and (ii) the existence of at least two H-bonds between paired bases irrespective of their being A:T or G:C.
RESULTS AND DISCUSSION
Progressive loss of WC base pairing in the duplexes
General considerations
MD simulations of DNA duplexes at room temperature routinely show fraying and reannealing of terminal base pairs independently of the force field used (39). In line with these findings, the A:T base pairs closest to the ends of the duplexes studied here were lost soon after initiation of the MD production runs (Figure 2). This strand dissociation process was progressively followed by ‘peeling’, that is, untwisting motions favoured by nucleic base competition with water molecules and ions, as well as by non-canonical hydrogen bonding of the purine and pyrimidine base edges with the sugar-phosphate backbone. Our choice of A:T base pairs surrounding the central CGG triplet clearly had an impact on the speed of base pair disruption (Figure 3) as only two hydrogen bonds need to be broken compared with three in the case of G:C base pairs, and they also present a smaller surface area for stacking interactions. Likewise, the selection of non-homopolymeric sequences prevented further base complementarity searching to yield a change of registry, as reported for d(A12)·d(T12) (21) or d(CGCG)2 duplexes (18).
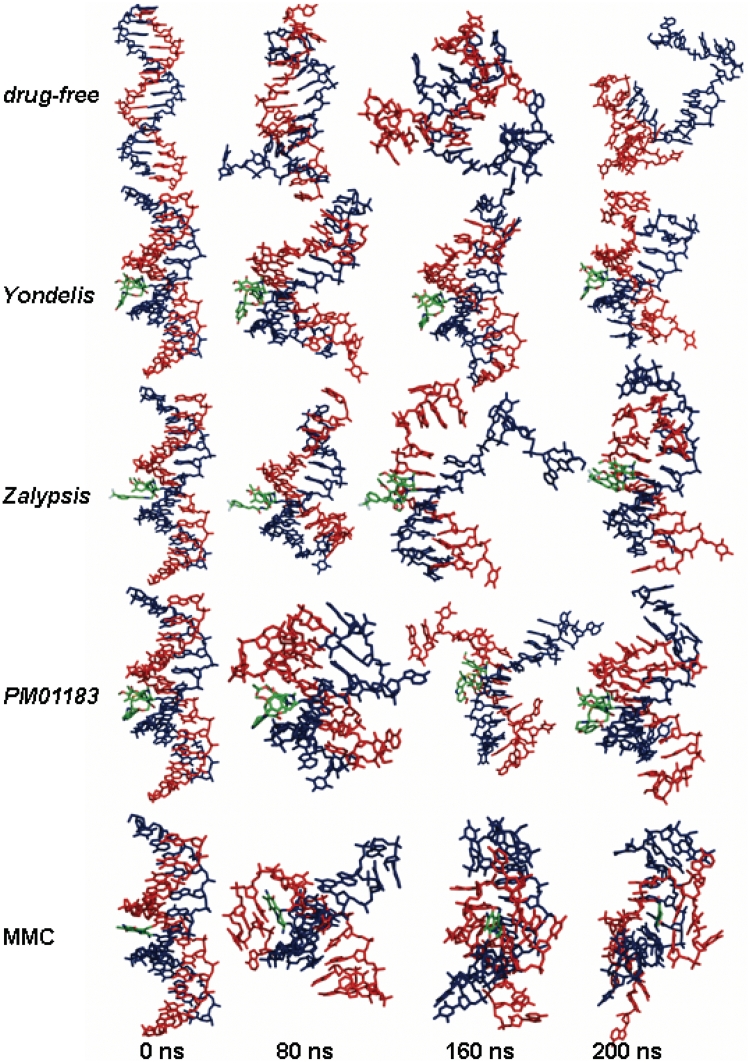
Figure 2.
Representative snapshots showing the time evolution of the DNA duplexes during the MD simulations in the absence of any bonded drug (top) and in the presence of Yondelis®, Zalypsis® or PM01183® covalently bonded to the blue strand, or MMC (bottom) covalently bonded to both blue and red strands. Drug carbon atoms are coloured in green. For more detailed visualization see the video in Supplementary Data.
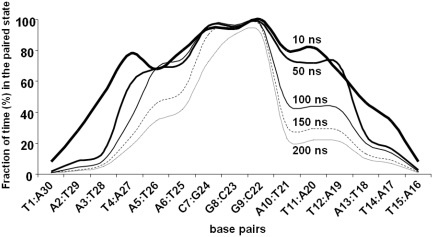
Figure 3.
Fraction of the MD simulation time (measured at 10, 50, 100, 150 and 200 ns) during which individual base pairs remained WC hydrogen bonded in the drug-free DNA 15-mer of sequence 5′-d(ATTATTCGGTAATAA)·5′-d(TTATTACCGAATAAT). Hydrogen bonds were rapidly lost for those base pairs found at the strand termini whereas those making up the central CGG triplet were the most durable. Note the asymmetry in the melting profiles of the DNA stretches located 5′ and 3′ to the central CGG triplet. This profile is to be compared with that obtained when these TAA and ATT stretches were swapped (Supplementary Figure S2).
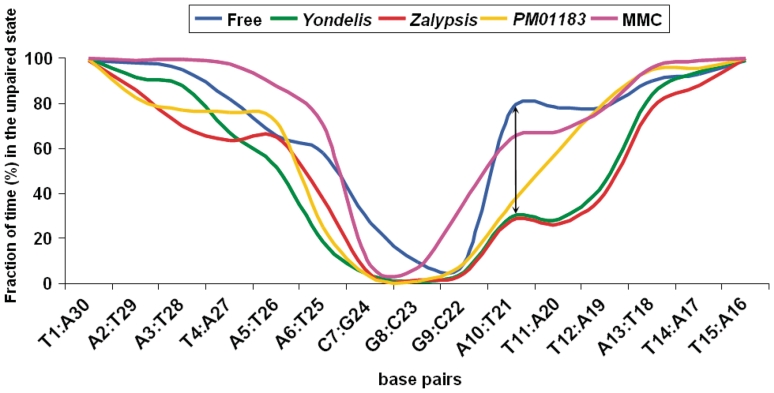
As the simulations progressed it became apparent that the stability of the central CGG triplet was crucial in the reaction coordinate of the melting process. Thus, for the drug-free DNA duplex this triplet remained WC hydrogen-bonded for not less than ~160 ns. At this point dehybridization took over rehybridization and at ~190 ns no standard WC hydrogen-bonded bases remained between the two strands. At 200 ns the simulation was stopped and we considered this time as the deadline for comparison with the other duplexes (Figure 4). Rather unexpectedly, the oligonucleotide containing the MMC bisadduct followed a very similar course, the main difference with respect to the drug-free DNA being that the two strands could not physically separate due to the existence of the ICL between the two guanines. But only the two G:C base pairs making up the CpG step remained WC hydrogen-bonded at the end of the simulation. In stark contrast, the duplexes containing the tetrahydroisoquinoline drug adducts in only one strand displayed a greatly increased stabilization in their central region, with up to 6 bp still hydrogen-bonded after 200 ns. Moreover, further dehybridization after ~110 ns was not only stopped but also reversed in the oligos containing Yondelis® or Zalypsis®. These reannealing events, which translate into a decreased percentage of time in the unpaired state for the base pairs on the 3′-side of the central CGG triplet relative to drug-free DNA (see shoulder in Figure 5), strongly suggest that the presence of the bulky drug in the minor groove helps stabilize the duplex on both sides of the covalent adduct.
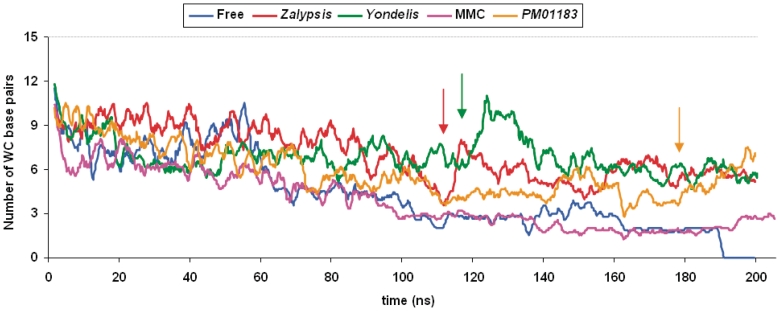
Figure 4.
DNA base-pair disruption along the production phase of the MD simulations of the free and drug-bonded DNA 15-mers. All the original base pairs were progressively lost in the drug-free DNA, the last ones belonging to the central CGG triplet. In all the drug-bonded complexes, however, this triplet remained H-bonded at the end of the simulation. Furthermore, in the DNA molecules containing the bonded tetrahydroisoquinolines, the WC base-paired region was extended further due to the occurrence of some conspicuous reannealing events (marked by a vertical arrow), which were more marked for Zalypsis® and Yondelis® than for PM01183®.
Figure 5.
Fraction of total MD simulation time during which a particular base pair was disrupted in the DNA 15-mer (note that for drug-free DNA this plot is the reverse of that shown in Figure 3). The lower frequency of the unpaired state in the central CGG triplet was extended to the ATT sequence 3′ to it (i.e. A10:T21 and T11:A20 base pairs) in the duplexes containing bonded Yondelis® and Zalypsis® (vertical double-headed arrow) as a consequence of the reannealing events shown in Figure 4.
Drug-free DNA complex
The rather dramatic decrease in the number of WC base pairs (from 15 down to 6) during the first ~12 ns of the MD production run at 400 K was mostly due to terminal base hydration, loss of stacking interactions (see below), and the existence of fast and reversible microevents of A:T base pair mismatching (Figure 4) following shearing of the canonical base pairs. This was followed by a period of ~40 ns during which an alternation of reversible dehybridization and rehybridization processes involving the central base pairs of the duplex maintained the double helical structure. The well-preserved central CGG triplet helped nucleate some extra dsDNA on both sides during this first part of the simulation forcing the closest AT base pairs to be temporarily reannealed (Figure 3). During the next 60 ns of simulation, the progressive and irreversible melting of all the A:T base pairs was observed. The result was that by 120 ns only the central and more stable CGG triplet remained WC hydrogen bonded. After this point, random hydrogen bonds between mismatched bases could be detected, but strong competition with the solvent prevented stabilization of any of these transition intermediates. Despite the occurrence of some occasional rehybridization microevents between 140 and 160 ns, the remaining two G:C base pairs were clearly not enough to promote further helicity and they broke up at 190 ns. At this time, we considered that the 15-mer was completely and irreversibly melted. Therefore, the three G:C base pairs making up the central triplet remained hydrogen bonded for a total length of about 190 ns.
The time evolution of the rmsd of the drug-free duplex with respect to the initial structure (Supplementary Figure S1) correlated strongly with the fraying-peeling process described above. The results of this simulation allowed us to understand the pathway of double helix disruption under denaturing conditions and were found to be in consonance with results from previous studies using different sequences (19,21). However, by introducing a central CGG triplet, we were able to assess more clearly the much higher stability of these three base pairs with respect to the surrounding AT pairs, which nicely highlights the base-pair dependence of the melting transition. Furthermore, examination of the evolution of the unpaired state of individual base pairs over the course of the simulation revealed some melting asymmetry. Thus, the ATT stretch located 3′ to the central CGG sequence behaved in an all-or-none fashion and melted as a block rather than progressively as did the TAA stretch located on the 5′ side (Figure 3). Interestingly, when TAA and ATT stretches on the 5′ and 3′ sides of the central CGG triplet were swapped, the asymmetry was even more accentuated (Supplementary Figure S2), with (ATT)2 on the 5′-side melting much faster and smoothly than the (TAA)2 block on the 3′-side, which was more resilient to melting and whose base pairs were also disrupted synchronously. Although more work is necessary to account for these differences, it seems clear that ATT triplets are more easily destabilized than TAA triplets, at least when located on the 5′-side of a CGG site. Our next goal was to examine the effect on this melting process of a drug-bonded guanine in the central triplet.
Drug-bonded DNA duplexes
The presence of the bonded drugs had a prominent effect on the denaturing profile of the DNA oligonucleotide (Figure 5). Most noticeable was the greater occurrence of reannealing events that took place over the course of the simulation, with some very dramatic examples showing up at ~120 ns (Figure 4). Indeed, six WC hydrogen-bonded base pairs still remained in the complexes containing the three tetrahydroisoquinolines midway the simulations and for Yondelis® this number went up to 11 at ~125 ns. In other words, the double helical structure was preserved at this time to virtually the same extent that it was at the beginning of the MD production run. These continuous dehybridization–rehybridization microevents kept taking place until the very end of the simulation. At this final time, the duplex structures were clearly maintained in the central region since the number of WC hydrogen-bonded base pairs was very close to six in the three complexes.
Remarkably, these critical reannealing events were not so perceptible in the MMC–DNA complex despite the fact that the two strands were physically linked through this drug, which effectively prevented strand separation (Figure 4). Therefore, the most likely explanation for this distinct behaviour is that the smaller size of this molecule, which fits into the minor groove causing only minimal distortion in the DNA (6), does not give rise to significant non-covalent (van der Waals and electrostatic) interactions with the DNA strands that could help stabilize the duplex. The two G:C base pairs making up the CpG step remained hydrogen bonded all along, with occasional extension to the remaining G:C base pair during the second half of the simulation, but this did not impinge on the stability of the neighbouring A:T base pairs. Interestingly, the last base pair to lose the WC hydrogen bonds in the simulation of the free DNA was G9:C22, whereas the most recalcitrant was C7:G24 when G8 and G24 were linked together by means of the bonded MMC.
Our results also showed, in common with previous evidence (19,21), that disrupted interstrand WC base pairing during the melting process, as described above, was accompanied by increased anomalous pairings in the duplex (Figure 2). Nonetheless, the solvent exposure of the polar atoms that make up the WC hydrogen bonds in a standard DNA double helix consistently increased during the simulated melting process of the drug-free duplex and reached a maximum after the two strands were separated (Supplementary Figure S3A). As expected, this increased exposure was much less marked in the duplexes containing the drug adducts, particularly in the case of Zalypsis®, in good correspondence with the better maintained WC base pairs discussed above.
Base stacking and hydrophobic effect
Base stacking plays an even greater role than hydrogen bonding in the stabilization of DNA duplexes in aqueous solutions due to a combination of the hydrophobic effect (40,41) and optimal dipolar interactions between the planar heteroaromatic systems (42). During the simulations, canonical intrastrand base stacking was largely maintained and, when lost, it was partially compensated by both non-contiguous intrastrand base stacking events and occasional interstrand face-to-face and edge-to-face interactions (data not shown). The overall result was that the individual strands showed a relatively high degree of compactness over the whole melting process (Figure 2). Nonetheless, despite the partial collapse of the individual strands, the DNA non-polar atoms were more exposed to the solvent at the end of the simulations than at the beginning (Supplementary Figure S3B) as a consequence of the increased disorder and loss of stacking interactions. Comparison of initial and final states for the different duplexes shows that those containing the adducts initially display a reduced solvent-accessible surface area (SASA) of their polar atoms relative to drug-free DNA due to partial occlusion by the bonded drug and also reduced solvent exposure at the end of the simulations as a consequence of the larger number of WC hydrogen-bonded base pairs. These differences are not as striking in the case of non-polar atoms because the extent of avoidance of interactions with water is much more similar in all the duplexes.
Implications for the mode of action of these drugs
The relevance of the present results is supported by (i) renewed interest in DNA minor groove binders as anti-cancer therapeutic agents (7,43), (ii) the established role of MMC in the therapy of bladder tumours (44) and other types of cancer, (iii) the recent approval of trabectedin (Yondelis®) (45) for the treatment of patients with advanced or metastatic soft tissue sarcoma and relapsed platinum-sensitive ovarian carcinoma and (iv) the ongoing clinical development of Zalypsis® (9) and PM01183® (10) for the treatment of solid tumours and haematological malignancies.
The three tetrahydroisoquinolines studied here have in common a fused pentacyclic skeleton that is mostly responsible for DNA recognition and binding to the amino group of a suitably embedded guanine (7). The sequence specificity of the three drugs depends on the establishment of highly specific hydrogen bonds with the nucleotides on both sides of the guanine that will be covalently bonded (46,47). Besides, non-covalent interactions within the minor groove help to stabilize the double helical structure of the duplex to which they bind, as inferred from experiments demonstrating notable increases (~20°C) in the melting temperature of dsDNA containing a single drug adduct (8–10) and the MD simulation results presented herein. Thus, although these three compounds are covalently bonded to only one strand of the DNA double helix, they have been viewed (8) as functional interstrand crosslinkers insofar as they prevent strand separation very effectively. To compare this action with that elicited by a true interstrand crosslinker, we chose MMC as a prototypical representative because of the similarities detected in cells exposed to all of these drugs regarding (i) phosphorylation of histone H2AX, a surrogate marker of double-strand breaks, (ii) Rad51 foci formation in the nucleus, which is known to facilitate DNA repair via homologous recombination and (iii) participation of the Fanconi anaemia (FA) pathway in the processing of the DNA damage (8). Nonetheless, a notable and yet unexplained difference regarding the effects of MMC and Yondelis® on the cell cycle of FA-deficient cells is that trabectedin, unlike MMC, did not induce any significant accumulation of cells in G2-M. Among the possible reasons for this differential effect on cell-cycle progression, the nature of the interaction of these drugs with DNA was pinpointed (8). The present results reinforce this possibility and provide a clear atomistic rationale for this interpretation even though more work will be necessary to clarify this point categorically.
CONCLUSIONS
DNA melting in the absence and presence of bonded drugs can provide information about the duplex stabilization brought about by drug binding. Performing the simulations under conditions similar to those used in the real-life experiments, i.e. progressively increasing the temperature until complete separation of the two strands is detected, is currently unfeasible given the time scale of this process. However, by employing a constant elevated temperature from the outset as a control parameter for overcoming energy barriers more easily and using high salt concentrations, we were able to detect complete thermal denaturation of the drug-free oligodeoxynucleotide in <200 ns, still a formidable time for simulating large macromolecular systems such as these but now made affordable by the combined use of optimized parallel algorithms and available supercomputing power.
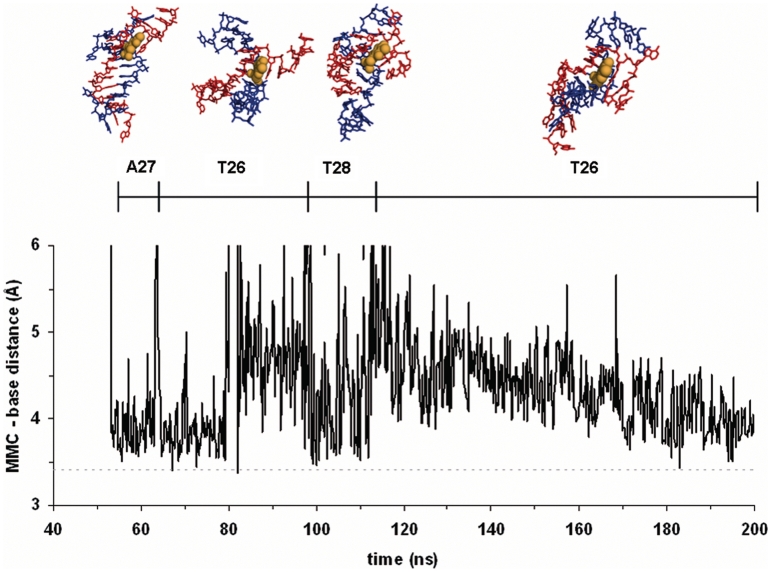
Initially, the duplex under study had fifteen WC hydrogen-bonded base pairs. During the initial stages of the simulation the frayed nucleobases at the ends started looping back to make transient hydrogen bonds with the sugar-phosphate backbone and this resulted in both untwisting and peeling apart of the DNA strands, essentially as described for similar duplexes (19,21). Although the number of intact base pairs consistently decreased with time, a striking asymmetry was observed on both sides of the central CGG triplet, as well as considerable fluctuations over the full course of the denaturation process. Furthermore, the progressive unzipping was clearly interspersed with some reannealing events, which were most noticeable in the oligonucleotides containing the tetrahydroisoquinoline monoadducts. After 200 ns of MD simulation, all the original WC hydrogen bonds were lost in the free oligonucleotide, whereas an extended WC-paired region embedding the central CGG triplet was still maintained in all drug–DNA complexes. This finding attests to the ability of these drugs to stabilize dsDNA and stall replication and transcription forks. On the other hand, in the MMC–DNA complex (Figure 6), but not in those with Yondelis®, Zalypsis® or PM01183®, several unique arrangements between the drug and different DNA bases in the ss region favoured the transient formation of distinct secondary structures in which MMC appeared pseudointercalated (Supplementary Figure S4). These hydrophobically collapsed intermediates, together with the existence of a true ICL physically connecting the two DNA strands, are likely to pose distinct challenges to the DNA repair machinery and may account for the differences in cellular response that have been detected experimentally (8).
Figure 6.
Distances between the ring centroids of MMC and different DNA nucleobases during the simulated melting process. The drug’s chromophore (CPK spheres coloured in orange) served as a platform for hydrophobic contacts with different nucleobases from one of the melted strands (that coloured in red), in the chronological order shown at the top of the figure. The dotted line given as a reference at 3.4 Å represents the canonical distance between stacked nucleobases in a standard B-DNA double helix.
Taken together, our results provide an atomistic view of the process of DNA melting in the presence and absence of several bonded anti-tumour drugs, and they are found to be in agreement with the large stabilization that single Yondelis®, Zalypsis® and PM01183® monoadducts have been shown to impose on the DNA double helix (8–10). On the contrary, the prototypical interstrand crosslinker MMC does prevent strand separation because of the fact that it forms a covalent bond with each DNA strand, but its effect on duplex stabilization does not appear to extend beyond the CpG step to which it is covalently attached. Therefore, this is another example of how theory and experiment can be successfully merged to help us improve our understanding in atomic detail of anti-tumour drugs acting on such a vital macromolecule as DNA.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
This work was partially supported by l’Association de la Recherche contre le Cancer (ARC n°3153), la Ligue contre le Cancer, INCA and an ERC Advanced grant (to J.M.E.), and Comisión Interministerial de Ciencia y Tecnología (SAF2009-13914-C02-02) (to F.G.). C.G. is a recipient of a fellowship from the Ministère de la Recherche and ARC. J.A.B.-C. enjoys a research fellowship from PharmaMar S.A.U. Funding for open access charge: Comisión Interministerial de Ciencia y Tecnología (SAF2009-13914-C02-02).
Conflict of Interest statement: C.M.G. is an employee and shareholder of PharmaMar S.A.U. No other potential conflicts of interest were disclosed.
Supplementary Material
ACKNOWLEDGEMENTS
We thankfully acknowledge the generous allowance of computer resources, technical expertise and assistance provided by the Red Española de Supercomputación at the Barcelona Supercomputing Center (MareNostrum).
REFERENCES
- 1.Mergny JL, Lacroix L. Analysis of thermal melting curves. Oligonucleotides. 2003;13:515–537. doi: 10.1089/154545703322860825. [DOI] [PubMed] [Google Scholar]
- 2.Stucki M, Stagljar I, Jónsson ZO, Hübscher U. A coordinated interplay: proteins with multiple functions in DNA replication, DNA repair, cell cycle/checkpoint control, and transcription. Prog. Nucleic Acid. Res. Mol. Biol. 2001;65:261–298. doi: 10.1016/s0079-6603(00)65007-9. [DOI] [PubMed] [Google Scholar]
- 3.Bustamante C, Cheng W, Meija YX. Revisiting the central dogma one molecule at a time. Cell. 2011;144:480–497. doi: 10.1016/j.cell.2011.01.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Negri A, Marco E, García-Hernández V, Domingo A, Llamas-Saiz AL, Porto-Sandá S, Riguera R, Laine W, David-Cordonnier MH, Bailly C, et al. Antitumor activity, X-ray crystal structure, and DNA binding properties of thiocoraline A, a natural bis-intercalating thiodepsipeptide. J. Med. Chem. 2007;50:3322–3333. doi: 10.1021/jm070381s. [DOI] [PubMed] [Google Scholar]
- 5.Lawley PD, Phillips DH. DNA adducts from chemotherapeutic agents. Mutat. Res. 1996;355:13–40. doi: 10.1016/0027-5107(96)00020-6. [DOI] [PubMed] [Google Scholar]
- 6.Tomasz M. Mitomycin C: small, fast and deadly (but very selective) Chem. Biol. 1995;2:575–579. doi: 10.1016/1074-5521(95)90120-5. [DOI] [PubMed] [Google Scholar]
- 7.Manzanares I, Cuevas C, García-Nieto R, Marco E, Gago F. Advances in the chemistry and pharmacology of ecteinascidins, a promising new class of anticancer agents. Curr. Med. Chem. Anti-Canc. Agents. 2001;1:257–276. doi: 10.2174/1568011013354561. [DOI] [PubMed] [Google Scholar]
- 8.Casado JA, Río P, Marco E, García-Hernández V, Domingo A, Pérez L, Tercero JC, Vaquero JJ, Albella B, Gago F, et al. The relevance of the Fanconi anemia pathway in the response of human cells to trabectedin. Mol. Cancer Ther. 2008;7:1309–1318. doi: 10.1158/1535-7163.MCT-07-2432. [DOI] [PubMed] [Google Scholar]
- 9.Leal JF, García-Hernández V, Moneo V, Domingo A, Bueren-Calabuig JA, Negri A, Gago F, Guillén-Navarro MJ, Avilés P, Cuevas C, et al. Molecular pharmacology and antitumor activity of Zalypsis in several human cancer cell lines. Biochem. Pharmacol. 2009;78:162–170. doi: 10.1016/j.bcp.2009.04.003. [DOI] [PubMed] [Google Scholar]
- 10.Leal JF, Martínez-Díez M, García-Hernández V, Moneo V, Domingo A, Bueren-Calabuig JA, Negri A, Gago F, Guillén-Navarro MJ, Avilés P, et al. PM01183, a new DNA minor groove covalent binder with potent in vitro and in vivo anti-tumour activity. Brit. J. Pharmacol. 2010;161:1099–1110. doi: 10.1111/j.1476-5381.2010.00945.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.McHugh PJ, Spanswick VJ, Hartley JA. Repair of DNA interstrand crosslinks: molecular mechanisms and clinical relevance. Lancet Oncol. 2001;2:483–490. doi: 10.1016/S1470-2045(01)00454-5. [DOI] [PubMed] [Google Scholar]
- 12.Pérez A, Marchán I, Svozil D, Sponer J, Cheatham TE, 3rd, Laughton CA, Orozco M. Refinement of the AMBER force field for nucleic acids: improving the description of alpha/gamma conformers. Biophys. J. 2007;92:3817–3829. doi: 10.1529/biophysj.106.097782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Schatz GC. Using theory and computation to model nanoscale properties. Proc. Natl. Acad. Sci. USA. 2007;104:6885–6892. doi: 10.1073/pnas.0702187104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Orozco M, Noy A, Pérez A. Recent advances in the study of nucleic acid flexibility by molecular dynamics. Curr. Opin. Struct. Biol. 2008;18:185–193. doi: 10.1016/j.sbi.2008.01.005. [DOI] [PubMed] [Google Scholar]
- 15.Drukker K, Schatz GC. A model for simulating dynamics of DNA denaturation. J. Phys. Chem. B. 2000;104:6108–6111. [Google Scholar]
- 16.Drukker K, Wu G, Schatz GC. Model simulations of DNA denaturation dynamics. J. Chem. Phys. 2001;114:579–590. [Google Scholar]
- 17.Tepper HL, Voth GA. A coarse-grained model for double-helix molecules in solution: spontaneous helix formation and equilibrium properties. J Chem. Phys. 2005;122:124906. doi: 10.1063/1.1869417. [DOI] [PubMed] [Google Scholar]
- 18.Linak MC, Dorfman KD. Analysis of a DNA simulation model through hairpin melting experiments. J. Chem. Phys. 2010;133:125101. doi: 10.1063/1.3480685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kannan S, Zacharias M. Simulation of DNA double-strand dissociation and formation during replica-exchange molecular dynamics simulations. Phys. Chem. Chem. Phys. 2009;11:10589–10595. doi: 10.1039/b910792b. [DOI] [PubMed] [Google Scholar]
- 20.Pérez A, Orozco M. Real-time atomistic description of DNA unfolding. Angew. Chem. Int. Ed. 2010;49:4805–4808. doi: 10.1002/anie.201000593. [DOI] [PubMed] [Google Scholar]
- 21.Hagan MF, Dinner AR, Chandler D, Chakraborty AK. Atomistic understanding of kinetic pathways for single base-pair binding and unbinding in DNA. Proc. Natl Acad. Sci. USA. 2003;100:13922–13927. doi: 10.1073/pnas.2036378100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wong KY, Pettitt BM. The pathway of oligomeric DNA melting investigated by molecular dynamics simulations. Biophys. J. 2008;95:5618–5626. doi: 10.1529/biophysj.108.141010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Piana S. Atomistic simulation of the DNA helix-coil transition. J. Phys. Chem. A. 2007;111:12349–12354. doi: 10.1021/jp0756552. [DOI] [PubMed] [Google Scholar]
- 24.Feuerhahn S, Giraudon C, Martínez-Díez M, Bueren-Calabuig JA, Galmarini CM, Gago F, Egly JM. XPF-dependent DNA breaks and RNA polymerase II arrest induced by antitumor DNA interstrand crosslinking-mimetic tetrahydroisoquinoline alkaloids. Chem. Biol. doi: 10.1016/j.chembiol.2011.06.007. (in preparation) [DOI] [PubMed] [Google Scholar]
- 25.de Laat WL, Appeldoorn E, Jaspers NGJ, Hoeijmakers JHJ. DNA structural elements required for ERCC1-XPF endonuclease activity. J. Biol. Chem. 1998;273:7835–7842. doi: 10.1074/jbc.273.14.7835. [DOI] [PubMed] [Google Scholar]
- 26.Rahn JJ, Adair GM, Nairn RS. Multiple roles of ERCC1-XPF in mammalian interstrand crosslink repair. Environ. Mol. Mutagen. 2010;51:567–581. doi: 10.1002/em.20583. [DOI] [PubMed] [Google Scholar]
- 27.Arnott S, Hukins DW. Optimised parameters for A-DNA and B-DNA. Biochem. Biophys. Res. Comm. 1972;47:1504–1509. doi: 10.1016/0006-291X(72)90243-4. [DOI] [PubMed] [Google Scholar]
- 28.Berman HM, Westbrook J, Feng Z, Gilliland G, Bhat TN, Weissig H, Shindyalov IN, Bourne PE. The Protein Data Bank. Nucleic Acids Res. 2000;28:235–242. doi: 10.1093/nar/28.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Frisch MJ, Trucks GW, Schlegel HB, Scuseria GE, Robb MA, Cheeseman JR, Zakrzewski VG, Montgomery JA, Jr, Stratmann RE, Burant JC, et al. Gaussian 03, Revision B. 04. Pittsburgh, PA: Gaussian, Inc.; 2003. [Google Scholar]
- 30.Bayly CI, Cieplak P, Cornell WD, Kollman PA. A well-behaved electrostatic potential based method using charge restraints for determining atom-centered charges: the RESP model. J. Phys. Chem. 1993;97:10269–10280. [Google Scholar]
- 31.Cornell WD, Cieplak P, Bayly CI, Gould IR, Merz KM, Ferguson DM, Spellmeyer DC, Fox T, Caldwell JW, Kollman PA. A second-generation force field for the simulation of proteins, nucleic acids and organic molecules. J. Am. Chem. Soc. 1995;117:5179–5197. [Google Scholar]
- 32.Jorgensen WL, Chandrasekhar J, Madura JD. Comparison of simple potential functions for simulating liquid water. J. Chem. Phys. 1983;79:926–935. [Google Scholar]
- 33.Åqvist J. Ion-water interaction potentials derived from free energy perturbation simulations. J. Phys. Chem. 1990;94:8021–8024. [Google Scholar]
- 34.Darden TA, York D, Pedersen LG. Particle mesh Ewald: an N*log(N) method for computing Ewald sums. J. Chem. Phys. 1993;98:10089–10092. [Google Scholar]
- 35.Ryckaert JP, Ciccoti G, Berendsen HJC. Numerical integration of the cartesian equations of motion of a system with constraints: molecular dynamics of n-alkanes. J. Comput. Phys. 1977;23:327–341. [Google Scholar]
- 36.Case DA, Darden TA, Cheatham TE, III, Simmerling CL, Wang J, Duke RE, Luo R, Crowley M, Walker W, Kollman PA, et al. AMBER 10. San Francisco: University of California; 2008. [Google Scholar]
- 37.Marco E, Negri A, Luque FJ, Gago F. Role of stacking interactions in the binding sequence preferences of DNA bis-intercalators: insight from thermodynamic integration free energy simulations. Nucleic Acids Res. 2005;33:6214–6224. doi: 10.1093/nar/gki916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lano WD. 2006. PyMOL version 0.99. DeLano Scientific LLC. http://www.pymol.org/ (10 May 2011, date last accessed) [Google Scholar]
- 39.Feig M, Pettitt BM. Structural equilibrium of DNA represented with different force fields. Biophys. J. 1998;75:134–149. doi: 10.1016/S0006-3495(98)77501-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Saenger W. Principles of Nucleic Acid Structure. NY: Springer-Verlag; 1984. [Google Scholar]
- 41.Friedman RA, Honig B. A free energy analysis of nucleic acid base stacking in aqueous solution. Biophys. J. 1995;69:1528–1535. doi: 10.1016/S0006-3495(95)80023-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gago F. Stacking interactions and intercalative DNA binding. Methods. 1998;14:277–292. doi: 10.1006/meth.1998.0584. [DOI] [PubMed] [Google Scholar]
- 43.Susbielle G, Blattes R, Brevet V, Monod C, Kas E. Target practice: aiming at satellite repeats with DNA minor groove binders. Curr. Med. Chem. 2005;5:409–420. doi: 10.2174/1568011054222355. [DOI] [PubMed] [Google Scholar]
- 44.Shelley MD, Mason MD, Kynaston H. Intravesical therapy for superficial bladder cancer: a systematic review of randomised trials and meta-analyses. Cancer Treat Rev. 2010;36:195–205. doi: 10.1016/j.ctrv.2009.12.005. [DOI] [PubMed] [Google Scholar]
- 45.D’Incalci M, Galmarini CM. A review of trabectedin (ET-743): a unique mechanism of action. Mol. Cancer Ther. 2010;9:2157–2163. doi: 10.1158/1535-7163.MCT-10-0263. [DOI] [PubMed] [Google Scholar]
- 46.García-Nieto R, Manzanares I, Cuevas C, Gago F. Bending of DNA upon binding of ecteinascidin 743 and phthalascidin 650 studied by unrestrained molecular dynamics simulations. J. Am. Chem. Soc. 2000;122:7172–7182. [Google Scholar]
- 47.Marco E, David-Cordonnier MH, Bailly C, Cuevas C, Gago F. Further insight into the DNA recognition mechanism of trabectedin from the differential affinity of its demethylated analog ecteinascidin ET729 for the triplet binding site CGA. J. Med. Chem. 2006;49:6925–6929. doi: 10.1021/jm060640y. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.