Abstract
The icsA gene of Shigella encodes a structural protein involved in colonization of the intestinal mucosa by bacteria. This gene is expressed upon invasion of the host and is controlled by a complex regulatory circuit involving the nucleoid protein H-NS, the AraC-like transcriptional activator VirF, and a 450 nt antisense RNA (RnaG) acting as transcriptional attenuator. We investigated on the interplay of these factors at the molecular level. DNase I footprints reveal that both H-NS and VirF bind to a region including the icsA and RnaG promoters. H-NS is shown to repress icsA transcription at 30°C but not at 37°C, suggesting a significant involvement of this protein in the temperature-regulated expression of icsA. We also demonstrate that VirF directly stimulates icsA transcription and is able to alleviate H-NS repression in vitro. According to these results, icsA expression is derepressed in hns- background and overexpressed when VirF is provided in trans. Moreover, we find that RnaG-mediated transcription attenuation depends on 80 nt at its 5′-end, a stretch carrying the antisense region. Bases engaged in the initial contact leading to sense–antisense pairing have been identified using synthetic RNA and DNA oligonucleotides designed to rebuild and mutagenize the two stem–loop motifs of the antisense region.
INTRODUCTION
Shigella is a highly adapted human pathogen that causes the bacillary dysentery. This enteric syndrome, mainly found in the developing world, is extremely contagious and, although self-limiting, may be life threatening and often fatal in children (1). Shigellosis is the result of the coordinated action of several genes encoded by both the large virulence plasmid (pINV) and the chromosome. Upon host invasion, the primary event consequent to the upshift of bacteria to the host temperature (37°C) is the synthesis of two plasmid-encoded regulators: VirF and VirB. First, VirF, an AraC-like activator, triggers a regulatory cascade involving the activation of the icsA and virB genes. Then, VirB activates several operons-encoding factors required to invade and colonize the intestinal mucosa (2,3).
At this stage, an important virulence factor is represented by the IcsA protein (also known as VirG), which sponsors the intra- and intercellular spread of bacteria (4). IcsA is an outer membrane protein, which asymmetrically accumulates at one pole of the bacterial cell, catalyzing the direct elongation of an actin tail which propels Shigella through the cytoplasm and facilitates the intercellular dissemination toward neighboring cells (5). Due to its prime role in the invasion process, it is reasonable to expect the regulation of the icsA gene to be a sophisticated multilayered process. VirF, the master activator of the plasmid-encoded virulence cascade, has been shown to induce the expression of icsA in vivo (2,6,7), although little is known on its molecular mechanism. Another player in the regulation of icsA is the nucleoid associated protein H-NS, implicated in chromosome organization and also acting as a global regulator (8–10). H-NS has been shown to repress icsA at low temperature (11) and to specifically interact with the icsA promoter (12). This protein also binds and represses the transcription of virF and virB as a function of temperature, playing a direct role in silencing the Shigella virulence regulon outside the host (13–16).
We have recently discovered a small non-coding antisense RNA of ~450 nt, termed RnaG, controlling icsA expression and transcribed in cis from the icsA sequence (17). Small RNAs are emerging as a powerful tool in gene regulation and particularly in modulating adaptive stress–responses that are important for bacteria to survive within the host (18–21). Different RNA–RNA-mediated mechanisms, entailing changes in processing and degradation of the target messenger and alterations of the efficiency of transcription and translation have been described (22–24). RnaG regulates icsA transcription by two independent mechanisms; transcriptional interference and transcriptional attenuation. As for the first mechanism, we have recently shown that silencing of the RnaG promoter results in a higher activity of the icsA promoter (17). On the other hand, we have also shown that RnaG represents the first evidence of a RNA-mediated transcriptional attenuation of a bacterial virulence gene. In fact, binding of RnaG to the nascent icsA transcript induces the formation of a stem–loop motif at the 5′-end of the mRNA, mimicking an intrinsic terminator and resulting in the synthesis of a truncated icsA mRNA (17).
Here, we report on the identification of the H-NS and VirF-binding sites on the icsA and RnaG promoter regions and on the in vivo and in vitro relevance of their interactions on the expression of icsA. VirF directly stimulates icsA transcription while H-NS contributes to the thermoregulation of icsA, severely reducing its transcription at 30°C. We also found that the RnaG-mediated premature termination of the icsA transcript depends on about 80 out of 120 nt of the antisense region of RnaG. In particular, by the use of synthetic RNA and DNA molecules and a site-specific mutagenesis approach, we have been able to identify the RnaG regions initially contacting with the icsA mRNA to form the so called kissing complex and needed for the transcriptional attenuation.
MATERIALS AND METHODS
Bacterial strains and general procedures
Escherichia coli K12 strain HMG9 is a HMG11 derivative defective in the hns gene. Strains ULS1127 and ULS1129 are P90C monolysogens for a λ transducing phage carrying a PicsA-lacZ transcriptional fusion with a functional or a silenced PRnaG convergent promoter, respectively (17). Proteins were expressed in E. coli strain BL21(DE3) containing the pLysS plasmid (Promega).
Plasmid pMYSH6601 is a pBR322-derivative containing the virA-icsA region of S. flexneri 2a pINV (31). A 478-bp icsA fragment (from positions −262 to +216) was amplified by PCR using the primer pair GZ24/ACC9 on the pMYSH6601 template and cloned into the BamHI site of pKK232-8, thus obtaining plasmid pKG450.
Plasmids pGT1127 is a pGEMT-easy derivative containing a 866 bp from the virA-icsA region (−448 to +419) of the S. flexneri 2a. Plasmids pGT1129 and pGT1083 contain the same fragment lacking the PRnaG or PicsA promoter, respectively.
Plasmid pGT80T contains the first 80 bp of the sequence-encoding RnaG followed by a transcription termination signal. It has been obtained by cloning into pGEMT-easy a fragment obtained by amplifying pGT1127 DNA with the oligo pair G+120H/G40T (or ACC9/G40T).
pMYSH6504 and pDIA510 plasmids are both pBR322 derivatives containing, respectively, the entire virF gene from S. flexneri 2a pINV (25) and the entire hns gene from E. coli (26).
β-galactosidase assays were performed with sodium dodecyl sulfate/chloroform-permeabilized cells grown in LB medium supplemented with ampicillin. β-galactosidase activity was determined as described by Miller (1992) (48) and the results were expressed as averages of three independent experiments.
Extraction of plasmid DNA, restriction digestion, electrophoresis, PCR and purification of DNA fragments were carried out as described previously (16). RNA extraction was carried out as previously described by La Teana et al. (27) except that hot phenol and chloroform:isoamyle alcohol (24:1) extractions were performed in place of purification in CsCl.
Radioactivity associated with DNA or RNA was detected and quantified by Molecular Imager (Bio-Rad, mod. FX) and oligos used in this study are listed in Supplementary Data and Table S1.
VirF purification
To obtain overexpression of VirF, we constructed pULS22F, a plasmid containing the entire virF gene of S. flexneri 2a. To this end, an amplicon obtained upon PCR amplification of the template plasmid pMYSH6504 DNA (25) with oligo pair FsinL and SW04 (modified to contain NdeI or XhoI sites, respectively) was introduced into the pET22b vector (Novagen) linearized by NdeI/XhoI digestion. pULS22F was then transformed into BL21(DE3) pLysS strain selecting for Apr. Expression of the C-terminally His-tagged VirF protein was induced with 1 mM IPTG in exponentially growing 500 ml of culture. Bacterial cells were then sonicated and the protein was purified under native conditions using immobilized-metal affinity chromatography (IMAC) with a Ni–nitrilotriacetic acid-agarose column according to the manufacturer protocol (Qiagen). The protein was finally eluted as 1-ml fractions with 8-ml elution buffer (50 mM Na2HPO4, 300 mM NaCl, 10% Glycerol, 0.1% Tween-20, 10 mM β-mercaptoethanol) containing from 50 to 500 mM imidazole. Fractions were analyzed by SDS–PAGE and those containing the VirF protein were pooled and dialyzed against 1 l of 20 mM Tris–HCl, pH 8, 30 mM Na2HPO4, 50 mM NaCl, 5% glycerol. The His-tagged wild type VirF protein was found to be fully functional in vivo (data not shown).
RnaG80 synthesis and purification
Plasmid pTZRnaG80 was obtained by cloning 80 nt of the RnaG gene (from positions +40 to +120 with respect to the icsA transcription start) into the HindIII/EcoRI restriction sites of pTZ19R, under the control of the T7 promoter. This DNA fragment was obtained by PCR using pGT1127 as DNA template and the primer G+120H and G+40E. To synthesize RnaG80 transcript, pTZRnaG80 was linearized with EcoRI and used as template in an in vitro transcription reaction with T7 RNA polymerase as described by Brandi et al. (28).
DNase I footprinting
Supercoiled plasmids pKG673 (17) and pKG450 (~150 ng/sample) were preincubated 5 min at 25°C with the indicated concentrations of H-NS or VirF in 30 µl of binding buffer (40 mM HEPES–HCl, pH 8.0, 100 mM KCl, 10 mM magnesium acetate and 0.5 mM dithiothreitol). The DNA–protein complex was incubated with 1 U of DNase I for 40 s. After stopping the reaction, the DNA was precipitated and extended as described by Giangrossi et al. (29).
In vitro transcription
In vitro transcription from supercoiled plasmids (~200 ng/sample) as DNA templates was carried out at 30 and at 37°C for 20 min. Each reaction mixture (40 µl) contained 40 mM Tris–HCl, pH 7.5, 150 mM KCl, 10 mM MgCl2, 10 mM dithiothreitol, 0.01% Triton X-100, 0.5 mM each of NTPs, 2 U of ribonuclease inhibitor and 0.2 U of E. coli RNA polymerase (USB). The reaction was stopped on ice and RNA was precipitated with ethanol in the presence of 1 µg tRNA as carrier. The transcription product was detected by primer extension using a [γ32 P]-labeled oligonucleotide essentially as described by Giangrossi et al. (17).
Chemical RNA probing
Chemical modification of RNA was performed as described by Giangrossi et al. (17) using the single-strand-specific reagents DMS (A and C specific) and CMCT (U and G specific). Control samples (−) were treated identically with the exception that no modifying reagents were added. The modified RNA was subjected to primer extension using the oligo G+40E.
RESULTS
H-NS and VirF modulate transcription of icsA and RnaG by a direct interaction at promoter regions
Although H-NS and VirF are known to be involved in the regulation of virulence genes in S. flexneri (2,30), very little is known on their role in the modulation of icsA expression, especially at the molecular level. While band shift assays indicate that H-NS recognizes the icsA promoter (12), no evidence supports a binding of VirF to the icsA promoter region.
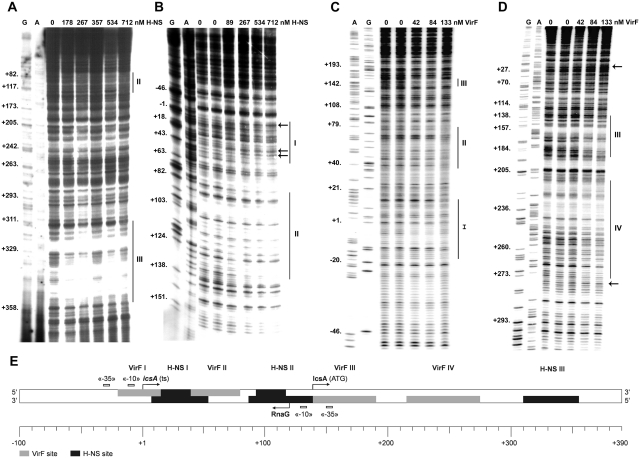
We investigated on the interaction of H-NS and VirF with the regulatory region of icsA by DNase I footprinting. H-NS protects three sites, numbered I, II and III and centered at positions +25, +115 and +330, respectively (Figure 1A and B), while VirF recognizes four regions, numbered I, II, III and IV and centered at positions −3, +62, +160 and +245, respectively (Figure 1C and D). DNase I protected regions are reported on the icsA sequence in Supplementary Figure S1 and are indicated in the schematic map shown in Figure 1E. H-NS-binding sites have an average length of ~50 bp and, within the limit of accuracy of this type of analysis, were bound with similar affinities by H-NS (~250–500 nM). As opposed to other H-NS regulated genetic systems, including the Shigella virulence genes virB, virF and virA, where H-NS-binding sites have been mapped on upstream regulatory regions (13–15), in the case of the icsA promoter H-NS binds downstream the transcriptional initiation site. Interestingly, H-NS site II includes the −10 conserved promoter element of RnaG whose occlusion might be relevant for transcriptional inhibition of PRnaG by H-NS (see below). VirF sites, spanning from ~40 to 60 bp in length, are detectable at low protein concentrations (~80–130 nM). VirF protection at site I covers the TATA box of PicsA and partially overlaps the left end of H-NS site I, while the right end of the latter site is superimposed on the VirF site II. Moreover, H-NS bound to site II and VirF bound to site III may contact the RnaG promoter (Figure 1E).
Figure 1.
H-NS and VirF show multiple binding sites on icsA regulatory region. Supercoiled plasmids pKG673 (A, C and D) and pKG450 (B), containing the promoter and part of the coding region of icsA, were incubated at the indicated concentrations of either H-NS (A and B) or VirF (C and D) expressed as dimer and monomer, respectively. Samples were processed as described in ‘Materials and methods’ section, using the oligos GBA (A), ACC9 (B), G–100 (C) and G+370 E (D) as primers. Lanes G, A and C represent the sequencing reactions using the same primers. Protections and sites hypersensitive to DNase I are indicated by vertical lines and arrows, respectively. The diagram shows the icsA promoter region from positions −100 to +390 (E). H-NS and VirF DNase I protections, −10 and −35 consensus elements, transcriptional start points of icsA and RnaG (ts) and the icsA initiation triplet (ATG) are indicated.
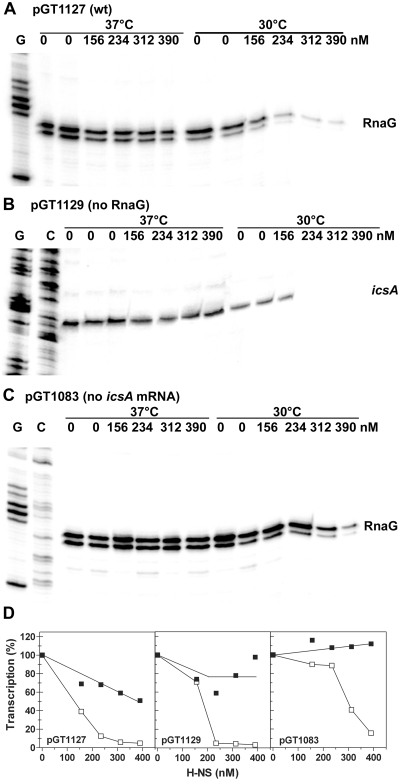
The position of H-NS and VirF-binding sites within the icsA promoter region suggests that both genes icsA and RnaG might be controlled by these regulators and provides a physical basis for a possible functional competition between the two proteins. Initially, we analyzed the effect of H-NS on the transcription of icsA and RnaG at 30 and 37°C in an in vitro system (Figure 2), using plasmids pGT1127 (wt), pGT1129 (inactive PRnaG) and pGT1083 (inactive PicsA) as DNA templates. The latter two constructs carry mutations in the TATA box of RnaG and icsA, respectively, therefore the transcription of these genes is abolished. This allowed us to study the transcription of these genes discriminating the effects caused by H-NS from those due to the transcriptional interference (TI) between the convergent promoters of icsA and RnaG (17). At 30°C, H-NS is able to inhibit transcription of both icsA and RnaG. Such repression is more pronounced on PicsA than on PRnaG. At ~200 nM H-NS, the icsA mRNA is no longer detectable and transcription attains <5% of the basal level obtained in the absence of protein (Figure 2B and D). Double amounts of H-NS (300–400 nM) are required to produce a comparable effect on RnaG transcription (Figure 2A and D). At 37°C, H-NS almost completely loses its capacity to act as transcriptional silencer, strongly suggesting that this protein plays a key role in the temperature-dependent regulation of icsA and RnaG genes. According to TI, the inactivation of icsA promoter stimulates the transcriptional activity of the convergent RnaG promoter. Under this condition of hyper-expression, RnaG seems less susceptible to H-NS repression even at 30°C. In fact, at 234 and 312 nM H-NS transcription is, respectively, 85 and 40% of the level without protein (Figure 2C and D).
Figure 2.
H-NS inhibits icsA and RnaG transcription. In vitro transcription was carried out at 30 and 37°C on ~200 ng of the supercoiled plasmid templates pGT1127 (A), pGT1129 (B) and pGT1083 (C) in the presence of the indicated concentrations of H-NS. The icsA and RnaG transcripts were detected by primer extension using the oligos G+110 and G+50, respectively, and quantified by imager (D). The transcription levels were normalized taking as 100%, the basal activity of the promoter measured at 30 and 37°C, in absence of H-NS. Values of the duplicated and/or tripled ‘0’ lanes were averaged and such value taken as 100%. All points of each individual set of samples were expressed as percentage of the corresponding ‘0’ samples. Lanes G and C represent the sequencing reactions using the same primer.
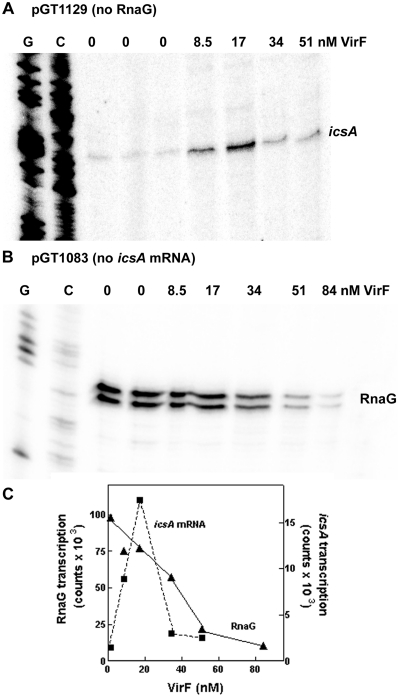
The VirF sites, localized on the icsA-RnaG locus, involve regions relevant for the regulation of both the icsA and the RnaG promoters (Figure 1E) and there is evidence that VirF is needed to activate icsA expression in vivo (7). By an in vitro assay, we succeeded in reproducing the VirF-mediated upregulation of icsA using the purified protein. As shown in Figure 3A and C, at low VirF concentrations the activity of the icsA promoter is strongly stimulated attaining an ~10-fold peak at 17 nM VirF. Transcription returns to basal levels when an excess of VirF is used (34 and 51 nM), possibly due to VirF occupying all its target sites and to the consequent formation of a transcriptionally inactive DNA–protein complex. As opposed to icsA, transcription of RnaG displays a progressive decrease with increasing VirF concentration: the activity of the RnaG promoter attains 58 and 22% in the presence of 34 and 51 nM VirF, respectively (Figure 3B and C). Taken together, these results indicate that VirF is able, depending on concentration, to stimulate PicsA and repress PRnaG.
Figure 3.
VirF stimulates icsA and represses RnaG transcription. In vitro transcription was carried out for 20 min at 37°C on the supercoiled plasmid templates pGT1129 (A) and pGT1083 (B) in presence of the indicated concentrations of VirF. The icsA and RnaG transcripts were detected essentially as described in Figure 2. Lanes G and C represent the sequencing reactions using the same primer. Radioactivity associated with icsA and RnaG transcripts is plotted versus VirF concentration (C).
Since both H-NS and VirF bind the icsA-RnaG locus (Figure 1), and antagonistically influence their transcription (Figures 2 and 3), an active competition between these regulators can be surmised. Therefore, this hypothesis has been tested in vitro by monitoring icsA promoter activity in the presence of both H-NS and VirF. The results indicate that VirF is able to significantly counteract the H-NS-dependent inhibition, thus almost completely relieving the transcriptional repression of icsA at the lowest H-NS concentration tested (Supplementary Figure S2).
H-NS and VirF modulate the in vivo expression of icsA and RnaG
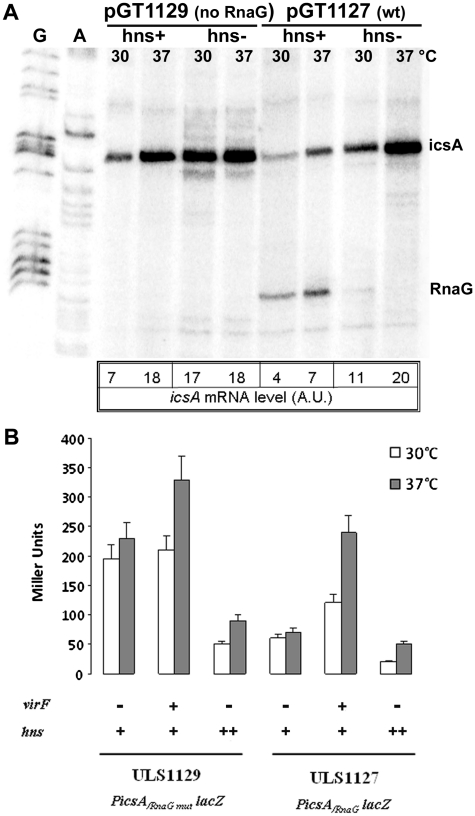
The in vitro functional assays (Figures 2 and 3) suggest that H-NS is able to inhibit transcription of both icsA and RnaG, as a function of temperature, while VirF behaves as an activator of icsA. In addition, the basal activities of PicsA and PRnaG (they are convergent and 120 nt apart) are not completely independent, since these promoters respond to transcriptional interference (TI) regulation (17). Hence, we investigated on the possible interplay among H-NS, VirF, temperature and TI in vivo. This was done by monitoring both the level of icsA mRNA and the expression of β-galactosidase from PicsA-lacZ transcriptional fusions carrying or lacking a functional PRnaG promoter (Figure 4A and B).
Figure 4.
Effect of H-NS and VirF on the in vivo expression of icsA. (A) Escherichia coli wt (HMG11) and its isogenic hns- strain (HMG9) harboring pGT1127 or pGT1129 plasmids were grown at 30 and 37°C in LB broth to A600 = 0.5. Aliquots of each culture were processed for total RNA extraction and 10 µg of each sample were analyzed by primer extension using the oligos G+50 and G+110 to detect RnaG and icsA mRNA, respectively. (B) Esherichia coli strains ULS1127 and ULS1129, carrying a wt hns gene (indicated with hns+) and a single-copy chromosomal PicsA-lacZ fusion with, respectively, a functional RnaG promoter (PicsA/RnaG-lacZ) or a defective one (PicsA/RnaGmut-lacZ), were used to assay β-galactosidase activity in the presence of pMYSH6504 (a plasmid harboring the S. flexneri virF gene indicated with virF+) or pDIA510 (a plasmid carrying extracopies of the E. coli hns gene indicated with hns++). Strains were grown in LB broth at 30 or 37°C to A600 = 0.4. The values represent the average of at least three independent experiments and the standard deviation is indicated.
The icsA promoter displays a natural thermoregulation and its transcription is higher at 37°C (20 A.U.) than at 30°C (11 A.U.) also in cells lacking an active H-NS. Moreover, a marked regulatory action can be attributed to this protein since mutations of the hns gene cause a further de-repression of icsA independently of temperature. Indeed, when both icsA and RnaG are transcribed (i.e. using plasmid pGT1127) the steady-state level of the icsA mRNA in hns defective strains increases about 3-fold as compared to the wt (from 4 to 11 A.U. at 30°C and from 7 to 20 A.U. at 37°C). In agreement with the transcriptional pattern of the icsA mRNA, the activity of the lacZ reporter gene in the E. coli strain ULS1127, carrying a single chromosomal copy of the PicsA-lacZ fusion, shows an ~3-fold reduction in cells overproducing H-NS from the pDIA510 plasmid. As expected, H-NS-mediated repression is more pronounced at 30°C than at 37°C (Figure 4B). Similar conclusions about the role of H-NS in the temperature-mediated regulation of icsA can be drawn using the pGT1129 construct, which carries a modified icsA regulatory region preventing RnaG synthesis. Under these conditions, the basal activity of PicsA is increased due to the absence of TI (compare hns+ cells transformed with pGT1129 or pGT1127 in Figure 4A). Consistently, TI also accounts for the higher expression of icsA in ULS1129 (an E. coli strain carrying a PicsA-lacZ fusion with an inactivated RnaG promoter) as compared to ULS1127. This is particularly evident in strains overproducing neither H-NS nor VirF (Figure 4B). The target of H-NS inhibition is mainly icsA rather than RnaG, in agreement with the results shown in Figure 2. Indeed, in an hns defective background, as an elevated synthesis of icsA mRNA is achieved (11–20 A.U.) RnaG becomes undetectable (Figure 4A). It is reasonable to assume that this is the outcome of TI, which leads to decreased accessibility of PRnaG to RNA polymerase when PicsA is overexpressed. Finally, as shown in Figure 4B, providing VirF in trans by introducing plasmid pMYSH6504, results in a stimulation of icsA expression. This activation mainly takes place at 37°C and in strain ULS1127, i.e. in the presence of a functional RnaG promoter.
Establishment of a kissing complex between sense and antisense RNAs mediates the transcriptional attenuation of icsA by RnaG
To gain deeper insight into the transcriptional attenuation mechanism governing the RnaG-mediated repression of icsA, we have studied how RnaG interacts with its target sequence on the icsA mRNA. RnaG is a 450 nt sRNA. We have demonstrated that its antisense region (RnaG120, i.e. nucleotides 1–120) still preserves the ability to repress icsA transcription in vitro, suggesting that this stretch constitutes a single functional domain. In particular, RNA probing showed that RnaG120 is highly structured and is characterized by three stem–loop motifs, GH1, GH2 and GH3 (17) (Supplementary Figure S3).
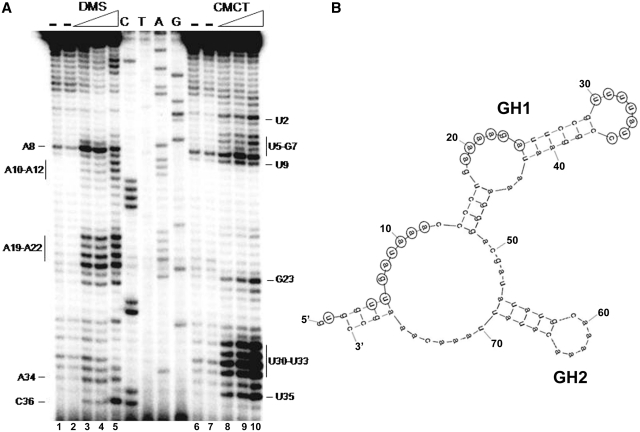
To identify the RnaG regions promoting the initial pairing with the icsA transcript, we first shortened RnaG120 by deleting 40 nt at its 3′-end, giving RnaG80. This smaller variant was then subjected to chemical probing using DMS and CMCT to verify whether the extended deletion had caused an extensive modification of the native structure. As seen in Figure 5A and B, the original structure of RnaG120 is essentially conserved in RnaG80: the GH1 and GH2 motifs are still present and structurally unchanged while the GH3 hairpin is lost.
Figure 5.
Secondary structure at 5′-end of RnaG. (A) Chemical probing of RnaG80 (nucleotides 1–80). Purified RnaG80 (2 pmol) was treated with increasing amounts of the single-strand specific reagents dimethyl sulfate (DMS) (0%, lanes 1 and 2; 0.22%, lane 3; 0.46%, lanes 4; 0.78%, lane 5) and 1-cyclohexyl-3-(2-morpholinoethyl) carbodiimide metho-p-toluene sulfonate (CMCT) (0 mg/ml, lanes 6 and 7; 1.5 mg/ml, lane 8; 3 mg/ml, lane 9; 6 mg/ml, lane 10) as described in ‘Materials and Methods’ section. Modified nucleotides were detected by primer extension using the oligo G+40H and accessible sites were evaluated comparing samples incubated in the absence (−) and in the presence of either DMS or CMCT. Lanes C, T, A and G correspond to DNA sequencing ladders made with the same primer. (B) Schematic representation of the secondary structure of RnaG80. Numbering is according to 5′-end of the antisense RNA corresponding to position +120 on icsA sequence. Nucleotides reactive to DMS or CMCT in Panel A are circled. Chemical probing data were superimposed on a computer prediction generated by the MFOLD program (47).
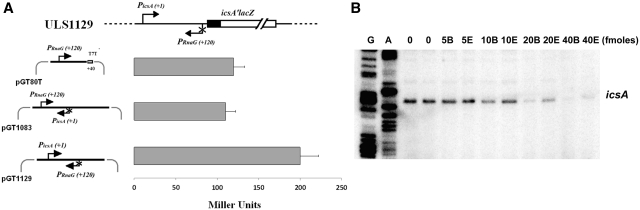
This result prompted us to examine the ability of RnaG80 to repress icsA transcription in vivo and in vitro. First, we compared the entire RnaG (expressed from pGT1083) to the shorter variant consisting of 80 nt (expressed from pGT80T) for their ability to inhibit the expression of a chromosomal single-copy PicsA-lacZ fusion (ULS119). As shown in Figure 6A both the full-length RnaG and RnaG80, induce a comparable decrease of β-galactosidase activity, supporting the repressive role of the first 80 nt at the 5′-end.
Figure 6.
RnaG80 is able to repress icsA expression. (A) The expression of a PicsA-lacZ fusion carried by E. coli strain ULS1129 was monitored in cells transformed with pGT80T, pGT1083 or pGT1129 as control. The data (±SD) represent the average of three independent experiments. (B) In vitro transcription was carried out at 37°C using 200 ng of supercoiled plasmid pGT1129 as template. Increasing amounts of RnaG80 were added to the transcription mixture either at the beginning of the reaction (prior to RNA polymerase addition, samples B) or at the end (immediately before RNA precipitation, samples E). The in vitro transcribed icsA mRNA was primer-extended with the oligo G+110.
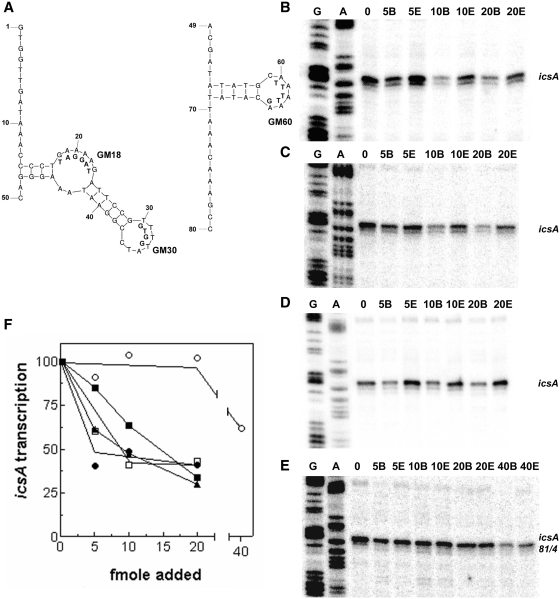
Next, an in vitro transcription assay was carried out in the presence of RnaG80 using a plasmid template (pGT1129) carrying the silenced RnaG promoter within the icsA locus. The icsA mRNA was detected by primer extension using an oligo recognizing only untruncated transcripts (Figure 6B). Since binding of RnaG to icsA mRNA can interfere with the elongation activity of the reverse transcriptase (17), RnaG80 was added to the transcription mixture either at beginning (samples B) or at the end (samples E) of the reaction: this strategy allowed us to discriminate the effect of the antisense RNA on transcription initiation from that on cDNA elongation. The assay showed that in the presence of low amounts of RnaG80 (up to 20 fmol), the icsA mRNA level is lower in samples B than in samples E. This result indicates that the shortened RnaG specifically causes termination of the icsA trascript. Higher amounts of RnaG80 (40 fmol) are required to interfere also with the detection of icsA mRNA by primer extension. By plotting icsA transcription, expressed as ‘sample B/sample E’ ratio, as a function of the amount of RnaG80 (Figure 7F), it can be seen that the icsA promoter activity attains 85, 64 and 34% at 5, 10 and 20 fmol of RnaG80, respectively. Since RnaG80 conserves its ability to properly function in vivo and in vitro, despite the loss of the GH3 hairpin, we can conclude that this motif is not required to interact with the icsA mRNA and that the first 80 nt confer RnaG the complete capability to teminate icsA transcription.
Figure 7.
Effects on icsA transcription of synthetic RNA and DNA oligonucleotides reproducing the structural motifs of RnaG. Secondary structure of DNA oligos used to construct the GH1 (left) and GH2 (right) stem–loops of RnaG (A). Mutations carried by oligos GM18, GM30 and GM60 are reported below the native sequence. In vitro transcription of icsA gene was performed, as described in the legend of Figure 6, using as templates the pGT1129 (B, C and D) and the pGT1129M (E) which synthesizes the mutated icsA81/4 mRNA (17). The increasing amounts of RNA oligo pair GR4–48/GR49–80 (B) and DNA oligo pairs G1–50/G49–80 (C and E) and G14–48/G49–80 (D) are indicated. After quantization, the radioactivity associated with icsA mRNA is expressed as ratio sample B/sample E and values are plotted as percentage assuming 100% the transcription of DNA template without oligos (F). Data from Figure 6B and from panels B, C, D and E are indicated with closed squares, open squares, closed triangles, closed circles and open circles, respectively.
We then investigated which regions within RnaG80 play a key role in establishing the initial contact with the icsA mRNA. For this purpouse, we designed a set of RNA and DNA oligonucletides partially or entirely reproducing the GH1 (oligos GR4–48, G1–50, G14–48 and G1–35) and GH2 (oligos GR49–80, G49–80) motifs which characterize the secondary structure of RnaG80 (Figures 5B and 7A). The ability of these oligos to block the synthesis of the icsA messenger was monitored by means of the previously illustrated in vitro transcription assay. While none of the oligos was individually able to affect icsA transcription (not shown), we found that the use of the RNA oligo pair GR4–48/GR49–80, which fully reconstructs RnaG80, inhibits trascription even more severely (60 and 41% at 5 and 10 fmol of oligos, respectively) than the purified RnaG80 (Figure 7B and F). Likewise, alternatively combining the DNA oligos G1–50 and G14–48 with G49–80 caused a reduction of icsA transcription, comparable to that observed with the RNA oligos (Figure 7C, D and F), suggesting that the secondary structure, rather than the nature of the molecule, is essential in determining the RnaG-mediated transcriptional repression. Recently, we showed that mutations on icsA gene (from positions 81 to 84) abolish the transcriptional attenuation by destabilizing the intrinsic terminator triggered on the nascent mRNA by the sense–antisense interaction (17). As expected, the oligo pair G1–50/G49–80 is almost totally inactive in repressing transcription from a plasmid carrying the icsA81/4 mutation (Figure 7E and F), which impedes the formation of the RnaG-mediated icsA mRNA terminator.
Altogether these results indicate that the minimum size necessary to maintain the RnaG attenuation activity on icsA is ~65 nt spanning from positions 14 to 80 and that both motifs, GH1 and GH2, are required. To further learn about which portions within this region primarily contribute to recognize the icsA mRNA, we used DNA oligos carrying base-exchanges localized at the level of unpaired nucleotides (apical loops and bulge) of the GH1 (oligos GM18 and GM30) and GH2 (oligo GM60) hairpins (Figure 7A). In vitro transcription assays were set up using each mutated GH1 or GH2 oligo in combination, respectively, with the wt GH2 (G49–80) or GH1 (G1–50) oligos, so as to cover the entire RnaG80 sequence. While the GM18 oligo conserves a residual repressive activity, GM30 and GM60 mutations completely abate the ability of these synthetic DNA molecules to terminate icsA transcription, suggesting that unpaired bases at the apical loops GH1 (nucleotides 30–33) and GH2 (nucleotides 60–64) provide the primary nucleation points, known as kissing complex, for sense–antisense RNAs pairing (Supplementary Figure S4 and Figure 8).
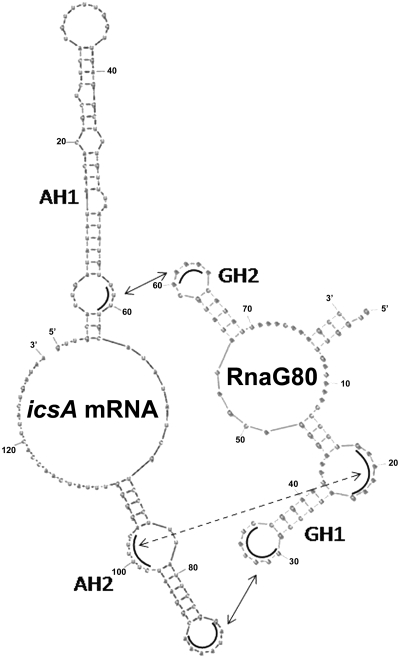
Figure 8.
Identification of stem–loop motifs involved in the formation of the kissing complex between RnaG and icsA mRNA. Schematic representation of the three regions (arrows) providing the initial nucleation points for the pairing of RnaG80 and icsA mRNA. Interactions occur between GH2 (positions 60–63) and AH1 (positions 59–62), as well as between GH1 (positions 18–23 and 30–36) and AH2 (positions 98–103 and 85–91).
DISCUSSION
The actin-based motility of Shigella, the causative agent of bacillary dysentery, depends on IcsA, an outer membrane protein that accumulates at one pole of the bacterial cell (4). IcsA is capable of directly recruiting and activating host cell factors, thus catalyzing the assembly of an actin tail which provides the propulsive force for intracellular movement and intercellular dissemination of bacteria through the human intestinal epithelium (5).
As is the case for other Shigella virulence genes, also the icsA gene is located on the large virulence plasmid (pINV) typically harbored by this pathogen (31). Over the years, evidence has built up indicating that the regulation of icsA depends on the nucleoid protein H-NS, which acts as a repressor, and on the AraC-like protein VirF, the major activator of the Shigella invasivity regulon (7,11,12). Nevertheless, the molecular details of this regulatory mechanism have remained elusive. We have recently shown that an additional player is involved: RnaG, a non-coding sRNA which acts as an antisense regulator on the icsA transcript (17). This study is an attempt to clarify the interplay among these factors in order to evaluate their contribution to the expression of icsA, not only as individual elements but also as components of a complex and integrated regulatory circuit.
H-NS modulates the expression of ~5% of the E. coli genome. Approximately one-third of these genes are linked to the adaptation of bacteria to stress conditions and to the regulation of pathogenicity processes (9,32–34). Actually, it is emerging that H-NS partecipates to the regulation of many genes whose expression is sensitive to temperature shifts and specifically to the virulence genes of Shigella. For example it has been found that H-NS represses transcription of virF, the primary regulator of the invasivity functions, at 30°C but not at 37°C (14,16). In the present study, we have shown that H-NS can also silence the virulence gene icsA in a temperature-dependent manner (Figures 2 and 4). This effect, exerted at the transcriptional level, is likely achieved through a temperature-mediated occupancy by H-NS of its binding sites on the icsA promoter region (Figure 1). This hypothesis is supported by the unsuccessful attemps to obtain clear protections by H-NS in footprints carried out at 37°C (Supplementary Figure S5).
Unlikely to other H-NS-regulated genes, in which binding sites have been mostly identified on the upstream regulatory regions (8,35), the three H-NS recognition sites on the icsA promoter are located downstream the icsA transcription start. Interstingly, two sites reside in the DNA tract between icsA and the RnaG promoters (Figure 1E). This arrangement likely favors the repressive action of H-NS on both icsA and RnaG genes. Indeed, the transcription of both genes is reduced by the presence of H-NS (Figure 2). The inhibition is more pronounced on icsA, suggesting that it may represent a preferential target as compared to RnaG. Nonetheless, the ability of H-NS to repress also a sRNA promoter further stresses the significance of this nucleoid protein as global regulator. It is not surprising that H-NS and sRNA molecules cooperate, since both are known to regulate bacterial gene expression in reponse to stress conditions (23,35). In this context, it is worth mentioning that the translation of the hns mRNA itself is controlled by DsrA, a sRNA (36). Moreover, DsrA can also counteract the H-NS silencing of rcsA, a regulatory gene involved in the synthesis of the capsular polysaccharide in E. coli (37). Recently, H-NS was also found to directly activate translation of several E. coli mRNAs with non-canonical Shine Dalgarno sequences (38), further supporting the view that significant connections exist between this nucleoid protein and the RNA world.
Besides icsA promoter, additional four H-NS-binding sites have been found on the regulatory region of virA, another important virulence gene responsible for destroying host cell microtubules (39). Notably, virA is located ~360 nt upstream icsA in the reverse orientation. These four H-NS binding sites, spanning from positions −220 to −350, possibly mediate the negative regulation of virA by H-NS (15). The icsA gene shows a high degree of intrinsic bending (12) and the center of curvature of the icsA-virA intergenic region has been located around position −70 (G. Micheli, personal comunication). According to the preferential interaction of H-NS with bent DNA (40), it is conceivable that the curvature flanked by multiple target sites might provide a nucleation point for protein oligomerization, resulting in the occlusion of the entire virA-icsA region to RNA polymerase. This hypothesis is supported by the fact that both icsA and virA are derepressed in hns- cells [Figure 4 and (15)]. The intrinsically curved DNA, localized between virA and icsA genes, might respond to temperature shifts, thereby controlling the binding of H-NS as demonstrated in the regulation of virF (14,16). Alternatively, it has been proposed that a thermal increase causes modifications in the quaternary structure of H-NS, leading to the decrease of high-order oligomers versus the appearance of discrete dimers. Such conformational change, in turn, would reduce the ability of H-NS to bind DNA (41). However, this issue has not been settled conclusively since a large number of H-NS-repressed genes remain silenced at 37°C, suggesting that a simple effect of temperature on the state of aggregation of H-NS cannot account for the temperature-dependent derepression of this subset of genes. Moreover, opposed to the temperature-mediated aggregation model, H-NS oligomerization has been reported to be constant in the range 28–42°C (33).
Our data reveal that, besides H-NS, also VirF is able to specifically interact with the icsA-RnaG region, by recognizing four biding sites (Figure 1) and it plays a direct role in activating icsA transcription (Figure 3). In vivo data indicate that VirF is able to stimulate icsA expression at 37°C and that this also occurs at 30°C, albeit to a much lesser extent (Figure 4). In most cases, VirF does not bind to only one site but rather simultaneously interacts with two sites. By different footprinting techniques, the 13-bp conserved sequence TTTaGYcTtTat (nucleotides with a frequency ≥60% in the consensus sequence are in uppercase and Y indicates pyrimidines) was found in VirF protected regions and it has been proposed that VirF tightly binds to this sequence when present as an inverted repeat and weakly when the sequence is present only once (49). Although the mechanism adopted by VirF to activate transcription is still questionable, VirF bound at its site I, which includes the −10 conserved element, presumably contacts the RNA polymerase thus facilitating its access to the icsA promoter. This condition can occur assuming that first VirF preferentially interacts with a high affinity site (i.e. site III). Subsequently, the occurrence of DNA looping, which is commonly involved in the AraC-mediated regulation (50), may favor the contact of VirF with the promoter proximal site I. This hypothesis is validated by some facts: (i) this two step binding model has been described in the regulation of yop genes of Yersinia by VirF (49); (ii) protection at VirF site III is quite pronounced even with the lowest concentrations of VirF used (42–84 nM VirF, Figure 1C and D), indicating that this site might be the first to be occupied by the protein; (iii) three regions, showing a high homology with the 13-bp consensus motif and inversely orientated, are present within VirF site III while only one or two have been found within sites I, II and IV (Supplementary Figure S1). This binding pathway can also explain repression of transcription by VirF. In fact, progressive filling-in of all available sites, as a function of protein amount, might cause the formation of a VirF–DNA complex covering a wide region which incorporates both the icsA and the RnaG promoters. This, in turn, would block transcription by trapping the RNA polymerase. Besides acting at the transcriptional level by directly stimulating PicsA, VirF increases IcsA expression by two additional strategies: (i) reduction of the intracellular level of RnaG, as observed in vivo by analyzing PRnaG-lacZ fusions (data not shown); (ii) lowering of the frequency of transcription initiation at PRnaG thus affecting the extent of transcriptional interference (TI) between RnaG and icsA. These mechanisms are direct consequences of the repressive action exerted by VirF on PRnaG. We have previously shown that TI occurs between the convergent RnaG and icsA promoters (17). Hence, VirF would limit the transit events and consequently the occupancy of PicsA by elongation complexes moving from the convergent PRnaG, thus enabling icsA mRNA synthesis.
VirF binding sites I and II are partially overlapping the H-NS box I while the VirF site III is adjacent to the H-NS box II (Figure 1E). Such an organization of target sites on the icsA promoter is consistent with a possible competition between the icsA-activating VirF and the repressing H-NS. Since virF expression rapidly increases when temperature is raised >32°C (14) and H-NS is able to recognize the icsA-RnaG region only at lower temperature, it could be argued that VirF and H-NS do not interfere with each other because their opposite action on icsA promoter is, somehow, spaced by a gap of temperature. On the contrary, transcriptional competition assays between VirF and H-NS reveal that, in the presence of H-NS, VirF is capable to restore, albeit not completely, the basal activity of PicsA (Supplementary Figure S2), thus proposing a more complex scenario. As the temperature raises, the interaction of VirF with sites I, II and III may disrupt the H-NS-stabilized complex by forming a putative H-NS-icsA-VirF intermediate, thus promoting a switch from a repressed to an activated state. In agreement with this observation, an antagonistic role between H-NS and VirF has been found in the regulation of virB (13). Regulatory antagonism between H-NS and members of the AraC-like protein family is emerging as a common strategy that bacterial pathogens adopt to control the virulence genes [reviewed in (42)], as exemplified, for instance, by (i) the V. cholerae TCP system, essential for the biogenesis of the toxin-co-regulated pilus (ToxT/H-NS) (43); (ii) the E. coli gad genes, involved in the adaptative response to acid stress (GadX/H-NS) (29); (iii) the ler gene of enteropathogenic E. coli, required to initiate the regulatory cascade activating the LEE locus (PerA/H-NS) (44). While the AraC-like protein represents the primary regulator, the involvement of H-NS might also reflect the need to fine-tune gene expression adapting it to the environmental changes that bacterial cells face upon host invasion (32,45).
The interplay between VirF and H-NS in the regulation of icsA is made more complex by the presence of RnaG, one of the largest (450 nt) non-coding RNAs in bacteria. We have shown that RnaG is a cis-encoded antisense RNA whose 120 nt at the 5′-end (RnaG120) are complementary to the 5′-UTR icsA transcript and behave as a functional domain inducing premature termination of the icsA mRNA by transcriptional attenuation (17). In general, the initial sense–antisense RNA contact, also known as kissing complex, takes place between two complementary loops (multistep pathway) or between a loop and a single-stranded region (one-step pathway). In both cases, such interactions, each one involving 4–7 exposed nucleotides, lead to a helix progression until a fully paired RNA is formed (22,46). We have recently described the secondary structure of RnaG120 and of the 5′-end (~150 nt) of the icsA mRNA (17); while the first is characterized by three hairpin structures, GH1, GH2 and GH3, the latter only shows two structural motifs, AH1 and AH2 (Figure 8 and Supplementary Figure S3). In the present study, we demonstrate that only GH1 and GH2 participate in establishing the kissing complex between RnaG and icsA mRNA, thus forming a functional domain responsible for icsA regulation by RnaG. This conclusion is supported by several lines of evidence. In vitro and in vivo experiments (Figure 6) indicate that out of 120 nt at the 5′-end of RnaG, the initial 80 nt (RnaG80) represent the minimum stretch able to repress icsA transcription. This finding is in agreement with the observation that a limited number of base pairs and not full-duplex formation between sense and antisense RNAs is usually required for control as shown in the case of R1, ColE1 and pIP501 (22). Despite the lack of GH3 in RnaG80, the secondary structures of GH1 and GH2 (Figure 5) are conserved in RnaG120 (Supplementary Figure S3), as well as at the 5′-end of the entire RnaG (not shown), stressing the relevance of the spatial organization of RnaG for its function. The need for both motifs, GH1 and GH2, is also evidenced by the finding that synthetic molecules carrying either GH1 or GH2 are unable to terminate icsA transcription and that this ability is restored as the native structure of RnaG80 is reconstituted by aligning the two motifs head-to-tail using suitable oligos (Figure 7). Termination of icsA mRNA, however, does not depend on the nature of the oligo molecule used (DNA or RNA).
The analysis of icsA mRNA and 5′-end of RnaG structures reveals the following potential points of interaction: (i) the apical loop of GH2 can pair with the basal bulge of AH1; (ii) the apical loop and the internal bulge of GH1 can anneal with the structurally similar motifs of AH2 (Figure 8). The relevance of contact points emerges from experiments using mutated RnaGs. Changing the exposed bases on the GH1 and GH2 loops to hinder their pairing with the complementary icsA mRNA regions, affects the repression activity of RnaG (Supplementary Figure S4). Complete inactivation occurs with the GM30 and GM60 mutations, indicating that these stretches of bases are essential for the formation of the kissing complex. Pairing might follow a two-step binding pathway, starting at the critical sequence of either the GH1 or the GH2 loop and using the remaining loop–loop interaction as nucleation site for RNA duplex propagation. Instead, GM18 mutation only partially alters RnaG functioning, suggesting that the internal bulge of GH1 may have an auxiliary role in establishing the early contact between the GH1 and AH2 structural motifs. Irrespective of the binding kinetics and loop–loop initiating system, the functional characterization of RnaG80 and its variants confirms the model based on the formation of an intrinsic terminator on the nascent icsA mRNA, triggered by the sense–antisense pairing (17). Indeed, both native and synthetic molecules carrying the 5′-end of RnaG fails to halt transcription of the icsA81/4 mRNA in which a four bases mutation in the stem of the terminator impedes the formation of an intramolecular RNA duplex (Figure 7 and data not shown).
Besides the relevance of IcsA as a virulence determinant, the fact that its expression is themoregulated and finely controlled by VirF, H-NS and the antisense RnaG makes this gene a very interesting candidate to comprehend how complicated regulatory circuits are functioning and how they have evolved. The aim of the present study has been to address some crucial points of this regulation, particularly those related to the antagonism between the two regulators VirF and H-NS and to their interplay with RnaG. Furthermore, icsA offers a chance to clarify several molecular aspects of sense–antisense pairing providing insights into the rules driving the formation of the kissing complex and, more generally, into the mechanisms of riboregulation.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Fondi Ateno Ricerca (FAR) and Progetti di ricerca di Rilevante Interesse Nazionale (PRIN) to M.F. and B.C. Funding for open access charge: FAR to M.F.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
The authors thank G. Micheli and R. Spurio for critical reading of the manuscript.
REFERENCES
- 1.Sansonetti PJ. Shigellosis: an old disease in new clothes? PLoS Med. 2006;3:e354. doi: 10.1371/journal.pmed.0030354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Prosseda G, Falconi M, Nicoletti M, Casalino M, Micheli G, Colonna B. Histone-like proteins and the Shigella invasivity regulon. Res. Microbiol. 2002;153:461–468. doi: 10.1016/s0923-2508(02)01346-3. [DOI] [PubMed] [Google Scholar]
- 3.Parsot C. Shigella spp. and enteroinvasive Escherichia coli pathogenicity factors. FEMS Microbiol. Lett. 2005;252:11–18. doi: 10.1016/j.femsle.2005.08.046. [DOI] [PubMed] [Google Scholar]
- 4.Bernardini ML, Mounier J, d'Hauteville H, Coquis-Rondon M, Sansonetti PJ. Identification of icsA, a plasmid locus of Shigella flexneri that governs bacterial intra- and intercellular spread through interaction with F-actin. Proc. Natl Acad. Sci. USA. 1989;10:3867–3871. doi: 10.1073/pnas.86.10.3867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brandon LD, Goehring N, Janakiraman A, Yan AW, Wu T, Beckwith J, Goldberg MB. IcsA, a polarly localized autotransporter with an atypical signal peptide, uses the Sec apparatus for secretion, although the Sec apparatus is circumferentially distributed. Mol. Microbiol. 2003;50:45–60. doi: 10.1046/j.1365-2958.2003.03674.x. [DOI] [PubMed] [Google Scholar]
- 6.Sakai T, Sasakawa C, Yoshikawa M. Expression of four virulence antigens of Shigella flexneri is positively regulated at the transcriptional level by the 30 kiloDalton VirF protein. Mol. Microbiol. 1988;2:589–597. doi: 10.1111/j.1365-2958.1988.tb00067.x. [DOI] [PubMed] [Google Scholar]
- 7.Le Gall T, Mavris M, Martino MM, Bernardini ML, Denamur E, Parsot C. Analysis of virulence plasmid gene expression defines three classes of effectors in the type III secretion system of Shigella flexneri. Microbiology. 2005;151:951–962. doi: 10.1099/mic.0.27639-0. [DOI] [PubMed] [Google Scholar]
- 8.Schroder Ö, Wagner R. The bacterial regulatory protein H-NS – a versatile modulator of nucleic acid structures. Biol. Chem. 2002;383:945–960. doi: 10.1515/BC.2002.101. [DOI] [PubMed] [Google Scholar]
- 9.Dorman CJ. H-NS: a universal regulator for a dynamic genome. Nat. Rev. Microbiol. 2004;2:391–400. doi: 10.1038/nrmicro883. [DOI] [PubMed] [Google Scholar]
- 10.Fang FC, Rimsky S. New insights into transcriptional regulation by H-NS. Curr. Opin. Microbiol. 2008;11:113–120. doi: 10.1016/j.mib.2008.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dagberg B, Uhlin BE. Regulation of virulence-associated plasmid genes in enteroinvasive Escherichia coli. J. Bacteriol. 1992;174:7607–7612. doi: 10.1128/jb.174.23.7606-7612.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Prosseda G, Fradiani PA, Di Lorenzo M, Falconi M, Micheli G, Casalino M, Nicoletti M, Colonna B. A role for H-NS in the regulation of the virF gene of Shigella and enteroinvasive Escherichia coli. Res Microbiol. 1998;149:15–25. doi: 10.1016/s0923-2508(97)83619-4. [DOI] [PubMed] [Google Scholar]
- 13.Tobe T, Yoshikawa M, Mizuno T, Sasakawa C. Transcriptional control of the invasion regulatory gene virB of Shigella flexneri: activation by VirF and repression by H-NS. J. Bacteriol. 1993;175:6142–6149. doi: 10.1128/jb.175.19.6142-6149.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Falconi M, Colonna B, Prosseda G, Micheli G, Gualerzi CO. Thermoregulation of Shigella and Escherichia coli EIEC pathogenicity. A temperature-dependent structural transition of DNA modulates accessibility of virF promoter to transcriptional repressor H-NS. EMBO J. 1998;17:7033–7043. doi: 10.1093/emboj/17.23.7033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Beloin C, Dorman CJ. An extended role for the nucleoid structuring protein H-NS in the virulence gene regulatory cascade of Shigella flexneri. Mol. Microbiol. 2003;47:825–838. doi: 10.1046/j.1365-2958.2003.03347.x. [DOI] [PubMed] [Google Scholar]
- 16.Prosseda G, Falconi M, Giangrossi M, Gualerzi CO, Micheli G, Colonna B. The virF promoter in Shigella: more than just a curved DNA stretch. Mol. Microbiol. 2004;51:523–537. doi: 10.1046/j.1365-2958.2003.03848.x. [DOI] [PubMed] [Google Scholar]
- 17.Giangrossi M, Prosseda G, Tran CN, Brandi A, Colonna B, Falconi M. A novel antisense RNA regulates at transcriptional level the virulence gene icsA of Shigella flexneri. Nucleic Acids Res. 2010;38:3362–3375. doi: 10.1093/nar/gkq025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Romby P, Vandenesch F, Wagner EG. The role of RNAs in the regulation of virulence-gene expression. Curr. Opin. Microbiol. 2006;9:229–236. doi: 10.1016/j.mib.2006.02.005. [DOI] [PubMed] [Google Scholar]
- 19.Toledo-Arana A, Repoila F, Cossart P. Small noncoding RNAs controlling pathogenesis. Curr. Opin. Microbiol. 2007;10:182–188. doi: 10.1016/j.mib.2007.03.004. [DOI] [PubMed] [Google Scholar]
- 20.Altuvia S. Identification of bacterial small non-coding RNAs: experimental approaches. Curr. Opin. Microbiol. 2007;10:257–261. doi: 10.1016/j.mib.2007.05.003. [DOI] [PubMed] [Google Scholar]
- 21.Papenfort K, Vogel J. Regulatory RNA in bacterial pathogens. Cell Host Microbe. 2010;8:116–127. doi: 10.1016/j.chom.2010.06.008. [DOI] [PubMed] [Google Scholar]
- 22.Brantl S. Regulatory mechanisms employed by cis-encoded antisense RNAs. Curr. Opin. Microbiol. 2007;10:102–109. doi: 10.1016/j.mib.2007.03.012. [DOI] [PubMed] [Google Scholar]
- 23.Waters LS, Storz G. Regulatory RNAs in bacteria. Cell. 2009;136:615–628. doi: 10.1016/j.cell.2009.01.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Repoila F, Darfeuille F. Small regulatory non-coding RNAs in bacteria: physiology and mechanistic aspects. Biol. Cell. 2009;101:117–131. doi: 10.1042/BC20070137. [DOI] [PubMed] [Google Scholar]
- 25.Sakai T, Sasakawa C, Makino S, Kamata K, Yoshikawa M. Molecular cloning of a genetic determinant for Congo red binding ability which is essential for the virulence of Shigella flexneri. Infect. Immun. 1986;51:476–482. doi: 10.1128/iai.51.2.476-482.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bertin P, Terao E, Lee EH, Lejeune P, Colson C, Danchin A, Collatz E. The H-NS protein is involved in the biogenesis of flagella in Escherichia coli. J. Bacteriol. 1994;176:5537–5540. doi: 10.1128/jb.176.17.5537-5540.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.La Teana A, Falconi M, Scarlato V, Lammi M, Pon CL. Characterization of the structural genes for the DNA-binding protein H-NS in Enterobacteriaceae. FEBS Lett. 1989;244:34–38. doi: 10.1016/0014-5793(89)81156-1. [DOI] [PubMed] [Google Scholar]
- 28.Brandi A, Pietroni P, Gualerzi CO, Pon CL. Post-transcriptional regulation of CspA expression in Escherichia coli. Mol. Microbiol. 1996;19:231–240. doi: 10.1046/j.1365-2958.1996.362897.x. [DOI] [PubMed] [Google Scholar]
- 29.Giangrossi M, Zattoni S, Tramonti A, De Biase D, Falconi M. Antagonistic role of H-NS and GadX in the regulation of the glutammate decarboxylase-dependent acid resistance system in Escherichia coli. J. Biol. Chem. 2005;280:21498–21505. doi: 10.1074/jbc.M413255200. [DOI] [PubMed] [Google Scholar]
- 30.Dorman CJ, Porter ME. The Shigella virulence gene regulatory cascade: a paradigm of bacterial gene control mechanisms. Mol. Microbiol. 1998;29:677–684. doi: 10.1046/j.1365-2958.1998.00902.x. [DOI] [PubMed] [Google Scholar]
- 31.Lett MC, Sasakawa C, Okada N, Sakai T, Makino S, Yamada M, Komatsu K, Yoshikawa M. virG, a plasmid-coded virulence gene of Shigella flexneri: identification of the VirG protein and determination of the complete coding sequence. J. Bacteriol. 1989;171:353–359. doi: 10.1128/jb.171.1.353-359.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hommais F, Krin E, Laurent-Winter C, Soutourina O, Malpertuy A, Le Caer JP, Danchin A, Bertin P. Large-scale monitoring of pleiotropic regulation of gene expression by the prokaryotic nucleoid-associated protein, H-NS. Mol. Microbiol. 2001;40:20–36. doi: 10.1046/j.1365-2958.2001.02358.x. [DOI] [PubMed] [Google Scholar]
- 33.Stella S, Falconi M, Lammi M, Gualerzi CO, Pon CL. Environmental control of the in vivo oligomerization of nucleoid protein H-NS. J. Mol. Biol. 2006;355:169–174. doi: 10.1016/j.jmb.2005.10.034. [DOI] [PubMed] [Google Scholar]
- 34.Navarre WW, McClelland M, Libby SJ, Fang FC. Silencing of xenogeneic DNA by H-NS—facilitation of lateral gene transfer in bacteria by a defense system that recognizes foreign DNA. Genes Dev. 2007;21:1456–1471. doi: 10.1101/gad.1543107. [DOI] [PubMed] [Google Scholar]
- 35.Atlung T, Ingmer H. H-NS, a modulator of environmentally regulated gene expression. Mol. Microbiol. 1997;24:7–17. doi: 10.1046/j.1365-2958.1997.3151679.x. [DOI] [PubMed] [Google Scholar]
- 36.Lease RA, Belfort M. A trans-acting RNA as a control switch in Escherichia coli: DsrA modulates funcion by forming alternative structures. Proc. Natl Acad. Sci. USA. 2000;97:9919–9924. doi: 10.1073/pnas.170281497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sledjeski D, Gottesman S. A small RNA acts as antisilencer of H-NS-silenced rcsA gene of Escherichia coli. Proc. Natl Acad. Sci. USA. 1995;92:2003–2007. doi: 10.1073/pnas.92.6.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Park HS, Ostberg Y, Johansson J, Wagner EGH, Uhlin BE. Novel role for a bacterial nucleoid protein in translation of mRNAs with suboptimal ribosome-binding sites. Genes Dev. 2010;24:1345–1350. doi: 10.1101/gad.576310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yoshida S, Handa Y, Suzuki T, Ogawa M, Suzuki M, Tamai A, Abe A, Katayama E, Sasakawa C. Microtubule-severing activity of Shigella is pivotal for intercellular spreading. Science. 2006;10:985–989. doi: 10.1126/science.1133174. [DOI] [PubMed] [Google Scholar]
- 40.Yamada H, Muramatsu S, Mizuno T. An Escherichia coli protein that preferentially binds to sharply curved DNA. J. Biochem. 1990;108:420–425. doi: 10.1093/oxfordjournals.jbchem.a123216. [DOI] [PubMed] [Google Scholar]
- 41.Ono AS, Goldberg MD, Olsson T, Esposito D, Hinton JCD, Ladbury JE. H-NS is a part of a thermally controlled mechanism for bacterial gene regulation. Biochem. J. 2005;391:203–213. doi: 10.1042/BJ20050453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Stoebel DM, Free A, Dorman CJ. Anti-silencing: overcoming H-NS-mediated repression of transcription in Gram-negative enteric bacteria. Microbiology. 2008;154:2533–2545. doi: 10.1099/mic.0.2008/020693-0. [DOI] [PubMed] [Google Scholar]
- 43.Yu RR, DiRita VJ. Regulation of gene expression in Vibrio cholerae by ToxT involves both antirepression and RNA polymerase stimulation. Mol. Microbiol. 2002;43:119–134. doi: 10.1046/j.1365-2958.2002.02721.x. [DOI] [PubMed] [Google Scholar]
- 44.Porter ME, Mitchell P, Roe AJ, Free A, Smith DGE, Gally DL. Direct and indirect transcriptional activation of virulence genes by an AraC-like protein, PerA from enteropathogenic Escherichia coli. Mol. Microbiol. 2004;54:1117–1133. doi: 10.1111/j.1365-2958.2004.04333.x. [DOI] [PubMed] [Google Scholar]
- 45.White-Ziegler CA, Davis TR. Genome-wide identification of H-NS-controlled, temperature-regulated genes in Escherichia coli K-12. J. Bacteriol. 2009;191:1106–1110. doi: 10.1128/JB.00599-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wagner EG, Altuvia S, Romby P. Antisense RNAs in bacteria and their genetic elements. Adv. Genet. 2002;46:361–398. doi: 10.1016/s0065-2660(02)46013-0. [DOI] [PubMed] [Google Scholar]
- 47.Zuker M, Mathews DH, Turner DH. Algorithms and thermodynamics for RNA secondary structure prediction: a practical guide in RNA biochemistry and biotechnology. In: Barciszewski J, Clark BFC, editors. NATO ASI Series. Dordrecht, the Netherlands: Kluwer Academic Publishers; 1999. [Google Scholar]
- 48.Miller JH. A short course in bacterial genetics. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1992. [Google Scholar]
- 49.Wattiau P, Cornellis GR. Identification of DNA sequences recognized by VirF, the transcriptional activator of the Yersinia yop regulon. J. Bacteriol. 1994;176:3878–3884. doi: 10.1128/jb.176.13.3878-3884.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Schleif R. AraC protein, regulation of the l-arabinose operon in Escherichia coli, and the light switch mechanism of AraC action. FEMS Microbiol. Rev. 2010;43:779–796. doi: 10.1111/j.1574-6976.2010.00226.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.