Abstract
Successful DNA repair within chromatin requires coordinated interplay of histone modifications, chaperones and remodelers for allowing access of repair and checkpoint machineries to damaged sites. Upon completion of repair, ordered restoration of chromatin structure and key epigenetic marks herald the cell’s normal function. Here, we demonstrate such a restoration role of H3K56 acetylation (H3K56Ac) mark in response to ultraviolet (UV) irradiation of human cells. A fast initial deacetylation of H3K56 is followed by full renewal of an acetylated state at ~24–48 h post-irradiation. Histone chaperone, anti-silencing function-1 A (ASF1A), is crucial for post-repair H3K56Ac restoration, which in turn, is needed for the dephosphorylation of γ-H2AX and cellular recovery from checkpoint arrest. On the other hand, completion of DNA damage repair is not dependent on ASF1A or H3K56Ac. H3K56Ac restoration is regulated by ataxia telangiectasia mutated (ATM) checkpoint kinase. These cross-talking molecular cellular events reveal the important pathway components influencing the regulatory function of H3K56Ac in the recovery from UV-induced checkpoint arrest.
INTRODUCTION
Genomic integrity is central to the successful survival and propagation of living organisms constantly challenged by exposures to endogenous and exogenous agents. Living organisms have evolved elaborate DNA repair mechanisms to overcome the deleterious effects of genotoxic exposures. Besides DNA repair, the deployment of DNA damage response (DDR) triggers signaling cascades for activating cell-cycle checkpoints to allow cells sufficient time to complete the repair (1,2). Failure of DDR and incomplete repair can result in the activation of apoptotic and other cell death pathways.
Nucleotide excision repair (NER) is the major DNA repair mechanism acting on diverse DNA lesions including UV damage from human exposure to sunlight. Defects in individual components of NER and resulting loss of repair capacity is the underlying cause of genetic disorders such as Xeroderma pigmentosum (XP) and Cockayne syndrome (CS) characterized by sensitivity to UV radiation and predisposition to skin cancers (3). Among the two major mutagenic photoproducts of UV exposure, (6-4)-pyrimidine pyrimidone photoproducts (6-4PPs) are readily detectable than cyclobutane pyrimidine dimers (CPDs) and excised at a 5-fold faster rate (4). For instance, 6-4PP in human cells are removed in about 1 h, while ~50% of the initial CPD are eliminated in ~24 h (5,6). Interestingly, CPD repair is faster in transcribed regions of the genome (7), and the repair microheterogeneity within discrete genomic regions is exquisitely controlled by additional layers of regulation including the native state and dynamics of chromatin structure.
Conventionally, basic steps of damage processing by NER are comprised of (i) initial recognition of the damaged DNA lesion; (ii) excision of a 24–32-bp oligonucleotide containing damaged lesion by a dual endonucleolytic incision; (iii) filling of the resulting gap by repair synthesis-specific DNA polymerase; and (iv) final ligation of the nick (8,9). The association of eukaryotic DNA with histone proteins, forming a highly compacted chromatin and higher-order structure has a major influence on all DNA-templated processes including repair (10–14). The chromatin organization poses an obvious challenge for the NER and DDR processes, requiring additional chromatin remodeling steps. Moreover, subsequent to successful repair, original conformation of chromatin must be restored to ensure normal function and propagation of cells. These additional steps form the basis for the ‘access–repair–restore’ model of NER (2).
DDR network is equally complex and composed of DNA damage sensors, signal transducers, and various effectors to ultimately invoke cellular checkpoints. Its central components are the phosphoinositide 3-kinase-related kinases (PIKKs), e.g. ataxia telangiectasia mutated (ATM), ATM and Rad3 related (ATR) and DNA-PK, whose substrates mediate cell-cycle arrest in G1, S or G2 phases (15). ATM appears to be the primary player in response to ionizing radiation, and the double-strand break (DSB) signal sensed by ATM is transduced to CHK2, while the UV damage signal sensed by ATR is transduced to CHK1. Some overlap and functional redundancy exist between ATM and ATR. Phosphorylation by activated CHK1 or CHK2 inactivates Cdc25A–C and prevents cells from the G1/S and G2/M transitions (16,17). Even though the ATR-CHK1 is the predominant pathway in response to UV damage, recent studies implicate a key role of the ATM-CHK2 pathway in UV damage repair (18,19). How the ATM-CHK2 pathway influences UV damage repair remains to be fully elucidated.
Post-translational modifications (PTMs) of histones (20,21), adenosine triphosphate (ATP)-dependent chromatin remodeling for nucleosome repositioning and reorganization (22), histone eviction and/or introduction by histone chaperones (23), and deposition of histone variants (24) constitute the key components of chromatin remodeling machinery. Efficient repair of diverse DNA damage require all or some of these chromatin-remodeling events. Therefore, any interference with chromatin remodeling results in impaired NER efficiency (25) and faulty checkpoint activation (19,26). PTMs such as methylation, ubiquitylation, phosphorylation, and acetylation, of both the histone N-terminal tails and histone core residues, regulate DNA repair and checkpoint activation. The best-known DDR-related histone modification is the ATM- and DNA-PKcs-mediated phosphorylation of Serine 139 of H2AX due to a variety of exposures including ionizing and UV irradiation (19,27). Another abundant and relevant histone PTM is the acetylation of histones H3 and H4. Induction of maximal core histone acetylation enhances NER following UV irradiation (28). Some studies indicate that acetylation of histones might signal nucleosome assembly following repair (29). Restoration of normal chromatin structure to allow re-entry into the cell cycle, referred as ‘checkpoint recovery’ (30) can be achieved by (i) the removal of introduced modifications by a specific enzymatic activity or (ii) the exchange of modified histones with an unmodified form. For example, in the case of γH2AX, PP2A was identified as the enzyme responsible for γH2AX dephosphorylation in mammalian cells (31). Reversal of other PTM remains a subject of active investigation.
Apart from histone PTM, histone chaperones such as anti-silencing function1 (ASF1) are important for nucleosome assembly and checkpoint recovery. Although the function of ASF1 in DNA transcription and replication is well established (32,33), its participatory role in DNA repair is only now beginning to emerge. For example, ASF1 is required for nucleosome assembly following in vitro NER in humans and DSB repair in yeast (34,35). ASF1 is also needed for the recovery and adaptation to the DNA damage checkpoint following repair by facilitating the acetylation of histone H3 on lysine 56 (H3K56Ac) by the histone acetyltransferase (HAT) rtt109 (36,37).
H3K56Ac, initially identified in Saccharomyces cerevisiae, is important in DNA-replication coupled repair and/or progression through the S phase (38,39). It is the abundant modification of newly synthesized histone H3 molecules incorporated into chromosomes during S phase (38,39). Defects in H3K56 acetylation result in sensitivity to genotoxic agents that cause strand breaks during replication. Increased level of H3K56Ac upon DNA damage is mediated by p300 HAT in association with ASF1A and deposited at sites of DNA damage in a CAF1-dependent manner (40–42). Nevertheless, constitutive H3K56 acetylation also results in poor growth, spontaneous DNA damage, and chromosome loss, suggesting that too much of this modified histone can negatively impact the chromosome structure (43,44). Therefore, H3K56 acetylation and deacetylation must be tightly regulated for maintaining genome stability. This study describes the nature and function of ASF1A-mediated H3K56 acetylation and its importance for restoration of chromatin and cell-cycle progression in mammalian cells.
MATERIALS AND METHODS
Cell lines
OSU-2 normal human fibroblasts were generated in our laboratory as described previously (45). GM04405 (AT) and GM18366 (Seckel) cells were obtained from Coriell Institute for Medical Research, Camden, NJ, USA. HeLa and OSU-2 cells were cultured in Dulbecco’s Modified Eagle’s Medium (DMEM) supplemented with 10% fetal bovine serum (FBS). AT cells were cultured in EMEM supplemented with Earle’s salts and nonessential amino acids and 15% FBS. Seckel cells were cultured in EMEM supplemented with Earle’s salts and nonessential amino acids and 10% FBS. H1299 cells expressing Flag-tagged histone H3.1 and Flag-tagged histone H3K56R were a kind gift from Dr. Zhenkun Lou, Division of Oncology Research, Department of Oncology, Mayo Clinic, Rochester, MN, USA, and are described by Yuan et al. (41). All cells were grown at 37°C in a humidified atmosphere with 5% CO2.
Antibodies
Antibodies toward H3K56Ac (21,34-1) were obtained from Epitomics, Burlingame, CA, USA. Antibodies against ASF1A (2,990) and γ-H2AX (2,5,77) were from Cell Signaling Technology, Danvers, MA, USA. Anti-H2AX (sc-54606), anti-ATM (sc-23921) and anti-ATR (sc-1887) antibodies were from Santa Cruz Biotechnology, Santa Cruz, CA, USA. Details of raising rabbit anti-CPD antibodies in our laboratory, and its specificity, are described earlier (46). Antibodies specific for 6-4PP were kindly provided by Dr. Toshio Mori (Nara Medical University, Nara, Japan).
UV irradiation, protein isolation and western blotting
Cells were washed with phosphate-buffered saline (PBS) and irradiated with various doses of UV using a germicidal lamp with UV-C light (254 nM) at a dose rate of 1.0 J m−2/s as measured with a Kettering model 65 radiometer (Cole-Palmer, Vernon Hills, IL, USA). Media was added to the cells, returned to 37°C incubator to allow repair and harvested at the indicated post-UV irradiation times. Total protein was extracted from the cells using sodium dodecyl sulfate (SDS) lysis buffer (62 mM Tris–HCl, pH 6.8, 2% SDS, 10% glycerol) with protease and phosphatase inhibitors followed by boiling for 5 min. Protein amount was estimated using Bio-Rad DCTM Protein assay kit and the whole cell lysates were resolved by SDS–polyacrylamide gel electrophoresis (PAGE) using Novex Tris-Glycine gels (Invitrogen, Carlsbad, CA, USA) followed by western blotting to detect specific proteins. The intensity of protein bands was determined using UN-SCAN-IT Gel 6.1 software and normalized to β-ACTIN levels.
Gene silencing
Small hairpin RNA (shRNA) toward ASF1A and ATM were obtained from Sigma Aldrich. ATR siRNA was from Dharmacon, Chicago, IL, USA. siRNA transfections were conducted using LipofectamineTM 2000 transfection reagent (Invitrogen) according to the manufacturer’s instructions. In brief, 0.3 × 106 HeLa cells were seeded in 60-mm tissue culture dishes and grown overnight in media without antibiotics. Lipofectamine 2000 reagent was diluted in Opti-MEM medium, siRNA added to it and incubated at room temperature (RT) for 20 min. The Lipofectamine–small interfering RNA (siRNA) complex was added to the cells and incubated for 48 h before UV irradiation.
shRNA transfections were performed using Fugene 6 transfection reagent (Roche, Mannheim, Germany) according to the manufacturer’s protocol. Briefly, cells were grown to 50% confluency in media containing serum but no antibiotics. Fugene 6 reagent was diluted in serum- and antibiotic-free medium. shRNA plasmid DNA was added to the diluted Fugene 6 reagent and incubated at RT for 20 min. The Fugene to DNA ratio was maintained at 6:1. The Fugene 6–DNA complex was added to cells in a drop-wise manner and returned to the incubator. Transfection was allowed to take place for 24 h prior to UV irradiation of the cells. shRNA transfections were performed again after UV irradiation to maintain the knockdown of the genes for a further 48 h.
HeLa cells stably transfected with ATM shRNA (HeLa–shATM cells) were generated by transfecting HeLa cells with ATM shRNA plasmids using the Fugene 6 transfection reagent. At 24 h post-transfection, 2 µg/ml of puromycin was added to the media to select for cells transfected with the shRNA plasmids. Selection with puromycin was continued further for 1 week. The surviving cells were trypsinized, diluted and re-seeded so as to achieve colonies from single cells under puromycin selection. Clones exhibiting knockdown of ATM were recovered and expanded for further experiments.
Localized micropore assay and immunofluorescence
Micropore UV irradiation was conducted by placing a 5-μm isopore polycarbonate filter (Millipore, Bedford, MA, USA) on top of the cell monolayer grown on glass coverslips followed by irradiation with UV light. UV-irradiated cells were maintained in a suitable medium for the desired period and processed thereafter. Immunofluorescence staining was conducted according to the method established in our laboratory. Briefly, cells were washed twice with cold PBS and fixed with 2% paraformaldehyde in 0.5% Triton X-100 at 4°C for 30 min. Cells were rinsed with PBS and blocked with 20% normal goat serum in PBS, stained with anti-γ-H2AX antibody followed by staining with Texas Red-conjugated secondary antibodies. The coverslips were mounted in Vectashield mounting medium with DAPI (Vector laboratories, Burlingame, CA, USA). Fluorescence images were obtained at RT with a Nikon fluorescence microscope E80i (Tokyo, Japan) equipped with a Nikon Plan Fluor objective lens with 40× magnification and numerical aperture of 0.75. The digital images were captured with a cooled SPOT RTKE charge-coupled-device camera (Diagnostic Instruments, Sterling Heights, MI) and processed with SPOT analysis software (Diagnostic Instruments). For CPD staining, cells were treated with 2 M HCL for 10 min at 37°C to denature the DNA, washed twice and stained with anti-CPD antibody followed by staining with Texas Red-conjugated secondary antibody.
Quantitation of CPD and 6-4PP by immuno-slot blot assay
CPD and 6-4PP were quantitated by a highly sensitive immuno-slot blot assay as described earlier (47). Briefly, cells were irradiated with 10 J m−2 UV and harvested at various time points. Cells were lysed and DNA isolated using phenol–chloroform method. DNA was quantitated using Quant iTTM PicoGreen DsDNA reagent (Invitrogen, Carlsbad, CA, USA). Equal amount of DNA was blotted onto nitrocellulose membranes and the amounts of CPD and 6-4PP were determined using anti-CPD and anti-6-4PP antibodies.
Cell-cycle arrest and fluorescence-activated cell sorting analysis
OSU-2 cells were arrested in G1 by serum starvation for 48 h before transfection with ASF1A shRNA. 24 h post-transfection, cells were UV irradiated, released into the cell-cycle by the addition of medium containing serum and collected by trypsinization at the indicated time points. H1299 cells expressing Flag-tagged H3.1 and Flag-tagged H3K56R were synchronized by serum starvation for 24 h (48) before UV irradiation, released into the cell-cycle by the addition of medium containing serum, and collected by trypsinization at the indicated time points.
For fluorescence-activated cell sorting (FACS) analysis cells were washed once with PBS and resuspended in 0.5 ml PBS. Care was taken to achieve a single-cell suspension. Cells were fixed by drop-wise addition of 5 ml of ice-cold 70% ethanol into the cell suspension. The fixed cells were either stored at 4°C or stained immediately with propidium iodide. For propidium iodide staining, the ethanol-suspended cells were centrifuged at 200g for 5 min and the ethanol decanted thoroughly. Cells were washed once with PBS and resuspended in 0.5 ml of propidium iodide-Triton X-100 staining solution containing RNase A (0.1% TritonX-100 in PBS, 0.2 mg/ml Dnase-free RNase A, 0.02 mg/ml propidium iodide) and incubated at 37°C for 15 min. DNA content analysis of the propidium iodide-stained cells was carried out in a BD FACSCalibur flow cytometer (BD Biosciences, San Jose, CA, USA). Cell-cycle analysis was performed using the Cell Quest Pro software from BD Biosciences.
RESULTS
Histone H3 is promptly deacetylated on lysine 56 in response to UV irradiation, but acetylation of H3K56 is also rapidly restored
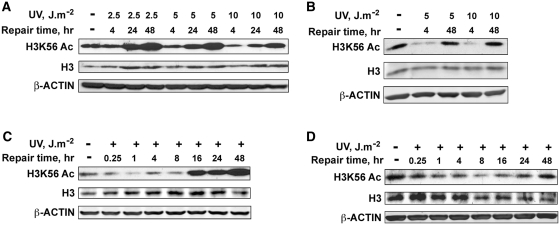
H3K56 in yeast chromatin has been reported to be acetylated in response to DNA damaging agents, whereas the acetylation status in response to DNA damage in mammalian system remains unclear. For instance, H3K56 acetylation is shown to increase in response to treatment with agents that mainly cause DNA breaks during replication (40–42). On the other hand, Tjeertes et al. and Miller et al. (49,50) have demonstrated that H3K56 is rapidly deacetylated at sites of DNA damage. To fully discern the state of H3K56 in chromatin and its role in DDR, we performed an extensive post-irradiation analysis of the levels of H3K56 acetylation following varying doses of UV irradiation of transformed HeLa and normal human fibroblast (NHF) cells. Western blot analysis of whole-cell protein lysates from UV-irradiated HeLa cells shows that H3K56 is consistently deacetylated in a dose-dependent manner at 4 h, but acetylation status was fully restored at 24 and 48 h of post-UV irradiation (Figure 1A). The restoration of H3K56 acetylation coincides with the well-established repair kinetics of UV-induced DNA lesions in human cells (25,51). Interestingly, the final levels of H3K56 acetylation restored at 24 and 48 h was significantly higher than the initial levels observed in the untreated cells (Figure 1A). Moreover, the H3K56 acetylation overshoot was distinctly related to the UV irradiation dose, i.e. a higher acetylation response at 2.5 than at 10 J m−2. A closer analysis indicates that greater deacetylation observed at higher doses might have interfered in the restoration of the acetylated state in HeLa cells. Effect of UV in NHF cells also exhibited a typical UV dose-dependent decrease followed by the post-irradiation recovery of H3K56 acetylation (Figure 1B). However, the levels of H3K56Ac at 48 h after irradiation were comparable to the untreated NHF cells. Therefore, unlike HeLa cells, the overshoot of H3K56 acetylation at later time points could not be seen in NHF cells. Although the reasons for enhanced H3K56 acetylation at later times in HeLa cells is not fully clear, it is possible that compared to normal cells rapidly dividing cancer cells require an efficient and faster recovery from the cell-cycle arrest. Therefore, this overshoot in H3K56Ac level could be attributable to cancerous cellular background. Interestingly, an identical increase in the H3K56Ac level has also been reported in several cancerous cell lines by Das et al. (40). Nevertheless, these data suggest that restoration of H3K56 acetylation in parallel with the repair of DNA damage is an integral component of maintaining genomic stability and normal cell function. A time course analysis of H3K56Ac deacetylation and its restoration in HeLa and NHF cells revealed a prompt and clear decrease in H3K56 deacetylation as early as 15 min after UV with a continuous steady decrease up to 8 h post-irradiation (Figure 1C and D). The beginning of restoration of the acetylation was evident from 16 h of post-UV irradiation in both cells lines. Since HeLa cells exhibited the typical overshoot of H3K56 acetylation beyond 16 h (Figure 1C), the completion of restoration occurring at around 48 h post-UV irradiation could be clearly discerned from the results of NHF cells (Figure 1D). As indicated above, the time course of restoration of H3K56Ac coinciding with the removal of UV-induced DNA lesions makes this recovery a potential mark for the completion of NER. In yeast, H3K56Ac is also shown to incorporate into chromatin during DNA replication (52). Therefore, to rule out the possibility that changes of the H3K56Ac observed in HeLa and NHF cells were not a result of actively cycling S phase cells, we specifically arrested NHF cells in the G1 phase of the cell-cycle before UV irradiation and determined the H3K56Ac levels by western blotting (Supplementary Figure S1). The initial decrease and restoration patterns of H3K56Ac levels in these G1-arrested cells were also similar to the asynchronous cells, indicating that H3K56 deacetylation and restoration of acetylation are UV damage-specific and DNA repair related responses.
Figure 1.
H3K56 is deacetylated in response to UV irradiation and acetylation is promptly restored after several hours of post-repair. (A) HeLa cells were exposed to increasing doses (2.5, 5 and 10 J m−2) of UV radiation, whole-cell lysates prepared immediately or at the indicated times, and the H3K56Ac levels determined by western blotting. (B) OSU-2 NHF cells were exposed to 5 and 10 J m−2 UV and cell lysates were prepared at the indicated post-repair times. Equal amounts of cell lysates were resolved on SDS–PAGE and probed for H3K56Ac levels by western blotting. (C) HeLa cells were exposed to 10 J m−2 UV irradiation, cell lysates prepared at various post-repair times and H3K56Ac levels determined by western blotting. (D) OSU-2 cells were exposed to 10 J m−2 UV irradiation and processed as described above for (C). Total H3 and β-actin levels were determined to ascertain equal protein loading.
Restoration of H3K56 acetylation is regulated by histone chaperone ASF1A
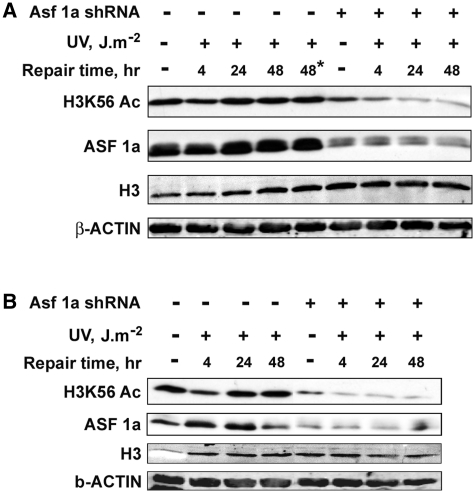
In yeast, ASF1 is important for the acetylation of H3K56 in the S phase (36,37). It also regulates H3K56 acetylation during DNA repair (35,40). Hence, we investigated the possible role of ASF1A in UV-induced H3K56 deacetylation and restoration of acetylation in mammalian cells. We quantitatively knocked down ASF1A in NHF cells by transfection of targeted shRNA plasmids (Supplementary Figure S2). As shown above, UV irradiation caused the typical loss of H3K56Ac levels and recovery following irradiation in NHF cells (Figure 2A). However, upon knockdown of ASF1A by specifically active shRNA plasmids, the recovery of H3K56 acetylation was drastically affected. As is clearly shown in Figure 2A, ASF1A depletion does not affect the deacetylation of H3K56 in response to UV irradiation and the expected reduction of H3K56 is apparent at 4 h. Nonetheless, the restoration of H3K56Ac at 24 and 48 h of post-UV repair is severely compromised in the absence of ASF1A (Figure 2A and Supplementary Figure S3) indicating that ASF1A is essential for H3K56 acetylation upon completion of NER, but is not required for the initial deacetylation process in UV-induced DNA damage. Interestingly, the decrease of H3K56Ac levels is exacerbated by ASF1A depletion even in the absence of UV damage (Supplementary Figure S3), suggesting that ASF1A is critical for maintaining the normal cellular levels of H3K56Ac. Previous studies have shown that yeast lacking ASF1 accumulate in metaphase of the cell-cycle due to the activation of the DNA damage checkpoint and are highly sensitive to DNA replication stress (53,54). In order to determine whether the defect in restoration of H3K56Ac in ASF1A-deficient NHF cells is specifically due to UV damage repair and not the potential replication stress, we knocked down ASF1A in G1-arrested NHF cells and determined the effect on the UV-induced H3K56 deacetylation–acetylation phenomenon (Figure 2B). ASF1A depletion had no effect on the deacetylation of H3K56 in the G1-arrested cells. However, as observed with the asynchronous cells, G1-arrested ASF1A-deficient cells exhibit the drastic defect in H3K56Ac restoration after the completion of NER. Hence, the data help conclude that ASF1A specifically mediates the restoration of H3K56 acetylation upon completion of UV damage repair in human cells.
Figure 2.
Restoration of H3K56 acetylation after UV damage repair is regulated by ASF1A histone chaperone. (A) ASF1A is required for H3K56Ac restoration in asynchronous NHF cells after UV damage repair. OSU-2 cells were transfected with ASF1A shRNA and after 24 h irradiated at 10 J m−2 UV. Cultures were again transfected with ASF1A shRNA to maintain the knockdown up to 48 h post-irradiation. Whole cell lysates were prepared at the indicated times and H3K56Ac levels determined by western blotting. Asterik indicates cells treated with Fugene transfection reagent alone to exclude any possible toxicity effects on H3K56Ac levels. (B) Regulation of H3K56Ac restoration by ASF1A in response to UV irradiation is independent of the effects of ASF1A during replication. OSU-2 cells were arrested in the G1 phase of the cell-cycle by serum starvation for 48 h, transfected with ASF1A shRNA and exposed to 10 J m−2 UV irradiation after 24 h of transfection. Cells were transfected again with ASF1A shRNA after UV irradiation to maintain the knockdown for a further 48 h. Cell lysates were prepared immediately or at the indicated post-UV repair times. H3K56Ac levels were determined by western blotting using anti-H3K56Ac antibody.
H3K56 acetylation is required for the dephosphorylation of γ-H2AX
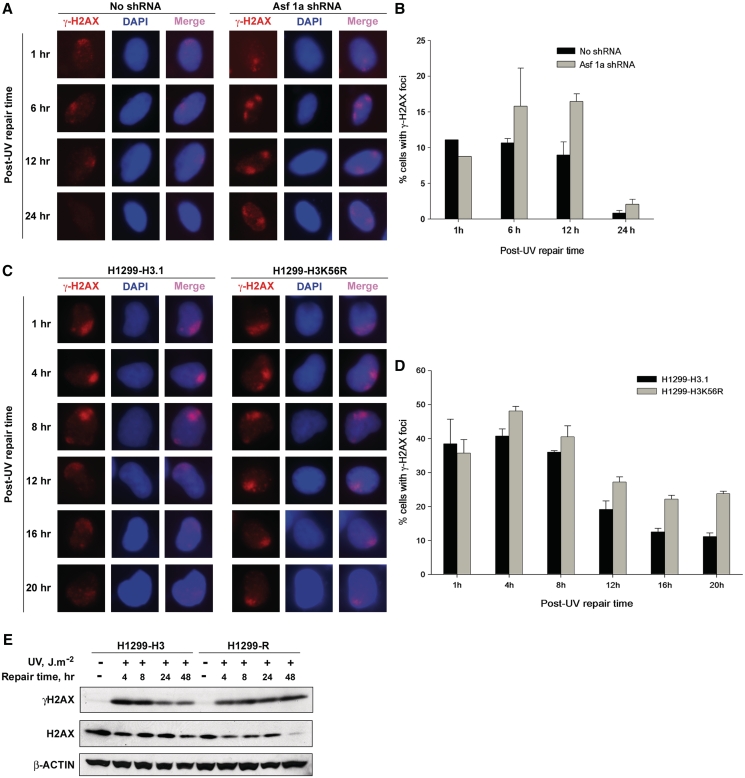
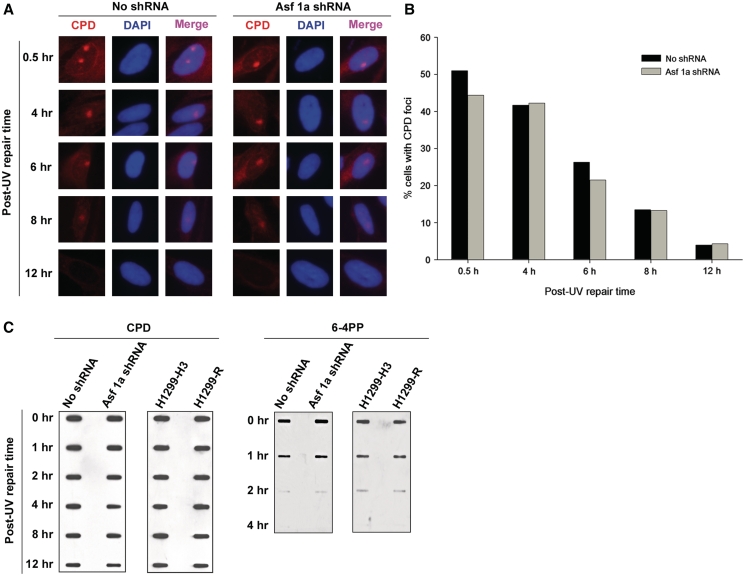
Deletion of ASF1 in yeast has been shown to cause spontaneous DNA damage, induction of DSB, and genomic instability (55). We wanted to find whether depletion of ASF1A in NHF cells can similarly result in spontaneous DNA damage. Formation of γ-H2AX is an established marker for DNA damage and presence of DSB (56), and H2AX is also phosphorylated upon processing of UV damage into DSB. Hence, we determined if γ-H2AX is induced in ASF1A-deficient cells. As usual, ASF1A was depleted in NHF cells by transfecting shRNA plasmids. Although we observed prompt phosphorylation of H2AX and spatial localization of γ-H2AX to the sites of DNA damage in UV-treated ASF1A-deficient and -proficient NHF cells (Figure 3A), we did not observe any γ-H2AX staining in the untreated cells (data not shown), indicating that the depletion of ASF1A does not lead to detectable spontaneous DNA damage. Nevertheless, we find that γ-H2AX in ASF1A-deficient cells is retained at sites of UV damage for longer times as compared to the control NHF cells (Figure 3A and Supplementary Figure S4A). About 10–15% of the cells formed γ-H2AX foci after UV irradiation (10 J m−2), which were retained up to 6 h post-UV irradiation in both control and ASF1A-deficient cells (Figure 3B). In control cells, however, ~50% of the cells with γ-H2AX foci lost their foci by 12 h post-UV irradiation, while complete loss of γ-H2AX foci was observed at 24 h post-UV irradiation. On the other hand, in shRNA transfected cells, the number of cells with γ-H2AX foci remained constantly higher than control up to 12 h post-UV irradiation. Besides, γ-H2AX foci were detectable even at 24 h post-UV irradiation in ASF1A-deficient cells. The prolonged retention of γ-H2AX foci at DNA damage sites could be a direct result of ASF1A deficiency or due to the observed defect in restoration of H3K56Ac in ASF1A-deficient cells. To determine the exact cause of γ-H2AX foci retention at the UV damage sites, we studied the kinetics of γ-H2AX foci formation and retention in H1299 cells harboring Flag-tagged H3K56R mutant (Figure 3C and Supplementary Figure S4B). As seen with ASF1A-deficient NHF cells, the H1299-K56R cells also retained γ-H2AX foci for longer times compared to the control H1299-H3.1 cells. Approximately, 40% of the cells formed γ-H2AX foci in response to UV irradiation in both H1299-3.1 and H1299-H3K56R cells (Figure 3D). These foci were maintained up to 8 h post-irradiation. At 12 h post-irradiation, only about 20% of H1299-H3.1 cells retained γ-H2AX foci and a steady decrease was observed at later times. However, in H1299-H3K56R cells, >20% of the cells still retained the foci even at 20 h of post-irradiation. These observations indicate that cells deficient in the acetylation of H3K56 show a delayed removal of γ-H2AX from the sites of DNA damage. Earlier reports indicate that suppression of PP2A, the enzyme that dephosphorylates γ-H2AX in mammalian cells, leads to the persistence of γ-H2AX foci (31). Hence, we wanted to determine if the persistence of γ-H2AX foci in H3K56 acetylation deficient cells was due to the delayed dephosphorylation of γ-H2AX. Western blot analysis of whole-cell protein lysates prepared from UV-treated H1299-H3.1 and H1299-H3K56R cells revealed that γ-H2AX was induced in both the cells after UV irradiation (Figure 3E and Supplementary Figure S7A) and levels of γ-H2AX were diminished at 24 and 48 h of post-irradiation in the control H1299-H3.1 cells, again coinciding with the kinetics of damage removal by NER. However, in the case of H1299-H3K56R cells, γ-H2AX levels remained unchanged and no discernable decrease could be observed even at 48 h of post-irradiation, revealing a defect in the dephosphorylation of γ-H2AX in these cells. To confirm whether NER was completed in ASF1A-depleted and H1299-H3K56R cells, we determined the kinetics of CPD and 6-4PP removal in these cells, using both immunofluorescence and immuno-slot blot assays. The results show that CPD and 6-4PP removal rate within the two cell lines is comparable at all the post-treatment time points (Figure 4 and Supplementary Figure S5). Thus, ASF1A-deficient and H1299-H3K56R cells are not defective in NER per se, and yet their retention of robust γ-H2AX signal further supports the conclusion that dephosphorylation of H2AX does not depend on NER but on the restoration of H3K56 acetylation.
Figure 3.
H3K56 acetylation is required for the dephosphorylation of γ-H2AX. (A) Depletion of ASF1A results in the retention of γ-H2AX for a longer time at sites of UV damage. OSU-2 cells were transfected with ASF1A shRNA and after 24 h irradiated with 10 J m−2 UV through micro-pore filters placed on the cell monolayers. Cells were recovered at indicated times and fixed with paraformaldehyde. Immunofluorescence was performed using anti-γ-H2AX antibody. Images shown are from a representative of multiple experiments. (B) Quantitation of γ-H2AX foci in ASF1A-depleted cells. Triplicate experiments were performed as described above in (A) and the number of cells containing γ-H2AX foci in shRNA-treated and -untreated cells were quantitated as follows. An average of 150 merged nuclear foci from at least five different microscopic fields were used for the quantitation of γ-H2AX foci for each experiment. The graph depicts the average number of cells containing γ-H2AX foci from the three independent experiments. The error bars show the mean standard deviation. (C) H3K56R mutation results in the retention of γ-H2AX foci at sites of UV damage. H1299-H3.1 and H1299-H3K56R cells were exposed to 10 J m−2 of UV radiation through micro-pore filters. Cells were fixed at the indicated intervals and immunofluorescence was performed to detect γ-H2AX foci. Images shown are from a representative experiment. (D) Quantitation of γ-H2AX foci in H1299 cells. Quantitation of γ-H2AX foci in H1299-H3.1 and H1299-H3K56R cells was essentially performed as described in (B) from two independent experiments. (E) H3K56 acetylation-deficient cells are defective in γ-H2AX dephosphorylation. H1299-H3.1 and H1299-H3K56R cells were irradiated at 10 J m−2 UV and whole cell lysates prepared at 4, 8, 24 and 48 h. Equal quantities of cell lysates were resolved on SDS–PAGE and the levels of γ-H2AX determined by western blotting. Total H2AX and β-ACTIN were used as loading controls.
Figure 4.
Acetylation of H3K56 is not required for NER. (A) Depletion of ASF1A does not affect CPD repair efficiency. OSU-2 cells were transfected with ASF1A shRNA and irradiated with 10 J m−2 UV after 24 h through micropore filters placed on the cell monolayers. Cells were fixed with paraformaldehyde at the indicated times and immunofluorescence was performed using anti-CPD antibody. Images shown are from a representative of duplicate experiments. (B) Quantitation of CPD foci in ASF1A-depleted cells. Duplicate experiments were performed as described above in (A) and the number of cells containing CPD foci in shRNA-treated and -untreated cells were quantitated as follows. An average of 150 merged nuclear foci from at least five different microscopic fields were used for the quantitation of CPD foci for each experiment. The graph depicts the average number of cells containing CPD foci from two independent experiments. (C) ASF1A-depleted and H1299-H3K56R cells are proficient in NER. ASF1A shRNA-treated cells and H1299-H3K56R cells were irradiated at 10 J m−2 UV and DNA isolated at the indicated post-irradiation times. Equal amounts of DNA was blotted on nitrocellulose membranes and probed for the presence of CPD and 6-4PP photoproducts with specific antibodies.
Restoration of H3K56 acetylation is necessary for recovery from UV-induced cell-cycle checkpoint
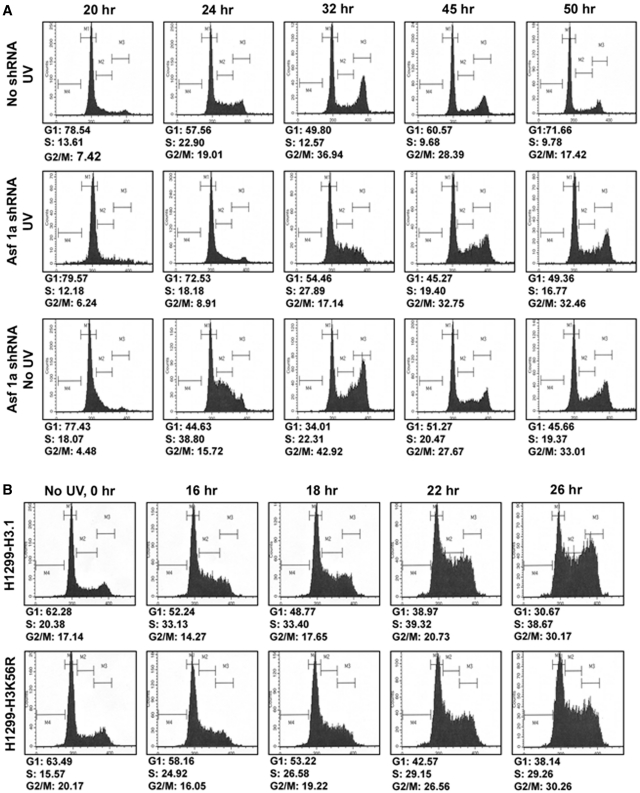
Removal of γ-H2AX from sites of DNA damage and its dephosphorylation is necessary for the inactivation of cell-cycle checkpoint (57). Moreover, ASF1 yeast mutants show activation of the DNA damage checkpoint during replication and ASF1 is required for the inactivation, recovery and adaptation of the checkpoint arrest after DSB repair (36,58). Since we observe the persistence of γ-H2AX foci in ASF1A-depleted cells and defect in γ-H2AX dephosphorylation in cells deficient in H3K56Ac restoration, we next determined if these cells were also deficient in the inactivation of UV-induced checkpoint. For this, NHF cells were arrested in G1 phase of the cell-cycle, transfected with ASF1A shRNA, UV irradiated (5 J m−2) and progression through the cell-cycle was exhaustively evaluated by flow cytometry (Figure 5A). In comparison to the control cells, ASF1A-deficient cells exhibited a significant delay in their progression from the G1 phase of the cell-cycle in response to UV irradiation (Figure 5A, compare panels 1 and 2). At 24 h of post-irradiation, only about 57% of the control cells remained in G1, while 72% of the ASF1A-deficient cells were present in the G1 phase. As reported earlier (41), a prominent S phase arrest was observed in ASF1A-deficient cells in the absence as well as the presence of UV damage (Figure 5A, panels 2 and 3). However, in the absence of DNA damage, ASF1A-deficient cells recovered from the S phase arrest, albeit slower than the ASF1A-proficient NHF cells (Figure 5A, compare panels 1 and 3). The S phase arrest in ASF1A-deficient cells was more prominent in the presence of UV damage and cells progressed slowly from the S phase as compared to nonirradiated cells (Figure 5A, compare panels 1, 2 and 3 at 32 h post-irradiation). Nevertheless, we did not observe any G2/M arrest in these cells.
Figure 5.
Restoration of H3K56 acetylation is necessary for recovery from UV-induced cell-cycle checkpoint arrest. (A) ASF1A-deficient cells exhibit cell-cycle checkpoint recovery defect. OSU-2 cells were arrested in G1 phase of the cell-cycle by serum starvation for 48 h. The arrested cells were transfected with ASF1A shRNA and after 24 h irradiated with 5 J m−2 UV and released into the cell-cycle by addition of media containing serum. Transfection with ASF1A shRNA was performed again to maintain the knockdown up to 50 h. Cells were harvested at the indicated times, fixed with 70% ethanol and the DNA content determined by FACS. (B) H3K56R mutant cells are defective in recovery from UV-induced cell-cycle checkpoint arrest. H1299-H3.1 and H1299-H3K56R cells were synchronized by serum starvation for 24 h. Cells were then irradiated with 10 J m−2 UV and released into the cell-cycle by addition of media containing serum. Cells were harvested at the indicated time points, fixed and DNA content determined by FACS.
Similar to the ASF1A-deficient NHF cells, UV-irradiated (10 J m−2) H1299-H3K56R cells also showed a delayed progression through the cell-cycle as compared to the control H1299-H3.1 cells (Figure 5B). These observations confirm that cells deficient in the acetylation of H3K56 show defects in the recovery from cell-cycle checkpoints activated in response to UV damage.
Recovery of H3K56 acetylation is regulated by the checkpoint kinase, ATM, but not ATR
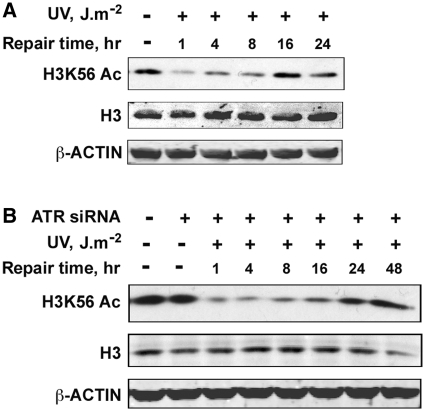
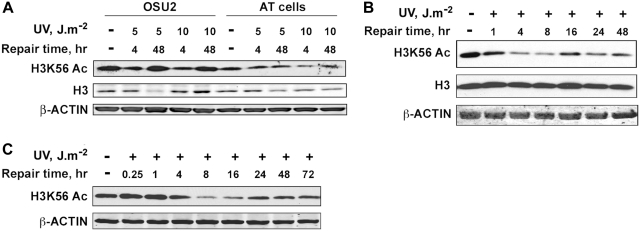
Since the primary kinases involved in checkpoint activation in response to UV damage are ATR and ATM, we were interested in finding out whether these kinases are involved in the regulation of H3K56Ac in response to UV-induced DNA damage. First, we analyzed H3K56Ac protein levels upon UV irradiation in ATR-deficient Seckel cells (Figure 6A, and Supplementary Figure S7B). We found that H3K56Ac levels decreased upon UV irradiation remained low up to 8 h and then started to return to normal at 16 h post-irradiation. These results are similar to those observed above with NHF and HeLa cells. Thus, the absence of functional ATR does not seem to affect either the deacetylation or the recovery of H3K56 acetylation in Seckel cells. To confirm these results under more controlled conditions, we quantitatively knocked down ATR in HeLa cells using targeted siRNA and determined H3K56Ac protein levels in response to UV irradiation (Figure 6B, and Supplementary Figure S7C). In these experiments, ATR knockdown was maintained at least up to 92 h (Supplementary Figure S2B). Importantly, the normal cellular levels of H3K56Ac were unaffected in the absence of ATR in HeLa cells. Upon UV irradiation of ATR-deficient HeLa cells, the H3K56Ac levels decreased predictably and promptly recovered to normal levels by 48 h post-UV irradiation (Figure 6B, and Supplementary Figure S7C). This again confirms the results with Seckel cells that ATR does not play any role in either the deacetylation or the restoration of acetylation of H3K56Ac in response to UV damage. We next investigated whether ATM has any regulatory effect on the H3K56 deacetylation and/or acetylation process. We irradiated the ATM-deficient AT cells with 5 and 10 J m−2 UV and determined H3K56Ac levels at 4 and 48 h post-irradiation (Figure 7A and Supplementary Figures S6 and S7D). We observed a decrease in H3K56Ac levels in response to UV irradiation up to 8 h similar to that observed in OSU-2 NHF cells ran in parallel, which is more discernible at a higher UV dose of 10 J m−2 compared to 5 J m−2. Surprisingly, the H3K56Ac levels failed to revert to normal levels at 48 h post-irradiation at the UV dose of 10 J m−2 (Figure 7A). In order to confirm this observation, we performed a detailed time course analysis of H3K56Ac protein levels in AT cells (Figure 7B, and Supplementary Figure S7E) and in HeLa–shATM cells (Figure 7C and Supplementary Figure S2C and S7 F) in response to 10 J m−2 UV irradiation. We observed a distinct decrease in H3K56Ac levels upon UV irradiation in AT cells, which is comparable to NHF cells (Figures 7B and 1D). HeLa–shATM cells also showed a decrease in H3K56Ac upon UV irradiation, becoming evident at 4 h and fully prominent at 8 h, which is comparable to parental HeLa cells (Figures 1A and 7 C). Notably, however, the H3K56Ac levels remained depressed up to 48 h in AT cells and even up to 72 h in HeLa–shATM cells as compared to the levels at these time points in NHF and HeLa cells, respectively. These observations show that ATM is crucial for the recovery of H3K56Ac levels in the UV DDR, but has no effect on the deacetylation of H3K56Ac immediately after UV irradiation.
Figure 6.
H3K56 deacetylation and acetylation is not regulated by ATR checkpoint kinase. (A) Seckel cells are proficient in the deacetylation and restoration of H3K56 acetylation in response to UV irradiation. Seckel cells were exposed to 10 J m−2 UV radiation and were harvested at the indicated times. Whole cell lysates prepared from the cells were resolved on SDS–PAGE and H3K56 acetylation levels determined by western blotting. (B) ATR deficiency does not affect either the deacetylation or restoration of H3K56 acetylation in response to UV irradiation. HeLa cells were transfected with 100 nM ATR siRNA using Lipofectamine tansfection reagent. At 48 h after transfection, cells were irradiated with 10 J m−2 UV and harvested at the indicated times. Whole-cell lysates were prepared, resolved on SDS–PAGE and H3K56Ac levels determined by western blotting with anti-H3K56Ac antibodies.
Figure 7.
Restoration of H3K56 acetylation is regulated by ATM checkpoint kinase. (A) AT cells are deficient in the restoration of H3K56 acetylation after UV repair. OSU-2 and AT cells were irradiated with 5 and 10 J m−2 UV, allowed to repair for 4 and 48 h and harvested to prepare whole-cell lysates. H3K56 acetylation levels were determined by western blotting. (B) AT cells are deficient in the restoration of H3K56 acetylation, but not the deacetylation in response to UV irradiation. AT cells were irradiated with 10 J m−2 UV and cells harvested at various intervals of post-UV repair. Whole-cell lysates were prepared and H3K56Ac levels analyzed by western blotting. (C) Depletion of ATM results in defective restoration of H3K56 acetylation. HeLa cells stably transfected with ATM shRNA were irradiated with 10 J m−2 UV, allowed to repair and harvested at various times. Whole-cell lysates were prepared and H3K56Ac levels determined by western blotting.
Taken together, above observations and the defects in cell-cycle progression observed in H3K56Ac-deficient H1299-H3K56R cells indicate that ASF1A-mediated H3K56Ac restoration is necessary for the recovery of UV-induced cell-cycle checkpoint arrest.
DISCUSSION
Acetylation of H3K56 is becoming increasingly important in unraveling the components and understanding the events orchestrating DDR in both yeast and mammalian systems. However, the nature of DNA damage-induced changes and the actual functional role of H3K56 acetylation and its regulation is in need of scientific clarity. The disparate observations about H3K56 acetylation reported in the literature could very well be due to the use of different damaging agents, their doses and the timing of the response measurements. In this study, we have conducted an extensive analysis of the functional role of H3K56 acetylation in regulating UV-induced DDR in mammalian cells. We report that H3K56 is deacetylated upon UV irradiation and represents an early upstream event of DNA damage signaling. In normal cells, acetylated state is promptly restored within a time course that parallels the completion of repair of UV lesions by NER. We further show that the restoration of H3K56 acetylation is intimately regulated by ASF1A histone chaperone and ATM checkpoint kinase. Lastly, H3K56Ac is required for the dephosphorylation of γ-H2AX and the recovery of checkpoint arrest.
Histone PTM are known to regulate the DDR in a number of ways including the recruitment of chromatin remodelers, DNA repair complexes, and checkpoint factors to the sites of damage. One of the best known examples is γ-H2AX, which is required for the recruitment of INO80 chromatin remodeling complex to facilitate recovery from checkpoint arrest (59,60). Similarly, it has been demonstrated that in H3K56R mutant yeast strains, Snf5 binding is reduced at the promoter as well as the coding regions of histone genes indicating that H3K56Ac is required for the recruitment of SWI/SNF complex to enable histone gene transcription (61). Moreover, deacetylation of H3K56 has been proposed in the closure of the entry–exit points of DNA on the histone octamer (62). This, however, might not be directly translatable to mammalian systems. Although the primary structure of yeast nucleosomes is similar to that of higher eukaryotes, subtle but meaningful differences have been observed in their crystal structures (63). For instance, unlike the higher eukaryotes, yeast nucleosomes contain hyperacetylated core histones (64). They also differ in physical properties such as thermal or salt stability (65,66). In addition, yeast is missing a conventional H1, which stabilizes nucleosomes in higher eukaryotes (67). These critical differences might explain why H3K56 is deacetylated in response to DNA damage in the mammalian system, unlike in yeast where it is acetylated. Moreover, species-specific differences in histone modifications have also been reported. Hence, the deacetylation rather than the acetylation of H3K56 in response to UV irradiation might very well serve as a signal for regulating the recruitment and/or exit of different chromatin remodeling factors in mammals. Additional experimentation is clearly needed to strengthen this concept. In support of this hypothesis, however, several other histones including H3K56 are also deacetylated in response to ionizing radiation (49), and the deacetylated state facilitates the recruitment of DSB repair factors. Therefore, it would also be worthwhile to determine whether H3K56 deacetylation could serve as a mark for the recruitment of NER and checkpoint factors, especially those involved early in the damage recognition phase.
Previous studies have shown the involvement of ASF1A in the acetylation of H3K56 in both the yeast and mammalian systems. In accordance with published reports, we also found that depletion of ASF1A severely affects the restoration of H3K56Ac in response to UV irradiation (Figure 2). However, our results clearly show that ASF1A depletion has no effect on the initial deacetylation of H3K56. Similarly, ATM and ATR were also not involved in regulating H3K56 deacetylation (Figures 6 and 7, and Supplementary Figure S6) indicating that H3K56 deacetylation is an upstream event or it may be regulated separately through an alternate pathway. Although ATR is an established checkpoint kinase acting primarily in UV damage response, we were surprised to find that the restoration of H3K56Ac is regulated by ATM, but not ATR checkpoint kinase. The role of ATM in the activation of checkpoint upon UV damage is now being increasingly appreciated due to the recent studies demonstrating a clear recruitment of ATM to UV damage sites and its impact on signaling cross-talk (18,19). Notably, Yajima et al. have reported that the downstream substrates of ATM, e.g. CHK2 and Kap1, were phosphorylated late in the UV damage response and that the late increase of ATM activity is needed to complement the decreasing ATR activity. Hence, it is possible that the late activation of ATM substrates such as CHK2 could influence the restoration of H3K56Ac brought about by ASF1A after the completion of repair. This assumption is further supported by studies demonstrating that various DNA damage checkpoint kinases regulate ASF1A and that CHK2 interacts directly with ASF1A in yeast (68–70).
We, for the first time, also report that the restoration of H3K56Ac is necessary for the removal of γ-H2AX from the sites of UV damage and for its dephosphorylation (Figure 3). Although earlier reports have observed spontaneous DNA damage in both ASF1A and H3K56 mutants, we could not observe any detectable γ-H2AX formation in ASF1A-depleted NHF cells and H1299-H3K56R that were not exposed to UV radiation. Furthermore, no defect in NER was observed in both ASF1A-depleted and H3K56 acetylation-deficient H1299-H3K56R cells (Figure 4), indicating that H3K56Ac is not required for the completion of repair. Dephosphorylation of γ-H2AX is an important step for the inactivation of checkpoint arrest (31). Accordingly, we found that cells depleted of ASF1A and H3K56 acetylation mutants remain arrested for a longer time compared to the control cells after UV irradiation (Figure 5). Hence, it can be concluded that the restoration of ASF1A-mediated H3K56 acetylation is required for the inactivation of UV-induced checkpoint arrest and for the progression of cells through the cell-cycle.
Our results show that the DDR is regulated by a dynamic H3K56 acetylation and deacetylation process for restoring the genomic integrity and successful progression of normal cell-cycle. Such a key role of H3K56Ac status in response to UV-damage could also occur in response to other kinds of genomic insults. H3K56 acetylation/deacetylation has already been implicated in chromatin assembly/disassembly in the yeast system (71). Therefore, it will be interesting to investigate whether the H3K56 acetylation/deacetylation plays a significant role in DDR and influence the transcription-coupled repair within active euchromatic as compared to the inactive heterochromatic regions of the genomes. Our current results cannot distinguish these specific epigenetic scenarios. Interestingly, the yeast acetylase Rtt109 requires ASF1 for stimulation of its enzymatic activity (72), further supporting a conserved role of ASF1 in regulating the H3K56 acetylation. Thus far, the combined results from other laboratories and ours strongly suggest that the modification of H3K56 plays a crucial conservatory role in maintaining different physiological processes in mammalian cells.
To summarize the overall observations, we propose that H3K56 deacetylation is an early event triggered by DNA damage upon UV irradiation of mammalian cells. According to our model (Figure 8), DNA damage results in the prompt deacetylation of H3K56, which in turn helps recruit different factors including chromatin remodelers to relax the chromatin structure for allowing easy access to complex NER and checkpoint machineries. Upon successful completion of repair, ASF1A is recruited in an ATM-dependent manner. At this point, the molecular mechanism of ASF1A recruitment is not clear, but it is possible that ASF1A interacts with ATM or ATM substrates to be recruited to the damage site. This is supported by the fact that ASF1A interacts with the ATM substrate, CHK2, in yeast. CHK2 protein, which is phosphorylated by ATM checkpoint kinase, allows the recruitment of ASF1A histone chaperone. In turn, ASF1A facilitates the recruitment of HATs needed for the restoration of native H3K56 acetylation status. Acetylation of H3K56 aids in the dephosphorylation of γ-H2AX and its removal from the damage sites resulting in the recovery from checkpoint arrest and cell-cycle progression.
Figure 8.
A model depicting the role of H3K56 deacetylation/acetylation in DNA damage response.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Institutes of Health grants (ES2388, ES12991 and CA93413 to A.A.W.); Pelotonia Postdoctoral Program (to A.B.). Funding for open access charges: National Institutes of Health.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
We acknowledge Dr. Zhenkun Lou, Division of Oncology Research, Department of Oncology, Mayo Clinic, Rochester, MN, for providing us Flag-tagged histone H3.1 and Flag-tagged histone H3K56R expressing cell lines.
REFERENCE
- 1.Harper JW, Elledge SJ. The DNA damage response: ten years after. Mol. Cell. 2007;28:739–745. doi: 10.1016/j.molcel.2007.11.015. [DOI] [PubMed] [Google Scholar]
- 2.Green CM, Almouzni G. When repair meets chromatin. First in series on chromatin dynamics. EMBO Rep. 2002;3:28–33. doi: 10.1093/embo-reports/kvf005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.De Boer J, Hoeijmakers JH. Nucleotide excision repair and human syndromes. car. 2000;21:453–460. doi: 10.1093/carcin/21.3.453. [DOI] [PubMed] [Google Scholar]
- 4.Reardon JT, Sancar A. Nucleotide excision repair. Prog. Nucleic Acid Res. Mol. Biol. 2005;79:183–235. doi: 10.1016/S0079-6603(04)79004-2. [DOI] [PubMed] [Google Scholar]
- 5.Mitchell DL, Haipek CA, Clarkson JM. Photoproducts are removed from the DNA of UV-irradiated mammalian cells more efficiently than cyclobutane pyrimidine dimers. Mutat. Res. 1985;143:109–112. doi: 10.1016/s0165-7992(85)80018-x. [DOI] [PubMed] [Google Scholar]
- 6.Kim JK, Choi BS. The solution structure of DNA duplex-decamer containing the (6-4) photoproduct of thymidylyl(3'–>5')thymidine by NMR and relaxation matrix refinement. Eur. J. Biochem. 1995;228:849–854. doi: 10.1111/j.1432-1033.1995.tb20331.x. [DOI] [PubMed] [Google Scholar]
- 7.Mellon I, Bohr VA, Smith CA, Hanawalt PC. Preferential DNA repair of an active gene in human cells. Proc. Natl Acad. Sci. USA. 1986;83:8878–8882. doi: 10.1073/pnas.83.23.8878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.De Laat WL, Jaspers NG, Hoeijmakers JH. Molecular mechanism of nucleotide excision repair. Genes Dev. 1999;13:768–785. doi: 10.1101/gad.13.7.768. [DOI] [PubMed] [Google Scholar]
- 9.Sancar A. DNA excision repair. Annu. Rev. Biochem. 1996;65:43–81. doi: 10.1146/annurev.bi.65.070196.000355. [DOI] [PubMed] [Google Scholar]
- 10.Wilkins RJ, Hart RW. Preferential DNA repair in human cells. Nature. 1974;247:35–36. doi: 10.1038/247035a0. [DOI] [PubMed] [Google Scholar]
- 11.Wang Z, Wu X, Friedberg EC. Nucleotide excision repair of DNA by human cell extracts is suppressed in reconstituted nucleosomes. J. Biol. Chem. 1991;266:22472–22478. [PubMed] [Google Scholar]
- 12.Evans DH, Linn S. Excision repair of pyrimidine dimers from simian virus 40 minichromosomes in vitro. J. Biol. Chem. 1984;259:10252–10259. [PubMed] [Google Scholar]
- 13.Hara R, Mo J, Sancar A. DNA damage in the nucleosome core is refractory to repair by human excision nuclease. Mol. Cell. Biol. 2000;20:9173–9181. doi: 10.1128/mcb.20.24.9173-9181.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Liu X, Smerdon MJ. Nucleotide excision repair of the 5 S ribosomal RNA gene assembled into a nucleosome. J. Biol. Chem. 2000;275:23729–23735. doi: 10.1074/jbc.M002206200. [DOI] [PubMed] [Google Scholar]
- 15.Lukas J, Lukas C, Bartek J. Mammalian cell cycle checkpoints: signalling pathways and their organization in space and time. DNA Repair. 2004;3:997–1007. doi: 10.1016/j.dnarep.2004.03.006. [DOI] [PubMed] [Google Scholar]
- 16.Ishikawa K, Ishii H, Saito T. DNA damage-dependent cell cycle checkpoints and genomic stability. DNA Cell Biol. 2006;25:406–411. doi: 10.1089/dna.2006.25.406. [DOI] [PubMed] [Google Scholar]
- 17.Harrison JC, Haber JE. Surviving the breakup: the DNA damage checkpoint. Annu. Rev. Genet. 2006;40:209–235. doi: 10.1146/annurev.genet.40.051206.105231. [DOI] [PubMed] [Google Scholar]
- 18.Yajima H, Lee KJ, Zhang S, Kobayashi J, Chen BP. DNA double-strand break formation upon UV-induced replication stress activates ATM and DNA-PKcs kinases. J. Mol. Biol. 2009;385:800–810. doi: 10.1016/j.jmb.2008.11.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ray A, Mir SN, Wani G, Zhao Q, Battu A, Zhu Q, Wang QE, Wani AA. Human SNF5/INI1, a component of the human SWI/SNF chromatin remodeling complex, promotes nucleotide excision repair by influencing ATM recruitment and downstream H2AX phosphorylation. Mol. Cell. Biol. 2009;29:6206–6219. doi: 10.1128/MCB.00503-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lee JS, Smith E, Shilatifard A. The language of histone crosstalk. Cell. 2010;142:682–685. doi: 10.1016/j.cell.2010.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.van AH, Gasser SM. The histone code at DNA breaks: a guide to repair? Nat. Rev. Mol. Cell Biol. 2005;6:757–765. doi: 10.1038/nrm1737. [DOI] [PubMed] [Google Scholar]
- 22.Bowman GD. Mechanisms of ATP-dependent nucleosome sliding. Curr. Opin. Struct. Biol. 2010;20:73–81. doi: 10.1016/j.sbi.2009.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Das C, Tyler JK, Churchill ME. The histone shuffle: histone chaperones in an energetic dance. Trends Biochem. Sci. 2010;35:476–489. doi: 10.1016/j.tibs.2010.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Altaf M, Auger A, Covic M, Cote J. Connection between histone H2A variants and chromatin remodeling complexes. Biochem. Cell Biol. 2009;87:35–50. doi: 10.1139/O08-140. [DOI] [PubMed] [Google Scholar]
- 25.Zhao Q, Wang QE, Ray A, Wani G, Han C, Milum K, Wani AA. Modulation of nucleotide excision repair by mammalian SWI/SNF chromatin-remodeling complex. J. Biol. Chem. 2009;284:30424–30432. doi: 10.1074/jbc.M109.044982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gong F, Fahy D, Liu H, Wang W, Smerdon MJ. Role of the mammalian SWI/SNF chromatin remodeling complex in the cellular response to UV damage. Cell Cycle. 2008;7:1067–1074. doi: 10.4161/cc.7.8.5647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hanasoge S, Ljungman M. H2AX phosphorylation after UV irradiation is triggered by DNA repair intermediates and is mediated by the ATR kinase. Carcinogenesis. 2007;28:2298–2304. doi: 10.1093/carcin/bgm157. [DOI] [PubMed] [Google Scholar]
- 28.Hasan S, Hassa PO, Imhof R, Hottiger MO. Transcription coactivator p300 binds PCNA and may have a role in DNA repair synthesis. Nature. 2001;410:387–391. doi: 10.1038/35066610. [DOI] [PubMed] [Google Scholar]
- 29.Brand M, Moggs JG, Oulad-Abdelghani M, Lejeune F, Dilworth FJ, Stevenin J, Almouzni G, Tora L. UV-damaged DNA-binding protein in the TFTC complex links DNA damage recognition to nucleosome acetylation. EMBO J. 2001;20:3187–3196. doi: 10.1093/emboj/20.12.3187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bartek J, Lukas J. DNA damage checkpoints: from initiation to recovery or adaptation. Curr. Opin. Cell Biol. 2007;19:238–245. doi: 10.1016/j.ceb.2007.02.009. [DOI] [PubMed] [Google Scholar]
- 31.Chowdhury D, Keogh MC, Ishii H, Peterson CL, Buratowski S, Lieberman J. gamma-H2AX dephosphorylation by protein phosphatase 2A facilitates DNA double-strand break repair. Mol. Cell. 2005;20:801–809. doi: 10.1016/j.molcel.2005.10.003. [DOI] [PubMed] [Google Scholar]
- 32.Krebs A, Tora L. Keys to open chromatin for transcription activation: FACT and Asf1. Mol. Cell. 2009;34:397–399. doi: 10.1016/j.molcel.2009.05.004. [DOI] [PubMed] [Google Scholar]
- 33.English CM, Adkins MW, Carson JJ, Churchill ME, Tyler JK. Structural basis for the histone chaperone activity of Asf1. Cell. 2006;127:495–508. doi: 10.1016/j.cell.2006.08.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mello JA, Sillje HH, Roche DM, Kirschner DB, Nigg EA, Almouzni G. Human Asf1 and CAF-1 interact and synergize in a repair-coupled nucleosome assembly pathway 6. EMBO Rep. 2002;3:329–334. doi: 10.1093/embo-reports/kvf068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chen CC, Carson JJ, Feser J, Tamburini B, Zabaronick S, Linger J, Tyler JK. Acetylated lysine 56 on histone H3 drives chromatin assembly after repair and signals for the completion of repair. Cell. 2008;134:231–243. doi: 10.1016/j.cell.2008.06.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Adkins MW, Carson JJ, English CM, Ramey CJ, Tyler JK. The histone chaperone anti-silencing function 1 stimulates the acetylation of newly synthesized histone H3 in S-phase. J. Biol. Chem. 2007;282:1334–1340. doi: 10.1074/jbc.M608025200. [DOI] [PubMed] [Google Scholar]
- 37.Recht J, Tsubota T, Tanny JC, Diaz RL, Berger JM, Zhang X, Garcia BA, Shabanowitz J, Burlingame AL, Hunt DF, et al. Histone chaperone Asf1 is required for histone H3 lysine 56 acetylation, a modification associated with S phase in mitosis and meiosis. Proc. Natl Acad. Sci. USA. 2006;103:6988–6993. doi: 10.1073/pnas.0601676103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Masumoto H, Hawke D, Kobayashi R, Verreault A. A role for cell-cycle-regulated histone H3 lysine 56 acetylation in the DNA damage response. Nature. 2005;436:294–298. doi: 10.1038/nature03714. [DOI] [PubMed] [Google Scholar]
- 39.Ozdemir A, Spicuglia S, Lasonder E, Vermeulen M, Campsteijn C, Stunnenberg HG, Logie C. Characterization of lysine 56 of histone H3 as an acetylation site in Saccharomyces cerevisiae. J. Biol. Chem. 2005;280:25949–25952. doi: 10.1074/jbc.C500181200. [DOI] [PubMed] [Google Scholar]
- 40.Das C, Lucia MS, Hansen KC, Tyler JK. CBP/p300-mediated acetylation of histone H3 on lysine 56. Nature. 2009;459:113–117. doi: 10.1038/nature07861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yuan J, Pu M, Zhang Z, Lou Z. Histone H3-K56 acetylation is important for genomic stability in mammals. Cell Cycle. 2009;8:1747–1753. doi: 10.4161/cc.8.11.8620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Vempati RK, Jayani RS, Notani D, Sengupta A, Galande S, Haldar D. p300 mediated acetylation of histone H3 lysine 56 functions in DNA damage response in mammals. J. Biol. Chem. 2010;285:28553–28564. doi: 10.1074/jbc.M110.149393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Celic I, Masumoto H, Griffith WP, Meluh P, Cotter RJ, Boeke JD, Verreault A. The sirtuins hst3 and Hst4p preserve genome integrity by controlling histone h3 lysine 56 deacetylation. Curr. Biol. 2006;16:1280–1289. doi: 10.1016/j.cub.2006.06.023. [DOI] [PubMed] [Google Scholar]
- 44.Maas NL, Miller KM, DeFazio LG, Toczyski DP. Cell cycle and checkpoint regulation of histone H3 K56 acetylation by Hst3 and Hst4. Mol. Cell. 2006;23:109–119. doi: 10.1016/j.molcel.2006.06.006. [DOI] [PubMed] [Google Scholar]
- 45.Venkatachalam S, Denissenko M, Wani AA. Modulation of (+/–)-anti-BPDE mediated p53 accumulation by inhibitors of protein kinase C and poly(ADP-ribose) polymerase. Oncogene. 1997;14:801–809. doi: 10.1038/sj.onc.1200890. [DOI] [PubMed] [Google Scholar]
- 46.Wani AA, Gibson-D'Ambrosio RE, D'Ambrosio SM. Antibodies to UV irradiated DNA: the monitoring of DNA damage by ELISA and indirect immunofluorescence. Photochem. Photobiol. 1984;40:465–471. doi: 10.1111/j.1751-1097.1984.tb04619.x. [DOI] [PubMed] [Google Scholar]
- 47.Wani AA, D'Ambrosio SM, Alvi NK. Quantitation of pyrimidine dimers by immunoslot blot following sublethal UV-irradiation of human cells. Photochem. Photobiol. 1987;46:477–482. doi: 10.1111/j.1751-1097.1987.tb04798.x. [DOI] [PubMed] [Google Scholar]
- 48.Yde CW, Olsen BB, Meek D, Watanabe N, Guerra B. The regulatory beta-subunit of protein kinase CK2 regulates cell-cycle progression at the onset of mitosis. Oncogene. 2008;27:4986–4997. doi: 10.1038/onc.2008.146. [DOI] [PubMed] [Google Scholar]
- 49.Miller KM, Tjeertes JV, Coates J, Legube G, Polo SE, Britton S, Jackson SP. Human HDAC1 and HDAC2 function in the DNA-damage response to promote DNA nonhomologous end-joining. Nat. Struct. Mol. Biol. 2010;17:1144–1151. doi: 10.1038/nsmb.1899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tjeertes JV, Miller KM, Jackson SP. Screen for DNA-damage-responsive histone modifications identifies H3K9Ac and H3K56Ac in human cells. EMBO J. 2009;28:1878–1889. doi: 10.1038/emboj.2009.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Emmert S, Kobayashi N, Khan SG, Kraemer KH. The xeroderma pigmentosum group C gene leads to selective repair of cyclobutane pyrimidine dimers rather than 6-4 photoproducts. Proc. Natl Acad. Sci. USA. 2000;97:2151–2156. doi: 10.1073/pnas.040559697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kaplan T, Liu CL, Erkmann JA, Holik J, Grunstein M, Kaufman PD, Friedman N, Rando OJ. Cell cycle- and chaperone-mediated regulation of H3K56ac incorporation in yeast. PLoS. Genet. 2008;4:e1000270. doi: 10.1371/journal.pgen.1000270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Yamaki M, Umehara T, Chimura T, Horikoshi M. Cell death with predominant apoptotic features in Saccharomyces cerevisiae mediated by deletion of the histone chaperone ASF1/CIA1. Genes Cells. 2001;6:1043–1054. doi: 10.1046/j.1365-2443.2001.00487.x. [DOI] [PubMed] [Google Scholar]
- 54.Ramey CJ, Howar S, Adkins M, Linger J, Spicer J, Tyler JK. Activation of the DNA damage checkpoint in yeast lacking the histone chaperone anti-silencing function 1. Mol. Cell Biol. 2004;24:10313–10327. doi: 10.1128/MCB.24.23.10313-10327.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Prado F, Cortes-Ledesma F, Aguilera A. The absence of the yeast chromatin assembly factor Asf1 increases genomic instability and sister chromatid exchange. EMBO Rep. 2004;5:497–502. doi: 10.1038/sj.embor.7400128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Mah LJ, El-Osta A, Karagiannis TC. gammaH2AX: a sensitive molecular marker of DNA damage and repair. Leukemia. 2010;24:679–686. doi: 10.1038/leu.2010.6. [DOI] [PubMed] [Google Scholar]
- 57.Keogh MC, Kim JA, Downey M, Fillingham J, Chowdhury D, Harrison JC, Onishi M, Datta N, Galicia S, Emili A, et al. A phosphatase complex that dephosphorylates gammaH2AX regulates DNA damage checkpoint recovery. Nature. 2006;439:497–501. doi: 10.1038/nature04384. [DOI] [PubMed] [Google Scholar]
- 58.Kim JA, Haber JE. Chromatin assembly factors Asf1 and CAF-1 have overlapping roles in deactivating the DNA damage checkpoint when DNA repair is complete. Proc. Natl Acad. Sci. USA. 2009;106:1151–1156. doi: 10.1073/pnas.0812578106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Morrison AJ, Kim JA, Person MD, Highland J, Xiao J, Wehr TS, Hensley S, Bao Y, Shen J, Collins SR, et al. Mec1/Tel1 phosphorylation of the INO80 chromatin remodeling complex influences DNA damage checkpoint responses. Cell. 2007;130:499–511. doi: 10.1016/j.cell.2007.06.010. [DOI] [PubMed] [Google Scholar]
- 60.Sarkar S, Kiely R, McHugh PJ. The Ino80 chromatin-remodeling complex restores chromatin structure during UV DNA damage repair. J. Cell Biol. 2010;191:1061–1068. doi: 10.1083/jcb.201006178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Xu F, Zhang K, Grunstein M. Acetylation in histone H3 globular domain regulates gene expression in yeast. Cell. 2005;121:375–385. doi: 10.1016/j.cell.2005.03.011. [DOI] [PubMed] [Google Scholar]
- 62.Xu F, Zhang Q, Zhang K, Xie W, Grunstein M. Sir2 deacetylates histone H3 lysine 56 to regulate telomeric heterochromatin structure in yeast. Mol. Cell. 2007;27:890–900. doi: 10.1016/j.molcel.2007.07.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.White CL, Suto RK, Luger K. Structure of the yeast nucleosome core particle reveals fundamental changes in internucleosome interactions. EMBO J. 2001;20:5207–5218. doi: 10.1093/emboj/20.18.5207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Davie JR, Saunders CA, Walsh JM, Weber SC. Histone modifications in the yeast S. cerevisiae. Nucleic Acids Res. 1981;9:3205–3216. doi: 10.1093/nar/9.13.3205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Morse RH, Pederson DS, Dean A, Simpson RT. Yeast nucleosomes allow thermal untwisting of DNA. Nucleic Acids Res. 1987;15:10311–10330. doi: 10.1093/nar/15.24.10311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Pineiro M, Puerta C, Palacian E. Yeast nucleosomal particles: structural and transcriptional properties. Biochemistry. 1991;30:5805–5810. doi: 10.1021/bi00237a025. [DOI] [PubMed] [Google Scholar]
- 67.Thoma F, Koller T, Klug A. Involvement of histone H1 in the organization of the nucleosome and of the salt-dependent superstructures of chromatin. J. Cell Biol. 1979;83:403–427. doi: 10.1083/jcb.83.2.403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Hu F, Alcasabas AA, Elledge SJ. Asf1 links Rad53 to control of chromatin assembly. Genes Dev. 2001;15:1061–1066. doi: 10.1101/gad.873201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Sharp JA, Rizki G, Kaufman PD. Regulation of histone deposition proteins Asf1/Hir1 by multiple DNA damage checkpoint kinases in Saccharomyces cerevisiae. Genetics. 2005;171:885–899. doi: 10.1534/genetics.105.044719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Emili A, Schieltz DM, Yates JR, III, Hartwell LH. Dynamic interaction of DNA damage checkpoint protein Rad53 with chromatin assembly factor Asf1. Mol. Cell. 2001;7:13–20. doi: 10.1016/s1097-2765(01)00150-2. [DOI] [PubMed] [Google Scholar]
- 71.Durairaj G, Chaurasia P, Lahudkar S, Malik S, Shukla A, Bhaumik SR. Regulation of chromatin assembly/disassembly by Rtt109p, a histone H3 Lys56-specific acetyltransferase, in vivo. J. Biol. Chem. 2010;285:30472–30479. doi: 10.1074/jbc.M110.113225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Driscoll R, Hudson A, Jackson SP. Yeast Rtt109 promotes genome stability by acetylating histone H3 on lysine 56 31. Science. 2007;315:649–652. doi: 10.1126/science.1135862. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.