Abstract
In the early ‘RNA world’ stage of life, RNA stored genetic information and catalyzed chemical reactions. However, the RNA world eventually gave rise to the DNA–RNA–protein world, and this transition included the ‘genetic takeover’ of information storage by DNA. We investigated evolutionary advantages for using DNA as the genetic material. The error rate of replication imposes a fundamental limit on the amount of information that can be stored in the genome, as mutations degrade information. We compared misincorporation rates of RNA and DNA in experimental non-enzymatic polymerization and calculated the lowest possible error rates from a thermodynamic model. Both analyses found that RNA replication was intrinsically error-prone compared to DNA, suggesting that total genomic information could increase after the transition to DNA. Analysis of the transitional RNA/DNA hybrid duplexes showed that copying RNA into DNA had similar fidelity to RNA replication, so information could be maintained during the genetic takeover. However, copying DNA into RNA was very error-prone, suggesting that attempts to return to the RNA world would result in a considerable loss of information. Therefore, the genetic takeover may have been driven by a combination of increased chemical stability, increased genome size and irreversibility.
INTRODUCTION
The RNA world theory posits that RNA fulfilled both of the major cellular functions, catalysis and information storage, during an early stage of life (1–3). RNA possesses the ability to store genetic information (e.g. in retroviruses), and RNA sequences can fold into complex structures, enabling enzymatic activity (ribozymes). The finding that the catalytic core of the ribosome comprises RNA lent considerable credence to the RNA world theory (4–6). This theory not only simplifies the origin of life by proposing a relatively uncomplicated replicating intermediate compared to a wholesale emergence of the transcription and translation machineries, but also implies that the RNA world then transitioned to the DNA–RNA–protein world. Our study is motivated by one of the central features of this transition: the ‘genetic takeover’ of RNA by DNA.
The transition to DNA has previously been considered primarily from a chemical perspective. In particular, the DNA backbone is less prone to hydrolysis, since it lacks the nucleophilic 2′-hydroxyl group, so it represents a more chemically stable genetic material. In this work, we consider a genetic perspective: does DNA replicate with greater intrinsic fidelity, thus allowing more information to be stored? Would information be lost during the genetic takeover or during a hypothetical reversion back to RNA? RNA viruses generally have higher mutation rates than DNA viruses (7,8), but it is unclear whether this is due to replication mechanisms in place today (e.g. DNA proofreading enzymes), natural selection [e.g. on evolvability (9)] or intrinsic properties of the nucleic acid backbones.
The mutation rate during replication places an important constraint on the amount of information that can be stored in the genome. Intuitively, the information degrades over subsequent generations if mutations are too frequent (an error ‘catastrophe’). In general, theoretical models indicate that the maximum genomic information content is inversely proportional to the mutation rate per base (10,11). The critical mutation rate above which the genomic information cannot survive is known as the error threshold (12). This relationship appears to hold for viruses, especially RNA viruses, which exist close to the error threshold of roughly one mutation per genome replication (13). Indeed, increasing the mutation rate to precipitate an error catastrophe appears to be a practical anti-retroviral strategy (14–16). The constraint on information is also a serious consideration for the early stages of life that were characterized by primitive replication mechanisms; mutation rates would have been quite high and would substantially limit the information content of the system (17–19).
Therefore, we sought to compare the intrinsic error susceptibility of RNA replication, DNA replication, copying RNA to DNA and copying DNA to RNA, using two strategies. First, we determined misincorporation rates in an experimental model of non-enzymatic polymerization. Although the polymerization chemistry present during the genetic takeover is not known, for these experiments we use 5′phosphorimidazolides as activated monomers and primers terminated by a 3′-amino-2′,3′-dideoxynucleotide; this system is capable of rapid polymerization, such that relatively slow rates of mis-incorporation can be measured (18,20–24). Similar systems have been previously used to demonstrate replication in model protocells (25,26).
Second, since experimental rates presumably depend on kinetic effects (e.g. the activation chemistry in non-enzymatic polymerization, or ribozyme mechanisms involved in catalyzed polymerization), we also attempted to estimate the lowest possible error rates achievable by any experimental system using equilibrium thermodynamic calculations (Figure 1). To our knowledge, this is the first comprehensive comparison of experimental and theoretical error rates and the first analysis of error rates in RNA/DNA hybrid duplexes. Both strategies showed that RNA is intrinsically error-prone compared to DNA. This effect can be attributed largely to the stability of G-U wobble pairs in RNA, which leads to a large increase in the frequency of mis-incorporation, particularly across U, in RNA compared to DNA. Characterization of the RNA–DNA hybrid systems showed that while RNA can be copied into the DNA complement fairly accurately, copying DNA back to an RNA complement would be quite inaccurate. These results suggest that information transfer to DNA would permit an increase in genomic information content as the mutation rate decreased, and that this transition would be essentially irreversible since copying back to RNA would be an error-ridden process.
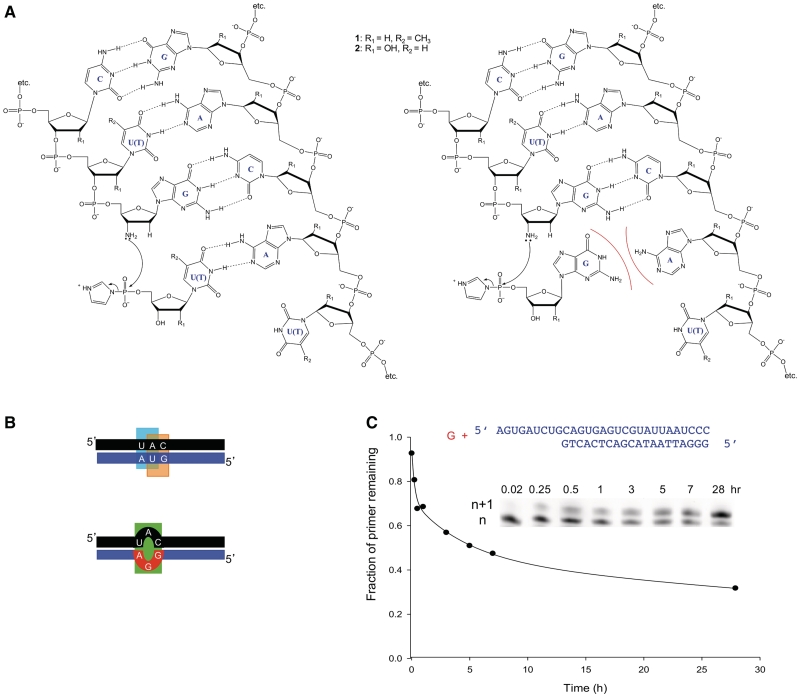
Figure 1.
Experimental and theoretical approaches to determining error rates. (A) Comparison of the experimental reaction rate of correct incorporation (left duplex) versus incorrect incorporation (right duplex). Template strand (right strand within duplex) is either DNA (1) or RNA (2). Primer strand (left strand within duplex) and activated nucleotide are either both DNA analogs (1) or both RNA analogs (2). (B) The free energy of the full-length duplex is calculated as a sum of independent contributions from stacking interactions and other simple structural elements (e.g. mismatches, bulges). Shown here is the comparison between a correctly matched product (top) using nearest–neighbor interactions (cyan and orange boxes) versus a product containing a single mismatch (bottom; green box). This comparison was used as a theoretical estimate for the lowest possible error rate. (C) Example of determination of experimental reaction rate: mis-incorporation of activated nucleotide (ImpdG; red) across template base G in RNA-templated DNA polymerization (blue). Gel image shows extension over time of the original primer (n) by one base (n + 1). Decrease of primer over time is plotted (initial rate = 0.24/h); the line is drawn to guide the eye. Initial rates were used because the reaction slows noticeably over time such that the yield is <100%; this is likely due to spontaneous hydrolysis of the activated monomer under the reaction conditions.
MATERIALS AND METHODS
Synthesis of activated nucleotides
All deoxynucleoside 5′-phosphorimidazolides (ImpdN) and nucleoside 5′ phosphorimidazolides (ImpN) were synthesized based on a previously published protocol (18,27,28) (Supplementary Data S4). ImpA, ImpC and ImpU were synthesized by GL Synthesis Inc. (Worcester, MA, USA). Activated nucleotides were verified by mass spectrometry and high-performance liquid chromatography (HPLC) as previously described (18) and were found to be >93% pure.
Oligonucleotides for non-enzymatic polymerization
The fluorescently labeled RNA primer was made by reverse synthesis in the W. M. Keck Biotechnology Resource Laboratory at Yale University (New Haven, CT, USA). The synthesis used 3′-O-tritylamino-N6-benzoyl-2′,3′-dideoxyguanosine-5′-cyanoethyl phosphoramidite (Metkinen Chemistry; Kuusisto, Finland) at the 3′-terminus and was labeled with Cy3 at the 5′-terminus. The primer was polyacrylamide gel electrophoresis (PAGE)-purified and its mass was verified by matrix-assisted laser desorption/ionization-time of flight (MALDI-TOF) (Supplementary Data S5). The fluorescently labeled DNA primer was synthesized as previously described (18). DNA oligonucleotides were synthesized and PAGE-purified by Sigma-Aldrich (St. Louis, MO, USA). RNA template sequences were from Dharmacon (Lafayette, CO, USA) and RNA excess primer was from UCDNA Services (Calgary, AB, Canada). See Supplementary Data S6 for oligonucleotide sequences.
Non-enzymatic polymerization
For observation on a laboratory timescale, the polymerization reaction required monomers activated at the 5′ position for incorporation. The activated monomer was a nucleoside 5′-phosphorimidazolide (ImpN) if the primer backbone was RNA, or a 2′-deoxynucleoside 5′-phosphorimidazolide (ImpdN) if the primer backbone was DNA. Templates were standard RNA or DNA, and primers were RNA or DNA with the exception that they were terminated by a single 3′-amino-2′,3′-dideoxynucleotide at the 3′ end. In all reactions, the template and primer were perfectly complementary at the beginning of the experiment, and each reaction was performed at least in duplicate. Extension was undetectable in the absence of template.
Primer extension reactions were carried out as previously described (18) (Supplementary Data S7), with the template and primer backbones varied to be either DNA or RNA. A primer (0.325 µM) and a template (1.3 µM) (1 µl each) were mixed in water, incubated at 95°C for 5 min, and annealed by cooling to room temperature on a benchtop for 5–7 min. In a reaction of 10 µl volume, 1 µl of 1 M Tris (pH 7) and 0.5 µl of 4 M NaCl were added to final concentrations of 100 mM Tris and 200 mM NaCl. For reactions with ImpdN/ImpN, where N = A, C, or G, the reaction was initiated by the addition of 1 µl of 100 mM ImpdN to a final concentration of 10 mM. For reactions involving ImpdT/ImpU, 1.38 µl of 289 mM stock solution was added to a final concentration of 40 mM. The reaction mixtures were incubated at room temperature, and aliquots were withdrawn at certain time points. For matched (Watson–Crick) reactions between the nucleotide and the template, time points were taken at 0.5 min, 1 min, 3 min, 7.5 min, 15 min, 30 min, 1 h, 2 h and 4 h. For mismatched reactions, time points were taken at 1 min, 7.5 min, 15 min, 30 min, 1 h, 2 h, 4 h, 8 h and 24 h. A negative control was taken before adding the ImpdN/ImpN in each reaction. For reactions using a DNA primer, time points were obtained by adding 1 µl of the reaction mixture to 9 µl of the loading buffer with 8 M urea, 100 mM ethylenediaminetetraacetic acid (EDTA), and 1.3 µM of a competitor DNA with the sequence: 5′ GG GAT TAA TAC GAC TCA CTN 3′, where N = A/T/G/C to match the primer employed in the reaction. For the RNA primer reactions, 65 µM of a competitor RNA with the sequence: 5′ GG GAU UAA UAC GAC UCA CUN 3′ was used instead of DNA. Time points were heated to 95°C for 5 min to disrupt primer–template complexes and were run on 20% denaturing PAGE. The initial rate of disappearance of primer was calculated. Examples of slow reactions are given in Supplementary Data (Supplementary Figure S2).
Calculation of experimental error rates
The frequency of incorporation (f) of a particular nucleotide a′ across a template base was calculated by dividing its rate of extension (r) by the sum of the rates of extension for all four ImpdNs or ImpNs across that template base:
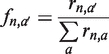 |
where n denotes the correct nucleotide complementary to the template base, and a ranges over all four nucleotides. If a′ = n, then fn,n is also called the fidelity. If a′ ≠ n, then fn, a′ is the error rate per site for a′ incorporated instead of n, or µn, a′. The mutation rate per site across a given template base B (μB) is Σ(µn, m), where m ≠ n. If the proportion of the genome composed of base X is PX, then the average mutation rate of a genome (μave) is Σ(PXμX). Since most aptamers and ribozymes have roughly even composition [22–28% of each nucleotide; (29,30)], we assume the genome has even composition. Therefore
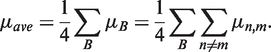 |
(1) |
Thermodynamic estimate of lower limit of the error rate
Using Equations 2 and 3 (Supplementary Data S8), the lower limit on the mutation rate is:
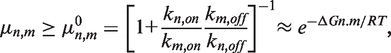 |
(2) |
where ΔGn,m is calculated as given below. Equation 2 can be generalized for four nucleotides (the correct nucleotide n and three erroneous nucleotides m) and non-equimolar nucleotide concentrations (10) as follows:
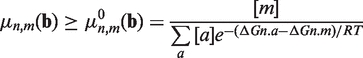 |
(3) |
where b is the vector of nucleotide concentrations and a ranges over all four nucleotides.
Calculation of nucleotide concentrations to minimize the mutation rate
The mutation rate is given by Equation 1, which is a function of b. The optimal nucleotide concentrations b* are obtained by simultaneous numerical minimization of Equation 1 with respect to b, given first-order kinetics with respect to the nucleotide concentration [i.e. rn, a′ is proportional to the concentration of (a′)] (Supplementary Data S9). To avoid large discrepancies among the optimized concentrations, we constrained all concentrations to be within a factor of 10 of each other. We calculated the optimal nucleotide supply using either the experimentally measured rates or the thermodynamic lower limits for the error rates, as given in Equation 3.
Thermodynamic calculation of free energy differences
The equilibrium probability of incorporating a certain nucleotide in a larger complex should include interactions with its 5′ and 3′ neighbors (Figure 1B). To estimate ΔGn,m, we use the nearest-neighbor model for predicting RNA and DNA duplex stabilities (31,32,33). For the example in Figure 1B,
We used energy parameters given in the literature for stacking and mismatches for the RNAt/RNAp and DNAt/DNAp systems, extrapolated to the experimental temperature of 22°C as described (34,32). These energy terms have an error of around 5–10% (32,33,35). Naïve error propagation implies that the relative uncertainties of the calculated mutation rates would be 30–60%; however, an accurate estimate of the uncertainty is also complicated by the fact that the energy parameters had been obtained by multivariate fitting of experimental data, so their values are likely to be highly correlated with each other. For the RNA/DNA hybrids, we used published stacking energies when available (35) or estimated them when not available (Supplementary Data S10).
RESULTS
To estimate the error rates (i.e. frequency of an error per residue) of non-enzymatic polymerization in the different nucleic acid systems, we used two approaches: measurements of mis-incorporation in an experimental model (Figure 1A) and calculations based on the thermodynamic differences between correct and incorrect incorporation (Figure 1B). Experimentally measured reaction rates yield straightforward estimates of error rates. However, we do not know whether the activation chemistry used in our experimental model is a good mimic of the chemistry of prebiotic replication. Therefore, we also sought to infer theoretical error rates based on the relative thermodynamic stability of correctly versus incorrectly paired complexes. These calculations estimate the theoretical lower limits for the error rates from the thermodynamics of RNA and DNA base pairing, which may or may not correlate with experimental rates that are kinetically determined. This combination of theory and experiment permitted us to cross-validate trends in the two separate sets of error rates and to quantitatively test how closely the experimental system approaches the theoretical limit. We first present the experimental results and then describe the theoretical calculations and their relationship to the experimental results.
We measured experimental reaction rates of non-enzymatic nucleic acid polymerization using a model system for template-directed replication (Figure 1C; Table 1; Supplementary Figure S1). Misincorporation rates were determined by comparing the rate of incorporation for the correct (Watson–Crick) base versus an incorrect base (Figure 1A; Table 2). The misincorporation rates and overall mutation rate for copying DNA into DNA had been previously determined (18); here, we determined the corresponding rates for copying RNA into RNA or DNA and for copying DNA into RNA using the same activation chemistry (5′-phosphorimidazolide nucleotides with primers terminated by a 2′,3′-dideoxy-3′-amino nucleoside at the 3′ end). The average mutation rate (µave) is given as the probability of a mutation (any error) per site. We refer to the different nucleic acid systems as ‘(template backbone)t/(primer backbone)p’, e.g. RNAt/DNAp designates an RNA template and DNA primer.
Table 1.
Experimental reaction rates (r, h−1) and standard deviation (σ)
| Template | ImpN or ImpdN | RNAt/RNAp |
RNAt/DNAp |
DNAt/RNAp |
|||
|---|---|---|---|---|---|---|---|
| r | σ | r | σ | r | σ | ||
| C | G | 2.3 | 0.08 | 1.6 | 1.2 | 3.8 | 0.47 |
| C | T or U | 0.0078 | 0.0021 | 0.0071 | 0.0057 | 0.021 | 0.0003 |
| C | C | 0.0035 | 0.001 | 0.00085 | 0.00007 | 0.022 | 0.0072 |
| C | A | 0.0075 | 0.0013 | 0.0080 | 0.0029 | 0.032 | 0.0049 |
| G | C | 20 | 2.2 | 2.5 | 0.7 | 1.1 | 0.42 |
| G | T or U | 0.91 | 0.23 | 0.75 | 0.13 | 0.29 | 0.11 |
| G | G | 0.25 | 0.02 | 0.25 | 0.003 | 0.11 | 0.0004 |
| G | A | 0.035 | 0.008 | 0.034 | 0.015 | 0.031 | 0.0036 |
| A | T or U | 1.1 | 0.16 | 1.6 | 0.13 | 0.47 | 0.023 |
| A | C | 0.078 | 0.029 | 0.022 | 0.0045 | 0.029 | 0.006 |
| A | G | 0.12 | 0.01 | 0.52 | 0.02 | 0.031 | 0.0005 |
| A | A | 0.016 | 0.0008 | 0.016 | 0.0075 | 0.029 | 0.005 |
| T or U | A | 0.55 | 0.26 | 0.30 | 0.052 | 0.093 | 0.026 |
| T or U | C | 0.011 | 0.004 | 0.0081 | 0.0037 | 0.019 | 0.0037 |
| T or U | G | 0.35 | 0.08 | 0.022 | 0.0011 | 0.065 | 0.0093 |
| T or U | T or U | 0.073 | 0.019 | 0.029 | 0.0004 | 0.064 | 0.0031 |
Table 2.
Theoretical (µtheory) and experimental (µexp) incorporation and mis-incorporation frequencies with standard deviation determined from replicates (σexp)
| Template | ImpN or ImpdN | DNAt/DNAp |
RNAt/RNAp |
RNAt/DNAp |
DNAt/RNAp |
||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| µtheory | µexp | σexp | µtheory | µexp | σexp | µtheory | µexp | σexp | µtheory | µexp | σexp | ||
| C | G | 0.9998 | 0.9939 | 2.4E-03 | 1.0000 | 0.9917 | 8.0E-04 | 0.9998 | 0.9939 | 1.0E-03 | 0.9999 | 0.9803 | 2.9E-03 |
| C | T or U | 1.6E-04 | 3.1E-03 | 1.6E-03 | 2.9E-05 | 3.4E-03 | 8.2E-04 | 1.6E-04 | 1.9E-03 | 4.0E-04 | 5.8E-05 | 5.5E-03 | 6.0E-04 |
| C | C | 3.6E-05 | 7.8E-04 | 2.3E-04 | 7.2E-06 | 1.6E-03 | 4.9E-04 | 3.8E-05 | 4.3E-04 | 1.7E-04 | 1.4E-05 | 5.7E-03 | 2.6E-03 |
| C | A | 1.7E-05 | 2.2E-03 | 6.1E-04 | 7.2E-06 | 3.3E-03 | 4.8E-04 | 2.7E-05 | 3.8E-03 | 4.6E-04 | 9.5E-06 | 8.4E-03 | 2.7E-04 |
| G | C | 0.9932 | 0.9487 | 1.5E-03 | 0.9628 | 0.9435 | 5.6E-03 | 0.9844 | 0.7057 | 3.4E-02 | 0.9486 | 0.7208 | 2.6E-02 |
| G | T or U | 4.1E-03 | 1.9E-02 | 1.1E-03 | 3.7E-02 | 4.3E-02 | 6.1E-03 | 1.5E-02 | 2.1E-01 | 1.2E-02 | 5.1E-02 | 1.8E-01 | 8.5E-03 |
| G | G | 2.4E-03 | 1.5E-02 | 4.2E-03 | 7.4E-06 | 1.2E-02 | 6.2E-04 | 1.7E-04 | 7.2E-02 | 1.6E-02 | 5.5E-04 | 7.4E-02 | 2.5E-02 |
| G | A | 2.0E-04 | 1.7E-02 | 6.8E-03 | 7.4E-06 | 1.7E-03 | 2.0E-04 | 4.8E-05 | 1.0E-02 | 6.6E-03 | 1.6E-04 | 2.1E-02 | 9.4E-03 |
| A | T or U | 0.9965 | 0.9058 | 4.7E-02 | 0.9997 | 0.8397 | 7.7E-03 | 0.9993 | 0.7458 | 1.8E-02 | 0.997 | 0.8422 | 2.3E-02 |
| A | C | 2.2E-04 | 1.8E-02 | 6.0E-03 | 8.4E-05 | 5.7E-02 | 1.4E-02 | 1.2E-04 | 1.0E-02 | 1.5E-03 | 4.9E-04 | 5.2E-02 | 1.2E-02 |
| A | G | 2.8E-03 | 4.6E-02 | 2.4E-02 | 8.4E-05 | 9.1E-02 | 2.0E-02 | 4.1E-04 | 2.4E-01 | 2.3E-02 | 1.7E-03 | 5.5E-02 | 2.8E-04 |
| A | A | 5.5E-04 | 3.1E-02 | 1.7E-02 | 8.4E-05 | 1.2E-02 | 1.0E-03 | 1.8E-04 | 7.1E-03 | 3.0E-03 | 7.8E-04 | 5.1E-02 | 1.1E-02 |
| T or U | A | 0.9921 | 0.8471 | 1.5E-02 | 0.8845 | 0.5515 | 6.7E-02 | 0.9323 | 0.8298 | 3.0E-02 | 0.955 | 0.3819 | 7.1E-02 |
| T or U | C | 5.2E-04 | 1.6E-02 | 9.3E-03 | 2.6E-04 | 1.3E-02 | 8.9E-03 | 1.2E-03 | 2.9E-02 | 6.7E-03 | 8.2E-04 | 7.9E-02 | 7.9E-03 |
| T or U | G | 2.7E-03 | 5.3E-02 | 3.4E-02 | 1.1E-01 | 3.6E-01 | 5.0E-02 | 5.9E-02 | 6.1E-02 | 1.1E-02 | 3.9E-02 | 2.7E-01 | 6.6E-02 |
| T or U | T or U | 4.7E-03 | 8.3E-02 | 2.9E-02 | 1.0E-03 | 7.5E-02 | 7.4E-03 | 7.5E-03 | 8.1E-02 | 1.2E-02 | 5.0E-03 | 2.7E-01 | 1.3E-02 |
Non-enzymatic replication of RNA has low intrinsic fidelity compared to DNA
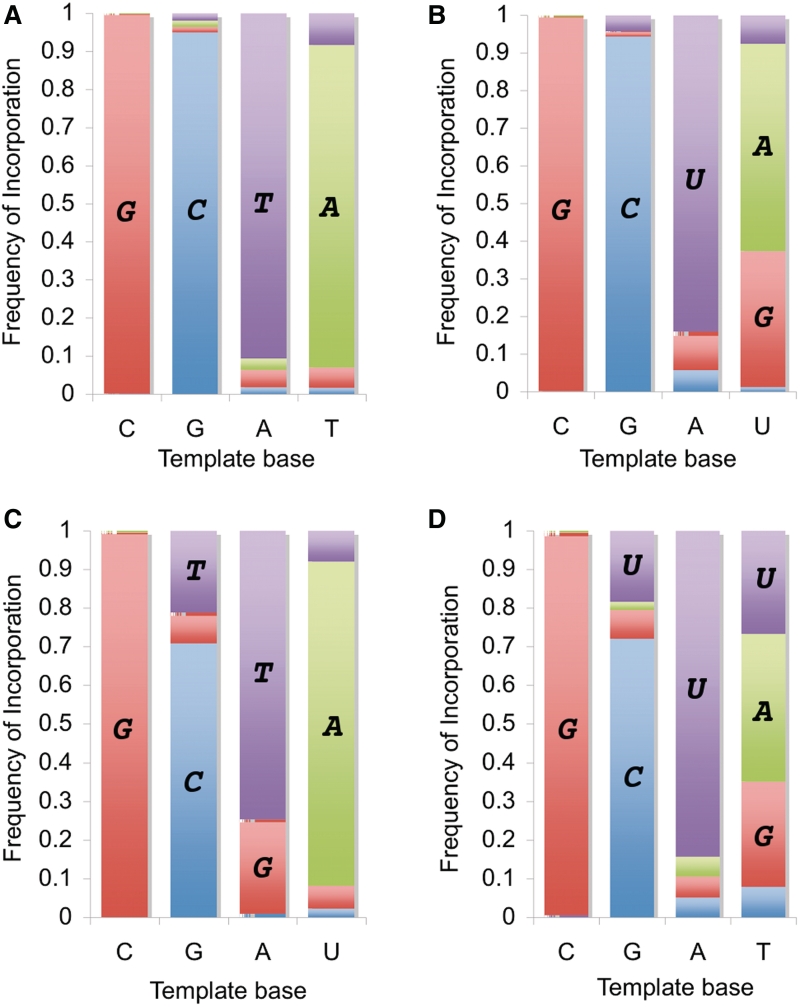
The experimental mutation rate of non-enzymatic polymerization on a DNA backbone with a DNA primer was measured in previous work [µave(DNAt/DNAp) = 7.6 ± 1.4%; Figure 2A] (18). We found that non-enzymatic polymerization on an RNA backbone with an RNA primer under analogous reaction conditions was more than twice as error-prone [Figure 2B; µave(RNAt/RNAp) = 16.8 ± 1.6%]. The correct Watson–Crick nucleotide incorporated across each template base with the highest rate. As in the DNA system, C and G templated with relatively high fidelity compared with A and U. However, mis-incorporation of G across U was a very prominent source of error in the RNA system, such that U templated with a fidelity of only 55 ± 7%, compared to 85 ± 2% for T templating in the DNA system. Mis-incorporation of U was also the dominant error across G in RNA, unlike G in DNA which did not have a dominant error; nevertheless, the absolute fidelity across G was relatively good (94.9 ± 0.2% in the DNA system; 94.5 ± 0.6% in the RNA system).
Figure 2.
Experimentally observed frequency of incorporation and mis-incorporation for copying (A) DNA into DNA, (B) RNA into RNA, (C) RNA into DNA, (D) DNA into RNA. Activated nucleotides (ImpN or ImpdN): red = G, blue = C, purple = U or T, green = A. Panel (A) is modified from (18). Substantial frequencies are labeled directly with the incorporated nucleotide in bold italic font within the bar graph.
RNA is copied into DNA complement with similar fidelity as RNA into RNA
As an experimental proxy for the transfer fidelity of genetic information from RNA to DNA, we measured the experimental misincorporation rate using an RNA template and DNA primer with 2′-deoxy activated nucleotides. The misincorporation rate of this system was similar to that for RNA/RNA replication: µave(RNAt/DNAp) = 18.1 ± 0.4% (Figure 2C). In contrast to the pure RNA and DNA systems, in this hybrid G templated with quite low fidelity (71 ± 3%), primarily from mis-incorporation of T. Also, in contrast to the dominant mis-incorporation of G across U in the pure RNA system, mis-incorporation of G across T did not occur at a high rate. Instead, most errors came from mis-incorporation of T across G and G across A, which correspond to non-Watson–Crick base pairs. In this system, only C templated relatively faithfully (fidelity of 99.4 ± 0.1%), while mutations across G, A, and U were quite frequent (fidelities of 71 ± 3, 75 ± 2, 83 ± 3%, respectively).
DNA is copied into RNA complement with low intrinsic fidelity
In the DNAt/RNAp experimental system, the misincorporation rate was found to be very high (27 ± 3%; Figure 2D). In particular, the misincorporation rate in this system was much higher (>3×) than that of pure DNA replication. As with the other systems, C templated with good fidelity (98 ± 0.3%). However, like the RNA-templated reactions, this system suffered from a high rate of mis-incorporation corresponding to the G:U or G:T wobble pair (mis-incorporation of G across T occurred at a rate of 27 ± 7%; mis-incorporation of U across G occurred at a rate of 18 ± 1%). In addition, the mis-incorporation of U across T was also a prominent error, occurring at a rate of 27 ± 1%. As a result, errors across T were especially frequent, such that the fidelity of copying T was only 38 ± 7%.
Correlation between misincorporation rates in non-enzymatic RNA replication and ribozyme-catalyzed RNA replication
The misincorporation rates of the experimental non-enzymatic RNAt/RNAp system correlate with those observed previously for a ribozyme-catalyzed system (r2 = 0.75) (17), although the ribozyme system had a lower overall µave of 3.3% (Supplementary Figure S2). As in the non-enzymatic system, wobble pairing is responsible for the bulk of the errors in the ribozyme system, indicating that this is a feature of the RNA backbone.
Theoretical basis for thermodynamic estimation of error rates
The conceptual basis for the theoretical approach is the well-established physico-chemical description of substrate discrimination and reading errors (10,36,37), in which the lower limit of the error rate is determined by the free energy difference between the correct versus incorrect products. This limitation is explicit in a Michaelis–Menten scheme describing the non-enzymatic elongation of an RNA or DNA primer across an RNA or DNA template:
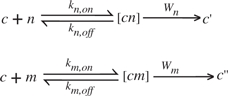 |
(4) |
where the primer–template complex c is either elongated by the correct Watson–Crick complementary nucleotide n or by a non-complementary base m. In this scheme, discrimination between n and m occurs in the nucleotide docking step to the reaction intermediates [cn] and [cm], as the incorrect base m has association (km, on) and dissociation (km, off) rates that differ from those of the correct base. The intermediates are converted to the correctly elongated template–primer complex c′ or the erroneous product c′′ via phosphodiester bond formation at rate Wn or Wm, respectively. Assuming that the different nucleotides are available at equal concentrations, Equation 4 results in an error ratio ϕ (rate of incorrect product formation divided by rate of correct product formation) given by:
The associated mutation rate µ is:
| (5) |
The error ratio is minimal when bond formation is much slower than unbinding. In this limit, the error ratio approaches the ratio of the equilibrium binding constants, resulting in the thermodynamic lower bound
| (6) |
where ΔGn,m is the free energy difference between the correct and incorrect product, R is the gas constant and T is the temperature. See ‘Materials and Methods’ section for details.
Thermodynamic estimates from nearest-neighbor interactions correlate with experimentally observed misincorporation rates
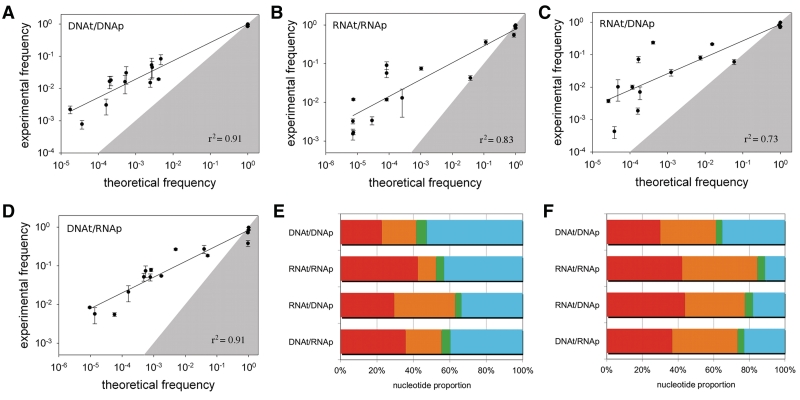
We estimated the lower theoretical limit for frequencies of each possible error in the four systems (DNAt/DNAp, RNAt/RNAp, RNAt/DNAp and DNAt/RNAp) using energetic calculations from a nearest-neighbor model based on the established free energy rules for DNA and RNA secondary structure formation (Figure 1B; Table 2) (34,31). The calculated frequencies correspond to a hypothetical situation in which the four possible fully elongated products of a given template (one correct and three erroneous primer–template complexes) would be allowed to reach thermodynamic equilibrium. The thermodynamic predictions correlated well with the experimentally observed error rates (Figure 3A–D). The thermodynamic calculations do appear to represent a lower limit to the experimentally observed rates, which were usually greater by one to three orders of magnitude. This discrepancy is probably due at least in part to the non-equilibrium conditions and lack of downstream incorporation in the experiments (see ‘Discussion’ section). Interestingly, the equilibrium model predicted the high frequency of mis-incorporation of G:U-type wobble pairs quite well (ratio of experimental to theoretical rates was <10).
Figure 3.
Experimental incorporation and mis-incorporation frequencies versus thermodynamic predictions for copying (A) DNA into DNA, (B) RNA into RNA, (C) RNA into DNA, (D) DNA into RNA. Lines: linear regression on the log values; r2 values as given. Error bars are from experimental replicates. The gray zones represent the areas in which the experimental frequency would be less than the theoretical frequency; correct incorporations (upper right corner) should lie within the gray zone (i.e. observed fidelity is less than the theoretical maximum) while mis-incorporations should lie outside the gray zone (i.e. observed error frequencies are greater than the theoretical minimum). Nucleotide supply for minimizing experimental (E) or theoretical (F) mutation rates. See Table 4 for values and experimental error bars. Systems are denoted as template/primer. Red = A, orange = C, green = G, blue = T or U.
Thermodynamic estimates follow the same trends as experimental mutation rates
To determine whether the trends we found comparing the DNA, RNA and hybrid systems in our experiments were intrinsic to the nucleic acid backbones versus heavily influenced by the activation chemistry, we looked for the same trends in our thermodynamic calculations. As in the experimental system, our thermodynamic calculations indicate that DNA replication (theoretical µave = 0.5%) is intrinsically more faithful than RNA replication (theoretical µave = 3.8%). In addition, copying RNA into DNA (theoretical µave = 2.1%) was about as faithful as RNA replication. Copying DNA into RNA was error-prone (theoretical µave = 2.5%) compared to pure DNA replication. These relationships verified the major trends observed in our experimental system. The error of these estimated mutation rates would be due to errors in the energy parameters on which these calculations are based, which have uncertainties of 5–10% (see ‘Materials and Methods’ section).
Alternative nucleotide ratios
Because polymerization is apparently first order with respect to nucleotide concentration, µave depends on the ratios of nucleotide concentrations. To match conditions for previously published data (18), actual experimental conditions were [A] = [C] = [G] = 10 mM, [T or U] = 40 mM, which were also used for the thermodynamic calculations above. We calculated the expected mutation rate in two additional conditions of interest: (i) equimolar nucleotide supply and (ii) an optimal nucleotide ratio that would minimize the mutation rate (Table 3). In (ii), we wondered whether very high rates of particular errors could be countered by adjustments in the nucleotide pool. We calculated the optimal nucleotide supply using either the experimentally determined error rates or the thermodynamic estimates (Figure 3E and F; Table 4). To avoid large discrepancies among the nucleotide concentrations, we also constrained them to be within 10-fold of each other. For all systems, reducing the concentration of G would improve fidelity, essentially because mis-incorporation of G tended to be a major source of error while the correct incorporation of G across template C was already very efficient (Figure 2).
Table 3.
Mutation rates (µave) predicted for equimolar or optimized nucleotide ratios, based on either experimental rates or thermodynamic lower bounds
| Equimolar ratios |
Optimized ratios |
|||||
|---|---|---|---|---|---|---|
| Rates from: | Experiment | Theory | Experiment | Theory | ||
| Template | Primer | |||||
| RNA | RNA | 22 ± 1% | 3% | 8.4 ± 0.9% | 0.5% | |
| DNA | DNA | 11 ± 4% | 0.6% | 5.8 ± 0.8% | 0.2% | |
| RNA | DNA | 21.5 ± 0.4% | 2% | 9.4 ± .05% | 0.4% | |
| DNA | RNA | 28 ± 4% | 2% | 21 ± 3% | 0.6% | |
Error bars given are standard deviations from calculations based on duplicate batches of experimental mutation rates.
Table 4.
Optimal relative concentrations of A, C, G, T or U nucleotides for minimization of mutation rate, based on rates from experimental system (ES) or thermodynamic calculation (TC)
| System | Template | Primer | Fraction of A | Fraction of C | Fraction of G | Fraction of T or U |
|---|---|---|---|---|---|---|
| ES | DNA | DNA | 0.227 ± 0.019 | 0.189 ± 0.047 | 0.0575 ± 0.0002 | 0.526 ± 0.066 |
| ES | RNA | RNA | 0.425 ± 0.009 | 0.101 ± 0.007 | 0.0432 ± 0.0001 | 0.432 ± 0.001 |
| ES | RNA | DNA | 0.295 ± 0.041 | 0.335 ± 0.021 | 0.034 ± 0.002 | 0.337 ± 0.018 |
| ES | DNA | RNA | 0.358 ± 0.011 | 0.1969 ± 0.0004 | 0.048 ± 0.004 | 0.397 ± 0.015 |
| TC | DNA | DNA | 0.30 | 0.31 | 0.04 | 0.35 |
| TC | RNA | RNA | 0.37 | 0.37 | 0.04 | 0.23 |
| TC | RNA | DNA | 0.44 | 0.34 | 0.04 | 0.18 |
| TC | DNA | RNA | 0.42 | 0.42 | 0.04 | 0.11 |
Standard deviations are calculated for optimization based on duplicate batches of experimental reaction rates.
The major trends, that RNA replication is error-prone compared to DNA replication, that RNA is copied into the DNA complement with similar fidelity as RNA replication and that DNA is copied into the RNA complement with a relatively high mutation rate, also held for equimolar conditions and optimized conditions (Table 3). Copying DNA into RNA was still very error-prone compared to the other systems, suggesting that this process could not be made as faithful as the other systems through optimization of the nucleotide supply alone.
DISCUSSION
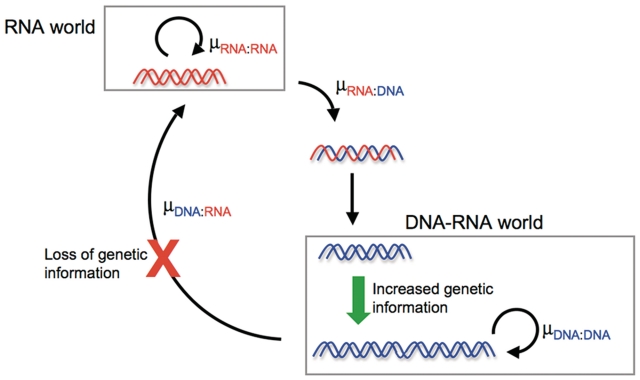
In our experiments, non-enzymatic RNA polymerization had about twice the misincorporation rate of DNA polymerization, suggesting that more information could be stably encoded after the switch to DNA as the genetic material. This might translate into roughly a doubling of genome information. In addition to the increased chemical stability of DNA, the potential to increase information content might present another selective advantage to an organism that made this transition. We also studied the RNAt/DNAp and DNAt/RNAp hybrids as exemplars of transitional forms, although the transitions could involve more complicated mixed backbones in reality. Using these exemplars, we found that copying RNA into DNA occurred with a mutation rate similar to RNA replication, suggesting that the genetic takeover itself would not cause much loss of information. In contrast, copying DNA back into RNA was a highly error-prone process, suggesting that an organism that attempted to switch from DNA back to RNA would be at an immediate disadvantage from the corruption of genetic information (Figure 4). It should be noted that the genetic takeover of the RNA world did not necessarily proceed directly to DNA, but might have proceeded through intermediate stages containing alternative nucleic acid backbones. If that were the case, one may not draw conclusions regarding the reversibility of the genetic takeover from our results, although the difference between RNA and DNA replication would still be relevant.
Figure 4.
Evolutionary consequences of the observed hierarchy of mutation rates. Expansion of genome upon genetic takeover and loss of information during reversion back to RNA.
The experimental system used here is only a laboratory model for nucleic acid polymerization without enzymes, and it is unclear what activation chemistries (and backbones) would have been present during the origin of life. The use of phosphorimidazolides in non-enzymatic, template-directed polymerization was pioneered by Orgel and others, who found that apparently minor substitutions on the leaving group led to large differences in reactivity with a 3′-OH nucleophile in an RNA primer; in particular, the 2-methylimidazole derivative resulted in much more efficient polymerization, which the authors proposed was due to improving the geometry of the reaction (38–43). Polymerization efficiency was further enhanced by use of a 3′-NH2 nucleophile, which reacted well even when the leaving group was relatively poor (e.g. imidazole) (44,45). While the 3'-amine nucleophile is not thought to be particularly prebiotically plausible, it is useful for laboratory study because of the fast rate of primer extension.
While the experimentally observed trends are suggestive, it would be difficult to draw conclusions about the genetic takeover from our experimental results alone. We therefore sought to validate the observed trends by a thermodynamic model, which is independent of the activation chemistry. To determine whether the trends in experimental error rates reflected underlying biophysical properties of the duplexes rather than specific properties of the activation chemistry, we calculated the error rates for a hypothetical system at thermodynamic equilibrium, i.e. if the four possible fully extended products (one perfectly matched and three mismatched duplexes) were allowed to equilibrate with one another. This analysis should give the thermodynamic error rates of the system, in contrast to error rates in an experimental implementation (which instead depend on the kinetic pathways, determined by activation chemistry and/or enzymes). Also, in our experimental system, we inferred error rates of non-enzymatic polymerization from the rates of misincorporation of single nucleotides, ignoring secondary effects such as stalling of polymerization downstream of a mismatch (see below). This is a simplification of replication of a full strand; in contrast, our theoretical calculations do estimate the lowest possible error rate for replication of full strands. An alternative approach might be to calculate the free energy difference between terminal matches and mismatches (i.e. after a single incorporation). Such an approach would assume that all of the potential discrimination is due to the energetics of a single incorporation. However, the presence of a terminal mismatch substantially decreases downstream polymerization speed (18), such that the mutation frequency of a single incorporation overestimates the frequency of mutations in fully extended products. To include such effects in an estimate of the thermodynamic bound on mutation rates, the equilibrium of fully extended products (not termini alone) must be calculated. Without enzymes or proofreading, substrate discrimination is limited by the thermodynamic free energy difference between the correct and incorrect final products. We used a nearest-neighbor model to estimate the equilibrium distribution of products and thus infer a lower bound on the rate for each possible mutation (Figure 1B). To our knowledge, this constitutes the first complete and quantitative exploration of the thermodynamic limit on error rates for RNA and DNA replication.
As expected, all our experimental error rates lie on or above the thermodynamic lower bound (Figure 3A–D). The fact that most experimental values lie substantially above the lower bound may be due to at least two factors. First, our experiments studied a single incorporation, but additional discrimination occurs when the primer is further extended because non-enzymatic polymerization slows after an incorrect vs. correct incorporation (18). Second, the thermodynamic lower bounds are approached in the equilibrium limit of very slow incorporation reactions [low W's in Equation (4); Supplementary Data S1], but any activation chemistry that gives reaction rates amenable to laboratory study is unlikely to be near this limit. An accurate prediction of the error rates in our experimental system would require a quantitative understanding of the microscopic kinetics of hybridization and chemical bond formation, which are currently not known (Supplementary Data S2).
Importantly, the thermodynamic limits on the average mutation rates, while lower than the experimental values, are also consistent with the trends represented in Figure 4, corroborating our qualitative conclusions on the evolutionary advantage of switching from RNA to DNA. Although not necessarily expected a priori, we also found a good correlation between the theoretical and experimental error rates in all of the four different template-primer systems (Figure 3A–D). While experimental trends may be strongly affected by the activation chemistry and presence of enzymes, the thermodynamic trends are presumably not affected by these kinetic considerations. Experimentally, the properties of the nucleophile and leaving group would affect the rate of bond formation [W's in Equation (4)]. Presumably, increasing W (e.g. 3′-amine nucleophile) implies a greater degree of kinetic rather than thermodynamic control, which may generally decrease fidelity. In other words, error rates should approach the thermodynamic limit as W decreases, because slow bond formation would allow more time to explore different conformations. One might further speculate that changing the activation chemistry would affect the Ws of different reactions in a similar way, such that experimental incorporation and mis-incorporation frequencies should correlate with the thermodynamic limits. Indeed, we found this to be the case (Figure 3A–D). A possible interpretation of this correlation is that bond formation is relatively independent of the properties of the base pair or mis-pair, such that the relative reaction rates reflect the binding equilibria of the monomers to the template–primer complex. The nucleotide triphosphates used as substrates in biological systems, including the RNA polymerase ribozyme, are kinetically stable (low W without enzymes), which might imply improved fidelity. In principle, this improvement may or may not be relevant because enzymes also change the reaction pathway. Regardless, we find a reasonable correlation between the incorporation and mis-incorporation rates of ribozyme-catalyzed RNA polymerization and non-enzymatic RNA polymerization (Supplementary Figure S2), suggesting some underlying similarity in the reaction pathways.
In both experiments and thermodynamic calculations, the major mispair that contributed to the higher mutation rate of RNA and the hybrids were the G:U(T) wobble pairs. This corroborates previous observations that the G:U wobble pair is a greater source of error when copying RNA compared to the G:T wobble pair in DNA (46). In principle, this difference may be due to the backbone structure or to the different structure of U versus T. Given that the hybrid duplexes tend to adopt conformations close to the A-form helix (47,48), our finding that G:T is a major source of error in the DNA/RNA hybrids suggests that the backbone may be more important than the additional methyl group of T in determining fidelity. Interestingly, the predominant mutations of the RNA polymerase ribozyme are also due to G:U mispairs (17), supporting the idea that this error is a feature of the RNA backbone rather than the activation chemistry. Why the A-form backbone might better accommodate this error is unclear; one may speculate that the possibilities include greater flexibility of the single-stranded template (49,50) or greater tolerance of non-canonical stacking interactions due to the presence of slide and roll in the helix (51).
We also observed the major trends summarized in Figure 4 using equimolar concentrations or concentrations that were optimized to minimize the overall mutation rate (a situation that might evolve under selective pressure to reduce errors). The consistent depletion of G to minimize the error rate (Figure 3E and F) suggests that practical implementations of non-enzymatic replication should decrease the relative concentration of G in order to improve overall fidelity. Interestingly, a similar depletion of GTP would also enhance the fidelity of an RNA polymerase ribozyme, reducing the error rate by more than a factor of two, as mis-incorporation of G across U is quite efficient (17). Furthermore, one may note that the concentrations of DNA precursors in the nucleus of various mammalian cells (~25% A, 20% C, 5–10% G, 45–50% T) (52) are surprisingly close to the values obtained from optimizing fidelity in non-enzymatic DNA replication (Figure 3E and F). While this correspondence may be coincidental, it is tempting to speculate that the observed dNTP supply might have evolved to control mutation rates (Supplementary Data S3).
In non-enzymatic RNA replication, like DNA replication, G and C templated with very good fidelity while A and U suffered the most errors. However, not all errors were equally likely, and in RNA the disparities among different errors were very pronounced, with G being incorporated across U as the dominant mutation. Over multiple generations of RNA replication, these disparities would bias toward a GC-rich genome, as U:A would tend to be replaced by C:G. This contrasts with DNA replication, in which no single type of error was particularly dominant. Therefore, a DNA genome might have tended toward a more even nucleotide composition, which could be advantageous since heavily GC-rich sequences pose practical problems (e.g. difficult strand separation), and indeed ribozymes and aptamers have relatively even composition compared to other RNAs (29).
The observed hierarchy of mutation rates also suggests that non-enzymatic transcription (e.g. of ribozymes encoded on a DNA genome) would be significantly more error-prone than genome replication. Error-prone transcription implies high phenotypic variability (53–55). Based on our non-enzymatic polymerization experiments, approximately one-quarter of nucleotides would be copied erroneously. Ribozyme-catalyzed polymerization could be more faithful, with a mutation rate <1% (56), but a significant proportion of transcripts would still have errors (e.g. for a 40-mer ribozyme, about one-third of transcripts would contain an error). In addition, a meta-analysis of two self-cleaving ribozymes showed that the majority of mutations (~75%) were deleterious (57), indicating that fitness may be greatly increased or decreased by a single mutation. Therefore, phenotypic variability could lead to an evolutionary ‘look-ahead’ effect (58): while each genotype specifies a particular ribozyme sequence, it also leads to a cloud of transcripts nearby in RNA sequence space. A given genotype may thus exhibit an overall phenotype influenced by its neighbors, resulting in a locally smoothened phenotypic fitness landscape (Supplementary Figure S3). The smoothing due to phenotypic variability may enhance evolvability by producing a selective benefit from relatively distant optima and facilitating evolutionary paths across low-fitness regions, although this advantage comes at the expense of decreased fitness for optimized sequences due to frequent transcription errors (53). This decreased fitness may have been a necessary cost of the transition to DNA as a more stable genetic material. Finally, phenotypic variability may also allow longer genomes because of a relaxed error threshold (59). Under prebiotic conditions, the evolutionary advantages of evolvability and larger genomes may have outweighed the cost.
The beginning of the RNA world would have been dominated by polymerization chemistry, so factors such as the geometry of the template–primer–nucleotide complex were important determinants of the reaction rates and the mutation rate. However, the later stages of the RNA world could have contained sophisticated ribozymes that could influence the mutation rate. For example, the RNA polymerase ribozymes (17,56) generally have lower mutation rates (0.88–4.3%) than the non-enzymatic polymerization systems, suggesting that the ribozyme imposes additional discrimination. Understanding the mechanistic basis of fidelity in these systems is an important goal for future research.
Mutation rates could be decreased by endergonic proofreading mechanisms. It is unclear whether protein enzymes evolved before or after the genetic takeover. Our results demonstrate that error rates for non-enzymatic polymerization are severely limiting, so one may speculate that the increase in fidelity accompanying a transition to DNA might have been required for the emergence of translation machinery. Regardless, the basal error ratios (ϕ) and their thermodynamic bounds (ϕ0) are also of fundamental significance for enzymes with proofreading capability. While these enzymes use chemical energy to drive one or more proofreading steps, the discrimination in each step is typically based on a scheme similar to Equation 1 and would be limited by an analogous thermodynamic bound. In theory, this could decrease the error ratio to ϕn+1, where ϕ is the basal error ratio of the interaction (which cannot be lower than the thermodynamic bound) and n is the number of proofreading steps (37). Therefore, while absolute fidelities could be improved by proofreading, the basal fidelity of the interaction is still an important factor; a replicator having greater basal fidelity would require less proofreading to achieve the same overall fidelity.
The genetic takeover of the RNA world by DNA may have been influenced by several factors, including chemical stability and multiple evolutionary considerations. Since the high mutation rate of non-enzymatic polymerization may have presented a serious limitation to information storage, our data and calculations suggest that the switch to DNA would allow an expansion of genomic information. The mutation profile of DNA appears to be relatively unbiased compared to RNA, so a DNA genome might be more conducive to ribozyme evolution. In addition, our results suggest that the switch from RNA to DNA would have been a one-way transition, as copying DNA back into RNA would cause loss of genomic information. At the same time, a high non-enzymatic ‘transcriptional’ error rate might present the advantage of greater evolvability. Based on the correspondence between our non-enzymatic data and equilibrium thermodynamic calculations, these trends appear to reflect intrinsic features of the different nucleic acid duplexes. However, while our results are suggestive, error rates should be investigated using different activation chemistries and nucleic acid backbones to determine the robustness of these conclusions about the genetic takeover. Eventually, the absolute mutation rates would change as ribozymes evolved in the RNA world and protein enzymes emerged; nevertheless, the basal fidelities may have played an important role early on as the first genetic systems became established and error correction mechanisms began to evolve.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
NIH grant GM068763 to the Center for Modular Biology at Harvard University; German Research Foundation grant GE1098/3-1 (Deutsche Forschungsgemeinschaft; to U.G.); IAC is a Bauer Fellow at Harvard University; German Academic Exchange Program (Deutscher Akademischer Austausch Dienst; to B.O.). Funding for open access charge: National Institutes of Health (grant GM068763).
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Jack Szostak, Bodo Stern, Allan Drummond, and Andrew Murray for comments, and Erin O’Shea for use of equipment.
REFERENCES
- 1.Woese CR, Dugre DH, Dugre SA, Kondo M, Saxinger WC. On the fundamental nature and evolution of the genetic code. Cold Spring Harb. Symp. Quant. Biol. 1966;31:723–736. doi: 10.1101/sqb.1966.031.01.093. [DOI] [PubMed] [Google Scholar]
- 2.Crick FH. The origin of the genetic code. J. Mol. Biol. 1968;38:367–379. doi: 10.1016/0022-2836(68)90392-6. [DOI] [PubMed] [Google Scholar]
- 3.Orgel LE. Evolution of the genetic apparatus. J. Mol. Biol. 1968;38:381–393. doi: 10.1016/0022-2836(68)90393-8. [DOI] [PubMed] [Google Scholar]
- 4.Ban N, Nissen P, Hansen J, Moore PB, Steitz TA. The complete atomic structure of the large ribosomal subunit at 2.4 Å resolution. Science. 2000;289:905–920. doi: 10.1126/science.289.5481.905. [DOI] [PubMed] [Google Scholar]
- 5.Wimberly BT, Brodersen DE, Clemons WM, Jr, Morgan-Warren RJ, Carter AP, Vonrhein C, Hartsch T, Ramakrishnan V. Structure of the 30S ribosomal subunit. Nature. 2000;407:327–339. doi: 10.1038/35030006. [DOI] [PubMed] [Google Scholar]
- 6.Yusupov MM, Yusupova GZ, Baucom A, Lieberman K, Earnest TN, Cate JH, Noller HF. Crystal structure of the ribosome at 5.5 Å resolution. Science. 2001;292:883–896. doi: 10.1126/science.1060089. [DOI] [PubMed] [Google Scholar]
- 7.Drake JW, Charlesworth B, Charlesworth D, Crow JF. Rates of spontaneous mutation. Genetics. 1998;148:1667–1686. doi: 10.1093/genetics/148.4.1667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Steinhauer DA, Domingo E, Holland JJ. Lack of evidence for proofreading mechanisms associated with an RNA virus polymerase. Gene. 1992;122:281–288. doi: 10.1016/0378-1119(92)90216-c. [DOI] [PubMed] [Google Scholar]
- 9.Pigliucci M. Is evolvability evolvable? Nat. Rev. Genet. 2008;9:75–82. doi: 10.1038/nrg2278. [DOI] [PubMed] [Google Scholar]
- 10.Eigen M. Selforganization of matter and the evolution of biological macromolecules. Naturwissenschaften. 1971;58:465–523. doi: 10.1007/BF00623322. [DOI] [PubMed] [Google Scholar]
- 11.Eigen M, Mccaskill J, Schuster P. The molecular quasi-species. Adv. Chem. Phys. 1989;75:149–263. [Google Scholar]
- 12.Nowak MA. Evolutionary Dynamics. Cambridge, Massachusetts: Harvard University Press; 2006. [Google Scholar]
- 13.Gago S, Elena SF, Flores R, Sanjuan R. Extremely high mutation rate of a hammerhead viroid. Science. 2009;323:1308. doi: 10.1126/science.1169202. [DOI] [PubMed] [Google Scholar]
- 14.Bull JJ, Sanjuan R, Wilke CO. Theory of lethal mutagenesis for viruses. J. Virol. 2007;81:2930–2939. doi: 10.1128/JVI.01624-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Anderson JP, Daifuku R, Loeb LA. Viral error catastrophe by mutagenic nucleosides. Annu. Rev. Microbiol. 2004;58:183–205. doi: 10.1146/annurev.micro.58.030603.123649. [DOI] [PubMed] [Google Scholar]
- 16.Eigen M. Error catastrophe and antiviral strategy. Proc. Natl Acad. Sci. USA. 2002;99:13374–13376. doi: 10.1073/pnas.212514799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Johnston WK, Unrau PJ, Lawrence MS, Glasner ME, Bartel DP. RNA-catalyzed RNA polymerization: accurate and general RNA-templated primer extension. Science. 2001;292:1319–1325. doi: 10.1126/science.1060786. [DOI] [PubMed] [Google Scholar]
- 18.Rajamani S, Ichida JK, Antal T, Treco DA, Leu K, Nowak MA, Szostak JW, Chen IA. Effect of stalling after mismatches on the error catastrophe in nonenzymatic nucleic acid replication. J. Am. Chem. Soc. 2010;132:5880–5885. doi: 10.1021/ja100780p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hagenbuch P, Kervio E, Hochgesand A, Plutowski U, Richert C. Chemical primer extension: efficiently determining single nucleotides in DNA. Angew. Chem. Int. Ed. Engl. 2005;44:6588–6592. doi: 10.1002/anie.200501794. [DOI] [PubMed] [Google Scholar]
- 20.Stutz JAR, Kervio E, Deck C, Richert C. Chemical primer extension: Individual steps of spontaneous replication. Chem. Biodivers. 2007;4:784–802. doi: 10.1002/cbdv.200790064. [DOI] [PubMed] [Google Scholar]
- 21.Kervio E, Hochgesand A, Steiner UE, Richert C. Templating efficiency of naked DNA. Proc. Natl Acad. Sci. USA. 2010;107:12074–12079. doi: 10.1073/pnas.0914872107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Stutz JAR, Richert C. A steroid cap adjusts the selectivity and accelerates the rates of nonenzymatic single nucleotide extensions of an oligonucleotide. J. Am. Chem. Soc. 2001;123:12718–12719. doi: 10.1021/ja011448i. [DOI] [PubMed] [Google Scholar]
- 23.Stutz JAR, Richert C. Tuning the reaction site for enzyme-free primer-extension reactions through small molecule substituents. Chem-Eur. J. 2006;12:2472–2481. doi: 10.1002/chem.200501008. [DOI] [PubMed] [Google Scholar]
- 24.Vogel SR, Deck C, Richert C. Accelerating chemical replication steps of RNA involving activated ribonucleotides and downstream-binding elements. Chem. Commun. 2005:4922–4924. doi: 10.1039/b510775j. [DOI] [PubMed] [Google Scholar]
- 25.Schrum JP, Ricardo A, Krishnamurthy M, Blain JC, Szostak JW. Efficient and rapid template-directed nucleic acid copying using 2′-amino-2′,3′-dideoxyribonucleoside-5′-phosphorimidazolide monomers. J. Am. Chem. Soc. 2009;131:14560–14570. doi: 10.1021/ja906557v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mansy SS, Schrum JP, Krishnamurthy M, Tobe S, Treco DA, Szostak JW. Template-directed synthesis of a genetic polymer in a model protocell. Nature. 2008;454:122–125. doi: 10.1038/nature07018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lohrmann R, Orgel LE. Preferential formation of (2′–5′)-linked internucleotide bonds in non-enzymatic reactions. Tetrahedron. 1978;34:853–855. [Google Scholar]
- 28.Prabahar KJ, Cole TD, Ferris JP. Effect of phosphate activating group on oligonucleotide formation on montmorillonite: the regioselective formation of 3′,5′-linked oligoadenylates. J. Am. Chem. Soc. 1994;116:10914–10920. doi: 10.1021/ja00103a006. [DOI] [PubMed] [Google Scholar]
- 29.Kennedy R, Lladser ME, Wu Z, Zhang C, Yarus M, De Sterck H, Knight R. Natural and artificial RNAs occupy the same restricted region of sequence space. RNA. 2010;16:280–289. doi: 10.1261/rna.1923210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lee JF, Hesselberth JR, Meyers LA, Ellington AD. Aptamer database. Nucleic Acids Res. 2004;32:D95–D100. doi: 10.1093/nar/gkh094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Freier SM, Kierzek R, Jaeger JA, Sugimoto N, Caruthers MH, Neilson T, Turner DH. Improved free-energy parameters for predictions of RNA duplex stability. Proc. Natl Acad. Sci. USA. 1986;83:9373–9377. doi: 10.1073/pnas.83.24.9373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mathews DH, Sabina J, Zuker M, Turner DH. Expanded sequence dependence of thermodynamic parameters improves prediction of RNA secondary structure. J. Mol. Biol. 1999;288:911–940. doi: 10.1006/jmbi.1999.2700. [DOI] [PubMed] [Google Scholar]
- 33.Xia T, SantaLucia J, Jr, Burkard ME, Kierzek R, Schroeder SJ, Jiao X, Cox C, Turner DH. Thermodynamic parameters for an expanded nearest-neighbor model for formation of RNA duplexes with Watson–Crick base pairs. Biochemistry. 1998;37:14719–14735. doi: 10.1021/bi9809425. [DOI] [PubMed] [Google Scholar]
- 34.SantaLucia J., Jr A unified view of polymer, dumbbell, and oligonucleotide DNA nearest-neighbor thermodynamics. Proc. Natl Acad. Sci. USA. 1998;95:1460–1465. doi: 10.1073/pnas.95.4.1460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sugimoto N, Nakano S, Katoh M, Matsumura A, Nakamuta H, Ohmichi T, Yoneyama M, Sasaki M. Thermodynamic parameters to predict stability of RNA/DNA hybrid duplexes. Biochemistry. 1995;34:11211–11216. doi: 10.1021/bi00035a029. [DOI] [PubMed] [Google Scholar]
- 36.Ninio J. A semi-quantitative treatment of missense and nonsense suppression in the strA and ram ribosomal mutants of Escherichia coli. Evaluation of some molecular parameters of translation in vivo. J. Mol. Biol. 1974;84:297–313. doi: 10.1016/0022-2836(74)90586-5. [DOI] [PubMed] [Google Scholar]
- 37.Hopfield JJ. Kinetic proofreading: a new mechanism for reducing errors in biosynthetic processes requiring high specificity. Proc. Natl Acad. Sci. USA. 1974;71:4135–4139. doi: 10.1073/pnas.71.10.4135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Inoue T, Orgel LE. Substituent control of the poly(C)-directed oligomerization of guanosine 5′-phosphoroimidazolide. J. Am. Chem. Soc. 1981;103:7666–7667. [Google Scholar]
- 39.Wu T, Orgel LE. Nonenzymatic template-directed synthesis on hairpin oligonucleotides .3. Incorporation of adenosine and uridine residues. J. Am. Chem. Soc. 1992;114:7963–7969. doi: 10.1021/ja00047a001. [DOI] [PubMed] [Google Scholar]
- 40.Lohrmann R, Bridson PK, Bridson PK, Orgel LE. Efficient metal-ion catalyzed template-directed oligonucleotide synthesis. Science. 1980;208:1464–1465. doi: 10.1126/science.6247762. [DOI] [PubMed] [Google Scholar]
- 41.Kozlov IA, Orgel LE. Nonenzymatic template-directed synthesis of RNA from monomers. Mol. Biol. 2000;34:781–789. [PubMed] [Google Scholar]
- 42.Orgel LE. Prebiotic chemistry and the origin of the RNA world. Crit. Rev. Biochem. Mol. 2004;39:99–123. doi: 10.1080/10409230490460765. [DOI] [PubMed] [Google Scholar]
- 43.Joyce GF. Nonenzymatic template-directed synthesis of informational macromolecules. Cold Spring Harb. Sym. 1987;52:41–51. doi: 10.1101/sqb.1987.052.01.008. [DOI] [PubMed] [Google Scholar]
- 44.Zielinski WS, Orgel LE. Oligomerization of activated derivatives of 3'-amino-3'-deoxyguanosine on poly(C) and poly(Dc) templates. Nucleic Acids Res. 1985;13:2469–2484. doi: 10.1093/nar/13.7.2469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tohidi M, Zielinski WS, Chen CHB, Orgel LE. Oligomerization of 3'-amino-3'-deoxyguanosine-5'-phosphorimidazolidate on a D(Cpcpcpcpc) template. J. Mol. Evol. 1987;25:97–99. doi: 10.1007/BF02101750. [DOI] [PubMed] [Google Scholar]
- 46.Zielinski M, Kozlov IA, Orgel LE. A comparison of RNA with DNA in template-directed synthesis. Helvet. Chim. Acta. 2000;83:1678–1684. doi: 10.1002/1522-2675(20000809)83:8<1678::AID-HLCA1678>3.0.CO;2-P. [DOI] [PubMed] [Google Scholar]
- 47.Wang AH, Fujii S, van Boom JH, van der Marel GA, van Boeckel SA, Rich A. Molecular structure of r(GCG)d(TATACGC): a DNA–RNA hybrid helix joined to double helical DNA. Nature. 1982;299:601–604. doi: 10.1038/299601a0. [DOI] [PubMed] [Google Scholar]
- 48.Egli M, Usman N, Rich A. Conformational influence of the ribose 2'-hydroxyl group: crystal structures of DNA–RNA chimeric duplexes. Biochemistry. 1993;32:3221–3237. [PubMed] [Google Scholar]
- 49.Smith SB, Cui YJ, Bustamante C. Overstretching B-DNA: the elastic response of individual double-stranded and single-stranded DNA molecules. Science. 1996;271:795–799. doi: 10.1126/science.271.5250.795. [DOI] [PubMed] [Google Scholar]
- 50.Seol Y, Skinner GM, Visscher K, Buhot A, Halperin A. Stretching of homopolymeric RNA reveals single-stranded helices and base-stacking. Phys. Rev. Lett. 2007;98:158103. doi: 10.1103/PhysRevLett.98.158103. [DOI] [PubMed] [Google Scholar]
- 51.Dickerson RE, Ng HL. DNA structure from A to B. Proc. Natl Acad. Sci. USA. 2001;98:6986–6988. doi: 10.1073/pnas.141238898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Mathews CK, Ji J. DNA precursor asymmetries, replication fidelity, and variable genome evolution. BioEssays. 1992;14:295–301. doi: 10.1002/bies.950140502. [DOI] [PubMed] [Google Scholar]
- 53.Ancel LW. Undermining the Baldwin expediting effect: does phenotypic plasticity accelerate evolution? Theor. Popul. Biol. 2000;58:307–319. doi: 10.1006/tpbi.2000.1484. [DOI] [PubMed] [Google Scholar]
- 54.Drummond DA, Wilke CO. The evolutionary consequences of erroneous protein synthesis. Nat. Rev. Genet. 2009;10:715–724. doi: 10.1038/nrg2662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Goldsmith M, Tawfik DS. Potential role of phenotypic mutations in the evolution of protein expression and stability. Proc. Natl Acad. Sci. USA. 2009;106:6197–6202. doi: 10.1073/pnas.0809506106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wochner A, Attwater J, Coulson A, Holliger P. Ribozyme-catalyzed transcription of an active ribozyme. Science. 2011;332:209–212. doi: 10.1126/science.1200752. [DOI] [PubMed] [Google Scholar]
- 57.Kun A, Santos M, Szathmary E. Real ribozymes suggest a relaxed error threshold. Nat. Genet. 2005;37:1008–1011. doi: 10.1038/ng1621. [DOI] [PubMed] [Google Scholar]
- 58.Whitehead DJ, Wilke CO, Vernazobres D, Bornberg-Bauer E. The look-ahead effect of phenotypic mutations. Biol. Direct. 2008;3:18. doi: 10.1186/1745-6150-3-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Sato K, Kaneko K. Evolution equation of phenotype distribution: general formulation and application to error catastrophe. Phys. Rev. E. 2007;75:061909–061918. doi: 10.1103/PhysRevE.75.061909. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.