Abstract
Autoimmune regulator (AIRE) is a transcription factor that induces the expression of a large subset of otherwise strictly tissue restricted antigens in medullary thymic epithelial cells, thereby enabling their presentation to developing T cells for negative selection. Mutations in AIRE lead to autoimmune-polyendocrinopathy-candidiasis-ectodermal dystrophy (APECED), a rare monogenetic disease. Although it has been reported that AIRE interacts with proteins involved in nuclear transport, DNA-damage response, chromatin remodeling, transcription and pre-mRNA-splicing, the precise mechanism of AIRE-induced tissue restricted antigen expression has remained elusive. In this study, we investigated an APECED patient mutation that causes the loss of the extreme C-terminus of AIRE and found that this mutant protein is transcriptionaly inactive. When tethered heterologously to DNA, this domain could stimulate transcription and splicing by itself. Moreover, the loss of this C-terminus disrupted interactions with the positive transcription elongation factor b (P-TEFb). Via P-TEFb, AIRE increased levels of RNA polymerase II on and enhanced pre-mRNA splicing of heterologous and endogenous target genes. Indeed, the inhibition of CDK9, the kinase subunit of P-TEFb, inhibited AIRE-induced pre-mRNA splicing of these genes. Thus, AIRE requires P-TEFb to activate transcription elongation and co-transcriptional processing of target genes.
INTRODUCTION
Tolerance to self is an essential element of the immune system and when tolerance is dysfunctional, autoimmune diseases can arise. Central tolerance is established by negative selection of developing T cells in the thymus (1). Most autoimmune diseases are dependent on many genetic loci, but a rare autoimmune disease, autoimmune-polyendocrinopathy-candidiasis-ectodermal dystrophy (APECED) is monogenetic. APECED patients have mutations in the AIRE gene (2,3), which encodes the autoimmune regulator (AIRE). AIRE is critical for the expression of a number of otherwise tissue restricted antigens (TRAs) in medullary thymic epithelial cells (mTECs) (4). Indeed, Aire−/− mice develop multi-organ autoimmunity with inflammatory infiltrates and auto-reactive antibodies, which is caused by dysregulated negative selection of developing T cells in the thymus (4), whereas the severity and organs affected are strain dependent (5–7). mTECs from these mice express a much smaller TRA repertoire than their wild-type (WT) counterparts.
Previous studies suggested a role for AIRE in transcription (8). AIRE is localized to the nucleus where it accumulates in distinct nuclear structures (9), and when expressed transiently, it activates the expression of endogenous genes as well as exogenous plasmid targets (10). In addition, AIRE interacts with the CREB-binding protein (11) and the positive transcription elongation factor b (P-TEFb) (10). AIRE contains several domains that are present in various transcription factors. First, it has a Sp100, Aire-1, NucP41/75 and DEAF-1 domain (SAND) that has been suggested to bind DNA (12). Second, AIRE contains two plant homeodomains (PHD1 and PHD2). PHD1 binds to histone-3 with non-methylated lysine at position 4 (H3K4me0) (13,14), which is abundant in transcriptionaly inactive chromatin. It was also proposed to be an E3-ubiquitin ligase (15). PHD2 might represent an accessory transcription activation domain (TAD) (16,17). Furthermore, AIRE can also associate with proteins involved with nuclear transport, DNA-damage response, chromatin remodeling, transcription and pre-mRNA splicing (18).
Protein coding genes are transcribed by RNA polymerase II (RNAPII) and their expression is regulated at many different steps (19). The C-terminal domain (CTD) of human Rbp1, the largest RNAPII subunit, consists of 52 Y1S2P3T4S5P6S7 heptapeptide repeats. Serine residues in these repeats are phosphorylated by different CTD-kinases. Thus, the pattern of CTD phosphorylation changes dynamically as the RNAPII transitions through different phases of transcription (20). CTD phosphorylated at serine-5 (S5P) is a hallmark of transcription initiation, and is accomplished by cyclin-dependent kinase 7 (CDK7), a component of the general transcription factor TFIIH (21). On the other hand, CTD phosphorylated at serine-2 (S2P) is a hallmark of transcription elongation, and is catalyzed by P-TEFb (22), composed of the CDK9 kinase and the regulatory cyclins T1 or T2a/b (23). Furthermore, pre-mRNA splicing occurs co-transcriptionaly, and the phosphorylated CTD is bridging these processes by binding components of pre-mRNA splicing and cleavage/polyadenylation machineries (24,25). Thus, pre-mRNA processing factors associate with the transcription elongation complex and the degree and type of CTD phosphorylation define the specificity and affinity of these interactions (26).
In this study, we examined the functional relevance of AIRE’s C-terminus by characterizing the molecular defect of an APECED patient mutation in AIRE. A previous report demonstrated that AIRE binds P-TEFb (10). We now mapped this interaction to the C-terminal 40 residues of AIRE and validated this finding with biochemical and functional assays. Indeed, our findings reveal that the C-terminal 40 residues of AIRE are critical for the recruitment of P-TEFb to target genes, leading to increased phosphorylation of the CTD, transcription elongation and pre-mRNA splicing.
MATERIALS AND METHODS
Cell lines
HeLa and 293T cells were obtained from ATCC and cultured in DMEM supplemented with 10% FBS and antibiotics. Mouse 1C6 mTECs were a kind gift from Dr M. Kasai [National Institute of Infectious Diseases, Tokyo, Japan (27)] and were cultured in S-MEM (Invitrogen) supplemented with 10% FBS (Invitrogen), 5 mM Glutamax (Invitrogen), 0.2 µM β-mercaptoethanol (Invitrogen), 0.5 µg/ml hydrocortisone hemisuccinate (Sigma-Aldrich) and 1 mM sodium pyruvate (Invitrogen).
Plasmids
Flag-AIRE was a kind gift from Dr M. Matsumoto [University of Tokushima, Kuramoto, Tokushima, Japan (15)] and GAL.AIRE was a kind gift from Dr P. Peterson (University of Tartu, Tartu, Estonia). hINSLuc was kind gift from Dr M. German (UCSF, San Francisco, CA, USA). G5HIV2SVEDA was a kind gift from Dr R. Kornbliht [Universidad de Buenos Aires, Buenos Aires, Argentina (28)] and G5HIV2dsx ± ESE were a kind gift from Dr J. Blencowe [University of Toronto, Toronto, Ontario, Canada (29)]. G5CAT has been previously described (30). HA-CDK9 and myc-cyclinT1 have been previously described (31,32).
The 505fsx520 mutation was generated in WT GAL.AIRE and WT Flag-AIRE by PCR mutagenesis using primers: (forward) AGGATGACACTCCAGTCACGAGC and (reverse) GCTCGTGACTGGAGTGTCATCCT. GAL.AIRE deletion mutants were generated with PCR mutagenesis deletion with QuikChange II XL Site-Directed Mutagenesis Kit (Stratagene) using primers:
425F TTGACTGTATCGCCGGAATTCCAGCAGAACCTGGCTCCTGGT and
425R ACCAGGAGCCAGGTTCTGCTGGAATTCCGGCGATACAGTCAA;
488F ACTGTATCGCCGGAATTCCTGGCCCCCAGCCCCGCC and
488R GGCGGGGCTGGGGGCCAGGAATTCCGGCGATACAGT.
Luciferase assay
1C6 mTECs were plated onto 6-well plates and transfected using Fugene 6 (Roche) to co-express empty plasmid vector, Flag-AIRE or Flag-AIREfsx, and hINSLuc and pRL-CMV (Promega). Cells were harvested after 24 h and luciferase assays were performed using the Dual Luciferase kit (Promega) according to the manufacturer’s instructions. Firefly luciferase activity of each sample was normalized to renilla luciferase readings. Averages were calculated from at least three separate transfections. Fold activation represents the ratio between the effector activated transcription and the activity of the reporter with empty vector. Error bars represent SEM.
CAT assay
1C6 cells were transfected with Fugene 6 (Roche) to co-express G5CAT and GAL-chimeras. CAT assays were performed as in Oven et al. (10) and are described in detail in Supplementary Methods.
RNA isolation and RT–PCR
Cells were harvested in Trizol (Invitrogen) and RNA was isolated according to manufacturer’s specifications. RNA was treated with DNAfree (Ambion) and reverse transcribed with M-MLV RT (Invitrogen) with random or specific primers. Detailed protocol and primers are described in Supplementary Methods.
Chromatin immunoprecipitation
Chromatin immunoprecipitation (ChIP) protocol was adapted from Carey et al. (33), and is described in detail in Supplementary Methods.
Co-immunoprecipitation
293T cells were transfected with plasmids encoding HA-CDK9, Myc-Cyc T1 and GAL, WT GAL.AIRE or mutant GAL.AIREfsx fusion proteins using calcium phosphate. Cells were harvested and resuspended in hypotonic buffer (10 mM HEPES pH 7.5, 1.5 mM MgCl2, 10 mM KCl, 5 mM EDTA, 0.05% NP-40, 1 mM DTT) supplemented with protease inhibitor cocktail (Sigma-Aldrich) and incubated on ice for 15 min. Cell lysates were centrifuged for 10 min at 1000g (4°C) and supernatant was removed as completely as possible and discarded. Nuclei were resuspended in sonication buffer (20 mM HEPES pH 7.5, 150 mM NaCl, 1.5 mM MgCl2, 0.2 mM EDTA, 0.1% NP-40, 1 mM DTT) supplemented with protease inhibitor cocktail and gently sonicated on a Bandelin sonoplus sonifier (2 × 10 s pulse at power setting 20%, on ice at all times). Nuclear lysates were centrifuged at maximum speed for 20 min to remove insoluble material and supernatants were used for immunoprecipitation. Two micrograms of antibodies were added and the samples were incubated 4 h at 4°C, then protein G Dynabeads (Invitrogen) were added and samples were incubated 1 additional hour. Precipitates were washed at room temperature four times with cold PBS supplemented with 0.05% NP-40 and protease inhibitor cocktail. Precipitated proteins were eluted by addition of 1× SDS–PAGE loading buffer and heating to 95°C for 5 min. Precipitates were separated by SDS–PAGE and proteins were detected with specific antibodies by western blotting.
Antibodies and chemicals
Antibodies used for western blotting, co-immunoprecipitation (co-IP), ChIP or immunofluorescence: anti-AIRE (sc-33188 Santa Cruz Biotechnology), anti-Flag (M2 Sigma-Aldrich), anti-CDK9 (sc-484 Santa Cruz Biotechnology, ab10874 Abcam), anti-RNAPII (ab5408 Abcam, MMS-126 R Covance), anti-S2P (ab5095 Abcam), anti-GAL (sc-510 Santa Cruz Biotechnology), anti-GAPDH (AM4300 Ambion), anti-α Tubulin (DM1A Sigma-Aldrich), anti-U5-116 kD (A300-957A Bethyl labs) rabbit and mouse control IgG (Santa Cruz Biotechnology). Flavopiridol was obtained from Sigma-Aldrich.
RESULTS
The frame-shift mutant AIRE protein cannot activate transcription
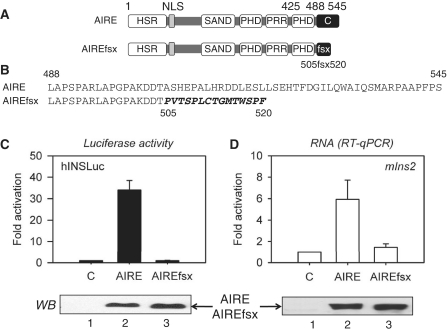
To reveal how AIRE induces the expression of TRAs, we focused on an APECED patient mutation in AIRE, which encodes the frame-shift mutant AIRE 505fsx520 (AIREfsx) protein with a truncated C-terminus, whose other domains important for homodimerization and recruitment to chromatin remain intact. The mutation was identified in two siblings with APECED by Ishii et al. (34). It represents a deletion of a guanosine at position 1640, which introduces a reading frame-shift at residue 505 and causes premature termination at residue 520 in the mutant AIREfsx protein. A schematic representation of AIRE protein domains and changes caused by the reading frame-shift mutation are depicted in Figure 1A and B. To characterize functionally the mutant AIREfsx protein, we used a co-transfection assay with plasmid effectors and a luciferase reporter target that was under the control of the human insulin promoter (hINSLuc) (Figure 1C). When the WT AIRE protein was co-expressed transiently, the luciferase activity of hINSLuc was 35-fold over that of the empty plasmid vector (Figure 1C, compare lanes 2 and 1), whereas the mutant AIREfsx protein was inactive (Figure 1C, compare lanes 3 and 1).
Figure 1.
The mutant AIREfsx protein does not activate transcription. (A) Schematic representation of WT AIRE and mutant AIREfsx proteins. From the N-terminus, domains are: homogenously staining region (HSR); nuclear localization signal (NLS), Sp100, Aire-1, NucP41/75 and DEAF-1 domain (SAND); plant homeodomains 1 and 2 (PHD); proline rich region (PRR); WT (C) and mutant alternative (fsx) C-termini. On top, numbers refer to borders of PHD2 and the full length Aire protein. Below numbers refer to the first frame-shifted residue and the end of the termination sequence (505fsx520). (B) Residues in the C-terminus of WT and mutant AIREfsx proteins. Numbers are as in (A). Changed residues are presented with italic letters. (C) Mutant AIREfsx protein does not activate transcription. Relative luciferase activity of plasmid target (hINSLuc) for WT AIRE and mutant AIREfsx proteins is presented as fold activation above the empty plasmid vector (C). Values represent the average of three independent transfections with standard errors of the mean (SEM) represented by error bars. Expression levels for AIRE proteins are presented below the bar graph (WB). (D) Mutant AIREfsx protein does not induce expression of the mouse Ins2 gene. mIns2 RNA levels from WT AIRE and mutant AIREfsx protein expressing 1C6 mTECs are presented as fold activation above the empty plasmid vector (C). Values represent the average of three independent transfections with SEM represented by error bars. Levels of Ins2 RNA were normalized to β-actin. Expression levels for AIRE proteins are presented below the bar graph (WB).
We also examined the expression of Ins2, an endogenous TRA gene (Figure 1D). Levels of mouse Ins2 transcripts from 1C6 mTECs transiently expressing WT AIRE or mutant AIREfsx proteins were quantified. While the WT AIRE protein induced Ins2 mRNA expression 6-fold (Figure 1D, compare lanes 2 and 1), the mutant AIREfsx protein had no effect (Figure 1D, compare lanes 3 and 1). Similar levels of WT AIRE and mutant AIRE proteins were expressed in these cells (Figure 1C, D).
AIRE is localized to the nucleus and has a typical punctate staining in AIRE-expressing mTECs in the thymus (35,36) as well as in transfected cells of various types (9). We also localized WT AIRE and mutant AIREfsx proteins in HeLa cells (Supplementary Figure S1). All examined AIRE proteins were found in the nucleus. However, while the WT AIRE protein showed a discrete punctuate staining, the mutant AIREfsx protein staining revealed a more dispersed pattern. Similar dispersed nuclear staining was also observed for a C-terminally deleted mutant AIRE (1–509) protein, confirming that it was indeed due to the loss of a functional C-terminus rather than mediated by the new C-terminal residues encoded by the frame-shifted reading frame. We conclude that the C-terminus of AIRE is required for the expression of TRAs and for the punctate nuclear distribution characteristic of the WT AIRE protein.
The C-terminus of AIRE is an autonomous TAD
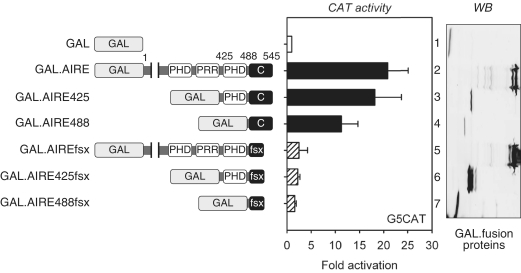
To examine if the C-terminus of AIRE can act as an independent TAD, we fused WT and different N-terminally truncated versions of AIRE as well as those bearing the 505fsx520 mutation to the GAL4 DNA-binding domain (GAL) (Figure 2, left panel). These fusion proteins were expressed transiently in 1C6 mTECs with G5CAT, consisting of a CAT reporter gene with five GAL4 upstream activating sequences (UASs) in its promoter. WT GAL.AIRE fusion protein activated G5CAT ~20-fold over GAL (Figure 2, middle panel, compare lanes 1 and 2). A similar activation was observed for the GAL. AIRE425 fusion protein (Figure 2, middle panel, lane 3), which included only the PHD2 and the C-terminus of AIRE. Importantly, the GAL.AIRE488 chimera, including only the C-terminus, also activated G5CAT 12-fold (Figure 2, middle panel, lane 4). In contrast, none of the mutant AIREfsx proteins (Figure 2, middle panel, lanes 5–7) activated this plasmid target. The expression and size of mutant fusion proteins was confirmed in cells (Figure 2, right panel). Of note, transcriptionaly inactive mutant proteins were more stable than their WT counterparts (Figure 2, right panel, compare lanes 2–4 and 5–7). Tethering AIRE to GAL enabled us to examine transcription activation independent of recruitment, thereby specifically mapping the core TAD to the 57 residues in the C-terminus of AIRE. The increased stability of our mutant proteins agrees with a previous observations that active transcription factors have shorter half-lives in cells (37). We conclude that the 57 residues of AIRE C-terminus represent a necessary and sufficient TAD of AIRE.
Figure 2.
The C-terminus of AIRE is an autonomous TAD. In the left panel is a schematic representation of GAL fusion proteins and their names. The CAT activity of the plasmid target (G5CAT) for the WT and mutant GAL.AIRE fusion proteins in 1C6 mTECs is presented as fold activation above that for GAL alone. Values represent the average of three independent transfections with SEM represented by error bars. Panel on the right presents expression levels of GAL and GAL.AIRE chimeras (WB).
The C-terminus of AIRE interacts with P-TEFb
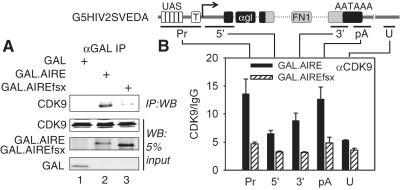
Given the functional importance of the C-terminus of AIRE, we wanted to determine if it mediates its interactions with P-TEFb (10). Therefore, we performed co-immunoprecipitations (co-IPs) from HeLa cells transiently expressing epitope-tagged CDK9 protein together with GAL, WT GAL.AIRE or mutant GAL.AIREfsx fusion proteins (Figure 3A). AIRE does not require a cell-specific co-factor (13,18). Since other cell lines beside mTECs were used in most publications, we used HeLa cells for our binding studies. As expected, CDK9 was co-precipitated with the WT GAL.AIRE fusion protein (Figure 3A, lane 2) but not with GAL (Figure 3A, lane 1). In contrast, no such interaction was observed with the mutant GAL.AIREfsx fusion protein (Figure 3A, lane 3).
Figure 3.
Mutant AIREfsx protein does not interact with P-TEFb. (A) Mutant AIREfsx protein does not interact with CDK9. GAL, GAL.AIRE and GAL.AIREfsx chimeras were expressed in HeLa cells. Proteins were immunoprecipitated with anti-GAL antibodies (αGAL) and the co-immunoprecipitation with CDK9 was detected with anti-CDK9 antibodies by western blotting (Top panel, IP:WB). The lower three panels contain 5% of input proteins for immunoprecipitations (WB). (B) Mutant AIREfsx protein cannot recruit CDK9 to DNA. Sonicated lysates from HeLa cells co-expressing GAL.AIRE or GAL.AIREfsx fusion proteins and G5HIV2SVEDA were immunoprecipitated with anti-CDK9 (αCDK9) antibodies for ChIP. Enrichment of amplicons is presented as fold enrichment over IgG control (CDK9/IgG). Black and striped bars represent values for the WT GAL.AIRE and mutant GAL.AIREfsx chimeras with SEM depicted by error bars. G5HIV2SVEDA and ChIP amplicons are depicted above and below the bar graph, respectively (promoter, Pr; 5′-end of gene, 5′; 3′-end of gene, 3′, polyadenylation signal, pA; untranscribed region, U). G5HIV2SVEDA contains five GAL binding sites (UAS) and the TATA box (T) upstream of the α-globin gene (αgl) with the inserted fragment from the fibronectin 1 gene (FN1).
To determine if the loss of P-TEFb binding abolished target gene recruitment of P-TEFb by the mutant AIREfsx protein, we performed ChIPs with anti-CDK9 antibodies. Recruitment of AIRE to its target genes depends on the binding between PHD1 and H3K4me0 (13,14) and as yet undetermined other interactions. Thus, to isolate our study from issues related to AIRE recruitment to DNA, we also chose the GAL-tethering approach for these experiments. We used G5HIV2SVEDA, containing from the 5′ direction 5 UASs, the TATA box from the HIV2 promoter, and a 1.5 kb fragment with the human α-globin gene, into whose third exon a 3 kb fragment from the human fibronectin 1 gene was inserted (Figure 3B) (28). This plasmid target has a long coding region (4 kb), which was optimal for our ChIP-qPCR analyses (promoter (Pr), 5′- and 3′-ends of the gene and polyadenylation site (pA), and a control untranscribed region (U)). Furthermore, this plasmid target had low basal levels of expression but could be induced potently by the co-expression with a GAL-fused TAD (28) such as the WT GAL.AIRE fusion protein (Supplementary Figure S2). The target gene analyzed in our ChIPs is depicted on top of Figure 3B. Below it are presented our amplicons for ChIP. Due to its artificial nature, primers designed to amplify different regions of G5HIV2SVEDA were specific for the plasmid target and did not amplify endogenous cellular genes. Indeed, we observed 1.5- to 2.5-fold enrichment of P-TEFb on this gene (Figure 3B, lower panel, black bars, Pr, 5′, 3′ and pA amplicons) above background levels (Figure 3B, lower panel, black bar, U amplicon) in cells expressing the WT GAL.AIRE fusion protein. In contrast, even though transiently expressed proteins were expressed at comparable levels (Supplementary Figure S3) the mutant GAL. AIREfsx fusion protein failed to recruit P-TEFb to G5HIV2SVEDA (Figure 3B, compare striped and black bars), thus establishing the functional role for interactions between P-TEFb and the C-terminus of AIRE in this process. Based on these results, we conclude that the AIRE TAD recruits P-TEFb to target genes and that the mutant AIREfsx protein has lost this property.
AIRE increases the phosphorylation of the CTD
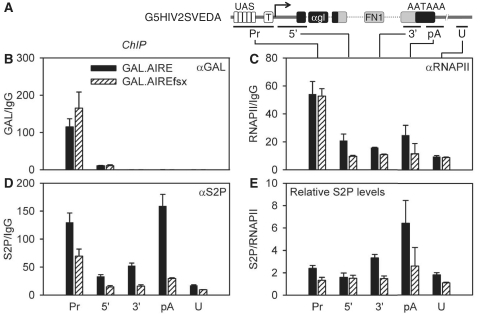
Since our assays defined the AIRE TAD and its interactions with P-TEFb, we wanted to analyze the phosphorylation of CTD on AIRE-induced genes. Since CDK9 phosphorylates S2P, we focused on this modification. G5HIV2SVEDA was co-expressed transiently with the WT GAL.AIRE or mutant GAL.AIREfsx fusion proteins in HeLa cells. Targeting of GAL fusion proteins to G5HIV2SVEDA was verified by ChIP with anti-GAL antibodies (Figure 4B). Indeed, the WT GAL.AIRE and mutant GAL.AIREfsx fusion proteins were recruited to DNA at comparable levels (Figure 4B, black and striped bars). Moreover, we observed a high occupancy of our GAL fusion proteins at the promoter (Pr) where the UASs are located (Figure 4B, black and striped bars, Pr). The 5′ amplicon was also enriched in the GAL ChIP, although these levels were approximately 20-fold lower than those for the Pr amplicon (Figure 4B, black and striped bars, compare Pr and 5′ amplicon). We conclude that our assay had a high resolution and the fusion proteins were recruited to DNA at comparable levels.
Figure 4.
AIRE increases S2P phosphorylation. (A) Schematic representation of the G5HIV2SVEDA minigene and ChIP amplicons is as in Figure 3B. (B–D) ChIP of G5HIV2SVEDA. Transfected cells were analyzed with specific antibodies as in Figure 3B. (B) ChIP with anti-GAL (αGAL) antibodies. (C) ChIP with anti-RNAPII (αRNAPII) antibodies. (D) ChIP with anti-S2P (αS2P) antibodies. (E) AIRE increases relative levels of S2P. To evaluate relative levels, levels of S2P were normalized to those of RNAPII (S2P/RNAPII) for each amplicon. Error bars represent SEM.
Whereas we found ~2-fold more RNAPII on the gene and at polyadenylation signal (Figure 4C, compare black and striped bars, 5′, 3′ and pA amplicons) in WT GAL.AIRE fusion protein-expressing cells compared to levels in mutant GAL.AIREfsx fusion protein-expressing cells or to levels in the untranscribed region (Figure 4C, compare black, U amplicon), its levels in the promoter region (Pr) did not differ significantly between the WT GAL.AIRE and mutant GAL.AIREfsx chimeras (Figure 4C, compare black and striped bars, Pr). Since the occupancy of RNAPII at the promoter in cells expressing the mutant GAL.AIREfsx fusion protein was above that at the untrancribed amplicon (U), this finding indicates that RNAPII is recruited to the promoter independently of its interaction with P-TEFb.
In contrast, levels of S2P were markedly higher on the gene in the presence of the WT GAL.AIRE fusion protein compared to the mutant GAL.AIREfsx fusion protein (Figure 4D, compare black and striped bars, 3′ and pA amplicons). Indeed, levels were ~2-fold above those from mutant GAL.AIREfsx fusion protein-expressing cells in the promotor (Pr) and 5′-end of the gene, 3-fold in the 3′-end of the gene and 8-fold at the polyadenylation site (pA) (Figure 4D, compare black and striped bars). To determine relative levels of S2P, we divided amounts of S2P by those of RNAPII for each amplicon. When they were examined in WT GAL.AIRE fusion protein-expressing cells and compared to the untranscribed region (U), 1.5-, 2- and 3-fold enrichment was observed at the promoter (Pr), at the 3′-end of the gene (3′) and at the polyadenylation site (pA), respectively (Figure 4E, black bars). In contrast, in mutant GAL.AIREfsx fusion protein-expressing cells, relative S2P levels throughout the gene were equivalent to those of the untranscribed region (U) (Figure 4E, striped bars). Thus, AIRE-induced activation of transcription is associated with elevated levels of RNAPII and S2P in the body and 3′-end of the gene, which depend on the AIRE TAD.
AIRE increases pre-mRNA splicing
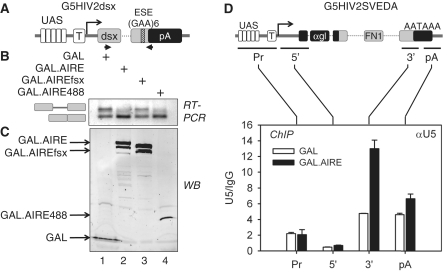
Since pre-mRNA splicing is a co-transcriptional process influenced by CTD phosphorylation, we wanted to examine its regulation by AIRE. We used G5HIV2dsx, a plasmid splicing reporter target containing a minigene of two exons with a weak splice site from the Drosophila dsx gene with or without an exonic splicing enhancer (ESE) (Figure 5A, Supplementary Figure S4A). ESEs are recognized by SR proteins, which interact with the splicing machinery to stimulate the inclusion of weak exons (38). Indeed, without a strong activator recruited to its 5′ UASs and CTD phosphorylation, G5HIV2dsx is transcribed at low levels and its transcripts are processed inefficiently (29,39). However, because it has a very short coding sequence (84 and 70 nucleotide exon fragments with a 114 nucleotide intron), we used the longer G5HIV2SVEDA for our ChIPs. As presented in Supplementary Figure S2, the WT GAL.AIRE fusion protein also activated G5HIV2dsx. To visualize AIRE-induced effects on co-transcriptional splicing of this minigene, we used a primer pair that amplified spliced and unspliced transcripts for RT-PCR. When the WT GAL.AIRE fusion protein was co-expressed with G5HIV2dsx, we detected more spliced (lower band) than unspliced transcripts (upper band) compared to GAL (Figure 5B, compare upper and lower bands in lanes 2 and 1). Interestingly, a similar trend was observed when we co-expressed just the AIRE TAD fused to GAL (GAL.AIRE488) (Figure 5B, compare lanes 2 and 4). In contrast, the mutant GAL.AIREfsx fusion protein did not have this effect (Figure 5B, compare lanes 1–3). Using the plasmid target lacking the ESE, we observed faint bands of spliced transcripts when the WT GAL.AIRE or GAL.AIRE488 fusion proteins were co-expressed, but these levels were much lower than those with the ESE-containing reporter (Supplementary Figure S4B, lanes 1–4). All GAL fusion proteins were expressed equivalently (Figure 5C and Supplementary Figure S4C). We conclude that AIRE increases the pre-mRNA splicing.
Figure 5.
AIRE increases pre-mRNA splicing. (A) Schematic representation of the G5HIV2dsx minigene with the RT–PCR primer pair. G5HIV2dsx contains 5 GAL-binding sites (UAS) upstream of the Drosophila doublesex (dsx) exons 3 and 4 and the intervening sequence (intron) with an exonic splicing enhancer (ESE) containing 6 GAA repeats and the SV40 poladenylation signal (pA). (B) AIRE affects splicing of dsx pre-mRNA. HeLa cells co-expressed GAL or GAL fusion proteins and G5HIV2dsx. Total mRNA was isolated and splicing was assayed by RT–PCR with a primer pair that amplified spliced (lower band) and unspliced (upper band) transcripts (RT–PCR). (C) Expression of fusion proteins was monitored with anti-GAL antibodies by western blotting (WB). GAL and GAL.AIRE fusion proteins are identified by arrows. (D) U5 snRNP is enriched on G5HIV2SVEDA in AIRE expressing cells. Sonicated lysates from HeLa cells co-expressing GAL or GAL.AIRE fusion protein and G5HIV2SVEDA were immunoprecipitated with anti-U5 (αU5) antibodies for ChIP. Increased levels of specific amplicons are presented as fold enrichment over the IgG control (U5/IgG). Black and white bars represent values for the GAL.AIRE chimeras and GAL, respectively, with SEM depicted by error bars. G5HIV2SVEDA with amplicons is diagrammed above the bar graph (promoter, Pr; 5′-end of gene, 5′; 3′-end of gene, 3′; polyadenylation signal, pA). It contains five GAL binding sites (UAS) upstream of α-globin gene (αgl) with inserted fragment of fibronectin 1 gene (FN1).
Prompted by these results and previous findings on co-transcriptional accumulation of cellular splicing factors (40), we examined if AIRE-induced increase of S2P stimulated the recruitment of spliceosome component U5 to our plasmid target. To this end, we examined the abundance of spliceosome subunits with anti-U5-116 kD antibodies by ChIP (Figure 5D, compare black and white bars). U5–116 kD (U5) is a subunit of U5 snRNP, which is a shared component of both the major U2 and minor U12 spliceosomes (41). Indeed, we observed an enrichment of U5 on the 3′-end of the gene in cells expressing the WT GAL.AIRE fusion protein. Thus, AIRE increases the recruitment of co-transcriptional processing factors to DNA that accumulate with RNAPII at the 3′-end of the gene.
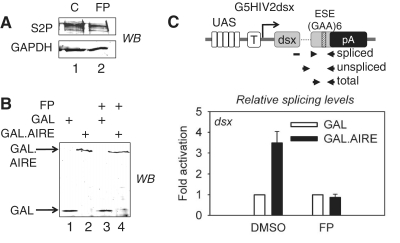
Inhibition of CDK9 abrogates effect of AIRE on pre-mRNA splicing
To examine if AIRE-induced increase of pre-mRNA splicing and increased S2P levels were linked, we made use of a pharmacological inhibitor of P-TEFb. Flavopiridol (FP) binds tightly to the ATP pocket of CDK9 and thereby inhibits its kinase activity (42). To this end, we co-expressed the WT GAL.AIRE fusion protein or GAL with G5HIV2dsx and compared splicing patterns between FP-treated and untreated cells. The reported half maximal inhibitory concentration of FP for the kinase activity CDK9 is 22 nM (43). Thus, we used a low concentration to minimize effects of FP on other CDKs. As presented in Figure 6A (compare lanes 2 and 1), when treating cells with FP at 25 nM, we observed a clear reduction in S2P levels in cells. We observed no decrease in cell proliferation after 24 h of treatment. The expression of the WT GAL.AIRE fusion protein or GAL in untreated as well as FP-treated cells was confirmed with specific antibodies by western blotting (Figure 6B). From these cells, we isolated RNA and used specific primers for spliced, unspliced and total dsx RNA (Figure 6C, upper panel) and RT–qPCR to measure activation of splicing by AIRE. Splicing efficiencies were calculated as ratios of normalized levels of spliced versus unspliced transcripts. In agreement with our earlier results, when we compared splicing efficiency in untreated cells, we detected a nearly 3.5-fold increased pre-mRNA splicing with the WT GAL.AIRE fusion protein compared to GAL (Figure 6C, compare black and white bars). By contrast, in FP-treated cells, no such AIRE-induced increase was observed (Figure 6C, compare black and white bars). Thus, by inhibiting the kinase activity of CDK9, AIRE-induced enhancement of pre-mRNA splicing is blocked.
Figure 6.
Flavopiridol inhibits effects of AIRE on splicing. (A) Flavopiridol decreases total levels of S2P. Lysates from Hela cells, which were treated with 25 nM flavopiridol (FP) or DMSO only (C) for 24 h, were analyzed with specific antibodies by western blotting (WB). The upper panel presents levels of S2P. Lower panel presents levels of GAPDH. (B) GAL fusion proteins and G5HIV2dsx are expressed in cells treated with FP. HeLa cells co-expressing G5HIV2dsx and GAL or the WT GAL.AIRE chimera were treated with 25 nM FP or DMSO only for 24 h as indicated. Expression of proteins was determined with anti-GAL antibodies by western blotting (WB). GAL and GAL.AIRE fusion proteins are indicated by arrows. (C) FP blocks AIRE-induced enhancement of pre-mRNA splicing. Upper panel presents schematic representation of G5HIV2dsx minigene with probes used for RT–qPCR that specifically amplified spliced, unspliced or total dsx transcripts. Relative splicing efficiency was determined and is presented for the WT GAL.AIRE chimera (black bars) as fold activation relative to GAL (white bars) for treated and untreated cells (FP, DMSO). Error bars represent SEM.
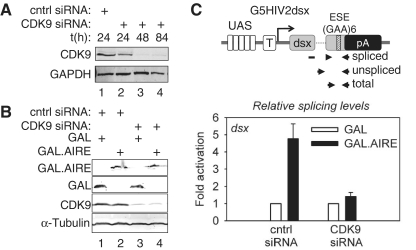
AIRE requires CDK9 to increase pre-mRNA splicing
Since FP may inhibit other CDKs, to validate further the role of CDK9 in AIRE-induced enhancement of splicing, we also knocked-down CDK9 with siRNA and examined AIRE-induced splicing. As presented in Figure 7A, CDK9 levels were decreased after 24 h and were depleted at 48 h. The corresponding siRNA and G5HIV2dsx were co-expressed with GAL or WT GAL.AIRE fusion proteins. The expression of protein effectors and CDK9 knock-downs were validated with specific antibodies by western blotting (Figure 7B). From these cells, RNA was isolated and levels of spliced and unspliced mRNAs were quantified as in Figure 6C. When comparing splicing efficiencies in cells transfected with control siRNA, we also observed 4.5-fold increased pre-mRNA splicing by AIRE (Figure 7C, compare black and white bars) which was comparable to effects in untreated cells (Figure 6C). Underlining the specific requirement for P-TEFb, AIRE had no effect when expressed in cells depleted of CDK9 (Figure 7C, compare black and white bars). Thus, AIRE requires P-TEFb to enhance pre-mRNA splicing.
Figure 7.
AIRE requires CDK9 for its effects on pre-mRNA splicing. (A) Time course of CDK9 siRNA knockdown. HeLa cells were transfected with CDK9 or control siRNAs (cntrl). Subsequently, cells were harvested at various time-points and protein levels were determined with specific antibodies by western blotting. In the upper and lower panels, levels of CDK9 and GAPDH are presented, respectively. (B) Protein effectors are expressed in CDK9-knockdown cells. HeLa cells were co-transfected with CDK9 or control siRNAs. GAL or the WT GAL.AIRE chimera and G5HIV2dsx were co-expressed as indicated. Protein levels of expressed proteins are presented in the upper two panels with those of CDK9 and tubulin in the lowest panels, respectively. (C) CDK9 siRNA knock-down inhibits effects of AIRE on splicing. Upper panel contains a schematic representation of the G5HIV2dsx minigene with amplicons used to amplify spliced, unspliced or total dsx transcripts by RT–qPCR. Relative splicing efficiency was determined and is presented for the WT GAL.AIRE chimera (black bars) as fold activation relative to GAL (white bars) for cells transfected with CDK9 or control siRNAs. Error bars represent SEM.
By recruiting P-TEFb, AIRE induces splicing of an endogenous TRA gene
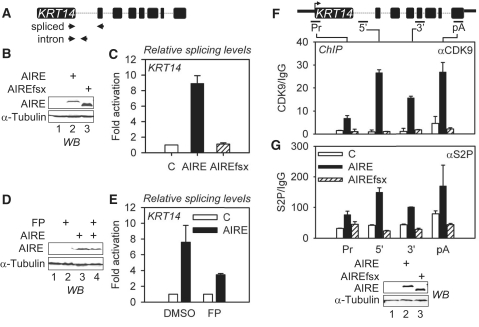
To validate our model, we examined the endogenous KRT14 gene, which is regulated by AIRE (18). Total RNA was isolated from 293T cells expressing transiently the empty plasmid vector, WT AIRE or mutant AIREfsx proteins (Figure 8B). Spliced KRT14 and KRT14 intron containing transcripts were amplified with specific primers by RT–qPCR (Figure 8A) and quantified relative to those from the endogenous GAPDH gene. As presented in Supplementary Figure S5A, the expression of KRT14 was induced by the WT AIRE but not mutant AIREfsx proteins. Splicing efficiencies were calculated as ratios of normalized levels of spliced versus unspliced transcripts and are presented as fold activation above the splicing observed in the presence of only the empty plasmid vector, which was set to 1. WT AIRE protein increased the splicing efficiency 9-fold above the control (Figure 8C, compare black to white bars). As expected, mutant AIREfsx protein did not affect the splicing efficiency of the KRT14 gene (Figure 8C, compare striped to white bars). Similar results were obtained for the S100A8 gene, which is also induced by AIRE (data not shown). We conclude that the mutant AIREfsx protein affects neither the transcription nor the splicing of endogenous TRA genes, which is in agreement with our observations with plasmid targets.
Figure 8.
By recruiting P-TEFb, AIRE induces splicing of an endogenous TRA gene. (A) Schematic representation of the KRT14 gene with primer pairs used to amplify spliced and unspliced transcripts by RT–qPCR. (B) WT AIRE and mutant AIREfsx proteins are expressed in 293T cells. Expression of proteins was determined with anti-AIRE and anti-α-tubulin antibodies by western blotting (WB). (C) Only the WT AIRE protein induces splicing of the KRT14 pre-mRNA. Relative splicing efficiency was determined and is presented for WT AIRE (black bar) and mutant AIREfsx (hashed bar) proteins as fold activation relative to the empty plasmid vector (C, white bar). Error bars represent SEM. (D) WT AIRE protein is expressed in 293T cells treated with FP. Expression of proteins was determined with anti-AIRE and anti-α-tubulin antibodies by western blotting (WB). (E) FP blocks AIRE-induced enhancement of KRT14 pre-mRNA splicing. Relative splicing efficiency was determined and is presented for the WT AIRE protein (black bars) as fold activation relative to the empty plasmid vector (C, white bars) for treated and untreated cells (FP, DMSO). Error bars represent SEM. (F) P-TEFb is enriched on the KRT14 gene in cells expressing the WT AIRE protein. ChIPs were performed with anti-CDK9 (αCDK9) and control IgG antibodies with lysates from 293T cells expressing the empty plasmid vector (C), WT AIRE or mutant AIREfsx proteins. Enrichment of specific KRT14 amplicons is presented as fold enrichment over IgG control (CDK9/IgG). White, black and striped bars represent values for the empty plasmid vector (C), WT AIRE and mutant AIREfsx proteins, respectively, with SEM depicted by error bars. The KRT14 gene and ChIP amplicons are depicted above the bar graph (promoter, Pr; 5′ of coding region, 5′; 3′ of coding region, 3′; polyadenylation signal, pA). (G) ChIP with anti-S2P (αS2P) antibodies. Expression levels for AIRE and α-tubulin proteins are presented below the bar graph (WB). Lysates from transfected cells were analyzed with specific antibodies by western blotting.
To examine further the role of P-TEFb in AIRE-induced expression of TRA genes, we compared levels of KRT14 mRNA from cells expressing the WT AIRE protein which were treated or not with FP (Figure 8D). Indeed, effects of AIRE were decreased by 85% in cells treated with FP compared to untreated cells (Supplementary Figure S5B, compare black bars). FP also inhibited AIRE-induced splicing of KRT14 transcripts (Figure 8E, FP, compare black to white bars). In these cells, AIRE was expressed at comparable levels (Figure 8D). Thus, AIRE requires P-TEFb to induce transcription and pre-mRNA splicing of endogenous TRA genes.
These results and a previous report of the recruitment of P-TEFb by AIRE to endogenous genes (10) led us to examine if the mutant AIREfsx protein fails to do the same. In 293T cells, we expressed transiently the empty plasmid vector, WT AIRE or mutant AIREfsx proteins. Subsequently, we performed ChIPs with anti-CDK9 antibodies and analyzed KRT14 DNA enrichment by qPCR. qPCR amplicons are presented in the schematic of the KRT14 gene above the bar graph (Figure 8F). Indeed, we observed a significant enrichment of CDK9 at the promoter (4.5-fold), in the coding region (26- and 14-fold) and at the polyadenylation site (6-fold) in WT AIRE protein-expressing cells compared to control cells (Figure 8F, compare black to white bars). Importantly, no significant enrichment was observed in mutant AIREfsx protein-expressing cells (Figure 8F, striped bars). With anti-S2P antibodies, we examined levels of S2P on the KRT14 gene. They were also enriched between 2- and 4-fold in WT AIRE protein-expressing cells compared to control and mutant AIREfsx protein-expressing cells (Figure 8G, compare black to white and striped bars). We conclude that by recruiting P-TEFb to endogenous TRA genes, AIRE also increases their levels of S2P.
DISCUSSION
By characterizing the functional defect caused by an APECED patient mutation, resulting in a frame-shift at position 505 in AIRE (AIREfsx), we elucidated the mechanisms of AIRE-induced activation of otherwise tissue-restricted genes. This mutation affects only the C-terminus of AIRE, which is highly conserved between mouse and human AIRE proteins. The mutant AIREfsx protein could not activate exogenous plasmid targets or representative endogenous TRA genes. This situation persisted even when AIRE proteins were targeted artificially to DNA via the GAL4 DNA-binding domain. This heterologous tethering also allowed us to map the minimal AIRE TAD, which was found to mediate interactions with P-TEFb. Next, the mutant AIREfsx protein could not recruit P-TEFb to plasmid DNA via heterologous DNA-tethering or to an endogenous TRA gene in chromatin when expressed transiently in cells. It also lost the ability of its WT counterpart to increase levels of RNAPII and CTD phosphorylation on the body and 3′-end of these target genes. These increased levels of S2P could be correlated with those of specific transcripts and improved co-transcriptional pre-mRNA processing of minigene plasmid targets and an endogenous TRA gene in chromatin. Of note, when the kinase activity of CDK9 was blocked by flavopiridol or its levels diminished by CDK9 siRNA, the WT AIRE protein also lost the ability to increase the pre-mRNA splicing of its target genes. In conclusion, we mapped the AIRE TAD, defined its interaction with P-TEFb, as well as characterized functional consequences of this binding on transcription of target genes and co-transcriptional processing of their nascent transcripts.
This study confirms and extends a previous observation on interactions between AIRE and P-TEFb (10). Importantly, we observed effects of increased levels of S2P on co-transcriptional processing of nascent transcripts. Moreover, enhanced splicing efficiencies and increased levels of steady state mRNAs correlated directly with CTD phosphorylation. Thus, our findings complement nicely a previous report, which reported that AIRE exerts most of its effects on gene expression at the level of pre-mRNA splicing (18). In their proteomic analysis, Abramson et al. identified 45 candidates that interacted with AIRE and were involved in nuclear transport, DNA-damage response, chromatin remodeling, transcription and pre-mRNA processing. Knockdowns of only 11 of these candidates impaired AIRE’s ability to regulate the expression of target genes in chromatin and on episomal plasmids. Of interest, except for the Myb binding protein 1 A (MYBBP1A), all of them were found associated with P-TEFb and/or RNAPII in another study (44). Since chromatin remodeling factors, splicing and polyadenylation machineries bind to the elongating transcription complex (26), associations between AIRE, P-TEFb and RNAPII can better explain these proteomic data than the ability of AIRE to bind all these proteins directly. Thus, AIRE enters a long list of transcription factors that recruit P-TEFb to target genes (22). Of interest, mono-ubiquitylation increases the affinity of VP16 and other TADs for P-TEFb (37,45). Since AIRE is ubiquitylated (15), this post-translational modification could also increase interactions between AIRE and CDK9.
How AIRE is targeted precisely to TRA genes in mTECs remains to be investigated. Since AIRE increases the expression of so many genes, it is clear that this binding has to be rather promiscuous. One aspect of this recruitment is the binding between PHD1 and histone H3K4me0 (13,14,46). However, AIRE activated exogenous plasmid targets in this and other studies, on which chromatin does not assemble efficiently (47). Furthermore, mutations outside the PHD1, e.g. in the SAND domain, prevent the recruitment of AIRE to target genes (35). Irrespective of this remaining conundrum, since P-TEFb does not initiate transcription, genes that are activated by AIRE will contain RNAPII already engaged on or near their promoters (48). The recruitment of P-TEFb will then modify these transcription complexes for elongation and co-transcriptional processing of target genes. Different levels of engaged but stalled transcription complexes on promoters could then explain why clones of primary mTECs isolated from the mouse thymus display different profiles of AIRE-induced TRAs (49).
Our study also finds resonance with whole genome analyses of P-TEFb target genes, for example those regulated by the proto-oncogene c-Myc (48). Similar to that situation, we find that AIRE does not increase levels of RNAPII at the promoter but rather in the body and especially at the 3′-end of susceptible genes. This finding suggests that P-TEFb phosphorylates preferentially subunits of negative elongation factors at the 5′-end of transcription units, which must be modified to allow RNAPII to elongate (22). Since P-TEFb remains associated with the transcription complex in the body and 3′-end of the gene, S2P levels will increase throughout elongation. Our ChIPs with anti-U5 antibodies revealed that spliceosome components were enriched at the 3′-end of the gene. This finding suggests that RNAPII accumulates at these 3′-ends to permit optimal co-transcriptional processing, i.e. pre-mRNA splicing and polyadenylation, which might not go to completion during the actual transcription of these sequences. In other words, transcription complexes might move too quickly for this maturation of primary transcripts to occur in situ (50). Consistent with this notion, two recent studies reported that RNAPII pauses for pre-mRNA splicing at the 3′-end of the gene in Saccharomyces cerevisiae (51,52). In summary, a single nucleotide mutation from an APECED patient leading to a mutant AIRE protein has allowed us to dissect these steps more precisely and to define how genes can be regulated in cells by a single protein, in this case for the specific purpose of directing central tolerance and prevent deleterious immune responses against self, which can result in autoimmunity.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Nora Eccles Treadwell Foundation and FiDiPro project funding by the Academy of Finland. Funding for open access charge: Nora Eccles Treadwell Foundation.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
We thank members of Peterlin and Saksela laboratories for their help with these studies and comments on the article; and Dr M. Kasai, Dr M. Matsumoto, Dr P. Peterson, Dr M. German, Dr R. Kornbliht and Dr J. Blencowe for provided reagents.
REFERENCES
- 1.Kyewski B, Klein L. A central role for central tolerance. Annu. Rev. Immunol. 2006;24:571–606. doi: 10.1146/annurev.immunol.23.021704.115601. [DOI] [PubMed] [Google Scholar]
- 2.Consortium F-GA. An autoimmune disease, APECED, caused by mutations in a novel gene featuring two PHD-type zinc-finger domains. Nat. Genet. 1997;17:399–403. doi: 10.1038/ng1297-399. [DOI] [PubMed] [Google Scholar]
- 3.Nagamine K, Peterson P, Scott HS, Kudoh J, Minoshima S, Heino M, Krohn KJ, Lalioti MD, Mullis PE, Antonarakis SE, et al. Positional cloning of the APECED gene. Nat. Genet. 1997;17:393–398. doi: 10.1038/ng1297-393. [DOI] [PubMed] [Google Scholar]
- 4.Anderson MS, Venanzi ES, Klein L, Chen Z, Berzins SP, Turley SJ, von Boehmer H, Bronson R, Dierich A, Benoist C, et al. Projection of an immunological self shadow within the thymus by the aire protein. Science. 2002;298:1395–1401. doi: 10.1126/science.1075958. [DOI] [PubMed] [Google Scholar]
- 5.Ramsey C, Winqvist O, Puhakka L, Halonen M, Moro A, Kampe O, Eskelin P, Pelto-Huikko M, Peltonen L. Aire deficient mice develop multiple features of APECED phenotype and show altered immune response. Hum. Mol. Genet. 2002;11:397–409. doi: 10.1093/hmg/11.4.397. [DOI] [PubMed] [Google Scholar]
- 6.Anderson MS, Venanzi ES, Chen Z, Berzins SP, Benoist C, Mathis D. The cellular mechanism of Aire control of T cell tolerance. Immunity. 2005;23:227–239. doi: 10.1016/j.immuni.2005.07.005. [DOI] [PubMed] [Google Scholar]
- 7.Kuroda N, Mitani T, Takeda N, Ishimaru N, Arakaki R, Hayashi Y, Bando Y, Izumi K, Takahashi T, Nomura T, et al. Development of autoimmunity against transcriptionally unrepressed target antigen in the thymus of Aire-deficient mice. J. Immunol. 2005;174:1862–1870. doi: 10.4049/jimmunol.174.4.1862. [DOI] [PubMed] [Google Scholar]
- 8.Peterson P, Org T, Rebane A. Transcriptional regulation by AIRE: molecular mechanisms of central tolerance. Nat. Rev. Immunol. 2008;8:948–957. doi: 10.1038/nri2450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bjorses P, Pelto-Huikko M, Kaukonen J, Aaltonen J, Peltonen L, Ulmanen I. Localization of the APECED protein in distinct nuclear structures. Hum. Mol. Genet. 1999;8:259–266. doi: 10.1093/hmg/8.2.259. [DOI] [PubMed] [Google Scholar]
- 10.Oven I, Brdickova N, Kohoutek J, Vaupotic T, Narat M, Peterlin BM. AIRE recruits P-TEFb for transcriptional elongation of target genes in medullary thymic epithelial cells. Mol. Cell Biol. 2007;27:8815–8823. doi: 10.1128/MCB.01085-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pitkanen J, Doucas V, Sternsdorf T, Nakajima T, Aratani S, Jensen K, Will H, Vahamurto P, Ollila J, Vihinen M, et al. The autoimmune regulator protein has transcriptional transactivating properties and interacts with the common coactivator CREB-binding protein. J. Biol. Chem. 2000;275:16802–16809. doi: 10.1074/jbc.M908944199. [DOI] [PubMed] [Google Scholar]
- 12.Purohit S, Kumar PG, Laloraya M, She JX. Mapping DNA-binding domains of the autoimmune regulator protein. Biochem. Biophys. Res. Commun. 2005;327:939–944. doi: 10.1016/j.bbrc.2004.12.093. [DOI] [PubMed] [Google Scholar]
- 13.Org T, Chignola F, Hetenyi C, Gaetani M, Rebane A, Liiv I, Maran U, Mollica L, Bottomley MJ, Musco G, et al. The autoimmune regulator PHD finger binds to non-methylated histone H3K4 to activate gene expression. EMBO Rep. 2008;9:370–376. doi: 10.1038/embor.2008.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Koh AS, Kuo AJ, Park SY, Cheung P, Abramson J, Bua D, Carney D, Shoelson SE, Gozani O, Kingston RE, et al. Aire employs a histone-binding module to mediate immunological tolerance, linking chromatin regulation with organ-specific autoimmunity. Proc. Natl Acad. Sci. USA. 2008;105:15878–15883. doi: 10.1073/pnas.0808470105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Uchida D, Hatakeyama S, Matsushima A, Han H, Ishido S, Hotta H, Kudoh J, Shimizu N, Doucas V, Nakayama KI, et al. AIRE functions as an E3 ubiquitin ligase. J. Exp. Med. 2004;199:167–172. doi: 10.1084/jem.20031291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bjorses P, Halonen M, Palvimo JJ, Kolmer M, Aaltonen J, Ellonen P, Perheentupa J, Ulmanen I, Peltonen L. Mutations in the AIRE gene: effects on subcellular location and transactivation function of the autoimmune polyendocrinopathy-candidiasis-ectodermal dystrophy protein. Am. J. Hum. Genet. 2000;66:378–392. doi: 10.1086/302765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pitkanen J, Vahamurto P, Krohn K, Peterson P. Subcellular localization of the autoimmune regulator protein. characterization of nuclear targeting and transcriptional activation domain. J. Biol. Chem. 2001;276:19597–19602. doi: 10.1074/jbc.M008322200. [DOI] [PubMed] [Google Scholar]
- 18.Abramson J, Giraud M, Benoist C, Mathis D. Aire's partners in the molecular control of immunological tolerance. Cell. 2010;140:123–135. doi: 10.1016/j.cell.2009.12.030. [DOI] [PubMed] [Google Scholar]
- 19.Weake VM, Workman JL. Inducible gene expression: diverse regulatory mechanisms. Nat. Rev. Genet. 2010;11:426–437. doi: 10.1038/nrg2781. [DOI] [PubMed] [Google Scholar]
- 20.Komarnitsky P, Cho EJ, Buratowski S. Different phosphorylated forms of RNA polymerase II and associated mRNA processing factors during transcription. Genes Dev. 2000;14:2452–2460. doi: 10.1101/gad.824700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shiekhattar R, Mermelstein F, Fisher RP, Drapkin R, Dynlacht B, Wessling HC, Morgan DO, Reinberg D. Cdk-activating kinase complex is a component of human transcription factor TFIIH. Nature. 1995;374:283–287. doi: 10.1038/374283a0. [DOI] [PubMed] [Google Scholar]
- 22.Peterlin BM, Price DH. Controlling the elongation phase of transcription with P-TEFb. Mol. Cell. 2006;23:297–305. doi: 10.1016/j.molcel.2006.06.014. [DOI] [PubMed] [Google Scholar]
- 23.Peng J, Zhu Y, Milton JT, Price DH. Identification of multiple cyclin subunits of human P-TEFb. Genes Dev. 1998;12:755–762. doi: 10.1101/gad.12.5.755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pandit S, Wang D, Fu XD. Functional integration of transcriptional and RNA processing machineries. Curr. Opin. Cell Biol. 2008;20:260–265. doi: 10.1016/j.ceb.2008.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Moore MJ, Proudfoot NJ. Pre-mRNA processing reaches back to transcription and ahead to translation. Cell. 2009;136:688–700. doi: 10.1016/j.cell.2009.02.001. [DOI] [PubMed] [Google Scholar]
- 26.Perales R, Bentley D. ‘Cotranscriptionality’: the transcription elongation complex as a nexus for nuclear transactions. Mol. Cell. 2009;36:178–191. doi: 10.1016/j.molcel.2009.09.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mizuochi T, Kasai M, Kokuho T, Kakiuchi T, Hirokawa K. Medullary but not cortical thymic epithelial cells present soluble antigens to helper T cells. J. Exp. Med. 1992;175:1601–1605. doi: 10.1084/jem.175.6.1601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kadener S, Cramer P, Nogues G, Cazalla D, de la Mata M, Fededa JP, Werbajh SE, Srebrow A, Kornblihtt AR. Antagonistic effects of T-Ag and VP16 reveal a role for RNA pol II elongation on alternative splicing. EMBO J. 2001;20:5759–5768. doi: 10.1093/emboj/20.20.5759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rosonina E, Bakowski MA, McCracken S, Blencowe BJ. Transcriptional activators control splicing and 3′-end cleavage levels. J. Biol. Chem. 2003;278:43034–43040. doi: 10.1074/jbc.M307289200. [DOI] [PubMed] [Google Scholar]
- 30.Kamine J, Subramanian T, Chinnadurai G. Sp1-dependent activation of a synthetic promoter by human immunodeficiency virus type 1 Tat protein. Proc. Natl Acad. Sci. USA. 1991;88:8510–8514. doi: 10.1073/pnas.88.19.8510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kanazawa S, Soucek L, Evan G, Okamoto T, Peterlin BM. c-Myc recruits P-TEFb for transcription, cellular proliferation and apoptosis. Oncogene. 2003;22:5707–5711. doi: 10.1038/sj.onc.1206800. [DOI] [PubMed] [Google Scholar]
- 32.Fujinaga K, Irwin D, Geyer M, Peterlin BM. Optimized chimeras between kinase-inactive mutant Cdk9 and truncated cyclin T1 proteins efficiently inhibit Tat transactivation and human immunodeficiency virus gene expression. J. Virol. 2002;76:10873–10881. doi: 10.1128/JVI.76.21.10873-10881.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Carey MF, Peterson CL, Smale ST. Chromatin immunoprecipitation (ChIP) Cold Spring Harb. Protoc. 2009;2009 doi: 10.1101/pdb.prot5279. doi:10.1101/pdb.prot5279. [DOI] [PubMed] [Google Scholar]
- 34.Ishii T. Novel mutations of the autoimmune regulator gene in two siblings with autoimmune polyendocrinopathy-candidiasis-ectodermal dystrophy. J. Clin. Endocrinol. Metab. 2000;85:2922–2926. doi: 10.1210/jcem.85.8.6726. [DOI] [PubMed] [Google Scholar]
- 35.Su MA, Giang K, Zumer K, Jiang H, Oven I, Rinn JL, Devoss JJ, Johannes KP, Lu W, Gardner J, et al. Mechanisms of an autoimmunity syndrome in mice caused by a dominant mutation in Aire. J. Clin. Invest. 2008;118:1712–1726. doi: 10.1172/JCI34523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Liston A, Gray DH, Lesage S, Fletcher AL, Wilson J, Webster KE, Scott HS, Boyd RL, Peltonen L, Goodnow CC. Gene dosage–limiting role of Aire in thymic expression, clonal deletion, and organ-specific autoimmunity. J. Exp. Med. 2004;200:1015–1026. doi: 10.1084/jem.20040581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Muratani M, Tansey WP. How the ubiquitin-proteasome system controls transcription. Nat. Rev. Mol. Cell Biol. 2003;4:192–201. doi: 10.1038/nrm1049. [DOI] [PubMed] [Google Scholar]
- 38.Wang Z, Burge CB. Splicing regulation: from a parts list of regulatory elements to an integrated splicing code. RNA. 2008;14:802–813. doi: 10.1261/rna.876308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rosonina E, Blencowe BJ. Analysis of the requirement for RNA polymerase II CTD heptapeptide repeats in pre-mRNA splicing and 3′-end cleavage. RNA. 2004;10:581–589. doi: 10.1261/rna.5207204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Listerman I, Sapra AK, Neugebauer KM. Cotranscriptional coupling of splicing factor recruitment and precursor messenger RNA splicing in mammalian cells. Nat. Struct. Mol. Biol. 2006;13:815–822. doi: 10.1038/nsmb1135. [DOI] [PubMed] [Google Scholar]
- 41.Patel AA, Steitz JA. Splicing double: insights from the second spliceosome. Nat. Rev. Mol. Cell Biol. 2003;4:960–970. doi: 10.1038/nrm1259. [DOI] [PubMed] [Google Scholar]
- 42.Baumli S, Lolli G, Lowe ED, Troiani S, Rusconi L, Bullock AN, Debreczeni JE, Knapp S, Johnson LN. The structure of P-TEFb (CDK9/cyclin T1), its complex with flavopiridol and regulation by phosphorylation. EMBO J. 2008;27:1907–1918. doi: 10.1038/emboj.2008.121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Biglione S, Byers SA, Price JP, Nguyen VT, Bensaude O, Price DH, Maury W. Inhibition of HIV-1 replication by P-TEFb inhibitors DRB, seliciclib and flavopiridol correlates with release of free P-TEFb from the large, inactive form of the complex. Retrovirology. 2007;4:47. doi: 10.1186/1742-4690-4-47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Jeronimo C, Forget D, Bouchard A, Li Q, Chua G, Poitras C, Therien C, Bergeron D, Bourassa S, Greenblatt J, et al. Systematic analysis of the protein interaction network for the human transcription machinery reveals the identity of the 7SK capping enzyme. Mol. Cell. 2007;27:262–274. doi: 10.1016/j.molcel.2007.06.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kurosu T, Peterlin BM. VP16 and ubiquitin; binding of P-TEFb via its activation domain and ubiquitin facilitates elongation of transcription of target genes. Curr. Biol. 2004;14:1112–1116. doi: 10.1016/j.cub.2004.06.020. [DOI] [PubMed] [Google Scholar]
- 46.Chignola F, Gaetani M, Rebane A, Org T, Mollica L, Zucchelli C, Spitaleri A, Mannella V, Peterson P, Musco G. The solution structure of the first PHD finger of autoimmune regulator in complex with non-modified histone H3 tail reveals the antagonistic role of H3R2 methylation. Nucleic Acids Res. 2009;37:2951–2961. doi: 10.1093/nar/gkp166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Jeong S, Stein A. Micrococcal nuclease digestion of nuclei reveals extended nucleosome ladders having anomalous DNA lengths for chromatin assembled on non-replicating plasmids in transfected cells. Nucleic Acids Res. 1994;22:370–375. doi: 10.1093/nar/22.3.370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Rahl PB, Lin CY, Seila AC, Flynn RA, McCuine S, Burge CB, Sharp PA, Young RA. c-Myc regulates transcriptional pause release. Cell. 2010;141:432–445. doi: 10.1016/j.cell.2010.03.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Derbinski J, Pinto S, Rosch S, Hexel K, Kyewski B. Promiscuous gene expression patterns in single medullary thymic epithelial cells argue for a stochastic mechanism. Proc. Natl Acad. Sci USA. 2008;105:657–662. doi: 10.1073/pnas.0707486105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Darzacq X, Shav-Tal Y, de Turris V, Brody Y, Shenoy SM, Phair RD, Singer RH. In vivo dynamics of RNA polymerase II transcription. Nat. Struct. Mol. Biol. 2007;14:796–806. doi: 10.1038/nsmb1280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Carrillo Oesterreich F, Preibisch S, Neugebauer KM. Global analysis of nascent RNA reveals transcriptional pausing in terminal exons. Mol. Cell. 2010;40:571–581. doi: 10.1016/j.molcel.2010.11.004. [DOI] [PubMed] [Google Scholar]
- 52.Alexander RD, Innocente SA, Barrass JD, Beggs JD. Splicing-dependent RNA polymerase pausing in yeast. Mol. Cell. 2010;40:582–593. doi: 10.1016/j.molcel.2010.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.